Abstract
Background:
Radio- and chemotherapy (RT/CT) resistance hampers success in combating small and non-small cell lung cancers (SCLC/NSCLC). The underlying molecular mechanisms of RT/CT resistance of LCs are multifactorial and have been understood in part hitherto. miRNAs, key regulators of mRNAs, are well-recognised oncomirs; however, their role in regulating RT response remains poorly understood.
Methods:
Six human NSCLC and five SCLC cell lines with different SF2 values were investigated. Using microarray we examined whether expression of miRNAs is linked to the RT resistance of NSCLCs or SCLCs. Obtained data were validated by quantitative real-time PCR. Apoptosis and senescence were analysed using siRNA transfection, western blot and flow cytometry.
Results:
miRNA-21, miRNA-1827, miRNA-214, miRNA-339-5p, miRNA-625, miRNA-768-3p, miRNA-523-3p, miRNA-1227, miRNA-324-5p, miRNA-423-3p, miRNA-1301 and miRNA-1249 are differentially expressed in LC cells. miRNA-214 is upregulated in RT-resistant NSCLC cells relative to radiosensitive counterparts. Considering miRNA-214 as a putative regulator of RT resistance, we demonstrate that knockdown of miRNA-214 in radioresistant NSCLCs sensitised them to RT by stimulation of senescence. Consistently, overexpression of miRNA-214 in radiosensitive NSCLCs protected against RT-induced apoptosis. Protection was mediated by p38MAPK, as downregulation of this kinase could reverse the miRNA-214 overexpression-induced resistance of NSCLC cells.
Conclusion:
miRNA profiling of LC revealed putative RT resistance signalling circuits, which might help in sensitisation of LC to RT.
Keywords: miRNA, microarray, lung cancer, small cell lung cancer, non-small cell lung cancer, radiotherapy
Lung cancer (LC) is the leading cause of cancer-related deaths worldwide (Jemal et al, 2008). On the basis of differences in histology, LC is roughly divided into small- and non-small cell LC (SCLC and NSCLC, respectively). The latter, which accounts for about 85% of all LC cases, can be further sub-classified into adeno-, squamous cell and large cell carcinomas. At early stage of disease development LC is asymptomatic and by the time of detection it often reaches an advanced stage where therapy response is limited and hence LC patients have poor prognosis. Radiotherapy (RT) has different roles at different stages of LC. In the treatment of early stages of NSCLC RT may be used as an adjunctive with or without chemotherapy (CT) even if this is still debated (Herbst et al, 2008; Hammerschmidt and Wirtz, 2009). In locally advanced inoperable LC, RT is delivered concomitant with CT. The CT regimen of NSCLC usually consists of platinum compounds, for example, cisplatin, carboplatin or oxaliplatin along with gemcitabine (Saha and Rudd, 2006). Regrettably, the response rate of NSCLC to first-line single-drug treatment is poor, about 10–15% (Edelman, 2004). In contrast, in SCLC, for which the CT course includes platinum compounds in combination with etoposide, the initial response rate is about 80–90% (Schmittel, 2011) but a large proportion of the cases recur with a CT refractory disease (Sher et al, 2008). In locally advanced SCLC limited to the thorax, RT is delivered concomitant with CT with a small fraction of patients (15–18%) reaching long-term survival, that is, 5 years. Efficacy of RT differs, however, between tumours among patients and understanding of the mechanisms behind this is highly warranted. Many biological factors have been suggested to be responsible for the treatment outcome after RT and among them tumour intrinsic cellular resistance remains an essential component. The underlying molecular mechanisms of CT and RT resistance of LC are multifactorial. We and others previously showed that a CT/RT refractory phenotype of LC may be explained by lack of proper apoptosis signalling, increased DNA-repair capacity and increased growth factor signalling circuits, for example, epidermal growth factor receptor and insulin growth factor 1 receptor (IGF-1R) (Dong et al, 2002; Arteaga, 2006; Cosaceanu et al, 2007; Viktorsson and Lewensohn, 2007). These altered circuits in SCLC and NSCLC require further exploration, and analysis of miRNAs expression pattern could be one avenue to identify putative novel targets with CT/RT-sensitising potential as well as to reveal biomarkers of CT/RT response.
miRNAs are small endogenous non-coding single-stranded RNA molecules of about 18–25 nucleotides. They negatively regulate gene expression at the post-transcriptional level by either degrading the target mRNA or inhibiting translation of the mRNA into functional protein (Wightman et al, 1993; Hutvagner and Zamore, 2002). The Sanger miRBase reports over 2000 miRNAs in the human genome and these miRNAs have indeed been shown to regulate multiple hallmarks of cancer, for example, increased proliferation and evasion of cell death (Ketting et al, 2001; He et al, 2005). Conceptually, miRNAs may have either tumour suppressor or oncogene potential within cells if they target oncogenes or tumour suppressor genes, respectively. The first description of the role of miRNAs in tumorigenesis was the identification of their localisation in close proximity to chromosomal breakpoints (Calin et al, 2004; Lu et al, 2005). Subsequent analysis revealed aberrant miRNA expression and function in many cancers, for example, miRNA-15a/16-1 and let-7 as tumour suppressors (Johnson et al, 2005; Bonci et al, 2008) and miRNA-17-92 cluster as oncogene (Hayashita et al, 2005). There are few studies supporting a link between miRNA expression and CT response of LC. It has been shown that cisplatin- and carboplatin-induced pro-apoptotic signalling pathways mediated by p53 are inhibited by miR-630 (Galluzzi et al, 2010). Moreover, certain miRNAs have been linked to RT response of some tumours, for example, miR-17-92 cluster, which impedes RT response of human mantle cell lymphoma (Jiang et al, 2010), miR-181 sensitising human gliomas to RT by targeting Bcl-2 (Chen et al, 2010), and miR-221/miR-222, which reported to regulate RT response of gastric carcinoma cells via phosphatase and tensin homologue (PTEN) (Chun-Zhi et al, 2010). Importantly, in LC, few studies have focused on the relationship between miRNA expression and RT responsiveness. It has been shown, however, that overexpression of miRNA-9 and let-7g in H1299 LC cells results in RT sensitisation by interfering with NF-κB pro-survival cascades (Arora et al, 2011). Furthermore, it has been shown that miRNA-101 sensitises LC cells to RT through targeting DNA-PKcs and ATM, thereby impairing the DNA-repair machinery (Yan et al, 2010). It has also been reported that upregulation of let-7a in NSCLC can sensitise these cells for RT through downregulation of K-RAS (Oh et al, 2010). Importantly, to our knowledge there are no studies which have, on a global scale, analysed miRNA expression in LC cells in relation to RT responsiveness. Therefore, we performed global miRNA expression profiling to find a link between miRNAs and RT resistance of SCLC and NSCLC. For this purpose, a panel of cell lines that represent both types of LC was used, displaying different radiosensitivity as measured by surviving cell fraction at 2 Gy (SF2) in clonogenic assays. Global profiling of SCLCs linked miRNA-324-5p to RR character of SCLC. In NSCLC cells, miRNA-214 was the only miRNA that was differentially expressed in the radioresistant (RR) cell lines vs the radiosensitive (RS) counterparts. We found that knockdown of miRNA-214 in RR NSCLC cells sensitised them to RT by increased senescence propensity. Along the same line, overexpression of miRNA-214 in RS NSCLC cells reversed the sensitivity of the cells through the regulation of p38MAPK pathway. Thus, our results suggest miRNA-214 to be a candidate biomarker of RT response of NSCLC and also propose it as a novel target with RT-sensitising potential.
