Abstract
The p53 tumor suppressor is highly responsive to different physiological stresses such as abnormal cell proliferation, nutrient deprivation, and DNA damage. Distinct signaling mechanisms have evolved to activate p53, which in turn modulate numerous pathways to enhance fitness and survival of the organism. Elucidating the molecular mechanisms of these signaling events is critical for understanding tumor suppression by p53 and development of novel therapeutics. Studies in the past decade have established that MDM2 and MDMX are important targets of signaling input from different pathways. Here, we focus our discussion on MDM2 and MDMX phosphorylation, which is important for p53 activation by DNA damage. Investigations in this area have generated new insight into the inner workings of MDM2 and MDMX and underscore the importance of allosteric communication between different domains in achieving an efficient response to phosphorylation. It is likely that MDM2 and MDMX regulation by phosphorylation will share mechanistic similarities to other signaling hub molecules. Phosphorylation-independent p53 activators such as ARF and ribosomal proteins ultimately achieve the same outcome as phosphorylation, suggesting that they may induce similar changes in the structure and function of MDM2 and MDMX through protein-protein interactions.
Keywords: MDM2, MDMX, p53, phosphorylation, ubiquitination
MDM2 and MDMX Are Key Regulators of p53
A unique feature of the p53 tumor suppressor is its stabilization after exposure to many stress signals. This leads to induction of numerous transcriptional targets that inhibit cell cycle progression, induce apoptosis, and regulate energy metabolism.1 The MDM2 and MDMX proteins are possibly responsible for establishing most of the dynamic features of the p53 pathway. It can be argued that the ability of p53 to act as a major tumor suppressor is in part due to its regulation by MDM2 and MDMX. MDM2 is best known as an ubiquitin E3 ligase for p53 that promotes p53 degradation.2,3 Although additional E3 ligases (Pirh2, Cop1) have also been shown to degrade p53 in cell culture,4,5 mouse models provided strong evidence that MDM2 function is indispensable for controlling p53 activity at all stages of life.6-8 The role of MDM2 as a major regulator of p53 stability is also validated by small molecule inhibitors that disrupt p53-MDM2 binding.9,10
The MDM2 homolog MDMX is also emerging as an important regulator of p53.11 The physiological significance of MDMX was revealed by the embryonic lethality of MDMX-null mice due to activation of p53.12-14 Tissue-specific knockout of MDMX generally results in mild phenotypes compared to MDM2,7,15,16 suggesting a supplemental function in p53 regulation. In fact, MDMX deficiency can be compensated by transgenic expression of additional MDM2 proteins in a mouse model.17 While MDM2 is a classic transcriptional target of p53, recent studies showed that MDMX is also a bona fide p53 target gene with its own (although weaker) p53-inducible P2 promoter.18,19 Therefore, both MDM2 and MDMX inducibility should be taken into consideration when analyzing and modeling the dynamics of the MDM2-negative feedback loop.
Ubiquitination of p53 by MDM2 and MDMX
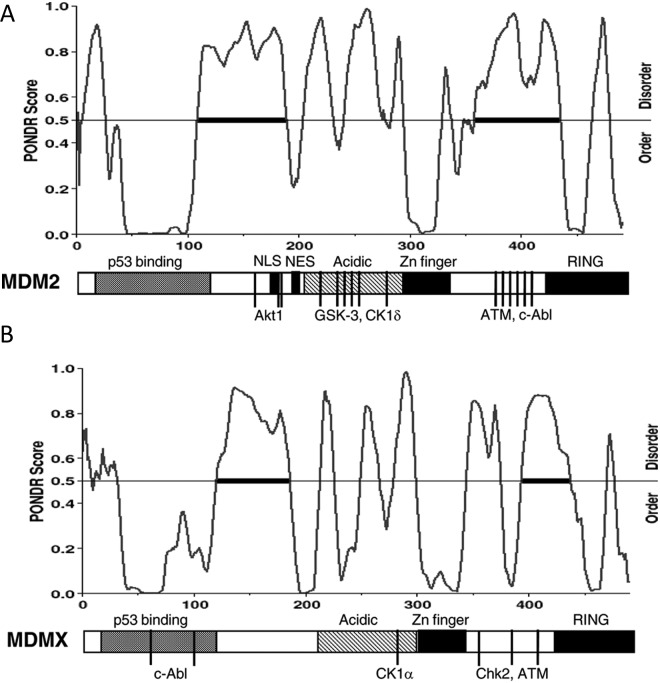
MDM2 promotes p53 degradation by forming a stable complex through N-terminal domains. The MDM2 C-terminal RING domain recruits ubiquitin-conjugating enzyme E2 that performs covalent modification of p53 lysine residues.20,21 In addition to E3 ligase activity, MDM2 also interacts with the proteasome subunit C8 and may deliver substrates directly to the proteasome.22 As expected, both the p53-binding domain and RING domains of MDM2 are critical for p53 degradation. However, the central acidic domain of MDM2 (residues 220-300) is also critical for ubiquitination of p53.23,24 The acidic domain has features of a partially unstructured region (Fig. 1A) that contains binding sites for most of the MDM2-binding proteins identified to date, including chromatin-modifying proteins (p300, YY1, KAP1, SUV39H1, EHMT1, etc.),25-27 de-ubiquitinating enzyme HAUSP,28 ribosomal proteins,29 and the tumor suppressor ARF.30 Furthermore, the MDM2 acidic domain has been shown to bind weakly to the p53 core domain and cause a conformational change.31-35 This interaction may be important for exposing the target lysines in the p53 core for ubiquitination or orienting the 2 proteins for efficient ubiquitin transfer.35 The disordered nature of the acidic domain probably provides the structural flexibility for interacting with multiple protein partners.36,37
Figure 1.
Diagrams of MDM2 (A) and MDMX domains (B) and the results of PONDR analysis, showing the unstructured sequences between domains (Predictors of Natural Disordered Regions, http://www.pondr.com). Short vertical lines indicate the location of phosphorylation sites and the corresponding kinases.
MDMX has weak intrinsic E3 ligase activity.38 The impact of MDMX expression on p53 stability is moderate in mouse embryonic fibroblasts (MEFs) derived from MDMX-null mice.39 Similarly, knockdown or overexpression of MDMX in tumor cells generally causes little change in the p53 level.40 Therefore, it has been proposed that MDMX regulates p53 mainly by formation of inactive p53-MDMX complexes.39-41 Early publications (pre-2004) showed that MDMX overexpression may even inhibit MDM2-mediated degradation of p53. It is noteworthy that some of these results may be due to the use of C-terminally epitope-tagged MDMX. Later work revealed that C-terminal epitope tagging is detrimental to MDMX ubiquitination by MDM2, suggesting an interference on MDM2 function.42 The structural basis of this phenomenon was only apparent after more recent studies revealed the importance of the extreme C-terminal sequences of MDM2 and MDMX in RING domain dimerization and E3 ligase function.43,44
The fact that MDMX has a strong tendency to form heterodimers with MDM2 through RING domains,45 the importance of RING domain dimerization for E3 activity, and the classic example of E3 ligase activation through BRCA1-BARD1 RING domain heterodimerization led many to hypothesize that MDM2-MDMX heterodimerization is important for p53 degradation. Indeed, biochemical experiments suggest that MDMX can stimulate the ability of MDM2 to ubiquitinate p53.46-50 The results of 2 recent mouse model experiments are consistent with a role of MDMX having at least a moderate role in regulating p53 stability in normal tissues.51 Therefore, the relatively insignificant role of MDMX as a regulator of p53 stability in tumor cell lines may reflect abnormalities in the p53 pathway in transformed cells.
