Abstract
While the presence, in the invertebrates, of genes related in sequence and function to the vertebrate p53 family has been known since the discovery of the fly Drosophila melanogaster Dmp53 and the worm Caenorhabditis elegans cep-1 gene, the failure to discover homologs of the essential vertebrate negative regulator of p53 Mdm2 in these species led to the false assumption that Mdm2 was only present in vertebrates. Very recently, clear homologs of Mdm2 have been discovered in a wide range of invertebrate species, raising a series of interesting questions about the evolution of the p53 pathway. Here, a personal account of the discovery of Mdm2-like genes in the Placozoa and Arthropoda is used to speculate on aspects of the evolution, structure, and function of the p53 pathway.
Keywords: Mdm2, evolution, invertebrate, p53 interaction, E3 ligase, RING domain
Introduction
Neither of us is an evolutionary biologist, so it was not our intent to look for homologs of Mdm2. One of us (C.V.), who is very computer literate, had just introduced the other (D.P.L.), who is not, to the UniProt program,1 which is very user friendly and maintained by the European Bioinformatics Institute (EBI) at the Sanger site in Cambridge. In exploring the use of the program on the morning of October 21, 2009, I (D.P.L.) used it to investigate the protein with which I was most familiar, namely p53. I noted that the program not only easily identified the p53 homologs from the entire vertebrate family tree but also the p53-like species from a number of invertebrates. Also noteworthy was the automatic display of the mutations that occur in human p53 in Li-Fraumeni syndrome as germline mutations and as somatic mutations in half of all human cancers. This recalled the early experience in the p53 field where puzzling sequence variations were ignored or dismissed as sequencing errors until the enormous significance of these variations was finally appreciated by the demonstration that these variants switched p53 from acting as a growth repressor to a growth activator in cell-based assays.2 I had been concerned for some time that mutations in human Mdm2 might be more frequent in cancers than had been appreciated so I decided to use the UniProt program to look for mutations in Mdm2. I was disappointed to see that mutations appeared to be quite rare in the database, and indeed, current cancer exome sequencing efforts confirm that initial conclusion, but I was fascinated by the number of homologs of Mdm2 that appeared in the search file. I began to look more closely at these new forms of Mdm2 and was amazed when a rather striking homolog was seen to be present in a species called Trichoplax adhaerens, annotated as an “unknown open reading frame B3RT05.” In retrospect, I did 3 very sensible things, although at the time, I was not really aware of how sensible they were. Firstly, I conducted a reverse search using the Trichoplax sequence to look for homologs to my putative Trichoplax Mdm2, and this clearly identified vertebrate Mdm2. Second, I read a little about Trichoplax and became fascinated by this little studied organism whose genome had nevertheless been completely sequenced.3 I found that it contains only 4 or 5 cell types, has very little structure, and can divide by splitting in two. Third and most importantly, I arranged to meet Chandra for coffee!
Trichoplax Mdm2
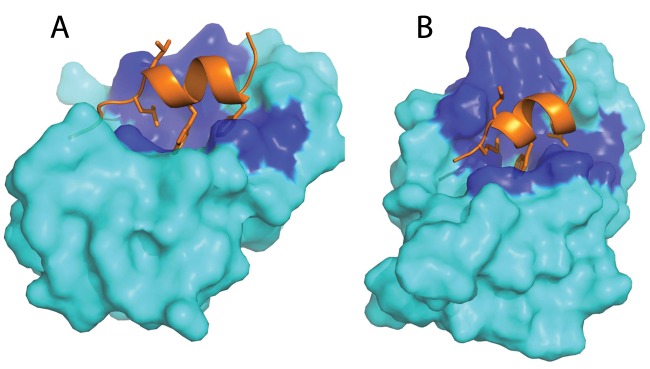
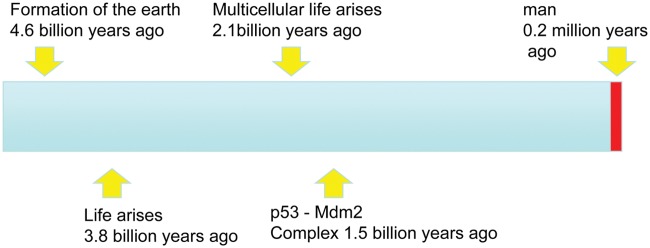