Materials and methods
Cell culture, siRNA transfection and irradiation
The human NSCLC cell lines H23, A549, H157, H661, H1299 and U-1810 and the SCLC cell lines H69, H82, U-1285, U-1690 and U-1906 were used (Bergh et al, 1982, 1985; Carney et al, 1983; Carmichael et al, 1989; Sirzen et al, 1998). All cells were maintained in RPMI 1640 medium (Invitrogen, Stockholm, Sweden) supplemented with 10% of heat-inactivated fetal bovine serum (FBS), ℒ-glutamine (2 mℳ), penicillin (100 U ml−1) and streptomycin (100 μg ml−1) (Gibco, Invitrogen, Paisley, UK) at 37 °C and 5% CO2. The cell lines were divided into RR and RS groups according to the values of their surviving fraction after radiation at 2 Gy (SF2) obtained in clonogenic assays (Carmichael et al, 1989; Sirzen et al, 1998; Franken et al, 2006). Cells were regarded as RR with SF2⩾0.3 and RS with SF2<0.3. To knockdown or overexpress miRNA-214, the cells were grown in complete medium for 24 h and then transfected with miRNA antagomir (cat#IH-300569-08-0005) or mimic (cat#C-300569-07-0005), respectively. After 48 h of transfection, cells were transferred to complete medium and irradiated on ice with 8 Gy by means of a Co60 source. p38MAPK-α was silenced using custom designed siRNA (5′-GAACUGCGGUUACUUAAACUU-3′) (Dharmacon, Thermo Fisher Scientific Inc., Lafayette, CO, USA).
RNA extraction, labelling and microarray analysis
Total RNA was extracted from 10 × 106 cells by using Trizol reagent (Invitrogen, Carlsbad, CA, USA), as previously described (Rio et al, 2010). The integrity of total RNA was examined by using 1.5% agarose gel electrophoresis. The Affymetrix Genechip miRNA array process was executed according to the manufacturer’s protocol (FlashTag Genisphere, Genisphere Inc., Hatfield, PA, USA). The Genechip used for the miRNA array consisted of 46 228 different probes comprising 7815 probe sets (Affymetrix Inc., Santa Clara, CA, USA). For this analysis, the FlashTag Biotin RNA Labelling Kit (FT30AFYB, Genisphere Inc.) was used to label 1 μg of purified RNA with biotin. After hybridisation, washing and staining steps were performed at Fluidics Station 450 in accordance with the manufacturer’s instructions (FlashTag Genisphere, Genisphere Inc.). The resulting hybridisation signal of biotin binding to a streptavidin–PE conjugate from the bound miRNA was detected with an Affymetrix scanner. To obtain statistically significant results with regard to miRNA expression, three independent samples from each cell line were examined.
Validation of miRNA microarray results by TaqMan miRNA real-time quantitative PCR analysis
Quantitative real-time PCR (q-RT–PCR) was used to validate the miRNA expression results. Eight differentially expressed miRNAs were selected for the validation in which the same RNA preparations, as used in the microarray, were subjected to q-RT–PCR. The selected miRNAs were reverse transcribed to cDNA by means of miRNA-specific primers: hsa-miRNA-214 (assay id: 002306), hsa-miRNA-324-5p (assay id: 000539), hsa-miRNA-1827 (assay id:002814), hsa-miRNA-532-3p (assay id:002355), hsa-miRNA-625 (assay id:002432), hsa-miRNA-1227 (assay id:002769), hsa-miRNA-1301 (assay id:002827), hsa-miRNA-1249 (assay id: 002868), and control miRNA Assay U6 snRNA (assay id: 001973) (Applied Biosystems, Carlsbad, CA, USA). q-RT–PCR was performed on the ABI (Applied Biosystems) 7900 HT Thermal cycler in standard mode for 40 cycles. Relative miRNA expression values (Target miRNA vs U6) were calculated with the 2−(ΔΔCt) method (Livak and Schmittgen, 2001).
Target prediction analysis
To find putative target genes of miRNA-214 and build hypothesis concerning its role in RT response of NSCLC, several bioinformatics algorithms were employed, that is, miRBase predicted mRNA targets of miRNAs (http://www.mirbase.org/), Targetscan (http://www.targetscan.org/), microrna.org (http://www.microrna.org/microrna/home.do), microcosm (http://www.ebi.ac.uk/enright-srv/microcosm/htdocs/targets/v5/) and PicTar (http://pictar.mdc-berlin.de/). The targets of miRNA-214, designated by at least three of the databases described above, were in the focus of further analysis and were also loaded into Ingenuity Pathway Analysis (IPA) (2011 Ingenuity Systems Inc., Redwood City, CA, USA).
Overexpression or downregulation of miRNA-214 expression by miRNA-214 mimic or antagomir
Cells (4 × 105 cells for H23 and 2 × 105 cells for U-1810) were seeded 24 h before 48 h transfection with miRNA-214 mimic or antagomir, respectively. In these experiments transfection reagents from Dharmacon (Thermo Scientific Inc.) were used and efficiency was confirmed by analysing miRNA-214 expression in 10 ng RNA samples from each reaction by means of the primers and methods described above.
PTEN expression analysis and validation of siRNA-mediated p38MAPK-α silencing by q-RT–PCR
PTEN mRNA level after overexpression or downregulation of miRNA-214, and p38MAPK-α mRNA level after its downregulation by siRNA transfection were analysed by SYBERGreen q-RT–PCR. The following primers: PTEN forward: 5′-AGTTCCCTCAGCCGTTACCT-3′, reverse: 5′-GAGGTTTCCTCTGGTCCTGGTA-3′, (Eurofins MWG Operon, Ebersberg, Germany), p38MAPK-α forward: 5′-GAAGAAGCTCTCCAGACCATTTC-3′, reverse: 5′-AACGTCCAACAGACCAATCAC-3′ and 18S ribosomal RNA,18S-1: 5′-CGCTACTACCGATTGGATGGTT-3′ and 18S-2: 5′-AGTCAAGTTCGACCGTCTTCTC-3′ (Invitrogen, Sweden) were consumed to carry out q-RT–PCR on a 7500 Real-Time PCR System (Applied Biosystems).