Stress Activation of p53
Many studies showed that different cellular stress and damage signals converge on MDM2 and MDMX to cause p53 activation. Oncogene-induced ARF expression induces p53 accumulation by binding MDM2 and inhibiting p53 ubiquitination.52 Inhibitors of rRNA transcription (such as actinomycin D, 5-FU, and growth factor deprivation) induce ribosomal stress, which stimulates MDM2 interaction with several ribosomal proteins (such as L5, L11, and L23) that also block p53 ubiquitination.53 These proteins interact with the MDM2 acidic domain, highlighting the importance of the acidic domain in sensing such growth-related stress signals.
DNA damage is probably the most extensively studied p53 activation signal. A critical player in p53 DNA damage response is the ATM kinase.54 ATM is activated within minutes after DNA double-strand breaks and phosphorylates numerous substrates involved in cell cycle regulation and DNA repair.55,56 ATM activation of Chk2 kinase further amplifies the signal and expands the number of target proteins. Loss of ATM prevents the rapid accumulation of p53 after Ionizing radiation (IR) and abrogates p53-mediated cell cycle arrest response.54 The importance of the ATM-mediated response is not limited to direct DNA damage by irradiation. A range of physiologically relevant stresses, including chemotherapy, oncogene activation, oxidative stress, and heat shock, can also trigger ATM activation either directly or by inducing DNA damage.
The Role of p53 Phosphorylation in DNA Damage Response
Although not the focus of this review, it is important to note that phosphorylation of p53 is an integral part of the signal transduction pathway that leads to its activation. MDM2-p53 binding has been extensively studied as a target of regulation by DNA damage. Several studies showed that DNA double-strand breaks induce phosphorylation of p53 S15 by ATM or DNA-PK.57,58 ATM also activates Chk2, which in turn phosphorylates p53 on S20, which is part of the MDM2-binding site.59,60 However, mutation of multiple phosphorylation sites including S15 and S20 does not abrogate p53 stabilization after DNA damage in cell culture.61-63 Mouse models showed that blocking p53 phosphorylation on S18 and S23 (equivalents of S15 and S20 in human p53) partially reduced p53 accumulation after DNA damage and caused partial defects in apoptosis and tumor suppression.64 S23 single-site mutation also caused partial defects in p53 stabilization by gamma irradiation and increased the incidence of B-cell lymphoma.65 Single-site mutation of S18 had no significant effect on p53 stabilization or tumor suppression but causes poor activation of certain p53 target genes after DNA damage.66 These studies showed that p53 phosphorylation contributes to its stabilization but also implicate the presence of additional signaling mechanisms.
DNA Damage–Induced Phosphorylation of MDM2
Since p53 phosphorylation is not sufficient to mediate its stabilization after DNA damage, its E3 ligase MDM2 is a logical target for regulation. DNA damage has been found to induce MDM2 phosphorylation on serine 395 by ATM,67 on serine 407 by ATR,68 and on tyrosine 394 by c-Abl.69 Phosphomimetic mutations of these sites inhibit MDM2-mediated degradation or nuclear export of p53.67,70 Recent mass spectrometric analysis of MDM2 purified from irradiated cells revealed the presence of additional phosphorylation sites (S386, T419, S425, and S429). Two of these sites were confirmed to be ATM targets and were strongly induced by DNA damage.71 As expected for most phosphorylation sites, they cluster in a region of MDM2 that is disordered (Fig. 1A).72 The MDM2 phosphorylation sites appear to have significant functional redundancy in regulating p53 degradation. Phosphomimetic substitution of a single site can strongly inhibit p53 degradation.71 Substitution of all phosphorylation sites with alanine results in an MDM2 protein that continues to degrade p53 after DNA damage.
If DNA damage inhibits the E3 ligase activity of MDM2 through phosphorylation, the ubiquitination level of stabilized p53 is expected to be lower in damaged cells. The literature contains 2 conflicting conclusions in this regard using 2 different definitions of ubiquitination level. Some studies concluded that p53 ubiquitination level does not reduce or even increase after DNA damage. This is generally referring to the total level of ubiquitinated p53 by comparing identical amounts of cell extract, not taking into consideration that there are much higher levels of p53 in the damaged cells. However, the turnover rate of the entire p53 population is dictated by the ratio of Ub-p53/free p53, not the total level of Ub-p53 in the cell. In fact, when p53 loading was equalized to offset the stabilization effect, the fraction of p53 present in ubiquitinated form was significantly lower after DNA damage.73 This is consistent with the observation that MDM2 purified from damaged cells has reduced E3 activity in ubiquitinating p53 in vitro.73 These recent findings favor the conclusion that DNA damage inhibits p53 ubiquitination.
ATM Regulates MDM2 Dimerization
DNA damage signaling is highly efficient in regulating MDM2 function; even cell lines overexpressing MDM2 undergo p53 stabilization after irradiation. Therefore, ATM phosphorylation of MDM2 may be targeting structural features critical for its E3 ligase activity. Although the understanding of E3 ligases is still incomplete and rapidly evolving, certain common features have emerged as potential general targets for regulation. It appears that dimerization is often required for the activation of RING domain E3 ligases or the structurally similar U-box E3 ligase.74-78 E2-conjugating enzymes also frequently form dimers, both charged and uncharged with ubiquitin.79 A secondary noncovalent ubiquitin-binding site on UbcH5 also allows the formation of oligomers, consisting of UbcH5 covalently charged with ubiquitin.80 Dimerization and oligomerization by proteins in the ubiquitination pathway may be important for promoting the synthesis of polyubiquitin chains by increasing the local concentration of E2, providing a scaffold to access the end of a growing ubiquitin chain, and increasing the probability of ubiquitin-ubiquitin conjugation over ubiquitin-substrate conjugation.