The coffee break was highly successful. Chandra agreed that he would create a homology model of the N-terminus of the Trichoplax Mdm2 in complex with p53. The initial result was very promising (Fig. 1) as the conservation of residues between the human N-terminal p53 binding domain of Mdm2 and the predicted Trichoplax protein was striking with a clear selective conservation of the residues involved in the interaction with p53. At this point, a very brief communication to Nature describing the discovery of Mdm2 in an invertebrate species was drafted and almost as promptly rejected. A second coffee break ensued and a slightly more thorough approach agreed upon. Arumugam Madhumualar from Chandra’s laboratory and Chit Fang Cheok, Christopher Brown, and Farid Ghadessy from my laboratory would join the team, and we all worked very closely together over an intensive period to produce the article that was accepted 2 weeks after the initial observation on November 4, 2009 and appeared in Cell Cycle on February 1, 2010.4,5 We first searched the Trichoplax genome sequence for p53 and, as had been noted in the original publication, discovered B3RZS6 as an excellent candidate for full annotation. The team then set out to fully annotate these candidate protein sequences by close comparison with p53 and Mdm2 proteins from other species. We were able to build homology models with the N-terminal domain, zinc finger domain, and RING finger domain of Mdm2 and for the DNA binding domain and oligomerization domain of the Trichoplax p53 protein. The analysis confirmed the close structural homology with the equivalent human protein, implying that both p53 and Mdm2 had been conserved from the Precambrian Era over 1 billion years ago (Research Highlights: Protein’s billion-year history. Nature. 2010;463(7280): 404). The N-terminus of the Trichoplax p53 contained a small peptide motif that showed clear homology to the very well-studied Mdm2 binding peptide of vertebrate p53, and indeed, homology modeling suggested that the 2 Trichoplax proteins would indeed interact with each other (Fig. 1). Mdm2 and p53 and their interaction have been a feature of the planet Earth for nearly a quarter of its total history of 4.54 billion years (Fig. 2).
Figure 1.
Molecular model of the human p53 Mdm2 complex (A) and predicted placozoan p53. The Mdm2 complex (B). p53 is shown as orange ribbon/sticks, and Mdm2 is shown in cyan; the dark blue regions are conserved Mdm2 residues in contact with p53. Reproduced from Research Highlights: Protein’s billion-year history. Nature. 2010;463(7280):404.
Figure 2.
Complete history of the earth, life, man, and p53/Mdm2.
Looking for Mdm2 in Other Invertebrates
Our growing familiarity with UniProt rapidly led to another discovery, indeed, that very afternoon: that of predicted Mdm2- and p53-like proteins in the arachnid Ixodes scapularis (Northern deer tick). The sequence B7QMD7 from the deer tick genome showed a striking alignment with vertebrate Mdm2 and Trichoplax Mdm2 with 23% overall amino acid identity over the entire sequence with an especially important 40% amino acid identity in the RING finger domain. Again, homology modeling strengthened the conclusion that these were highly related proteins with striking conservation of the predicted structure in the N-terminal p53 binding domain, zinc finger domain, and RING finger domain. The discovery of Mdm2 in the arachnids strengthened the conclusion that the absence of the gene from Drosophila and Caenorhabditis might be an exception among the invertebrate phyla. Two p53 genes were identified in Ixodes scapularis; one of these genes, Isp53-A (B7PEY7), aligns with the full-length human p53 protein and indeed contains a predicted N-terminal Mdm2 binding peptide sequence. The second gene, Isp53-B (B7QF52), shows 32% amino acid identity to human p53 but is a truncated form of the protein that initiates within the DNA binding domain at precisely the same region where one of the recently identified isoforms of human p53 (the delta 133 form) initiates.5