Assessment of apoptosis
To evaluate the percentage of apoptosis (cellular shrinkage, chromatin condensation and DNA fragmentation), cells were harvested 48 h post irradiation with cell dissociation solution (Sigma Aldrich, Stockholm, Sweden). Attached and floating cells were pooled and fixed in cold ethanol (70%). Fixed cells were centrifuged at 2000, r.p.m. for 5 min and washed twice with 1 × PBS. Nuclear morphology was assessed under a fluorescent microscope (Carl Zeiss Inc., Thornwood, NY, USA). Cell nuclei, which showed chromatin condensation and DNA fragmentation, were considered as apoptotic and the percentage of cells with apoptotic nuclei among 200 cells examined was quantified.
Caspase-3 activity and cell cycle analysis
Cells were fixed in ethanol and after washing in 1 × PBS the amount of active caspase-3 was assessed by staining digitonin-permeabilised cells with a FITC-conjugated antibody recognising active caspase-3 (BD Pharmingen, Franklin Lakes, NJ, USA). A mixture of digitonin (100 μg ml−1) and FITC-conjugated antibody (1 : 20 dilution) was prepared in 100 μl of 1 × PBS and cells were stained at room temperature for 1 h in the dark with mild agitation. After washing twice in 1 × PBS the amount of active caspase-3 was quantified in the FL1 channel on a FACS-Calibur flow cytometer (BD Biosciences, San Jose, CA, USA). Ten thousand cells were analysed for each sample with the Cell Quest Program and the percentage of cells with active caspase-3 was taken as readout and, subsequently, compared with scrambled (non-targeting, NT) and siRNA transfected samples with or without irradiation.
For cell cycle analysis, ethanol-fixed cells were, after washing with 1 × PBS, treated with RNaseA (100 μg ml−1) for 1 h at 37 °C and then stained with propidium iodide (50 μg ml−1) for 30 min at room temperature under mild agitation. The distribution of cells in different phases of the cell cycle (G1, S and G2/M) was analysed on a FACS-Calibur cytometer (BD Biosciences) and calculated with ModFitLT V2.0 software (Verity Software House Inc., Topsham, ME, USA).
Assessment of senescence
To assess senescence, β-galactosidase activity was measured with a histochemical staining kit (Sigma Aldrich; cat#CS0030) and performed according to the manufacturer’s instructions. Briefly, NT or miRNA-214 antagomir-transfected cells at 72 h post irradiation were washed twice with 1 × PBS, fixed in 1 × fixation buffer for 5 min, washed three times with 1 × PBS and incubated with the staining mixture overnight at 37 °C. β-Galactosidase-positive cells were visualised as blue cells, when examined by a light microscope. For both NT and miRNA-214 antagomir-treated samples a total of 100 cells were counted from different fields of the plate and the percentage of blue cells was calculated.
Western blot analysis
Protein extraction from 1 × 106 cells was performed with either complete lysis-M reagent (Roche Diagnostic GmbH, Mannheim, Germany) or urea buffer (containing 6 ℳ urea, 200 mℳ ammonium bicarbonate, 2% SDS) supplemented with protease inhibitor cocktail (PIC, Complete mini, Roche Diagnostic GmbH). Protein concentration was measured by BCA protein assay (Pierce, Thermo Scientific, Rockford, IL, USA) and lysates were mixed with 5 × Laemlli’s loading buffer. Samples were preheated for 10 min at 95 °C and 30 μg of protein from each sample were subjected to SDS–polyacrylamide gel electrophoresis followed by transfer to nitrocellulose membrane (Trans-Blot, Bio-Rad, Hercules, CA, USA) at 0.25 A for 2 h at 4 °C in transfer buffer (25 mℳ Tris, 200 mℳ glycine, 20% methanol). Membranes were blocked for 1 h in 5% non-fat dry milk or 5% BSA in Tris Buffer saline Tween-20 (TBS-T) at room temperature and probed with primary antibodies overnight at 4 °C and with secondary antibody for 2 h at room temperature. The following antibodies were applied: anti-Akt, anti-Akt-p (Ser473) (Thr308), anti PTEN, anti-PTEN-p (Ser380), anti-PI3K, anti-PI3K-p (p85 (Tyr458)/p55 (Tyr199)), anti-p38MAPK, anti-p38MAPK-p (Thr180/Tyr182), anti-HSP27-p (Ser82), anti-FoxO4, anti-CDK6, anti-cleaved PARP (Asp214) (all from Cell Signaling Technology, Inc., Trask Lane, Danvers, MA, USA), anti-PARP (BD Pharmingen, Stockholm, Sweden) and anti-FoxO4-p (Thr-451) (Abcam, Cambridge, UK). Equal loading was verified with Ku80 (XRCC5) (Abcam), anti-actin (Sigma Aldrich) or anti-G3PDH (Trevingen, Gaithersburg, MD, USA). In blots of H23 cells, the binding of primary antibodies was detected by horseradish peroxidase-labelled secondary antibodies (anti-mouse IgG and anti-rabbit IgG). For HRP detection, an ECL chemiluminescence kit (GE Healthcare, Little Chalfont, Buckinghamshire, UK) was used according to the manufacturer’s instructions and exposed to X-ray film. Protein densitometry was executed by using Image J software (http://rsbweb.nih.gov/ij/). For the detection of proteins in U-1810 cell lysates, after incubation with primary antibody membranes were exposed to Alexa fluor 680-conjugated antibodies (anti-mouse IgG or anti-rabbit IgG, Invitrogen), or Alexa fluor 800-labelled counterparts (goat anti-mouse or goat anti-rabbit, Odyssey). An Odyssey (LI-COR Biosciences, Lincoln, NE, USA) scanner was used to detect the signal from the Alexa fluor-labelling and Odyssey Sa-infrared imaging system application software (version 1.0.12) was used to analyse the images by examining near infrared light of the corresponding protein bands.
Statistical analysis
The results from three independent experiments were presented as mean of ±s.d. One-tailed paired t-test was used for statistical evaluation.
