Abstract
Hedgehog (Hh) signaling plays pivotal roles in embryonic development and adult tissue homeostasis in species ranging from Drosophila to mammals. The Hh signal is transduced by Smoothened (Smo), a seven-transmembrane protein related to G protein coupled receptors. Despite a conserved mechanism by which Hh activates Smo in Drosophila and mammals, how mammalian Hh signal is transduced from Smo to the Gli transcription factors is poorly understood. Here, we provide evidence that two ciliary proteins, Evc and Evc2, the products of human disease genes responsible for the Ellis-van Creveld syndrome, act downstream of Smo to transduce the Hh signal. We found that loss of Evc/Evc2 does not affect Sonic Hedgehog-induced Smo phosphorylation and ciliary localization but impedes Hh pathway activation mediated by constitutively active forms of Smo. Evc/Evc2 are dispensable for the constitutive Gli activity in Sufu−/− cells, suggesting that Evc/Evc2 act upstream of Sufu to promote Gli activation. Furthermore, we demonstrated that Hh stimulates binding of Evc/Evc2 to Smo depending on phosphorylation of the Smo C-terminal intracellular tail and that the binding is abolished in Kif3a−/− cilium-deficient cells. We propose that Hh activates Smo by inducing its phosphorylation, which recruits Evc/Evc2 to activate Gli proteins by antagonizing Sufu in the primary cilia.
Keywords: Evc, Evc2, Hedgehog, Smo, Gli
Introduction
The Hedgehog (Hh) pathway is a major developmental signaling pathway essential for the growth and patterning of various tissues during embryonic development and adult tissue homeostasis1,2. Malfunction of this pathway has been implicated in numerous human disorders including congenital anomalies and cancers1,2. The hedgehog (hh) gene was first identified as an embryonic segment polarity gene in Drosophila3. Three Hh family members have been identified in mammals including Sonic Hedgehog (Shh), Indian Hedgehog (Ihh) and Desert Hedgehog. Key components of the Hh signaling pathway include the 12-transmembrane receptor Patched (Ptc) that functions as the Hh receptor, the seven-transmembrane protein Smoothened (Smo) that functions as the obligated Hh signal transducer, and the Zinc-finger transcription factor Cubitus interruptus (Ci)/Gli4. In the absence of Hh ligands, Ptc blocks the activity of Smo and full-length Ci/Gli is phosphorylated by multiple kinases including PKA, CK1 and GSK3β, which targets it for Slimb/β-TRCP-mediated proteolysis to generate its repressor form (CiR/GliR)5. In the presence of Hh, the Ptc inhibition of Smo is released through Hh binding to Ptc; Smo is phosphorylated by multiple kinases including PKA (Drosophila only), CK1 and Gprk2 kinases, which promotes its active conformation and changes its subcellular localization with Drosophila Smo accumulating on the cell surface and mammalian Smo accumulating in the primary cilium6,7,8,9,10,11. Ci/Gli phosphorylation and proteolysis is blocked, leading to diminished CiR/GliR activity12,13,14. In addition, full-length Ci/Gli is converted into its activator form (CiA/GliA)15.
Although Smo is activated through a conserved mechanism in Drosophila and mammals7,9, the mechanisms by which activated Smo transduces the signal to the downstream signaling components appear to diverge. In Drosophila, Smo forms a complex with the kinesin-like protein Costal2 (Cos2) and the Ser/Thr kinase Fused (Fu) in a manner regulated by Hh and Smo phosphorylation16,17,18,19,20,21. Hh-induced Smo conformational switch and C-tail dimerization lead to Fu dimerization, phosphorylation and activation20,21,22. Activated Fu regulates both the activator and repressor form of Ci, likely by phosphorylating Cos2 and Sufu18,20,21,22,23. Both Sufu and the mammalian homolog of Cos2, Kif7, are involved in Hh signaling24,25,26,27,28. By contrast, the mammalian homolog of Fu is not required for Hh signaling during development29,30. Moreover, there is no evidence that Kif7 physically interacts with mammalian Smo. Hence, the signaling component acting immediately downstream of mammalian Smo has remained unknown.
EVC and EVC2 are adjacent genes identified in the recessive skeletal dysplasia Ellis-van Creveld syndrome (EvC; MIM: 225500), and loss-of-function mutation in either gene leads to the same condition31,32,33,34. Recent studies indicate that Evc promotes chondrocyte proliferation, hypertrophy and the differentiation of osteoblasts in the perichondrium, and localizes to the primary cilia of osteoblasts to mediate Hh signaling in the osteoblast lineage35. EVC2, which is in close proximity to EVC, was also implicated as a positive regulator of Hh signaling. Evc and Evc2 associate with each other and co-localize at primary cilia in a mutually dependent manner36. The mechanism by which Evc/Evc2 regulate Hh signaling has remained unknown. Here, we provide evidence that Evc/Evc2 act downstream of Smo to promote Gli activation. We find that knockdown of Evc/Evc2 attenuates Hh signaling without affecting Smo phosphorylation and ciliary localization. In contrast, knockdown of Evc/Evc2 does not affect Hh pathway activation caused by Gli overexpression or loss of Sufu, indicating that Evc/Evc2 function upstream of Sufu and Gli to regulate Hh signaling. We provide evidence that Evc/Evc2 are required for Gli activation and inhibition of Gli processing to generate the truncated Gli repressors. We show that Hh induces a physical interaction between Smo and Evc/Evc2 and that this association depends on phosphorylation of the Smo C-terminal intracellular tail. Together, our results suggest that Evc/Evc2 transduce Hh signal downstream of Smo activation and promote Gli activation by antagonizing Sufu.
Results
EVC/EVC2 knockdown attenuates Hh signaling
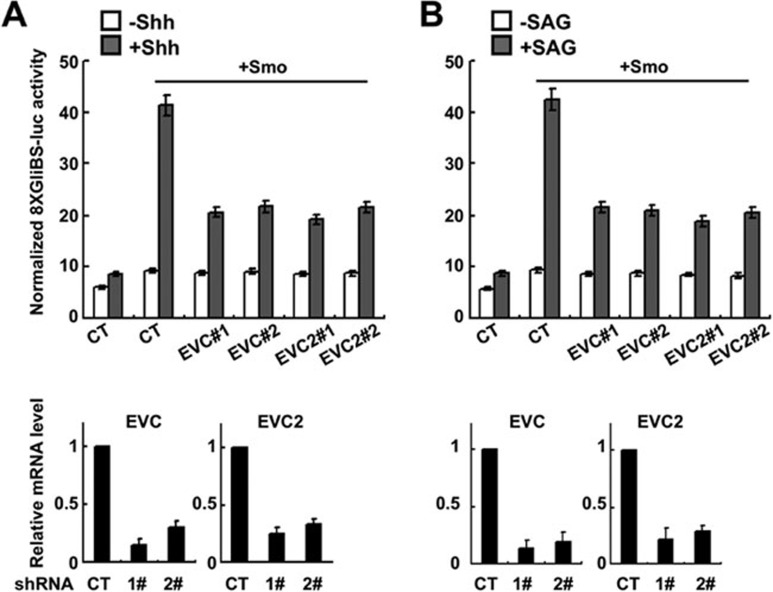
Previous studies demonstrated that Evc is essential for Ihh signaling in the cartilage growth plate, and Evc2, a close homolog of Evc, is a positive modulator of Shh signaling in cultured cells36,37. To define the underlying mechanism by which Evc/Evc2 modulate Hh signaling, we employed the RNAi approach to inactivate the two genes. We designed two independent shRNAs each against EVC and EVC2 (Supplementary information, Figure S1). We first confirmed the positive role of Evc/Evc2 in the Hh signaling pathway using 8× GliBS luciferase (Gli-luc) assay in NIH3T3 cells. In line with previous findings36, knockdown of Evc/Evc2 using two independent shRNAs in NIH3T3 cells attenuated the activation of Gli-luc reporter gene induced with Shh or a Smo agonist SAG (Figure 1A and 1B, top). The knockdown efficiency of Evc/Evc2 by these shRNAs was determined by real-time PCR and over 70% reduction was achieved for each shRNA (Figure 1A and 1B, bottom).