Since this analysis was published, other examples of Mdm2 in the invertebrates have been discovered, as will be described below. It became very important to be certain that the Mdm2 gene really was absent from the Drosophila and Caenorhabditis genomes. With our colleague Sebastian Maurer-Stroh from the Bioinformatics Institute in Singapore, an exhaustive search of all published sequences from these organisms was conducted, and the conclusion that they lack an Mdm2 gene was confirmed. Interestingly, their p53-like genes are less like human p53 than the p53 genes that we identified in the Placozoa and Arachnida, suggesting that the p53 pathway in these organisms had been subject to considerable evolutionary variation.4,5
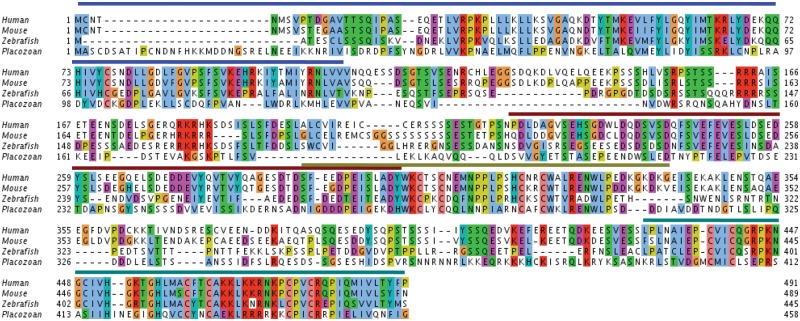
A recently published review of the invertebrate Mdm2 genes has indentified Mdm2-like predicted proteins in a variety of invertebrate species.6 The most detailed work to date has been the studies reported by Muttray et al.,7 who discovered an Mdm2 homolog in the mussel Mytilus trossulus and showed that it could bind to the p53 homolog from Mytilus trossulus in both yeast 2-hybrid assays and in vitro “pull-down” assays in which in vitro translated His-tagged Mdm2 protein was shown to interact with 35S-methionine–labeled p53 and vice versa. In unpublished studies, we have similarly found that the Trichoplax Mdm2 protein can bind to human p53. It is clear that further studies are warranted to establish the degree of conservation of the p53 pathway in the invertebrates. To date, 6 invertebrate Mdm2 species have been annotated from Trichoplax adhaerens, Lottia gigantea, Mytilus trossulus, Strongylocentrotus purpuratus, Nematostella vectensis, and Ixodes scapularis. An alignment of several of these invertebrate Mdm2 predicted proteins is published by Muttray et al.7 and is represented in Figure 3. Does the invertebrate Mdm2 function as E3 ligases with specificity for p53, does it induce the proteasomal degradation of p53, is it induced by p53, and finally, is its E3 ligase activity towards p53 inhibited by DNA damaging signals and nucleolar stress signals?
Figure 3.
Clustal comparison of human, mouse, zebrafish, and placozoan Mdm2. Alignment of the Mdm2 sequences from the human, mouse, zebrafish, and Trichoplax. The bars above the sequence represent the various identified domains in humans. Blue denotes the N-terminal domain, brown denotes the acidic domain, light green/gray denotes the zinc finger domain, and dark green denotes the RING finger domain. Reproduced from Lane et al.5
In order to be able to carry out these critical studies on the endogenous proteins, new antibody reagents that recognize the invertebrate species of p53 and Mdm2 will have to be produced, and a system that allows in vitro culture, labeling, and exposure to drugs and radiation must be established. Here, the Trichoplax system seems most attractive in that the organism can be grown in vitro and as it represents the most evolutionarily distant system. If the pathway functioned in this organism as it does in mammalian cells, it would be strong evidence that the entire pathway has been conserved.
Mdm2 and Mdm4 in the Vertebrates
Whole genome sequencing studies in Singapore inspired by the insights of Dr. Sydney Brenner have focused on extremely careful and deep sequencing of vertebrate species of special interest. The projects started with the sequencing of the pufferfish Fugu rubripes, notable for its compact genome, and have now proceeded to the elephant shark and the lamprey. One benefit of these studies has been the ability to examine the evolution of the p53 pathway in the vertebrates. While the data for the lamprey are still in final analysis, the results for the elephant shark have been published.8 This model vertebrate contains all 3 p53 family members and genes for both Mdm2 and Mdm4. Analysis of the genome structure and intron-exon positions confirms that all 3 p53 family members are derived from a common ancestral gene via gene duplication and that the same is true for Mdm2 and Mdm4.