Results
To analyse whether expression of certain miRNAs can be linked to RT resistance of LC cells, miRNA expression profiling was performed in a panel of NSCLC and SCLC cell lines. Three biological replicates for each cell line were applied in Affymetrix miRNA microarray and the data were further analysed in relation to the SF2 values (Figure 1A). Four probes for each miRNA were on the microarray chip. The detected signal intensities of the probes were normalised to the background signal and the P-value of each miRNA were calculated. Those miRNAs showing signal intensity of four or more and, importantly, P-value of <0.06 were regarded as expressed, whereas the others were under detection level and excluded from the subsequent analyses. Total profiling of miRNA in different NSCLC and SCLC cell lines revealed that each cell line expressed ∼100–200 different miRNAs (Figure 1B). For each LC subtype, SCLC and NSCLC, the cell lines were grouped into RS (SF2<0.3) and RR (SF2⩾0.3) on the basis of their clonogenic survival data (Bergh et al, 1982, 1985; Carney et al, 1983; Carmichael et al, 1989; Sirzen et al, 1998). The expression of the different individual miRNAs within each of these cohorts was examined. In total, eight miRNAs in NSCLC were found to be expressed in the RR cell lines cohort and in either none or only one of the RS counterparts (Table 1). Moreover, two miRNAs (miRNA-1260 and miRNA-558) were differentially detected in the RS NSCLC cell lines counterpart. Similarly, five miRNAs were found to be expressed in all the RR SCLC cell lines and either in one or no RS counterpart, whereas no miRNA could be linked to an RS phenotype in the SCLC cell line panel (Table 1).
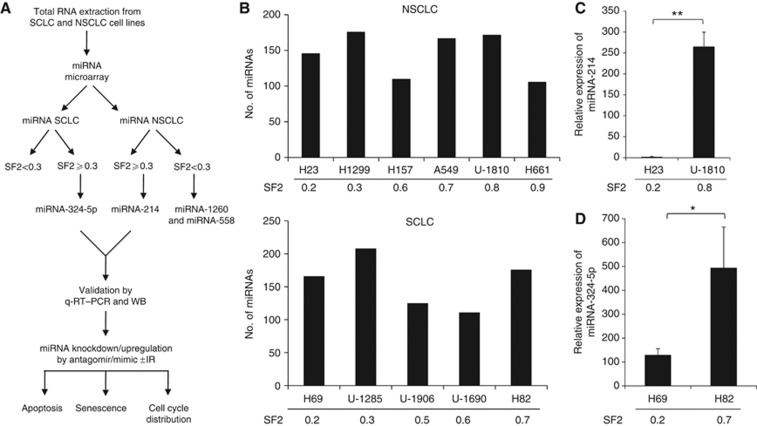
Figure 1.
Outline of miRNA expression profiling in LC cells. (A) Eleven lung cancer cell lines (five SCLC and six NSCLC) were analysed. Total RNA was isolated and subjected to miRNA expression profiling using Affymetrix miRNA chips. Both SCLC and NSCLC were each divided into RS and RR groups on the basis of SF2 values. For validation of the microarray results real-time quantitative PCR was performed on the differentially expressed miRNAs. miRNA-214 and NSCLC cells were in focus of subsequent analysis presented in this study as it was upregulated in all the RR and downregulated in all the RS NSCLC cell lines. The miRNA-214 was knocked down in U-1810 cells by transfecting with miRNA-214 antagomir and conversely the miRNA-214 was overexpressed in H23 cells by using the mimic of miRNA-214. After the transfection, apoptosis, senescence and cell cycle distribution were assessed. Validation of the predicted targets of the miRNA-214 was performed by western blot. (B) Total RNA was extracted from three biological replicates of each cell line and hybridised with microarray chips. The total number of miRNAs detected in each cell line is shown. The numbers below the bars in B, C and D, represent the mean of SF2 value of each cell line. (C, D) Validation of miRNA-214 in NSCLC and miRNA-324-5p in SCLC cell lines with different radiosensitivity/radioresistance using a real-time quantitative PCR. The expression levels of the miRNAs were quantified by q-RT–PCR. The Ct value of each sample was compared with internal control (snU6) and delta value was calculated and normalised to one. Data represent the mean±s.d. of three biological replicates of each cell line tested. Paired t-test was used to analyse statistics (*P<0.05, **P<0.01). (C) miRNA-214 in NSCLC cell lines, H23 and U-1810. (D) miRNA-324-5p in SCLC cell lines, H69 and H82. Abbreviation: WB=western blot.
Table 1. miRNAs expressed in RR and RS NSCLC or SCLC cell lines with SF2⩾0.3 or <0.3, respectively.
| miRNA in RR cell lines | miRNA in RS cell lines | |
|---|---|---|
| NSCLC | miRNA-214 | miRNA-1260 |
| miRNA-1827 | miRNA-558 | |
| miRNA-21 | ||
| miRNA-339-5p | ||
| miRNA-625 | ||
| miRNA-768-3p | ||
| miRNA-532-3p | ||
| miRNA-1301 | ||
| SCLC | miRNA-324-5p | |
| miRNA-1227 | ||
| miRNA-625 | ||
| miRNA-423-3p | ||
| miRNA-1249 |
Abbreviations: NSCLC=non-small cell lung cancer; RR=radioresistant; RS=radiosensitive; SCLC=small cell lung cancer; SF2=surviving cell fraction at 2 Gy.
Note, no miRNAs were found to be common among the RS SCLC cell lines.
miRNA-214 and miRNA-324-5p showed higher expression in RR than RS LC cells
Among the differentially expressed miRNAs, miRNA-214 and miRNA-324-5p were found to be expressed only in the RR NSCLC and SCLC cell lines, respectively (Table 1). These results were validated with q-RT–PCR. Indeed, the validation revealed that miRNA-214 had a higher expression (about 250-fold) in the U-1810 cells than in the H23 cells (representatives of RR and RS NSCLC, respectively) (Figure 1C). Similarly, miRNA-324-5p expression was examined in the H82 cells and H69 cells (representatives of RR and RS SCLC, respectively). As demonstrated, H82 cells had about five-fold higher expression of miRNA-324-5p compared with H69 cells (Figure 1D). Thus, the miRNA findings from the microarray were validated by q-RT–PCR.