The MDM2 C-terminal fragment containing the RING domain behaves as a high molecular weight oligomeric complex in gel filtration chromatography81,82 and can be cross-linked into dimers and oligomers by chemical cross-linking.73 MDM2 phosphorylation by ATM or phosphomimetic substitution inhibits RING domain oligomer formation during gel filtration and blocks RING domain dimerization in cross-linking assays.71,73 These findings suggest that ATM-mediated phosphorylation inhibits MDM2 RING domain homodimerization and oligomerization, preventing the formation of a scaffold for synthesis of polyubiquitin chains on p53. Consistent with an important role of MDM2 oligomerization, artificially promoting MDM2 oligomerization in vivo by FKBP fusion or in vitro by GST fusion significantly stimulates the E3 ligase function of MDM2, particularly favoring the synthesis of long ubiquitin chains on p53.48,73
How does phosphorylation near the RING domain inhibit dimerization? Currently, the atomic structure of only the RING domain of MDM2 in isolation has been determined by nuclear magnetic resonance (NMR) and crystallography. The ATM sites located in a region adjacent to the RING domain that is unstructured thus were removed in structural studies in order to improve the solubility of recombinant proteins.43,83 It appears that the RING domain itself is sufficient for dimerization and even oligomerization.81,83 Recent analysis showed that inclusion of adjacent sequences reduces RING domain dimerization efficiency, suggesting that the ATM sites are located in a region that negatively regulates RING domain dimerization.73 It is conceivable that after phosphorylation by ATM, the regulatory sequence adopts a conformation that can more efficiently conceal the dimerization surface of the RING domain through intramolecular binding, thus blocking dimerization.
Other Potential Effects of MDM2 Phosphorylation by ATM
In addition to direct regulation of RING domain dimerization, phosphorylation of MDM2 may also regulate interactions with other proteins. Several factors have been shown to cooperate with MDM2 in p53 polyubiquitination, such as p300/CBP and UBE4B.84,85 These proteins are thought to have E3 ligase activity of their own and interact with preformed ubiquitin conjugate on the substrate to further extend the ubiquitin chains. When recruited by MDM2, they may play a role in chain elongation on p53. The de-ubiquitinase HAUSP is an important regulator of p53 and MDM2 stability.86,87 MDM2 interaction with HAUSP is stimulated by DNA damage, which promotes p53 de-ubiquitination,88 possibly also contributing to p53 stabilization.
DNA damage has been shown in many studies to cause transient down-regulation of MDM2 levels. Recent findings suggest that data related to this phenomenon should be treated with caution. This is due to the fact that many studies used SMP14 or 2A10 to detect MDM2 because of the availability and sensitivity of these antibodies. However, the 2A10 epitope on MDM2 contains S395 and can be masked by ATM-mediated phosphorylation.67,89 SMP14 reactivity to MDM2 is also blocked by phosphorylation of an unknown site in its epitope. In fact, it has been shown in a previous study,90 and confirmed in recent experiments, that the MDM2 level does not undergo a significant decrease after irradiation when detected using other antibodies.91 In certain publications, the MDM2 level decreased after high-dose ultraviolet (UV) irradiation (>20 J/m2) and was interpreted as protein degradation, disregarding the fact that high-dose UV strongly down-regulates MDM2 mRNA.92 Therefore, whether MDM2 stability is regulated by phosphorylation remains to be demonstrated.
Phosphorylation of the MDM2 Acidic Domain Promotes p53 Degradation
While most of the phosphorylation sites stimulated by DNA damage are located near the C-terminal region of MDM2, the central acidic domain of MDM2 (220-300) also contains multiple serine phosphorylation sites that are constitutively modified in the absence of stress but down-regulated by DNA damage.93 GSK3 and CK1δ kinases have been shown to phosphorylate these sites.94-96 Down-regulation of GSK3 levels by DNA damage may explain the reduction in acidic domain phosphorylation.95 Alanine substitution of some of the phosphorylation sites in the acidic domain significantly inhibits p53 degradation without abrogating ubiquitination. A recent study suggests that the acidic domain phosphorylation sites regulate the ability of MDM2 to interaction with the 19S proteasome regulatory subunit, thus regulating MDM2 delivery of ubiquitinated p53 for degradation by the proteasome.97
C-Terminal ATM Sites Regulate the MDM2 Acidic Domain
Deletion analysis showed that the MDM2 acidic domain is important for ubiquitination and degradation of p53.23,24 Acidic domain interaction with ARF and ribosomal proteins also inhibits p53 ubiquitination.53 The mechanism by which the acidic domain participates in p53 ubiquitination remains poorly defined. Several groups showed that the MDM2 acidic domain binds to the p53 core domain.31,32,98 The binding affinity between the acidic domain peptides and the p53 core domain (Kd = 1,000 nM) is significantly weaker than the canonical N-terminal interaction (Kd = 130 nM) but may be important for stabilizing the complex and proper orientation of p53 for ubiquitination reactions to occur.32 The acidic domain–p53 core interaction is also responsible for inducing conformational change in p53, which may help expose the target lysine residues in the core.34,35 As expected, CK1δ phosphorylation of the MDM2 acidic domain stimulates binding to the p53 core.99 However, recent experiments showed that the acidic domain of MDM2 is also regulated by the C-terminal ATM sites. The weak binding between the acidic domain and the p53 core is inhibited by ATM after DNA damage, and the ability of MDM2 to induce p53 conformational change is also enhanced by ATM site mutations.73 Therefore, the ATM sites control the conformation and functions of multiple domains on MDM2.
MDMX Phosphorylation by ATM, Chk2, and c-Abl
MDM2 undergoes self-ubiquitination and has a very short half-life in cell culture (15-30 minutes). In contrast, MDMX has a significantly longer half-life (>3 hours). The MDMX level is controlled by MDM2-mediated ubiquitination in a stress-dependent fashion.42,100,101 A fraction of MDMX from cells with DNA damage has delayed migration consistent with phosphorylation in an ATM-dependent fashion.102 Candidate approach and mass spectrometry analysis have identified several phosphorylation sites near the C-terminal RING domain (S342, S367, S403).102,103 S342 and S367 were phosphorylated by Chk2, whereas S403 was modified by ATM (Fig. 1B). UV irradiation also induces S367 phosphorylation through activation of Chk1 kinase.104 In vivo metabolic labeling experiments showed that S367 phosphorylation is the most prominent modification site.105 This is consistent with the functional significance of S367 on MDMX ubiquitination: S367A substitution significantly reduced ubiquitination by MDM2, whereas S342A and S403A substitutions have negligible effects.102 In addition to the C-terminal phosphorylation sites, it has been reported that the p53-binding domain of MDMX is phosphorylated by c-Abl on Tyr-99 and Tyr-55. Phosphorylation of Tyr-99 interferes with p53 binding, presumably facilitating the activation of p53.106
MDMX Phosphorylation Controls Subcellular Localization
MDM2 has both Nuclear Localization Signal (NLS) (179-185, RQRKRHK) and Nuclear Export Signal (NES) (197-199, LSFDESLAL) sequences and undergoes nuclear-cytoplasmic shuttling. Akt1-mediated phosphorylation (S166, S186, S188) near the NLS has been shown to promote MDM2 nuclear translocation and enhance its ability to inactivate p53 in response to growth factor signaling.107-110 In contrast, MDMX appears to lack the NLS and NES signals in the corresponding locations, and phosphorylation in this region has not been reported. In the absence of stress, MDMX is predominantly cytoplasmic but shows strong nuclear accumulation after DNA damage.111 Although nuclear proteins such as p53 and MDM2 can bind to MDMX and promote MDMX nuclear import, DNA damage–induced MDMX nuclear import also occurs in p53- and MDM2-null cells.111,112 It has been shown that phosphorylated S367 becomes a high-affinity docking site for 14-3-3, suggesting that 14-3-3 binding may promote conformational change in the RING domain to expose a cryptic NLS.104,105,113 Although MDMX nuclear translocation is a long-established phenomenon, the biological function of this shift remains speculative. It may serve to accelerate MDMX degradation because most of MDM2 is in the nucleus. Alternatively, it may be an active mechanism to suppress p53 activity in the nucleus.