Careful examination of the invertebrate Mdm2 predicted proteins suggests that they most closely resemble the vertebrate Mdm2 rather than Mdm4. This is especially clear in a completely conserved sequence motif (the 249 FEVE 252 motif human numbering) found at the N-terminal end of the acidic domain, which is present in all Mdm2 species reported but is absent in Mdm4. Its function is not yet clear. The Mdm2 proteins all contain a structurally, very highly conserved zinc finger motif, and finally, at the C-terminus, a tightly defined and most conserved RING finger domain is present. Importantly, the exact length of the C-terminus of Mdm2 is completely conserved, being exactly 13 amino acids from the last Cys residue of the RING finger. Experiments in vertebrate systems have shown that all of these features are essential for the correct functioning of Mdm2.9 The basis of this conservation of structure and implied conservation of function is well understood in structural terms for all of these motifs except the acidic domain and its FEVE motif, whose activity is not yet fully resolved.
Analysis of the aligned sequences of all of the invertebrate Mdm2 genes and a selection of representative vertebrate Mdm2 supports the conclusions originally reached in the analysis of the Trichoplax and deer tick proteins. All of the Mdm2 proteins contain a structurally conserved N-terminal p53 binding domain in which the key contact residues responsible for p53 BOX 1 binding have been especially conserved. The interaction between the N-terminal domains of Mdm2 and p53 appears to be largely conserved in evolution. Three residues of p53 that appear critical for this interaction are F19, W23, and L26 (human numbering) as they are sequestered within a hydrophobic pocket of Mdm2.10 Recent studies further show that there is a kinetic component to the feature whereby a complex stepwise association between the 2 proteins witnesses an initial capture by residues in the Mdm2 pocket that grasp at F19 followed by a gradual induction of opening of the binding site associated with encapsulation of the rest of p53.11 It was demonstrated that the residues in Mdm2 are conserved in their ability to hydrophobically capture p53 upon initial encounter. Although the identity of residues is somewhat lost as one nears the arachnids and placozoans, nevertheless, the general features of the capture mechanism are probably conserved. The changes in amino acids are only likely to lead to differing kinetics of association.
The study further demonstrated that this stepwise process modulates the motion of gatekeeper Y100 (human numbering) that in turn appears to be responsible for triggering an allosteric signal. This signal possibly modulates the interactions between the acidic domains of Mdm2 and the DNA binding domain of p53, which in turn lead to ubiquitination of p53.11
The importance of the p53 Mdm2 axis in controlling the p53 response has been underlined recently by 2 remarkable studies in which subtle point mutations in the Mdm2 gene when introduced into the germline of mice have been shown to have the most profound and differential effects on the p53 response. The first of these mice contains a point mutation in the zinc finger domain C305F.12 These mice are able to respond to radiation with a normal p53 response and are not tumor prone. However, they are prone to myc- but not ras-induced tumorigenesis, are defective in their p53 response to nucleolar disrupting agents and pol 1 inhibitors such as actinomycin D, and most remarkably show defects in the normal metabolic response of mice to calorific restriction. Biochemically, the mutation prevents the ability of the ribosomal protein L11 to bind to and inhibit Mdm2’s activity as an E3 ligase for p53. This suggests that the Mdm2/p53 pathway is able to monitor metabolic effects through measuring aspects of ribosomal synthesis and assembly. Two lines of evidence suggest that this may be the primordial function of the p53 Mdm2 axis. Firstly, the interaction is likely completely conserved, all the Mdm2s examined in vertebrates and invertebrates contain this critical zinc finger interaction site, and the C305 position is completely conserved. Second experiments in mice have shown that acetylation mutants of p53 that are defective in inducing cell cycle arrest and apoptotic responses are still able to be tumor suppressive, able to induce Mdm2, and regulate metabolism.13
The second very striking point mutation is S394A.14 In mice homozygous for this mutation, which removes an ATM phosphorylation site in the RING domain, the response to ionizing radiation is profoundly ablated. Examination of the putative phosphorylation sites in the RING domains of vertebrate and invertebrate Mdm2 proteins does not show the remarkable conservation seen in the RING finger domains.