miRNA-214 antagomir sensitises RR NSCLC cells to IR
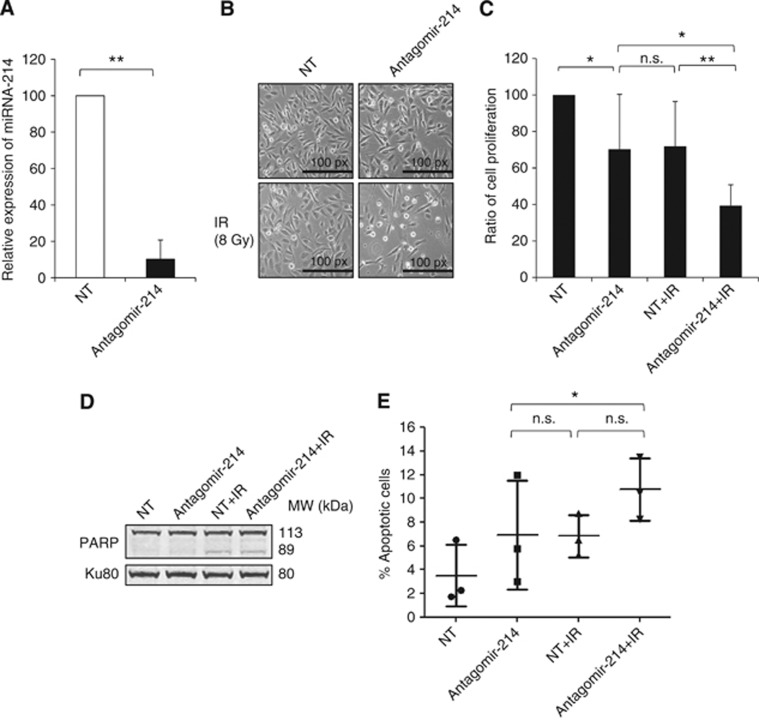
To assess whether miRNA-214 downregulation could sensitise NSCLC cells to ionising radiation (IR), its expression was knocked down by transfecting the U-1810 cells with miRNA-214 antagomir. The efficiency of antagomir-induced miRNA-214 downregulation was confirmed at 24 and 48 h of transfection and this effect lasted for at least 5 subsequent days (Supplementary Figure S1). To verify the role of miRNA-214 in RT resistance of NSCLC, miRNA-214 antagomir-transfected U-1810 cells were irradiated and their response was analysed. Under these conditions, the miRNA-214 expression level was reduced by about 90% relative to NT miRNA-antagomir (NT)-transfected U-1810 cells (Figure 2A). The effect of miRNA-214 on cell proliferation was analysed at 48 h post IR (Figure 2B). Downregulation of miRNA-214 expression per se was found to reduce cell proliferation, as demonstrated by the lower density of cells transfected with miRNA-214 antagomir compared with NT-antagomir counterparts (Figure 2B). This result was also confirmed by cell counting experiments in which a reduction in cell viability of about 30% was observed (Figure 2C). Importantly, a more pronounced growth inhibition of U-1810 cells (about 60%) was found when the miRNA-214 antagomir was knocked down in combination with irradiation (Figures 2B and C). This inhibition of cell growth in the combined treatment (miRNA-214 antagomir+IR) was found to be significantly higher compared with the two individual treatments (P=0.008). This indicates that knockdown of miRNA-214 sensitises RR NSCLC U-1810 cells to radiation.
Figure 2.
Downregulation of miRNA-214 in NSCLC U-1810 cells results in their sensitisation to IR-induced cell death. (A) miRNA-214 expression was knocked down by transfecting (48 h) NSCLC U-1810 cells with miRNA-214 antagomir. The knockdown of miRNA-214 was confirmed by q-RT–PCR. Data show the relative expression of miRNA-214 in miRNA-214 antagomir-transfected samples as compared with an NT-antagomir-transfected samples in which miRNA-214 Ct value was subtracted with internal control (snU6), as described in the Materials and Methods section. The data represent the mean±s.d. of three experiments. Paired t-test was used to analyse the statistics (**P<0.01). (B) Cell morphology of U-1810 cells transfected with miRNA-214 antagomir or NT-antagomir was analysed at 48 h post IR by light microscopy ( × 10 magnification). Representative images from three biological replicates are shown. The bar indicates the magnification power of the image. (C) U-1810 cells were treated as in B and the ratio between the total number of cells plated in the beginning of the experiment and the total number of cells harvested 48 h post IR was calculated. The NT-antagomir-transfected sample was normalised to 100% cell viability and the effect of the different treatments was calculated accordingly. The data represent the mean±s.d. of three separate experiments. Paired t-test was used to analyse the statistics (*P<0.05 and **P<0.01). (D) U-1810 cells were treated as in B and 72 h post IR, PARP cleavage was analysed by western blot. The upper and the lower bands represent the full-length and cleaved PARP, respectively. Ku80 was used as a loading control. (Abbreviations: NT=non-targeting antagomir, antagomir-214=miRNA-214 antagomir, IR=ionising radiation). (E) Apoptotic nuclear morphology of U-1810 cells was examined after treatment, as indicated in B, by staining the cell nuclei with DAPI and analysis with a fluorescence microscope. For each sample, 200 cells were counted and the percentage of cells with apoptotic nuclear morphology was calculated. The data represent the mean±s.d. of three separate experiments. Paired t-test was used to analyse the statistics (Abbreviation: n.s.=non-significant).
To examine the mode of cell death induced by the combined effect of miRNA-214 antagomir and IR, several apoptosis-related markers were analysed, that is, number of cells with active caspase-3 and cleaved poly(ADP-ribose) polymerase (PARP), as well as the percentage of cells with apoptotic nuclear morphology (Figures 2D and E and Supplementary Figure S2A). Knockdown of miRNA-214 expression with or without IR did not increase the percentage of cells with active caspase-3 by more than 10% and no more than what was observed in NT-antagomir or combined IR and NT-antagomir treatment (Supplementary Figure S2A). In line with the lack of caspase-3 activity, there was no increase in PARP cleavage, when miRNA-214 antagomir and IR were combined compared with a combination of NT-antagomir and IR (Figure 2D). Moreover, only a minor fraction of cells showed apoptotic morphology in response to either of the treatments applied (Figure 2E). We also analysed whether miRNA-214 antagomir influenced IR-induced cell cycle progression (Supplementary Figure S2B). As expected, IR caused an accumulation of U-1810 cells, in which p53 is mutated, in G2 phase at 48 h, but no difference was observed between miRNA-214 and NT-antagomir-transfected irradiated cells in this respect (Supplementary Figure S2B). Thus, our data show that miRNA-214 antagomir sensitises RR NSCLC U-1810 cells to radiation by a mechanism other than apoptosis and without influencing IR-induced cell cycle perturbations.