MDMX Phosphorylation Promotes Ubiquitination by MDM2
A validated function of MDMX phosphorylation near the RING domain is to regulate degradation by MDM2. MDMX phosphorylation increases its binding affinity for MDM2, independent of nuclear import. In a cell free–binding reaction, GST-MDM2 binds to phosphorylated MDMX more efficiently than nonphosphorylated MDMX.102 Transfection/Immunoprecipitation (IP) assay also showed that MDM2-MDMX co-precipitation efficiency is increased after DNA damage.114,115 Therefore, ATM-mediated phosphorylation of MDM2 near the RING domain inhibits homodimerization to suppress p53 ubiquitination, whereas Chk2-mediated phosphorylation near the MDMX RING domain promotes heterodimerization with MDM2 and enhances MDMX ubiquitination. Another effect of MDMX phosphorylation is the inhibition of MDMX-HAUSP binding, which may also contribute to increased MDMX ubiquitination.116,117 Dephosphorylation of MDMX by the WIP1 phosphatase also results in higher HAUSP binding and MDMX stabilization.114
The biological relevance of MDMX phosphorylation sites has been verified in mouse knockin experiments. Alanine substitution of all 3 phosphorylation sites near the MDMX RING domain causes stabilization and resistance to DNA damage–mediated degradation in the MEFs and tissue of the mutant mice.118 Furthermore, the animals have reduced p53 activation after irradiation and increased tumor incidence when introduced into a Myc transgenic background. These findings nicely mirror the cell culture results obtained mostly by overexpression and knockdown assays. Furthermore, p53 accumulation after DNA damage appears to be partially deficient in the 3A mice.118 This suggests that MDMX phosphorylation may play a role in regulating p53 stability in vivo. This is consistent with the increased MDM2-MDMX heterodimer formation after DNA damage114,115 and p53 accumulation in MDMX RING domain mutant mice.51
MDMX Phosphorylation by CK1α
MDM2 typically co-purifies with large amounts of ribosomal proteins (mainly L5, L11, and L23). In contrast, MDMX co-purifies mainly with 14-3-3 and near stoichiometric amounts of casein kinase 1 alpha (CK1α). CK1α interacts with the central region of MDMX including the acidic domain and zinc finger (150-350). S289 of MDMX has been identified as the major phosphorylation site by CK1α.119 CK1α expression stimulates the binding between MDMX and p53 and leads to inhibition of p53 activity. As expected, pharmacological inhibition or knockdown of CK1α leads to p53 activation and cooperates with DNA-damaging drugs to activate p53 in culture.119
Is endogenous CK1α important for regulating p53? CK1α is an abundant serine/threonine kinase that regulates multiple cellular pathways, such as Wnt/β-catenin signaling and circadian rhythm.120 Circumstantial but supportive evidence has recently been reported in a mouse model with tissue-specific conditional knockout of CK1α in the intestine.121 As expected from its role in the Wnt pathway, CK1α knockout leads to β-catenin stabilization and increased cell proliferation. CK1α knockout also leads to significant increases of p53 and p21 levels in vivo, which was interpreted as a secondary result of the proliferative stress from β-catenin stabilization. However, the data are also consistent with a role of CK1α through MDMX interaction. Mutation of the CK1α phosphorylation site on MDMX in mouse models will be needed to test the direct mechanism from CK1α.
Several questions need to be addressed in future studies. 1) How does CK1α binding and phosphorylation of the central domain of MDMX promote p53 binding? This effect implicates an interdomain communication between different parts of MDMX through conformational change or direct binding. 2) If the ATM sites on the MDM2 C-terminus can regulate the conformation and function of the acidic domain, it is possible that the ATM/Chk2 sites on the MDMX C-terminus also regulate acidic domain–CK1α binding. 3) The near stoichiometric binding between MDMX and CK1α is unusual for a kinase substrate relationship. This suggests that MDMX may also recruit CK1α to modify other proteins or regulate CK1α function.
Phosphorylation-Independent Mechanisms of MDMX Degradation
Interestingly, other types of stress that activate p53 without triggering DNA damage signaling also promote MDMX degradation. For example, ribosomal stress resulting from inhibition of rRNA transcription promotes MDMX degradation through L11-MDM2 interaction40; oncogenic stress promotes MDMX degradation through ARF expression.122 L11 binds to the zinc finger of MDM2 to promote MDMX ubiquitination, although detailed mechanisms remain undetermined. ARF also binds to MDM2 and stimulates ubiquitination of MDMX. A recent study showed that ARF promotes MDM2-MDMX heterodimer formation by mediating a second site interaction between the MDM2 and MDMX central domains.122 This highlights the importance of MDM2 and MDMX homodimerization/heterodimerization as a common target for different stress signals, producing the same outcome of p53 stabilization and MDMX degradation.
Potential Intramolecular Interactions in MDM2 and MDMX
There is growing evidence that different domains of MDM2 and MDMX are functionally and allosterically connected. It has been suggested that p53 binding to the MDM2 N-terminus stimulates acidic domain–p53 core binding. This second site interaction is critical for p53 ubiquitination.123 The ATM sites on the MDM2 C-terminus control both RING domain dimerization and acidic domain–p53 core interaction.73 These findings suggest that the N-terminal p53-binding domain and C-terminal RING domain may communicate directly or indirectly. In fact, it has been shown that a point mutation in the MDM2 RING domain can cause conformational change in the acidic domain, which in turn increases p53 binding by the N-terminal domain.124 There is also biochemical evidence that a C-terminal fragment of MDM2 binds weakly to the acidic domain,125 suggesting the presence of an intramolecular interaction. MDMX also presents a similar picture: the ATM/Chk2 sites regulate RING domain heterodimerization and, by analogy to MDM2, may also regulate acidic domain function. CK1α binding to the acidic domain of MDMX promotes p53 binding, suggesting an allosteric connection to the N-terminal p53-binding domain.
Although static atomic structures of full-length MDM2 and MDMX may not be obtainable because some parts of these proteins lack rigid structures, it is likely that the flexible peptide sequences interact frequently with the stably folded domains and influence their conformation and function. In such a model, ligand binding or phosphorylation of one region may alter the intramolecular interaction between different domains. Such a mechanism plays a prominent role in the regulation of pRb-E2F1 binding by Cdk-mediated phosphorylation.126 In the case of MDM2 and MDMX, intramolecular domain coupling may provide the structural basis for different signaling mechanisms to achieve an identical biological outcome, which is inhibiting p53 ubiquitination and promoting MDMX ubiquitination by MDM2.