Figure 1.
Evc and Evc2 are positive regulators in Hh signaling pathway. (A, B) Representative Gli-luciferase assay in NIH3T3 cells transfected with Smo and shRNAs against Evc/Evc2, respectively, and treated with or without Shh-conditioned medium (A) or SAG (B). Individual Evc/Evc2 shRNA knockdown efficiency was verified by real-time PCR indicated by the bottom panel.
Smo phosphorylation and ciliary localization are not blocked by Evc/Evc2 inactivation
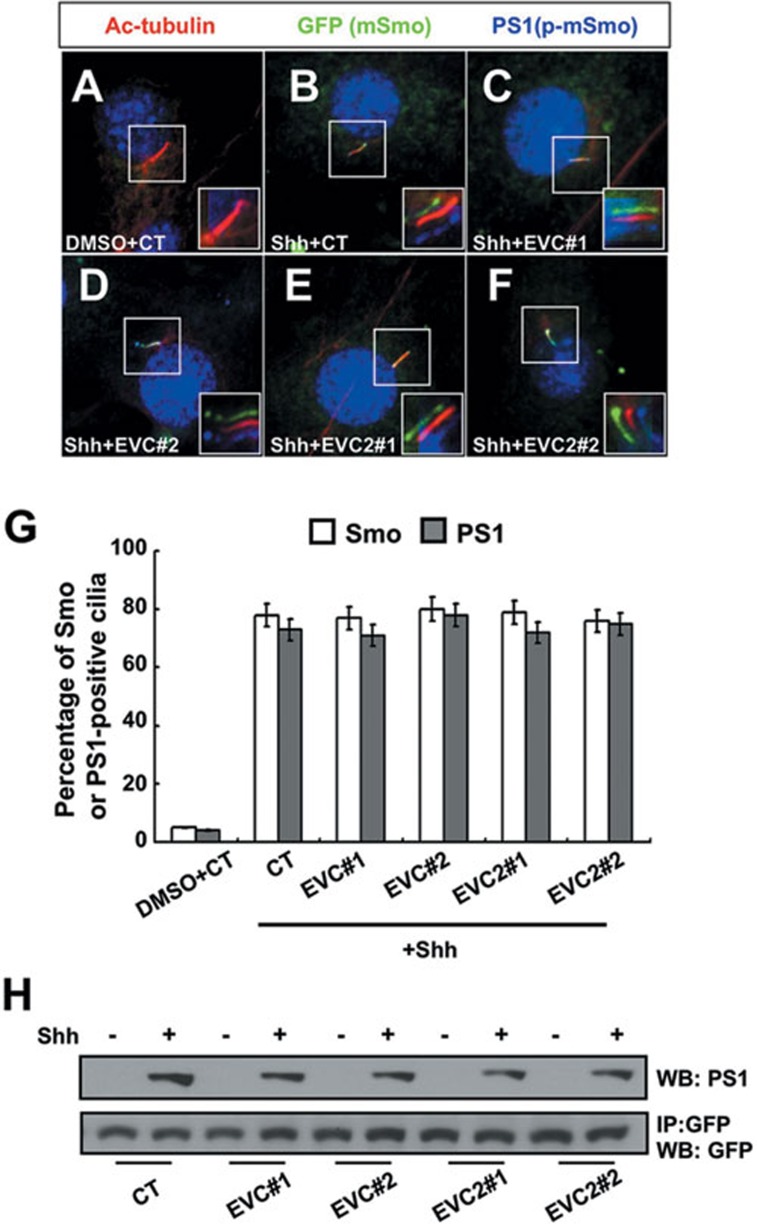
Our previous studies have demonstrated that Smo is hyperphosphorylated at multiple sites in its C-terminal tail by CK1α and GRK2 kinases upon Shh stimulation and that these phosphorylation events activate Smo through promoting its open active conformation and ciliary accumulation9. Evc/Evc2 knockdown could attenuate Shh signaling by downregulating Shh-induced Smo activation. Alternatively, Evc/Evc2 could act at a step downstream of Smo after it is activated by Hh. To distinguish these two possibilities, we examined whether inactivation of Evc/Evc2 affected Smo phosphorylation and ciliary accumulation. To monitor Smo phosphorylation, we took advantage of a phospho-specific antibody (PS1) that recognizes the S1 phosphorylation cluster (pS615ADVpS619pS620) in Smo C-tail, which plays a major role in Smo activation9. To monitor Smo ciliary localization, we made use of an NIH3T3 cell line that stably expresses CFP-tagged Smo (NIH3T3Smo-CFP)9. Consistent with our pervious findings, Smo-CFP did not exhibit significant ciliary localization in the absence of Shh, but accumulated in the primary cilia upon Shh stimulation (Figure 2A and 2B). By immunostaining with the PS1 antibody, we found that PS1 signal was not detected in primary cilia of NIH3T3Smo-CFP cells in the absence of Shh (Figure 2A). After Shh stimulation, PS1 was detected in primary cilia that also accumulated Smo-CFP (Figure 2B), suggesting that ciliary localized Smo is in its phosphorylated form. We found that Evc/Evc2 knockdown by shRNAs had little if any effect on Hh-induced ciliary accumulated Smo-CFP and PS1 signal (Figure 2C-2F, compared with Figure 2B and 2G). We confirmed that Evc/Evc2 knockdown did not block Hh-induced Smo phosphorylation by western blot analysis using the PS1 antibody (Figure 2H). Since Smo phosphorylation and primary cilia accumulation are indicative for its activation, these data suggest that Evc and Evc2 do not participate in Hh-induced Smo activation.
Figure 2.