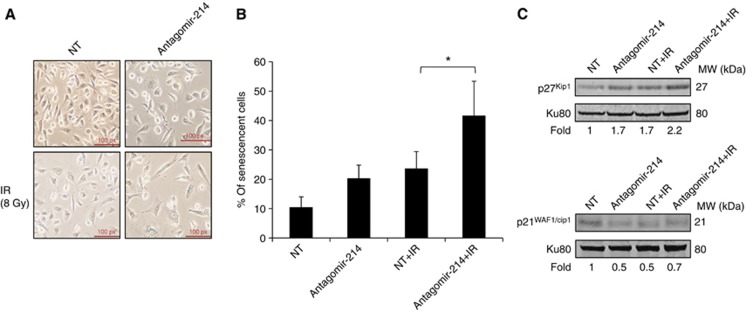
Suppression of miRNA-214 expression sensitises NSCLC cells to IR through induction of senescence
Morphological evaluation of U-1810 cells after knockdown of miRNA-214 expression in combination with IR revealed an elongated phenotype resembling cellular senescence (Figure 2B). Therefore, β-galactosidase, an immuno-histochemical marker of senescent cells, was examined in NT- or miRNA-214 antagomir-transfected cells with or without IR. It was found that 10 to 20% of the cells became senescent when treated with either the miRNA-214 or NT-antagomir separately or when IR was combined with NT-antagomir (Figures 3A and B). In contrast, when miRNA-214 antagomir and IR were applied together, a significant proportion of the cells, about 40%, became positive for β-galactosidase (P=0.04). Next, the expression level of p21WAF1/Cip1 and p27Kip1, two regulators of senescence, was analysed (Figure 3C). A 1.7-fold upregulation of p27Kip1 was found after downregulation of the miRNA-214 and after transfection with NT-antagomir with IR. The combination of miRNA-214 downregulation with IR further increased the expression of p27Kip1 2.2-fold compared with NT-treated cells (Figure 3C). In contrast, a reduction in the expression level of p21WAF1/Cip1 was observed after transfection with miRNA-214 antagomir with or without IR and after NT-transfection combined with IR. These data demonstrate that miRNA-214 downregulation followed by IR enhances upregulation of p27Kip1 and drives these NSCLC U-1810 cells to senescence rather than to apoptosis.
Figure 3.
miRNA-214 antagomir combined with ionising radiation (IR) leads to increased senescence in NSCLC U-1810 cells. U-1810 cells were treated as in Figure 2B and at 72 h post IR, cells were stained for β-galactosidase activity. (A) Examples of β-galactosidase blue-stained cells. (B) The percentage of senescent cells out of 100 cells examined was assessed in three different fields under a light microscope ( × 40 magnification). The data represent the mean±s.d. of three separate experiments. Paired t-test was used to analyse the statistics (*P<0.05). (C) p27Kip1 and p21WAF1/Cip1 were analysed by western blot at 48 h post IR in samples shown in A. Ku80 was used as a loading control (Abbreviations: NT=non-targeting antagomir, antagomir-214=miRNA-214 antagomir, IR=ionising radiation). One representative out of three different experiments is shown.
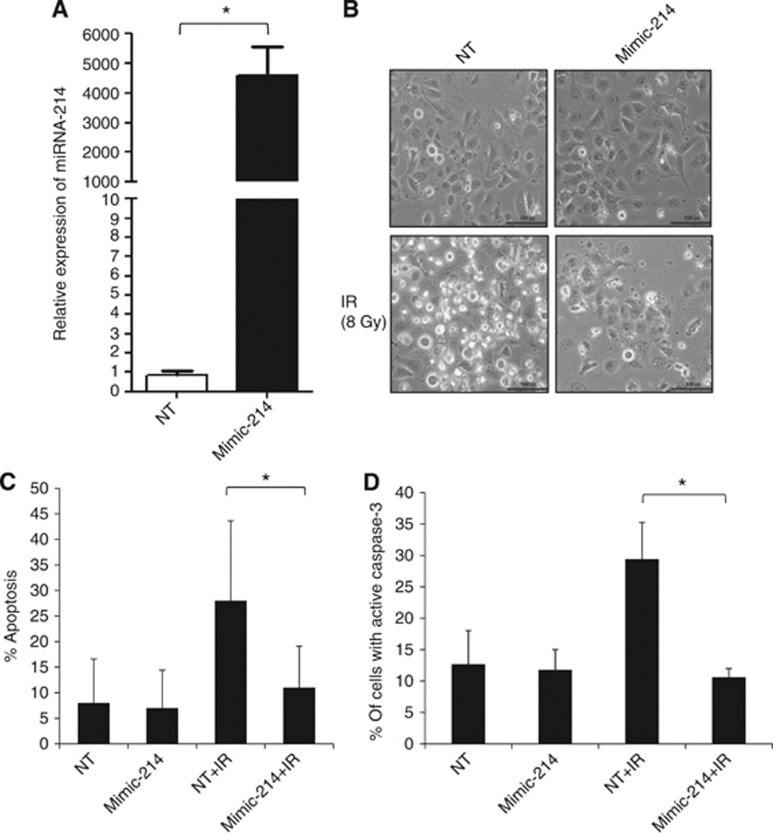
Overexpression of miRNA-214 in NSCLC cells confers IR resistance by blocking IR-induced apoptotic signalling
As downregulation of miRNA-214 was found to sensitise RR NSCLC cells to IR, we next examined whether increasing the low level of miRNA-214 could make RS NSCLC H23 cells more resistant to IR (Figure 1C). First, an increased level of miRNA-214 in H23 cells was confirmed by q-RT–PCR after miRNA-214 mimic transfection, with a relative expression value of ∼5000-fold (P-value <0.05) compared with H23 cells transfected with NT mimic (Figure 4A). Then, the response to IR in miRNA-214 or NT-mimic-transfected H23 cells was examined. Very few floating/dead cells were observed in samples with miRNA-214 overexpression and upon IR, which was in marked contrast to NT-mimic and IR-treated H23 cells where cell death was apparent (Figure 4B). Then, we studied whether increased level of miRNA-214 could block the appearance of IR-induced apoptotic morphology and caspase-3 activation (Figures 4C and D). Importantly, whereas the numbers of H23 cells with apoptotic morphology rose to ⩾25% among NT-mimic-transfected H23 cells upon irradiation, overexpression of miRNA-214 inhibited IR-induced apoptosis and only 10% of the cells showed apoptotic morphology. This percentage of cells with apoptotic phenotype was almost the same in H23 cells overexpressing miRNA-214 before and after IR (Figure 4C), illustrating a complete block of IR-induced apoptosis by miRNA-214 overexpression. In the subsequent experiments, we checked whether augmented miRNA-214 expression also blocked IR-induced caspase-3 activation (Figure 4D). In accordance with the block of changes in IR-induced apoptotic morphology by miRNA-214 overexpression, a significantly reduced number of H23 cells with active caspase-3 were found after increased expression level of the miRNA-214 and subsequent irradiation of the cells (Figure 4D). Thus, the number of H23 cells with active caspase-3 was elevated to >30% in NT samples after irradiation, which is in marked contrast to cells with miRNA-214 overexpression, where only 10% of the cells were positive for active caspase-3 staining after IR (Figure 4D). These results suggest that miRNA-214 mimic enhances radiation resistance of NSCLC by inhibiting caspase-3-mediated apoptosis and thus confirm a role for miRNA-214 in regulation of RT response of NSCLC.