Footnotes
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by grants from the National Institutes of Health (CA109636 and CA141244).
References
- 1. Vousden KH, Lane DP. p53 in health and disease. Nat Rev Mol Cell Biol. 2007;8:275-83 [DOI] [PubMed] [Google Scholar]
- 2. Kubbutat MH, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature. 1997;387:299-303 [DOI] [PubMed] [Google Scholar]
- 3. Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296-9 [DOI] [PubMed] [Google Scholar]
- 4. Leng RP, Lin Y, Ma W, et al. Pirh2, a p53-induced ubiquitin-protein ligase, promotes p53 degradation. Cell. 2003;112:779-91 [DOI] [PubMed] [Google Scholar]
- 5. Dornan D, Wertz I, Shimizu H, et al. The ubiquitin ligase COP1 is a critical negative regulator of p53. Nature. 2004;429:86-92 [DOI] [PubMed] [Google Scholar]
- 6. Montes de Oca Luna R, Wagner DS, Lozano G. Rescue of early embryonic lethality in Mdm2-deficient mice by deletion of p53. Nature. 1995;378:203-6 [DOI] [PubMed] [Google Scholar]
- 7. Grier JD, Xiong S, Elizondo-Fraire AC, Parant JM, Lozano G. Tissue-specific differences of p53 inhibition by Mdm2 and Mdm4. Mol Cell Biol. 2006;26:192-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jones SN, Roe AE, Donehower LA, Bradley A. Rescue of embryonic lethality in Mdm2-deficient mice by absence of p53. Nature. 1995;378:206-8 [DOI] [PubMed] [Google Scholar]
- 9. Vassilev LT, Vu BT, Graves B, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844-8 [DOI] [PubMed] [Google Scholar]
- 10. Shangary S, Qin D, McEachern D, et al. Temporal activation of p53 by a specific MDM2 inhibitor is selectively toxic to tumors and leads to complete tumor growth inhibition. Proc Natl Acad Sci U S A. 2008;105:3933-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shvarts A, Steegenga WT, Riteco N, et al. MDMX: a novel p53-binding protein with some functional properties of MDM2. EMBO J. 1996;15:5349-57 [PMC free article] [PubMed] [Google Scholar]
- 12. Finch RA, Donoviel DB, Potter D, et al. Mdmx is a negative regulator of p53 activity in vivo. Cancer Res. 2002;62:3221-5 [PubMed] [Google Scholar]
- 13. Migliorini D, Lazzerini Denchi E, Danovi D, et al. Mdm4 (Mdmx) regulates p53-induced growth arrest and neuronal cell death during early embryonic mouse development. Mol Cell Biol. 2002;22:5527-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Parant JM, Reinke V, Mims B, Lozano G. Organization, expression, and localization of the murine Mdmx gene and pseudogene. Gene. 2001;270:277-83 [DOI] [PubMed] [Google Scholar]
- 15. Xiong S, Van Pelt CS, Elizondo-Fraire AC, Liu G, Lozano G. Synergistic roles of Mdm2 and Mdm4 for p53 inhibition in central nervous system development. Proc Natl Acad Sci U S A. 2006;103:3226-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Maetens M, Doumont G, Clercq SD, et al. Distinct roles of Mdm2 and Mdm4 in red cell production. Blood. 2007;109:2630-3 [DOI] [PubMed] [Google Scholar]
- 17. Steinman HA, Hoover KM, Keeler ML, Sands AT, Jones SN. Rescue of Mdm4-deficient mice by Mdm2 reveals functional overlap of Mdm2 and Mdm4 in development. Oncogene. 2005;24:7935-40 [DOI] [PubMed] [Google Scholar]
- 18. Li B, Cheng Q, Li Z, Chen J. p53 inactivation by MDM2 and MDMX negative feedback loops in testicular germ cell tumors. Cell Cycle. 2010;9:1411-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Phillips A, Teunisse A, Lam S, et al. HDMX-L is expressed from a functional p53-responsive promoter in the first intron of the HDMX gene and participates in an autoregulatory feedback loop to control p53 activity. J Biol Chem. 2010;285:29111-27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fang S, Jensen JP, Ludwig RL, Vousden KH, Weissman AM. Mdm2 is a RING finger-dependent ubiquitin protein ligase for itself and p53. J Biol Chem. 2000;275:8945-51 [DOI] [PubMed] [Google Scholar]
- 21. Honda R, Tanaka H, Yasuda H. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 1997;420:25-7 [DOI] [PubMed] [Google Scholar]
- 22. Sdek P, Ying H, Chang DL, et al. MDM2 promotes proteasome-dependent ubiquitin-independent degradation of retinoblastoma protein. Mol Cell. 2005;20:699-708 [DOI] [PubMed] [Google Scholar]
- 23. Kawai H, Wiederschain D, Yuan ZM. Critical contribution of the MDM2 acidic domain to p53 ubiquitination. Mol Cell Biol. 2003;23:4939-47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Meulmeester E, Frenk R, Stad R, et al. Critical role for a central part of Mdm2 in the ubiquitylation of p53. Mol Cell Biol. 2003;23:4929-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang C, Ivanov A, Chen L, et al. MDM2 interaction with nuclear corepressor KAP1 contributes to p53 inactivation. EMBO J. 2005;24:3279-90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sui G, Affar el B, Shi Y, et al. Yin Yang 1 is a negative regulator of p53. Cell. 2004;117:859-72 [DOI] [PubMed] [Google Scholar]
- 27. Chen L, Li Z, Zwolinska AK, et al. MDM2 recruitment of lysine methyltransferases regulates p53 transcriptional output. EMBO J. 2010;29:2538-52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hu M, Gu L, Li M, Jeffrey PD, Gu W, Shi Y. Structural basis of competitive recognition of p53 and MDM2 by HAUSP/USP7: implications for the regulation of the p53-MDM2 pathway. PLoS Biol. 2006;4:e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhang Y, Wolf GW, Bhat K, et al. Ribosomal protein L11 negatively regulates oncoprotein MDM2 and mediates a p53-dependent ribosomal-stress checkpoint pathway. Mol Cell Biol. 2003; 23:8902-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Midgley CA, Desterro JM, Saville MK, et al. An N-terminal p14ARF peptide blocks Mdm2-dependent ubiquitination in vitro and can activate p53 in vivo. Oncogene. 2000;19:2312-23 [DOI] [PubMed] [Google Scholar]
- 31. Ma J, Martin JD, Zhang H, et al. A second p53 binding site in the central domain of Mdm2 is essential for p53 ubiquitination. Biochemistry. 2006;45:9238-45 [DOI] [PubMed] [Google Scholar]
- 32. Yu GW, Rudiger S, Veprintsev D, Freund S, Fernandez-Fernandez MR, Fersht AR. The central region of HDM2 provides a second binding site for p53. Proc Natl Acad Sci U S A. 2006;103:1227-32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Burch LR, Midgley CA, Currie RA, Lane DP, Hupp TR. Mdm2 binding to a conformationally sensitive domain on p53 can be modulated by RNA. FEBS Lett. 2000;472:93-8 [DOI] [PubMed] [Google Scholar]