Evc/Evc2 are not required for Smo activation upon Shh stimulation. (A-F) NIH3T3Smo-CFP cells infected with control (CT) or the indicated Evc/Evc2 shRNA viruses and treated with or without Shh-conditioned medium were immunostained to show the expression of acetylated (Ac)-tubulin (Red) that labels the primary cilium, GFP (green) that labels the CFP-tagged Smo proteins and PS1 (blue) that labels the phosphorylated Smo. More than 50 cells were analyzed for each experiment and representative images were shown. The insets show enlarged views of the selected regions with shifted overlays. (G) The percentage of Smo-CFP (GFP) or phosphorylated Smo (PS1) positive primary cilia in cells that were infected by different shRNA-expression viruses and treated with or without Shh. Over 100 ciliated cells were counted for each time point, n = 3. (H) NIH3T3Smo-CFP cells treated as in A-F were collected for immunoprecipitation with GFP antibody, followed by representative western blot analysis with PS1 or GFP antibodies, respectively.
Inactivation of Evc/Evc2 inhibits Hh pathway activity downstream of activated Smo
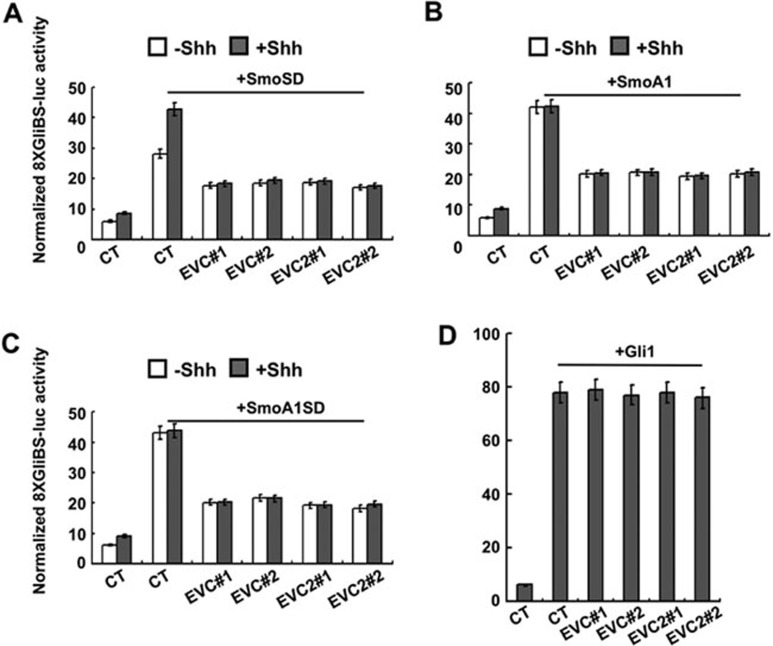
Previous work has implicated that Evc/Evc2 are required for Hh- and Smo agonist-induced pathway activation, suggesting that they act at the level of Smo in Hh signal transduction; however, it did not distinguish whether Evc/Evc2 are required for Smo activation or signal transduction downstream of activated forms of Smo36,37. Our above observations suggest that Evc/Evc2 act at a step downstream of Smo after it is phosphorylated in response to Hh. To further test this hypothesis, we asked whether inactivation of Evc/Evc2 affects Hh signal transduction from constitutively active forms of Smo: SmoSD0-5 (SmoSD for short) in which all six phosphorylation clusters were converted to acidic residues to mimic phosphorylation; SmoA1 (also known as SmoM2), which is an oncogenic form of Smo; and SmoA1SD0-5 (SmoA1SD), which combines both the A1/M2 and SD0-5 mutations9. SmoSD, SmoA1, and SmoA1SD were transfected into NIH3T3 cells with the Gli-luc reporter gene in the presence or absence of Evc/Evc2 shRNAs. The transfected cells were treated with or without Shh-conditioned media, followed by dual luciferase assay. In line with our previous findings9, SmoSD exhibited high basal activities compared with wild-type (WT) Smo; and Shh further increased the activity of SmoSD (Figure 3A), while both SmoA1 and SmoA1SD exhibited maximal activity regardless of the presence or absence of Shh (Figure 3B and 3C). We found that knockdown of Evc/Evc2 reduced both high basal activities and Shh-induced activities of SmoSD as well as the constitutive activity of SmoA1 and SmoA1SD (Figure 3A-3C), indicating that Evc/Evc2 function downstream of Smo activation. By contrast, we found that loss of Evc/Evc2 did not affect Gli-luc activity induced by overexpression of Gli1 or Gli2 (Figure 3D and Supplementary information, Figure S2), suggesting that Evc/Evc2 act upstream of Gli transcription factors.
Figure 3.
Evc/Evc2 function downstream of Smo but upstream of Gli. (A-D) Representative Gli-luciferase assay in NIH3T3 cells transfected with SmoSD, SmoA1, SmoA1SD, Gli1 and shRNAs against Evc/Evc2, respectively, and treated with or without Shh-conditioned medium. Individual Evc/Evc2 knockdown efficiency was verified by real-time PCR (data not shown).
Evc/Evc2 act upstream of Sufu to regulate Gli activity
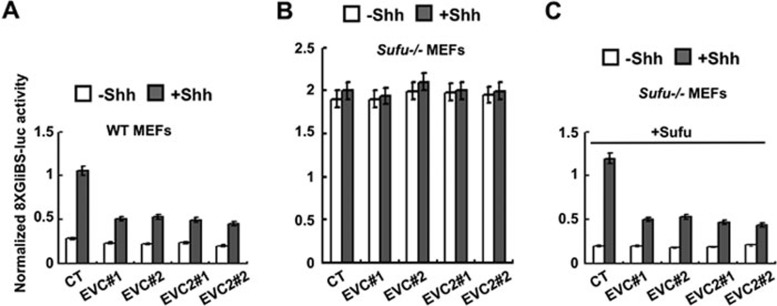
Sufu is a major inhibitory component of the mammalian Hh pathway downstream of Smo as loss of Sufu leads to ectopic Hh signaling activation similar to loss of Ptc28. We next asked whether Evc/Evc2 are required for the constitutive Hh signaling activity in the absence of Sufu, and we made use of Sufu−/− MEF cells28. We compared the Gli-luc response between WT MEF and Sufu−/− MEF cells, and found that loss of Evc/Evc2 by shRNAs attenuated Shh-induced Gli-luc activation in WT MEFs (Figure 4A), but did not affect the constitutive Gli-luc activity in Sufu−/− cells regardless of the presence or absence of Shh (Figure 4B). Introducing a Sufu expression construct back into Sufu−/− cells suppressed the high basal Gli-luc activity and restored the dependency on Evc/Evc2 for Shh-induced activation of Gli-luc (Figure 4C), suggesting that Evc/Evc2 are dispensable for the constitutive Gli activity in Sufu−/− cells.
Figure 4.
Evc/Evc2 are not required for constitutive Gli activity in Sufu−/− cells. Representative Gli-luciferase assay in WT and Sufu−/− MEF cells transfected with the indicated constructs and treated with or without Shh-conditioned medium. Individual Evc/Evc2 shRNA knockdown efficiency was verified by real-time PCR (data not shown).