Figure 4.
Overexpression of miRNA-214 in NSCLC H23 cells confers resistance to IR-induced cell death. (A) miRNA-214 expression was increased in NSCLC H23 cells by transfection with mimic miRNA-214 for 48 h, as confirmed by q-RT–PCR. Data are relative expression of miRNA-214 in a mimic miRNA-214-transfected sample as compared with a mock-transfected sample (NT) in which miRNA-214 Ct value has been subtracted with internal control (snU6), as described in the Materials and Methods. The data represent the mean±s.d. of three separate experiments. Paired t-test was used to analyse the statistics (*P<0.05). (B) Morphology of H23 cells transfected with non-targeting (NT) or mimic miRNA-214 was examined at 48 h post IR. Images shown are representatives of three independent experiments. (C) Apoptotic nuclear morphology of H23 cells was examined at 48 h post IR as described in Figure 2E. Data shown are the mean±s.d. from three independent experiments. Paired t-test was used to analyse the statistics (*P<0.05). (D) Caspase-3 activity was examined using an FITC-conjugated antibody against active caspase-3 in flow cytometric analysis of samples treated as in Figure 4B. The percentage of cells with active caspase-3 was quantified in three independent experiments. Data shown are the mean±s.d. Paired t-test was used to analyse the statistics (*P<0.05) (Abbreviations: NT=non-targeting mimic, mimic-214=mimic miRNA-214, IR=ionising radiation).
Identification of miRNA-214 targets and their validation
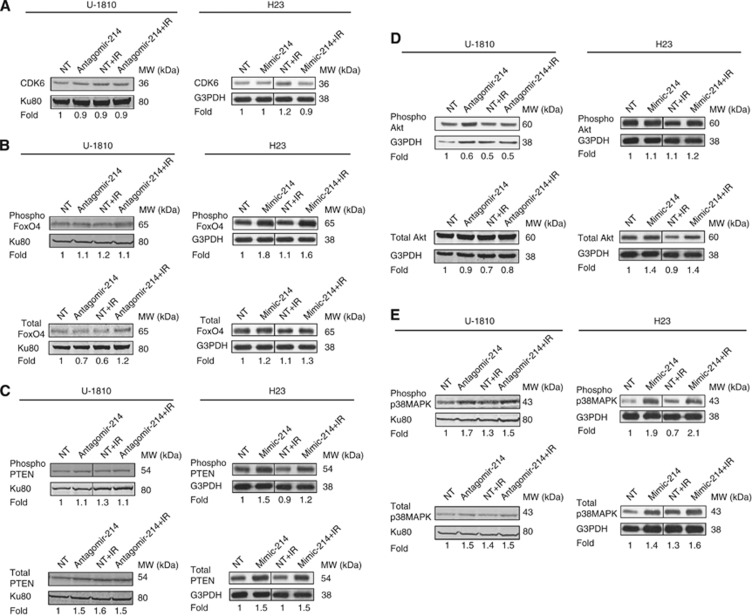
To reveal putative targets for miRNA-214, which might have a role in radiation-induced signalling circuits and hence explain its role in confining sensitivity, several different target prediction tools were used (see Materials and Methods). By using these tools, we were able to identify ∼400 different target mRNAs for miRNA-214 and further elaborated on, using IPA. This analysis of the targets created a map of putative signalling components (Supplementary Figure S5), which was further investigated with respect to cell proliferation, cell death and senescence using PUBMED annotations, as well as the IPA knowledge database (Table 2). Interestingly, CDK6 (cyclin-dependent kinase 6), which holds a vital role in cell cycle progression, and FoxO4 (Forkhead box protein O4), a regulator of a multitude of cellular signalling events including cell cycle arrest, senescence and apoptosis, were among the potential predicted targets of miRNA-214 (Brunet et al, 1999; Dijkers et al, 2000, 2002; Hui et al, 2008). As downregulation of miRNA-214 was observed to cause increased IR-induced senescence in NSCLC U-1810 cells, the subsequent protein expression of these targets was examined by western blot (Figure 5A). No changes in the expression level of CDK6 were found in U-1810 cells, when miRNA-214 expression was blocked, as an almost equal amount of CDK6 was found in all samples (Figure 5A). Similarly, neither alteration in FoxO4 expression was revealed, when miRNA-214 expression was blocked, nor did we observe any difference in FoxO4 phosphorylation (Figure 5B). Thus, although our data showed that downregulation of miRNA-214 could sensitise RR U-1810 cells to IR by driving them into senescence, the targets of miRNA-214 remain to be identified. Cell cycle progression and CDK6 expression level were also analysed in H23 cells after overexpression of miRNA-214 in combination with IR (Figure 5A, Supplementary Figure S3). In line with the miRNA-214 antagomir results, overexpression of miRNA-214 in H23 cells did not influence IR-induced cell cycle distribution, as no difference in G2 and S phase cell populations was observed upon miRNA-214 mimic transfection alone or in combination with IR (Supplementary Figure S3). Accordingly, no change in CDK6 level was observed among miRNA-214 overexpressing and control H23 cells, with or without IR treatment (Figure 5A). Thus, although our data show that increasing the miRNA-214 level in NSCLC confers IR resistance, it probably does not involve altered IR-induced cell cycle regulation. However, overexpression of miRNA-214 in H23 cells resulted in a 1.8-fold increase in phosphorylation of FoxO4 at Thr-451, which also remained high even after IR (Figure 5B). Moreover, a 1.3-fold increase in FoxO4 total level was observed when miRNA-214 was combined with IR in H23 cells (Figure 5B). In contrast, in U-1810 cells, downregulation of miRNA-214 did not affect FoxO4 phosphorylation or expression (Figure 5B), suggesting that FoxO4 is probably an indirect target of miRNA-214 in the NSCLC H23 cells.