- 34. Cross B, Chen L, Cheng Q, Li B, Yuan ZM, Chen J. Inhibition of p53 DNA binding function by the MDM2 protein acidic domain. J Biol Chem. 2011;286:16018-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sasaki M, Nie L, Maki CG. MDM2 binding induces a conformational change in p53 that is opposed by heat-shock protein 90 and precedes p53 proteasomal degradation. J Biol Chem. 2007;282:14626-34 [DOI] [PubMed] [Google Scholar]
- 36. Dunker AK, Silman I, Uversky VN, Sussman JL. Function and structure of inherently disordered proteins. Curr Opin Struct Biol. 2008;18:756-64 [DOI] [PubMed] [Google Scholar]
- 37. Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol. 2005;6:197-208 [DOI] [PubMed] [Google Scholar]
- 38. Badciong JC, Haas AL. MdmX is a RING finger ubiquitin ligase capable of synergistically enhancing Mdm2 ubiquitination. J Biol Chem. 2002;277:49668-75 [DOI] [PubMed] [Google Scholar]
- 39. Francoz S, Froment P, Bogaerts S, et al. Mdm4 and Mdm2 cooperate to inhibit p53 activity in proliferating and quiescent cells in vivo. Proc Natl Acad Sci U S A. 2006;103:3232-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gilkes DM, Chen L, Chen J. MDMX regulation of p53 response to ribosomal stress. EMBO J. 2006;25:5614-25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Toledo F, Krummel KA, Lee CJ, et al. A mouse p53 mutant lacking the proline-rich domain rescues Mdm4 deficiency and provides insight into the Mdm2-Mdm4-p53 regulatory network. Cancer Cell. 2006;9:273-85 [DOI] [PubMed] [Google Scholar]
- 42. Pan Y, Chen J. MDM2 promotes ubiquitination and degradation of MDMX. Mol Cell Biol. 2003;23:5113-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Linke K, Mace PD, Smith CA, Vaux DL, Silke J, Day CL. Structure of the MDM2/MDMX RING domain heterodimer reveals dimerization is required for their ubiquitylation in trans. Cell Death Differ. 2008;15:841-8 [DOI] [PubMed] [Google Scholar]
- 44. Uldrijan S, Pannekoek WJ, Vousden KH. An essential function of the extreme C-terminus of MDM2 can be provided by MDMX. EMBO J. 2007;26:102-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tanimura S, Ohtsuka S, Mitsui K, Shirouzu K, Yoshimura A, Ohtsubo M. MDM2 interacts with MDMX through their RING finger domains. FEBS Lett. 1999;447:5-9 [DOI] [PubMed] [Google Scholar]
- 46. Linares LK, Hengstermann A, Ciechanover A, Muller S, Scheffner M. HdmX stimulates Hdm2-mediated ubiquitination and degradation of p53. Proc Natl Acad Sci U S A. 2003;100:12009-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gu J, Kawai H, Nie L, et al. Mutual dependence of MDM2 and MDMX in their functional inactivation of p53. J Biol Chem. 2002;277:19251-4 [DOI] [PubMed] [Google Scholar]
- 48. Wang X, Wang J, Jiang X. MdmX protein is essential for Mdm2 protein-mediated p53 polyubiquitination. J Biol Chem. 2011;286:23725-34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Okamoto K, Taya Y, Nakagama H. Mdmx enhances p53 ubiquitination by altering the substrate preference of the Mdm2 ubiquitin ligase. FEBS Lett. 2009;583:2710-4 [DOI] [PubMed] [Google Scholar]
- 50. Kawai H, Lopez-Pajares V, Kim MM, Wiederschain D, Yuan ZM. RING domain-mediated interaction is a requirement for MDM2’s E3 ligase activity. Cancer Res. 2007;67:6026-30 [DOI] [PubMed] [Google Scholar]
- 51. Huang L, Yan Z, Liao X, et al. The p53 inhibitors MDM2/MDMX complex is required for control of p53 activity in vivo. Proc Natl Acad Sci U S A. 2011;108:12001-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sherr CJ. Divorcing ARF and p53: an unsettled case. Nat Rev Cancer. 2006;6:663-73 [DOI] [PubMed] [Google Scholar]
- 53. Zhang Y, Lu H. Signaling to p53: ribosomal proteins find their way. Cancer Cell. 2009;16:369-77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kastan MB, Zhan Q, el-Deiry WS, et al. A mammalian cell cycle checkpoint pathway utilizing p53 and GADD45 is defective in ataxia-telangiectasia. Cell. 1992;71:587-97 [DOI] [PubMed] [Google Scholar]
- 55. Shiloh Y. ATM and related protein kinases: safeguarding genome integrity. Nat Rev Cancer. 2003;3:155-68 [DOI] [PubMed] [Google Scholar]
- 56. Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell. 2010;40:179-204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Banin S, Moyal L, Shieh S, et al. Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science. 1998;281:1674-7 [DOI] [PubMed] [Google Scholar]
- 58. Shieh SY, Ikeda M, Taya Y, Prives C. DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell. 1997;91:325-34 [DOI] [PubMed] [Google Scholar]
- 59. Chehab NH, Malikzay A, Appel M, Halazonetis TD. Chk2/hCds1 functions as a DNA damage checkpoint in G(1) by stabilizing p53. Genes Dev. 2000;14:278-88 [PMC free article] [PubMed] [Google Scholar]
- 60. Shieh SY, Ahn J, Tamai K, Taya Y, Prives C. The human homologs of checkpoint kinases Chk1 and Cds1 (Chk2) phosphorylate p53 at multiple DNA damage-inducible sites. Genes Dev. 2000;14:289-300 [PMC free article] [PubMed] [Google Scholar]
- 61. Ashcroft M, Kubbutat MH, Vousden KH. Regulation of p53 function and stability by phosphorylation. Mol Cell Biol. 1999;19:1751-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Blattner C, Tobiasch E, Litfen M, Rahmsdorf HJ, Herrlich P. DNA damage induced p53 stabilization: no indication for an involvement of p53 phosphorylation. Oncogene. 1999;18:1723-32 [DOI] [PubMed] [Google Scholar]
- 63. Dumaz N, Meek DW. Serine15 phosphorylation stimulates p53 transactivation but does not directly influence interaction with HDM2. EMBO J. 1999;18:7002-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Chao C, Herr D, Chun J, Xu Y. Ser18 and 23 phosphorylation is required for p53-dependent apoptosis and tumor suppression. EMBO J. 2006;25:2615-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. MacPherson D, Kim J, Kim T, et al. Defective apoptosis and B-cell lymphomas in mice with p53 point mutation at Ser 23. EMBO J. 2004;23:3689-99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Chao C, Hergenhahn M, Kaeser MD, et al. Cell type- and promoter-specific roles of Ser18 phosphorylation in regulating p53 responses. J Biol Chem. 2003;278:41028-33 [DOI] [PubMed] [Google Scholar]