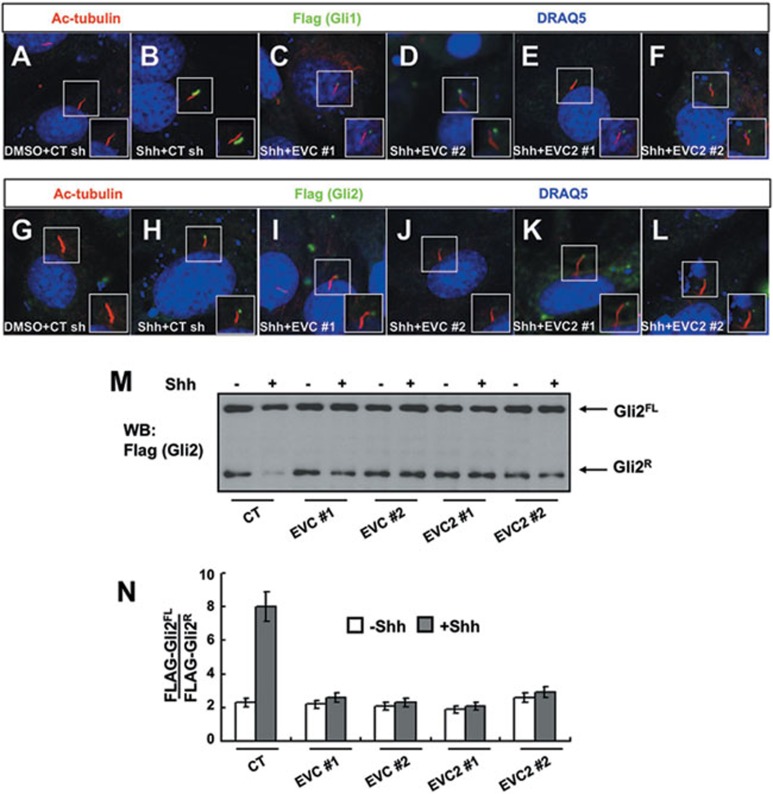
Hh signaling activates the Gli transcription factors by inhibiting the production of GliR and promoting the formation of GliA. We therefore determined whether these processes are affected by inactivation of Evc/Evc2. We made use of cell lines that stably express Flag-tagged Gli2 (Shh-EGFPFLAG-Gli2) or Gli1 (Shh-LIGHT2FLAG-Gli1)38. Consistent with a previous study38, Shh induced accumulation of Flag-Gli1 and Flag-Gli2 at the tip of primary cilia, which is indicative of Gli activation (Figure 5B, 5H and Supplementary information, Figure S3). When cells are infected with high-titer Evc/Evc2 shRNA expression viruses, the accumulation of Flag-Gli1 and Flag-Gli2 at the tip of primary cilia was significantly reduced (Figure 5C-5F, 5I-5L and Supplementary information, Figure S3), suggesting that Gli activation is attenuated by inactivation of Evc/Evc2. The knockdown efficiencies for shRNA viruses were verified in the same batch of cells used in the above experiments (data not shown). Furthermore, we collected the cells for western blot analysis to determine whether Evc/Evc2 are required for Shh to inhibit the formation of GliR. In the absence of Shh, Flag-Gli2 was processed into a truncated repressor form (Gli2R) while treating cells with Shh-conditioned medium inhibits Gli2R formation (Figure 5M), resulting in a 4-fold increase in the ratio of Gli2FL/Gli2R (Figure 5N). Consistent with a previous finding37, we found that Evc/Evc2 RNAi had little if any effect on Gli2R formation in the absence of Shh, suggesting that Evc/Evc2 are not required for Gli processing to generate the repressor forms. However, we found that Shh did not effectively block the production of Gli2R in the absence of Evc/Evc2 (Figure 5M and 5N), suggesting that Evc/Evc2 are required for Hh to inhibit Gli2R formation in cultured cells. A previous study indicated that Shh can still block Gli repressor formation in the absence of Evc function in vivo37. It is possible that Hh-mediated inhibition of Gli processing is more dependent on Evc/Evc2 in vitro, whereas redundant or context-dependent mechanisms may exist in vivo. In addition, only a fraction of cells are exposed to Hh in vivo and a partial change in Gli processing in Evc/Evc2 mutants could have been “masked”, whereas in cell-based assays, all cells are exposed to Hh, and as a consequence, it might be more sensitive to detect the effect of Evc/Evc2 inactivation on Shh-regulated Gli processing. Taken together, these data suggest that Evc/Evc2 regulate the balance of GliA and GliR by promoting GliA activation while inhibiting GliR formation.
Figure 5.
Evc/Evc2 modulate GliA ciliary accumulation and inhibit GliR formation in response to Hh. (A-L) Shh-LIGHT2FLAG-Gli1 and Shh-EGFPFLAG-Gli2 cells were infected with control (CT) or the indicated Evc/Evc2 shRNA viruses twice; 2 days later, the cells were split for additional treatment with or without Shh-conditioned medium or real-time PCR analysis for knockdown efficiency. For Gli cilia localization test, cells were immunostained to show the expression of acetylated (Ac)-tubulin (Red) that labels the primary cilium, Flag (green) that labels the Flag-tagged Gli1 (A-F) or Gli2 (G-L) proteins and DRAQ5 (blue) that labels the nucleus. More than 50 cells were analyzed for each experiment and representative images were shown. The insets show enlarged views of the selected regions with shifted overlays. (M, N) Shh-EGFPFLAG-Gli2cells treated as in G-L were collected and probed with anti-Flag primary antibody. Representative western blot was shown in M, and the ratio for Flag-Gli2FL/Flag-Gli2R was quantified with three independent western blot results using Image J software (N).