Conclusions
The study or evolution of the p53 Mdm2 axis is in its infancy but has already raised many profound uncertainties about our knowledge of this most important of all human tumor suppressor loci. Why would a pair of proteins that appear to have been conserved for 1.5 billion years have been lost selectively by the 2 model eukaryotes most studied by academic scientists? Why have C. elegans and D. melanogaster evolved new ways to control p53 that nevertheless allow it to respond to the very same damage and developmental clues that the lost system of Mdm2-based control has perfected? How can such highly conserved genes be dispensable for all aspects of development in a mouse? We have much to learn!
Footnotes
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) received following financial support for the research, authorship, and/or publication of this article:
Funding was provided by BMSI (A*STAR) Singapore.
References
- 1. The Uniprot Consortium Reorganizing the protein space at the Universal Protein Resource (UniProt). Nucleic Acid Res. 2012; 40(D1):D71-D75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lane DP, Benchimol S. p53: oncogene or anti-oncogene? Genes Dev. 1990;4(1):1-8 [DOI] [PubMed] [Google Scholar]
- 3. Srivastava M, Begovic E, Chapman J, et al. The Trichoplax genome and the nature of placozoans. Nature. 2008;454(7207):955-60 [DOI] [PubMed] [Google Scholar]
- 4. Lane DP, Cheok CF, Brown C, Madhumalar A, Ghadessy FJ, Verma C. Mdm2 and p53 are highly conserved from placozoans to man. Cell Cycle. 2010;9(3):540-7 [DOI] [PubMed] [Google Scholar]
- 5. Lane DP, Cheok CF, Brown CJ, Madhumalar A, Ghadessy FJ, Verma C. The Mdm2 and p53 genes are conserved in the Arachnids. Cell Cycle. 2010;9(4):748-54 [DOI] [PubMed] [Google Scholar]
- 6. Momand J, Villegas A, Belyi VA. The evolution of MDM2 family genes. Gene. 2011;486(1-2):23-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Muttray AF, O’Toole TF, Morrill W, Van Beneden RJ, Baldwin SA. An invertebrate mdm homolog interacts with p53 and is differentially expressed together with p53 and ras in neoplastic Mytilus trossulus haemocytes. Comp Biochem Physiol B Biochem Mol Biol. 2010;156(4):298-308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lane DP, Madhumalar A, Lee AP, et al. Conservation of all three p53 family members and Mdm2 and Mdm4 in the cartilaginous fish. Cell Cycle. 2011;10(24):4272-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dolezelova P, Cetkovska K, Vousden KH, Uldrijan S. Mutational analysis of Mdm2 C-terminal tail suggests an evolutionarily conserved role of its length in Mdm2 activity toward p53 and indicates structural differences between Mdm2 homodimers and Mdm2/MdmX heterodimers. Cell Cycle. Epub 2012. March 1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kussie PH, Gorina S, Marechal V, et al. Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science. 1996;274(5289):948-53 [DOI] [PubMed] [Google Scholar]
- 11. Dastidar SG, Lane DP, Verma CS. Why is F19Ap53 unable to bind MDM2? Simulations suggest crack propagation modulates binding. Cell Cycle. 2012;11(12):2239-47 [DOI] [PubMed] [Google Scholar]
- 12. Macias E, Jin A, Deisenroth C, et al. An ARF-independent c-MYC-activated tumor suppression pathway mediated by ribosomal protein-Mdm2 interaction. Cancer Cell. 2010;18(3):231-43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li T, Kon N, Jiang L, et al. Tumor suppression in the absence of p53-mediated cell-cycle arrest, apoptosis, and senescence. Cell. 2012;149(6):1269-83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gannon HS, Woda BA, Jones SN. ATM phosphorylation of Mdm2 Ser394 regulates the amplitude and duration of the DNA damage response in mice. Cancer Cell. 2012;21(5):668-79 [DOI] [PMC free article] [PubMed] [Google Scholar]