- 67. Maya R, Balass M, Kim ST, et al. ATM-dependent phosphorylation of Mdm2 on serine 395: role in p53 activation by DNA damage. Genes Dev. 2001;15:1067-77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Shinozaki T, Nota A, Taya Y, Okamoto K. Functional role of Mdm2 phosphorylation by ATR in attenuation of p53 nuclear export. Oncogene. 2003;22:8870-80 [DOI] [PubMed] [Google Scholar]
- 69. Sionov RV, Coen S, Goldberg Z, et al. c-Abl regulates p53 levels under normal and stress conditions by preventing its nuclear export and ubiquitination. Mol Cell Biol. 2001;21:5869-78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Goldberg Z, Vogt Sionov R, Berger M, et al. Tyrosine phosphorylation of Mdm2 by c-Abl: implications for p53 regulation. EMBO J. 2002;21:3715-27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Cheng Q, Chen L, Li Z, Lane WS, Chen J. ATM activates p53 by regulating MDM2 oligomerization and E3 processivity. EMBO J. 2009;28: 3857-67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Iakoucheva LM, Radivojac P, Brown CJ, et al. The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res. 2004;32:1037-49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Cheng Q, Cross B, Li B, Chen L, Li Z, Chen J. Regulation of MDM2 E3 ligase activity by phosphorylation after DNA damage. Mol Cell Biol. 2011;31:4951-63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Dueber EC, Schoeffler AJ, Lingel A, et al. Antagonists induce a conformational change in cIAP1 that promotes autoubiquitination. Science. 2011;334:376-80 [DOI] [PubMed] [Google Scholar]
- 75. Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399-434 [DOI] [PubMed] [Google Scholar]
- 76. Nikolay R, Wiederkehr T, Rist W, Kramer G, Mayer MP, Bukau B. Dimerization of the human E3 ligase CHIP via a coiled-coil domain is essential for its activity. J Biol Chem. 2004;279:2673-8 [DOI] [PubMed] [Google Scholar]
- 77. Mace PD, Linke K, Feltham R, et al. Structures of the cIAP2 RING domain reveal conformational changes associated with ubiquitin-conjugating enzyme (E2) recruitment. J Biol Chem. 2008;283:31633-40 [DOI] [PubMed] [Google Scholar]
- 78. Vander Kooi CW, Ohi MD, Rosenberg JA, et al. The Prp19 U-box crystal structure suggests a common dimeric architecture for a class of oligomeric E3 ubiquitin ligases. Biochemistry. 2006;45:121-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. David Y, Ziv T, Admon A, Navon A. The E2 ubiquitin-conjugating enzymes direct polyubiquitination to preferred lysines. J Biol Chem. 2010;285:8595-604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Brzovic PS, Lissounov A, Christensen DE, Hoyt DW, Klevit RE. A UbcH5/ubiquitin noncovalent complex is required for processive BRCA1-directed ubiquitination. Mol Cell. 2006;21:873-80 [DOI] [PubMed] [Google Scholar]
- 81. Poyurovsky MV, Priest C, Kentsis A, et al. The Mdm2 RING domain C-terminus is required for supramolecular assembly and ubiquitin ligase activity. EMBO J. 2007;26:90-101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Cheng Q, Chen L, Li Z, Lane WS, Chen J. ATM activates p53 by regulating MDM2 oligomerization and E3 processivity. EMBO J. 2009;28: 3857-67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Kostic M, Matt T, Martinez-Yamout MA, Dyson HJ, Wright PE. Solution structure of the Hdm2 C2H2C4 RING, a domain critical for ubiquitination of p53. J Mol Biol. 2006;363:433-50 [DOI] [PubMed] [Google Scholar]
- 84. Shi D, Pop MS, Kulikov R, Love IM, Kung AL, Grossman SR. CBP and p300 are cytoplasmic E4 polyubiquitin ligases for p53. Proc Natl Acad Sci U S A. 2009;106:16275-80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Wu H, Pomeroy SL, Ferreira M, et al. UBE4B promotes Hdm2-mediated degradation of the tumor suppressor p53. Nat Med. 2011;17:347-55 [DOI] [PubMed] [Google Scholar]
- 86. Li M, Brooks CL, Kon N, Gu W. A dynamic role of HAUSP in the p53-Mdm2 pathway. Mol Cell. 2004;13:879-86 [DOI] [PubMed] [Google Scholar]
- 87. Cummins JM, Rago C, Kohli M, Kinzler KW, Lengauer C, Vogelstein B. Tumour suppression: disruption of HAUSP gene stabilizes p53. Nature. 2004;428:1 p following 486. [DOI] [PubMed] [Google Scholar]
- 88. Tang J, Qu LK, Zhang J, et al. Critical role for Daxx in regulating Mdm2. Nat Cell Biol. 2006;8:855-62 [DOI] [PubMed] [Google Scholar]
- 89. Maya R, Oren M. Unmasking of phosphorylation-sensitive epitopes on p53 and Mdm2 by a simple Western-phosphatase procedure. Oncogene. 2000;19:3213-5 [DOI] [PubMed] [Google Scholar]
- 90. de Toledo SM, Azzam EI, Dahlberg WK, Gooding TB, Little JB. ATM complexes with HDM2 and promotes its rapid phosphorylation in a p53-independent manner in normal and tumor human cells exposed to ionizing radiation. Oncogene. 2000;19:6185-93 [DOI] [PubMed] [Google Scholar]
- 91. Cheng Q, Chen J. The phenotype of MDM2 auto-degradation after DNA damage is due to epitope masking by phosphorylation. Cell Cycle. 2011;10:1162-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Wu L, Levine AJ. Differential regulation of the p21/WAF-1 and Mdm2 genes after high-dose UV irradiation: p53-dependent and p53-independent regulation of the Mdm2 gene. Mol Med. 1997;3:441-51 [PMC free article] [PubMed] [Google Scholar]
- 93. Blattner C, Hay T, Meek DW, Lane DP. Hypophosphorylation of Mdm2 augments p53 stability. Mol Cell Biol. 2002;22:6170-82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Winter M, Milne D, Dias S, et al. Protein kinase CK1delta phosphorylates key sites in the acidic domain of murine double-minute clone 2 protein (MDM2) that regulate p53 turnover. Biochemistry. 2004;43:16356-64 [DOI] [PubMed] [Google Scholar]