Hh-induced phosphorylation of Smo promotes its binding to Evc/Evc2
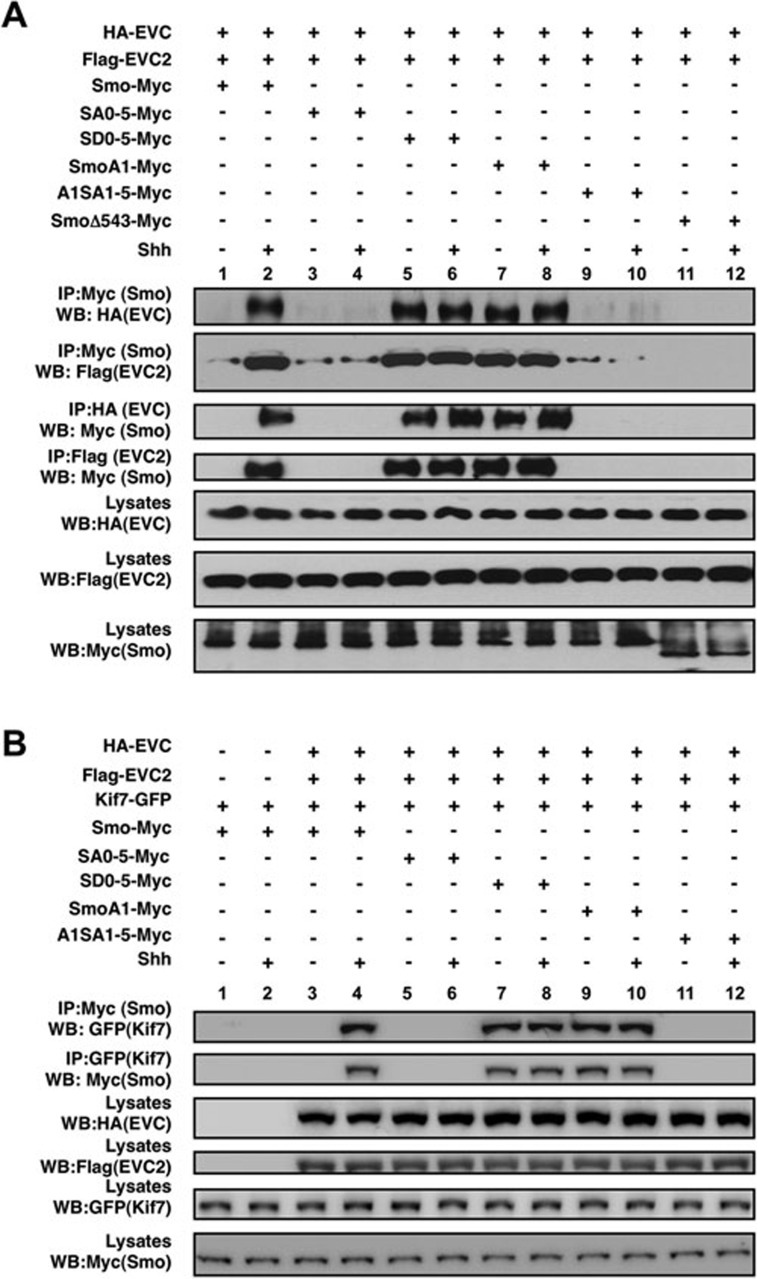
Evc and Evc2 associate with each other and their localization in primary cilia appears to be mutually dependent36. Smo is also accumulated in primary cilia in response to Shh, which depends on its phosphorylation by CK1 and GRK29. Our epistasis experiments suggest that Evc/Evc2 act downstream of Smo activation to regulate Hh pathway activity (Figures 2 and 3). To gain further insight into the mechanism by which Evc and Evc2 exert their function, we decided to test the possibility that they form a complex with Smo after Smo is activated by Hh. To do this, we determined the binding between HA-tagged EVC (HA-EVC)/Flag-tagged EVC2 (Flag-EVC2) and Myc-tagged WT (Smo-Myc) or mutant forms of Smo (SmoSA0-5-Myc, SmoSD0-5-Myc, SmoA1-Myc, SmoA1SA1-5-Myc or SmoΔ543-Myc) by coimmunoprecipitation experiments after transient transfection of these constructs into NIH3T3 cells9. In the absence of Shh, little if any HA-EVC or Flag-EVC2 were coimmunoprecipitated with Smo-Myc and similarly, Smo-Myc was not coimmunoprecipitated with HA-EVC or Flag-EVC2 (Figure 6, lane 1). By contrast, Shh induced a robust association between Smo-Myc and HA-EVC/Flag-EVC2 (Figure 6, lane 2). Shh-induced complex formation between Smo-Myc and HA-EVC/Flag-EVC2 depends on Smo phosphorylation because the phospho-deficient form SmoSA0-5-Myc failed to form a complex with HA-EVC/Flag-EVC2 even in the presence of Shh (Figure 6, lanes 3 and 4). On the other hand, the phospho-mimetic form of Smo, SmoSD0-5-Myc, exhibited constitutive binding to HA-EVC/Flag-EVC2, which is not further stimulated by Shh (Figure 6, lanes 5 and 6). Similarly, the oncogenic form of Smo, SmoA1-Myc, also formed a constitutive complex with HA-EVC/Flag-EVC2 (Figure 6, lanes 7 and 8) and complex formation between SmoA1 and EVC/EVC2 was abolished by mutating multiple phosphorylation clusters in its C-tail (SmoA1SA1-5-Myc, Figure 6, lanes 9 and 10). Shh-induced association between activated forms of Smo and Evc/Evc2 was abolished by deleting the Smo C-terminal tail (SmoΔ543-Myc, Figure 6, lanes 11 and 12) or by treating cells with Smo antagonist cyclopamine (data not shown). Finally, Shh-induced Smo/Evc/Evc2 complex formation is lost in Kif3a−/− MEF cells that lack primary cilia (Supplementary information, Figure S4). Taken together, these observations suggest Shh-induced phosphorylation of Smo is both necessary and sufficient to recruit Evc/Evc2, and the binding between Smo and Evc/Evc2 is mediated by Smo C-tail and is likely dependent on primary cilia.
Figure 6.
Hh promotes Smo binding to Evc/Evc2 and recruitment of Kif7. (A) Coimmunoprecipitation assays to determine the interaction between HA-tagged EVC (HA-EVC)/Flag-tagged EVC2 (Flag-EVC2) and the indicated forms of Myc-tagged Smo. NIH3T3 cells were cotransfected with the indicated Smo and Evc/Evc2 constructs, followed by immunoprecipitation and western blot analysis with the indicated antibodies. Cell lysates were also directly immunoblotted by the indicated antibodies. (B) Hh promotes the interaction between activated forms of Smo with Kif7, depending on Evc/Evc2 coexpression. Coimmunoprecipitation assays to determine the interaction between GFP-tagged Kif7 (Kif7-GFP) and the indicated forms of Myc-tagged Smo. NIH3T3 cells were cotransfected with the indicated Smo, Kif7 and Evc/Evc2 constructs, followed by immunoprecipitation and western blot analysis with the indicated antibodies. Cell lysates were also directly immunoblotted by the indicated antibodies.
Several studies have shown that Kif7, the mammalian homolog of Drosophila Cos2, is involved in Hh signaling24,25,26,27. In Drosophila, Hh-induced Smo phosphorylation and conformational change promotes Smo/Cos2 association20,21. Interestingly, we found that Hh and oncogenic Smo mutation can promote the recruitment of Kif7 to Smo in a manner depending on Evc/Evc2 and Smo phosphorylation (Figure 6B), suggesting that Kif7 is recruited to the activated Smo/Evc/Evc2 complex.
Discussion
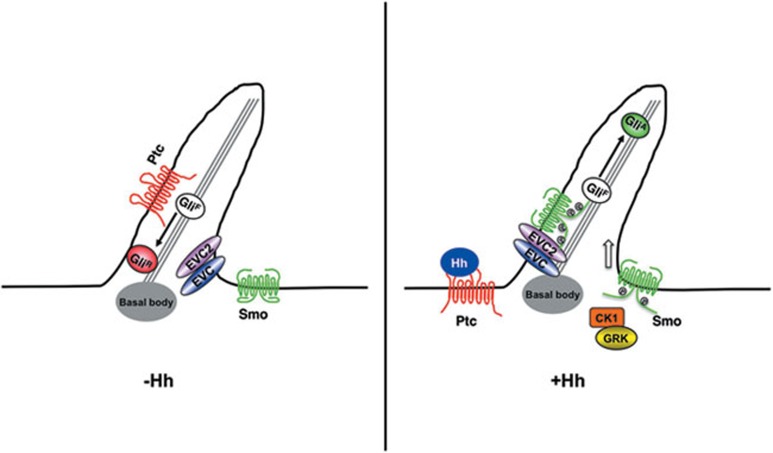
Despite that great efforts have been devoted to understand how Hh signaling works in the past two decades, the mechanism by which mammalian Hh signals are transduced from Smo to the Gli transcription factors is still poorly understood1. Most notably, the component acting immediately downstream of mammalian Smo has remained elusive, and thus represents a major gap in the mammalian Hh signaling pathway. Here, we demonstrated that the Evc/Evc2 complex, the products of two human disease genes responsible for the Ellis-van Creveld syndrome, mediates Hh signal transduction from Smo to the Gli transcription factors. We demonstrated that Shh induces binding of Evc/Evc2 to Smo, depending on phosphorylation of its C-tail. Furthermore, we provide evidence that Evc and Evc2 act upstream of Sufu to regulate Gli activity. We propose that Hh-induced phosphorylation of Smo promotes its ciliary accumulation and open conformation, leading to an association between Smo and Evc/Evc2. Smo/Evc/Evc2 complex further transduces the Hh signal to activate Gli by antagonizing Sufu (Figure 7). Our findings thus provide a novel insight into how activated Smo relays the Hh signal to downstream signaling components in the primary cilium.
Figure 7.
A working model for Evc/Evc2 involvement in Hh signaling. Hh promotes the activation of Smo and its association with Evc/Evc2 complex in the primary cilia to transduce the signal intracellularly, leading to Gli activation.
The microtubule-based organelle primary cilium has been implicated as essential for mammalian Hh signal transduction, and most of Hh signaling components including Ptc, Smo and Gli proteins exhibit dynamic ciliary localization39. For example, Hh-induced Smo phosphorylation promotes its ciliary localization and active conformation9. Evc and Evc2 form a complex localized in the primary cilia and both proteins have been implicated as positive regulators of mammalian Hh signaling36,37. We found that inactivation of Evc/Evc2 does not significantly affect Hh-induced Smo phosphorylation and ciliary accumulation in cultured cells, suggesting that Evc and Evc2 are unlikely to be involved in Smo activation. Furthermore, we demonstrated that inactivation of Evc/Evc2 blocks the activity of phospho-mimetic and constitutively active forms of Smo, suggesting that Evc and Evc2 function at a step downstream of activated Smo. Importantly, we demonstrated that Hh promotes an association between Evc/Evc2 and Smo, suggesting that Evc/Evc2 may act immediately downstream of Smo. Interestingly, we found that complex formation between Evc/Evc2 and Smo is regulated by Smo phosphorylation. Because Evc and Evc2 are localized in the primary cilia, Hh-induced phosphorylation of Smo may facilitate its interaction with Evc/Evc2 by promoting its ciliary accumulation. Consistent with this, we found that Hh-induced association between Smo and Evc/Evc2 is abolished in Kif3a−/− cells that lack primary cilia. In addition, we speculate that phosphorylation-induced conformational change in Smo may expose a binding pocket in Smo C-tail for Evc/Evc2 interaction. Indeed, a Smo-deletion mutant that lacks the C-tail failed to bind Evc/Evc2. Finally, we provide evidence that binding of Evc/Evc2 to Smo further recruits Kif7 (Figure 6B). We propose that Hh induces the formation of an active Smo/Evc/Evc2 signaling complex in the primary cilia to transduce the signal intracellularly, leading to the activation of Gli proteins (Figure 7).
It has been shown that Sufu retains Gli in the cytoplasm and promotes its proteolysis into a truncated repressor whereas activation of Smo in primary cilia upon Hh stimulation leads to recruitment of endogenous Sufu-Gli complexes to the cilia and causes the dissociation of Sufu-Gli complex, allowing activated Gli proteins to enter the nucleus40,41,42. We found that Evc and Evc2 function upstream of Sufu to promote Gli activation. Hence, it is possible that the activated Smo/Evc/Evc2 complex may modulate the association between Sufu and Gli proteins in the primary cilia.
We have obtained evidence that Hh stimulates the binding of Kif7 to Smo, which is facilitated by Evc/Evc2. Increased binding of Cos2 to activated Smo has also been observed in Drosophila20,21. It has been shown that binding of Cos2 to activated Smo promotes Fu dimerization, phosphorylation and activation, and activated Fu then regulates Ci by inhibiting its repressor form and stimulating its activator form20,21,22. The mammalian Fu homolog is dispensable for Hh signaling29,30; however, it is possible that another kinase(s) may substitute for the Fu function in the mammalian Hh pathway. It has been shown that Hh stimulates Gli3 phosphorylation, which correlates with formation of Gli3A40. Therefore, it would be interesting to determine whether Hh-induced Gli3 phosphorylation is affected by loss of Evc/Evc2. It is tempting to speculate that the activated Smo/Evc/Evc2 complex may recruit one or more kinases to phosphorylate Gli proteins and promote their activation.
The data from mouse Evc KO experiments and human patients clearly established a role for Evc proteins in Ihh signaling37. The “extra toe” phenotype in human patient suggests that Evc/Evc2 may also play a role in Shh pathway although they might not be absolutely required for Shh signaling. Our in vitro study also suggested that Evc/Evc2 are only required for maximal Hh pathway activity because residual pathway activities persisted when Evc and Evc2 were knocked down. It remains to be determined whether Evc mutants exhibit any defects in Shh responses in vivo.
Materials and Methods
Constructs, cell culture, transfection, immunoprecipitation, western blot and small molecule treatment
WT or mutant Smo DNA fragments were amplified by PCR and inserted between HindIII and SalI digestion sites into the pGE vector; Myc tag was PCR amplified and inserted in frame after Smo coding sequence; all the constructs were sequence verified; Smo mutants have been described previously9. Independent shRNAs against EVC and EVC2 were constructed using pLKO.1 vector, and constructs were sequence verified. Lentiviruses were generated according to the manufacturer's protocol; supernatants containing different lentiviruses were collected 48 and 72 h post transfection, cells were infected twice with 48 h and 72 h viruses with 4 μg/ml polybrene, respectively. 21-bp targeting sequences are indicated in Supplementary information, Figure S1. NIH3T3 cells were obtained from ATCC; HEK-293T were kindly provided by Dr Alec Zhang; and Sufu−/− MEFs cells and Kif7-GFP construct were kindly provided by Dr Chi-Chuang Hui. All the mammalian cells were cultured in DMEM supplemented with 10% fetal bovine serum and incubated at 37 °C. For protein expression, the mammalian cells were transfected with X-tremeGENE transfection reagent (Roche) according to the manufacturer's instructions, harvested and lysed in RIPA buffer (50 mM Tris-Cl pH 7.9, 150 mM NaCl, 5 mM EDTA, 1% NP-40 supplemented with protease inhibitors (Roche)). Lysates were frozen and thawed 2∼3 times. Immunoprecipitation experiments were performed as previously described14. Primary and second antibodies used are: mouse anti-Myc (Sigma), mouse anti-Flag (Santa Cruz Biotechnology), monoclonal anti-acetylated tubulin (Sigma, #T7451), PS1 phospho-specific antibody (1:509), goat anti-mouse IgG HRP, goat anti-rabbit IgG HRP. Reagents used were: Recombinant Mouse Sonic Hedgehog N-terminus (ShhNp, R&D Systems, Cat # 464-SH), 293-Shh-conditioned medium (1:6 v/v43), and SAG (200 nM).