- 95. Kulikov R, Boehme KA, Blattner C. Glycogen synthase kinase 3-dependent phosphorylation of Mdm2 regulates p53 abundance. Mol Cell Biol. 2005;25:7170-80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Inuzuka H, Tseng A, Gao D, et al. Phosphorylation by casein kinase I promotes the turnover of the Mdm2 oncoprotein via the SCF(beta-TRCP) ubiquitin ligase. Cancer Cell. 2010;18:147-59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Kulikov R, Letienne J, Kaur M, Grossman SR, Arts J, Blattner C. Mdm2 facilitates the association of p53 with the proteasome. Proc Natl Acad Sci U S A. 2010;107:10038-43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Shimizu H, Burch LR, Smith AJ, et al. The conformationally flexible S9-S10 linker region in the core domain of p53 contains a novel MDM2 binding site whose mutation increases ubiquitination of p53 in vivo. J Biol Chem. 2002;277:28446-58 [DOI] [PubMed] [Google Scholar]
- 99. Kulikov R, Winter M, Blattner C. Binding of p53 to the central domain of Mdm2 is regulated by phosphorylation. J Biol Chem. 2006;281: 28575-83 [DOI] [PubMed] [Google Scholar]
- 100. de Graaf P, Little NA, Ramos YF, Meulmeester E, Letteboer SJ, Jochemsen AG. Hdmx protein stability is regulated by the ubiquitin ligase activity of Mdm2. J Biol Chem. 2003;278:38315-24 [DOI] [PubMed] [Google Scholar]
- 101. Kawai H, Wiederschain D, Kitao H, Stuart J, Tsai KK, Yuan ZM. DNA damage-induced MDMX degradation is mediated by MDM2. J Biol Chem. 2003;278:45946-53 [DOI] [PubMed] [Google Scholar]
- 102. Chen L, Gilkes DM, Pan Y, Lane WS, Chen J. ATM and Chk2-dependent phosphorylation of MDMX contribute to p53 activation after DNA damage. EMBO J. 2005;24:3411-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Pereg Y, Shkedy D, de Graaf P, et al. Phosphorylation of Hdmx mediates its Hdm2- and ATM-dependent degradation in response to DNA damage. Proc Natl Acad Sci U S A. 2005;102:5056-61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Jin Y, Dai MS, Lu SZ, et al. 14-3-3gamma binds to MDMX that is phosphorylated by UV-activated Chk1, resulting in p53 activation. EMBO J. 2006;25:1207-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. LeBron C, Chen L, Gilkes DM, Chen J. Regulation of MDMX nuclear import and degradation by Chk2 and 14-3-3. EMBO J. 2006;25:1196-206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Zuckerman V, Lenos K, Popowicz GM, et al. c-Abl phosphorylates Hdmx and regulates its interaction with p53. J Biol Chem. 2009; 284:4031-9 [DOI] [PubMed] [Google Scholar]
- 107. Mayo LD, Donner DB. A phosphatidylinositol 3-kinase/Akt pathway promotes translocation of Mdm2 from the cytoplasm to the nucleus. Proc Natl Acad Sci U S A. 2001;98:11598-603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Zhou BP, Liao Y, Xia W, Zou Y, Spohn B, Hung MC. HER-2/neu induces p53 ubiquitination via Akt-mediated MDM2 phosphorylation. Nat Cell Biol. 2001;3:973-82 [DOI] [PubMed] [Google Scholar]
- 109. Milne D, Kampanis P, Nicol S, et al. A novel site of AKT-mediated phosphorylation in the human MDM2 onco-protein. FEBS Lett. 2004;577: 270-6 [DOI] [PubMed] [Google Scholar]
- 110. Ashcroft M, Ludwig RL, Woods DB, et al. Phosphorylation of HDM2 by Akt. Oncogene. 2002;21:1955-62 [DOI] [PubMed] [Google Scholar]
- 111. Li C, Chen L, Chen J. DNA damage induces MDMX nuclear translocation by p53-dependent and -independent mechanisms. Mol Cell Biol. 2002;22:7562-71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Migliorini D, Danovi D, Colombo E, Carbone R, Pelicci PG, Marine JC. Hdmx recruitment into the nucleus by Hdm2 is essential for its ability to regulate p53 stability and transactivation. J Biol Chem. 2002;277:7318-23 [DOI] [PubMed] [Google Scholar]
- 113. Okamoto K, Kashima K, Pereg Y, et al. DNA damage-induced phosphorylation of MdmX at serine 367 activates p53 by targeting MdmX for Mdm2-dependent degradation. Mol Cell Biol. 2005;25:9608-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Zhang X, Lin L, Guo H, et al. Phosphorylation and degradation of MdmX is inhibited by Wip1 phosphatase in the DNA damage response. Cancer Res. 2009;69:7960-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Waning DL, Lehman JA, Batuello CN, Mayo LD. c-Abl phosphorylation of Mdm2 facilitates Mdm2-Mdmx complex formation. J Biol Chem. 2011;286:216-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Meulmeester E, Maurice MM, Boutell C, et al. Loss of HAUSP-mediated deubiquitination contributes to DNA damage-induced destabilization of Hdmx and Hdm2. Mol Cell. 2005;18: 565-76 [DOI] [PubMed] [Google Scholar]
- 117. Pereg Y, Lam S, Teunisse A, et al. Differential roles of ATM- and Chk2-mediated phosphorylations of Hdmx in response to DNA damage. Mol Cell Biol. 2006;26:6819-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Wang YV, Leblanc M, Wade M, Jochemsen AG, Wahl GM. Increased radioresistance and accelerated B cell lymphomas in mice with Mdmx mutations that prevent modifications by DNA-damage-activated kinases. Cancer Cell. 2009;16:33-43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Chen L, Li C, Pan Y, Chen J. Regulation of p53-MDMX interaction by casein kinase 1 alpha. Mol Cell Biol. 2005;25:6509-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Knippschild U, Gocht A, Wolff S, Huber N, Lohler J, Stoter M. The casein kinase 1 family: participation in multiple cellular processes in eukaryotes. Cell Signal. 2005;17:675-89 [DOI] [PubMed] [Google Scholar]
- 121. Elyada E, Pribluda A, Goldstein RE, et al. CKIalpha ablation highlights a critical role for p53 in invasiveness control. Nature. 2011;470:409-13 [DOI] [PubMed] [Google Scholar]
- 122. Li X, Gilkes D, Li B, et al. Abnormal MDMX degradation in tumor cells due to ARF deficiency. Oncogene. Epub 2011. November 28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Wallace M, Worrall E, Pettersson S, Hupp TR, Ball KL. Dual-site regulation of MDM2 E3-ubiquitin ligase activity. Mol Cell. 2006;23: 251-63 [DOI] [PubMed] [Google Scholar]
- 124. Wawrzynow B, Pettersson S, Zylicz A, et al. A function for the RING finger domain in the allosteric control of MDM2 conformation and activity. J Biol Chem. 2009;284:11517-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Dang J, Kuo ML, Eischen CM, Stepanova L, Sherr CJ, Roussel MF. The RING domain of Mdm2 can inhibit cell proliferation. Cancer Res. 2002;62:1222-30 [PubMed] [Google Scholar]
- 126. Burke JR, Deshong AJ, Pelton JG, Rubin SM. Phosphorylation-induced conformational changes in the retinoblastoma protein inhibit E2F transactivation domain binding. J Biol Chem. 2010;285:16286-93 [DOI] [PMC free article] [PubMed] [Google Scholar]