8× GliBS-luciferase assay
Luciferase assay was performed following standard protocol described in9. Briefly, the day before transfection, cells were seeded at a density of 1∼2 × 105 cells/ml, and transfected with a 4:1 ratio of 8× GliBS-luciferase vector:pRL-TK and other transgene or shRNA constructs with X-tremeGENE (Roche) according to the manufacturer's instructions. 2 days after transfection, cells were further cultured with or without additional treatments as indicated in different figures and harvested. Luciferase activities were determined. Individual sample was performed in triplicate and all the assays were repeated for at least 3 times. shRNA knockdown efficiency against Evc/Evc2 was verified each time by real-time PCR for each transfection.
Immunofluorescence
Cells were seeded on poly-𝒟-Lysine coated LAB-TEK chamber slides and infected twice with Evc/Evc2 shRNA viruses, followed by treating with the indicated reagents for the indicated time. Cells were washed 3 times with 1× PBS and fixed with 4% PFA, stained and observed using Zeiss LSM510 confocal microscope.
Acknowledgments
We thank Drs Alec Zhang, L Lum (University of Texas Southwestern Medical Center at Dallas, USA), CC Hui (the Hospital for Sick Children, Canada), James Chen (Stanford University, USA) and Pao-Tien Chuang (University of California, San Francisco, USA) for providing reagents. J Jiang was supported by grants from NIH (GM61269 and GM067045), Welch Foundation (I-1603) and CPRIT (RP100561). Y Chen was supported by American Heart Association postdoctoral fellowship (10POST3640046) and the National Natural Science Foundation of China (31271579). C Yang and Y Chen were both supported by Kunming Institute of Zoology startup fund and Chinese Academy of Sciences.
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
Diagrams show the sequences of the corresponding shRNAs targeting EVC and EVC2.
Evc/Evc2 knockdown does not affect Gli2 overexpression-induced 8×Gli-luciferase activity in NIH3T3 cells.
Evc/Evc2 knockdown reduces Shh-induced Gli1/2 ciliary accumulation.
The Shh-induced association between Smo and Evc/Evc2 depends on Kif3a or primary cilium.
References
- Jiang J, Hui CC. Hedgehog signaling in development and cancer. Dev Cell. 2008;15:801–812. doi: 10.1016/j.devcel.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingham PW, McMahon AP. Hedgehog signaling in animal development: paradigms and principles. Genes Dev. 2001;15:3059–3087. doi: 10.1101/gad.938601. [DOI] [PubMed] [Google Scholar]
- Nusslein-Volhard C, Wieschaus E. Mutations affecting segment number and polarity in Drosophila. Nature. 1980;287:795–801. doi: 10.1038/287795a0. [DOI] [PubMed] [Google Scholar]
- Jia J, Jiang J. Decoding the Hedgehog signal in animal development. Cell Mol Life Sci. 2006;63:1249–1265. doi: 10.1007/s00018-005-5519-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J. Regulation of Hh/Gli signaling by dual ubiquitin pathways. Cell Cycle. 2006;5:2457–2463. doi: 10.4161/cc.5.21.3406. [DOI] [PubMed] [Google Scholar]
- Jia J, Tong C, Wang B, Luo L, Jiang J. Hedgehog signalling activity of Smoothened requires phosphorylation by protein kinase A and casein kinase I. Nature. 2004;432:1045–1050. doi: 10.1038/nature03179. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Tong C, Jiang J. Hedgehog regulates smoothened activity by inducing a conformational switch. Nature. 2007;450:252–258. doi: 10.1038/nature06225. [DOI] [PubMed] [Google Scholar]
- Chen Y, Li S, Tong C, et al. G protein-coupled receptor kinase 2 promotes high-level Hedgehog signaling by regulating the active state of Smo through kinase-dependent and kinase-independent mechanisms in Drosophila. Genes Dev. 2010;24:2054–2067. doi: 10.1101/gad.1948710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Sasai N, Ma G, et al. Sonic Hedgehog dependent phosphorylation by CK1alpha and GRK2 is required for ciliary accumulation and activation of smoothened. PLoS Biol. 2011;9:e1001083. doi: 10.1371/journal.pbio.1001083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbit KC, Aanstad P, Singla V, Norman AR, Stainier DY, Reiter JF. Vertebrate Smoothened functions at the primary cilium. Nature. 2005;437:1018–1021. doi: 10.1038/nature04117. [DOI] [PubMed] [Google Scholar]
- Rohatgi R, Milenkovic L, Scott MP. Patched1 regulates hedgehog signaling at the primary cilium. Science. 2007;317:372–376. doi: 10.1126/science.1139740. [DOI] [PubMed] [Google Scholar]
- Wang B, Fallon JF, Beachy PA. Hedgehog-regulated processing of Gli3 produces an anterior/posterior repressor gradient in the developing vertebrate limb. Cell. 2000;100:423–434. doi: 10.1016/s0092-8674(00)80678-9. [DOI] [PubMed] [Google Scholar]
- Pan Y, Bai CB, Joyner AL, Wang B. Sonic hedgehog signaling regulates Gli2 transcriptional activity by suppressing its processing and degradation. Mol Cell Biol. 2006;26:3365–3377. doi: 10.1128/MCB.26.9.3365-3377.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Zhao Y, Tong C, et al. Hedgehog-regulated costal2-kinase complexes control phosphorylation and proteolytic processing of cubitus interruptus. Dev Cell. 2005;8:267–278. doi: 10.1016/j.devcel.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Ohlmeyer JT, Kalderon D. Hedgehog stimulates maturation of Cubitus interruptus into a labile transcriptional activator. Nature. 1998;396:749–753. doi: 10.1038/25533. [DOI] [PubMed] [Google Scholar]
- Ruel L, Rodriguez R, Gallet A, Lavenant-Staccini L, Therond PP. Stability and association of Smoothened, Costal2 and Fused with Cubitus interruptus are regulated by Hedgehog. Nat Cell Biol. 2003;5:907–913. doi: 10.1038/ncb1052. [DOI] [PubMed] [Google Scholar]
- Jia J, Tong C, Jiang J. Smoothened transduces Hedgehog signal by physically interacting with Costal2/Fused complex through its C-terminal tail. Genes Dev. 2003;17:2709–2720. doi: 10.1101/gad.1136603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lum L, Zhang C, Oh S, et al. Hedgehog signal transduction via Smoothened association with a cytoplasmic complex scaffolded by the atypical kinesin, Costal-2. Mol Cell. 2003;12:1261–1274. doi: 10.1016/s1097-2765(03)00426-x. [DOI] [PubMed] [Google Scholar]
- Stegman MA, Goetz JA, Ascano M, Jr, Ogden SK, Nybakken KE, Robbins DJ. The Kinesin-related protein Costal2 associates with membranes in a Hedgehog-sensitive, Smoothened-independent manner. J Biol Chem. 2004;279:7064–7071. doi: 10.1074/jbc.M311794200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Q, Li S, Jia J, Jiang J. The Hedgehog-induced Smoothened conformational switch assembles a signaling complex that activates Fused by promoting its dimerization and phosphorylation. Development. 2011;138:4219–4231. doi: 10.1242/dev.067959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Mao F, Lu Y, Wu W, Zhang L, Zhao Y. Transduction of the Hedgehog signal through the dimerization of Fused and the nuclear translocation of Cubitus interruptus. Cell Res. 2011;21:1436–1451. doi: 10.1038/cr.2011.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Kalderon D. Hedgehog activates Fused through phosphorylation to elicit a full spectrum of pathway responses. Dev Cell. 2011;20:802–814. doi: 10.1016/j.devcel.2011.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranieri N, Ruel L, Gallet A, Raisin S, Therond PP. Distinct phosphorylations on kinesin costal-2 mediate differential hedgehog signaling strength. Dev Cell. 2012;22:279–294. doi: 10.1016/j.devcel.2011.12.002. [DOI] [PubMed] [Google Scholar]
- Liem KF, Jr, He M, Ocbina PJ, Anderson KV. Mouse Kif7/Costal2 is a cilia-associated protein that regulates Sonic hedgehog signaling. Proc Natl Acad Sci USA. 2009;106:13377–13382. doi: 10.1073/pnas.0906944106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu SH, Zhang X, Yu C, et al. Kif7 promotes hedgehog signaling in growth plate chondrocytes by restricting the inhibitory function of Sufu. Development. 2011;138:3791–3801. doi: 10.1242/dev.069492. [DOI] [PubMed] [Google Scholar]
- Endoh-Yamagami S, Evangelista M, Wilson D, et al. The mammalian Cos2 homolog Kif7 plays an essential role in modulating Hh signal transduction during development. Curr Biol. 2009;19:1320–1326. doi: 10.1016/j.cub.2009.06.046. [DOI] [PubMed] [Google Scholar]
- Cheung HO, Zhang X, Ribeiro A, et al. The kinesin protein Kif7 is a critical regulator of Gli transcription factors in mammalian hedgehog signaling. Sci Signal. 2009;2:ra29. doi: 10.1126/scisignal.2000405. [DOI] [PubMed] [Google Scholar]
- Svard J, Heby-Henricson K, Persson-Lek M, et al. Genetic elimination of Suppressor of fused reveals an essential repressor function in the mammalian Hedgehog signaling pathway. Dev Cell. 2006;10:187–197. doi: 10.1016/j.devcel.2005.12.013. [DOI] [PubMed] [Google Scholar]
- Merchant M, Evangelista M, Luoh SM, et al. Loss of the serine/threonine kinase fused results in postnatal growth defects and lethality due to progressive hydrocephalus. Mol Cell Biol. 2005;25:7054–7068. doi: 10.1128/MCB.25.16.7054-7068.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen MH, Gao N, Kawakami T, Chuang PT. Mice deficient in the fused homolog do not exhibit phenotypes indicative of perturbed hedgehog signaling during embryonic development. Mol Cell Biol. 2005;25:7042–7053. doi: 10.1128/MCB.25.16.7042-7053.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Perez VL, Ide SE, Strom TM, et al. Mutations in a new gene in Ellis-van Creveld syndrome and Weyers acrodental dysostosis. Nat Genet. 2000;24:283–286. doi: 10.1038/73508. [DOI] [PubMed] [Google Scholar]
- Ruiz-Perez VL, Tompson SW, Blair HJ, et al. Mutations in two nonhomologous genes in a head-to-head configuration cause Ellis-van Creveld syndrome. Am J Hum Genet. 2003;72:728–732. doi: 10.1086/368063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tompson SW, Ruiz-Perez VL, Blair HJ, et al. Sequencing EVC and EVC2 identifies mutations in two-thirds of Ellis-van Creveld syndrome patients. Hum Genet. 2007;120:663–670. doi: 10.1007/s00439-006-0237-7. [DOI] [PubMed] [Google Scholar]
- Valencia M, Lapunzina P, Lim D, et al. Widening the mutation spectrum of EVC and EVC2: ectopic expression of Weyer variants in NIH 3T3 fibroblasts disrupts Hedgehog signaling. Hum Mutat. 2009;30:1667–1675. doi: 10.1002/humu.21117. [DOI] [PubMed] [Google Scholar]
- Pacheco M, Valencia M, Caparros-Martin JA, Mulero F, Goodship JA, Ruiz-Perez VL. Evc works in chondrocytes and osteoblasts to regulate multiple aspects of growth plate development in the appendicular skeleton and cranial base. Bone. 2012;50:28–41. doi: 10.1016/j.bone.2011.08.025. [DOI] [PubMed] [Google Scholar]
- Blair HJ, Tompson S, Liu YN, et al. Evc2 is a positive modulator of Hedgehog signalling that interacts with Evc at the cilia membrane and is also found in the nucleus. BMC Biol. 2011;9:14. doi: 10.1186/1741-7007-9-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Perez VL, Blair HJ, Rodriguez-Andres ME, et al. Evc is a positive mediator of Ihh-regulated bone growth that localises at the base of chondrocyte cilia. Development. 2007;134:2903–2912. doi: 10.1242/dev.007542. [DOI] [PubMed] [Google Scholar]
- Hyman JM, Firestone AJ, Heine VM, et al. Small-molecule inhibitors reveal multiple strategies for Hedgehog pathway blockade. Proc Natl Acad Sci USA. 2009;106:14132–14137. doi: 10.1073/pnas.0907134106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggenschwiler JT, Anderson KV. Cilia and developmental signaling. Annu Rev Cell Dev Biol. 2007;23:345–373. doi: 10.1146/annurev.cellbio.23.090506.123249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humke EW, Dorn KV, Milenkovic L, Scott MP, Rohatgi R. The output of Hedgehog signaling is controlled by the dynamic association between Suppressor of Fused and the Gli proteins. Genes Dev. 2010;24:670–682. doi: 10.1101/gad.1902910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tukachinsky H, Lopez LV, Salic A. A mechanism for vertebrate Hedgehog signaling: recruitment to cilia and dissociation of SuFu-Gli protein complexes. J Cell Biol. 2010;191:415–428. doi: 10.1083/jcb.201004108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen MH, Wilson CW, Li YJ, et al. Cilium-independent regulation of Gli protein function by Sufu in Hedgehog signaling is evolutionarily conserved. Genes Dev. 2009;23:1910–1928. doi: 10.1101/gad.1794109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JK, Taipale J, Cooper MK, Beachy PA. Inhibition of Hedgehog signaling by direct binding of cyclopamine to Smoothened. Genes Dev. 2002;16:2743–2748. doi: 10.1101/gad.1025302. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Diagrams show the sequences of the corresponding shRNAs targeting EVC and EVC2.
Evc/Evc2 knockdown does not affect Gli2 overexpression-induced 8×Gli-luciferase activity in NIH3T3 cells.
Evc/Evc2 knockdown reduces Shh-induced Gli1/2 ciliary accumulation.
The Shh-induced association between Smo and Evc/Evc2 depends on Kif3a or primary cilium.