Table 2. Some of the predicted targets for miRNA-214 involved in apoptosis, senescence and/or cell cycle regulation.
| Predicted target | Cellular processes |
|---|---|
| CDK6 | Senescence, cell cycle, apoptosis |
| Foxo4 | Senescence, cell cycle, apoptosis |
| BCL2L2 | Senescence, apoptosis |
| BCL2L11 | Senescence, apoptosis |
| CBL | Senescence |
| ACVR1B | Senescence, apoptosis |
| ADORA2A | Senescence, apoptosis |
| ASXL1 | Senescence, |
| BMF | Senescence, apoptosis |
| CADM1 | Senescence, apoptosis |
| CBL | Senescence |
| CD320 | Senescence |
| CD47 | Senescence, apoptosis |
| CDCA7L | Senescence |
| CSF1 | Cell cycle, apoptosis |
| CTNNB1 | Cell cycle, apoptosis |
| ESRRG | Cell cycle |
| ING4 | Cell cycle, apoptosis |
| MFN2 | Cell cycle, apoptosis |
| MITF | Cell cycle, apoptosis |
| NOTCH2 | Cell cycle, apoptosis |
| RUNX1 | Cell cycle, apoptosis |
Figure 5.
Western blot validation of proteins upon both downregulation of miRNA-214 in U-1810 cells and its upregulation in H23 cells. The expression level of CDK6 and the phosphorylated or total level of FoxO4, PTEN, Akt and p38MAPK were analysed in U-1810 cells transfected with miRNA-214 antagomir or NT miRNA for 48 h with or without IR (8 Gy) and in H23 cells transfected with mimic miRNA-214 or NT mimic for the same period with or without IR (8 Gy). Ku80 or G3PDH were used as a loading control for U-1810 and H23 cells, respectively. Fold data are given relative to NT after correction for loading differences. (A) CDK6, (B) Phospho FoxO4 (Thr-451) and total FoxO4, (C) phospho PTEN (Ser380) and total PTEN, (D) phospho Akt (Ser473, Thr308) and total Akt. (E) Phospho p38MAPK (Thr180/Tyr182) and total p38MAPK.
Downregulation of miRNA-214 expression alters p38MAPK and PI3K signalling networks in NSCLC cells
miRNA-214 target prediction and IPA revealed PTEN to be a putative target of miRNA-214, and a role for miRNA-214 in regulating PTEN expression has also been documented in some ovarian cancer cells (Yang et al, 2008). Therefore, PTEN expression at both mRNA and protein level was examined in U-1810 cells after miRNA-214 and NT-antagomir transfection (Figure 5C and Supplementary Figure S4A). Following both downregulation of miRNA-214 and NT-antagomir transfection with IR, the expression level of total PTEN was increased 1.5-fold, whereas combination of miRNA-214 downregulation with IR did not further increase PTEN level (Figure 5C). In addition, miRNA-214 antagomir did not affect the phosphorylation status of PTEN. As PTEN is a known negative regulator of the PI3K/Akt pathway, the expression level of both phospho- and total Akt upon miRNA-214 antagomir transfection was examined by western blot (Figure 5D). No major alteration in total Akt was observed, whereas a reduction of almost 50% of Akt phosphorylation (Thr308) was found when miRNA-214 antagomir was applied as compared with NT antagomir (Figure 5D). However, NT-antagomir combined with IR did not further reduce Akt Thr308 phosphorylation. Altogether, these results show that downregulation of miRNA-214 leads to upregulation of PTEN, which subsequently reduces Akt phosphorylation.
We previously showed that IR in U-1810 cells stimulates the IGF-1R and downstream p38MAPK signalling, leading to cell survival and resistance to IR (Cosaceanu et al, 2007). Therefore, both total and phospho-p38MAPK were examined by western blot (Figure 5E). A 1.7-fold increase in p38MAPK phosphorylation was observed after miRNA-214 downregulation, whereas no difference in phosphorylation was detected when the combined treatment with antagomir-214 and IR was compared with NT antagomir and IR (Figure 5E). Moreover, a non-specific increase in the total p38MAPK was found after treatment with NT-antagomir and after miRNA-214 downregulation with or without IR (Figure 5E). This suggests that albeit downregulation of miRNA-214 in U-1810 cells increases the p38MAPK phosphorylation, it does not have a significant role in sensitisation of U-1810 cells to radiation.
miRNA-214 overexpression increases p38MAPK but not PI3K signalling in H23 cells
To understand the potential targets for miRNA-214 in NSCLC H23 cells, we focused on analysis of signalling pathways previously shown to influence survival or IR-induced cell death. As the influence of Akt pathway on IR-induced apoptotic propensity is well documented, so alike in U-1810, we first examined whether overexpressing miRNA-214 affected Akt phosphorylation at Ser473 and/or its total expression (Figure 5D). No change was observed in Akt phosphorylation between these treatments. Analysis of total Akt expression, however, revealed a 1.4-fold increase in H23 cells overexpressing miRNA-214, an effect which was not corroborated further when cells were irradiated (Figure 5D).
As binding of miRNA-214 to the 3′-UTR of PTEN in other tumour cell types has been shown to inhibit PTEN translation, subsequently resulting in activation of the Akt pathway (Yang et al, 2008), PTEN expression was analysed, with the assumption that it was indeed regulated after overexpressing miRNA-214 in H23 cells and could corroborate altered cell survival of NSCLC cells upon IR (Figure 5C). However, miRNA-214 overexpression increased total PTEN expression, which remained stable after IR (Figure 5C). A slight decrease in PTEN phosphorylation at Ser380 was observed after combined treatment with miRNA-214 mimic and IR (Figure 5C). As in U-1810, regulation of PTEN expression at mRNA level was analysed by q-RT–PCR upon miRNA-214 manipulation in H23 cells as well (Supplementary Figure S4). It was found that overexpression of miRNA-214 did not influence PTEN mRNA level profoundly. Thus, in these NSCLC cells the effect of miRNA-214 on IR response probably does not involve direct deregulation of PTEN.
Next we examined whether altered expression and/or activation of p38MAPK could be involved in the conferred radioresistance of H23 cells after increasing their low level of miRNA-214 expression (Figure 5E). A significant 1.9-fold increase in p38MAPK (Thr180/Tyr182) phosphorylation was observed after the introduction of miRNA-214 in H23 cells as compared with NT mimic (Figure 5E). Moreover, whereas IR reduced p38MAPK phosphorylation in NT mimic-expressing H23 cells, a further increase in p38MAPK phosphorylation was observed in miRNA-214 mimic-expressing H23 cells after irradiation (Figure 5E). In line with this, upon miRNA-214 overexpression the already increased level of total p38MAPK was further augmented to 1.6-fold after radiation (Figure 5E). Our results suggest that overexpression of miRNA-214 leads to upregulation of p38MAPK and FoxO4 expression, and their enhanced phosphorylation upon IR may account at least in part for increased cell survival through inhibition of apoptosis.
Knockdown of p38MAPK resensitises NSCLC cells to IR after miRNA-214 overexpression
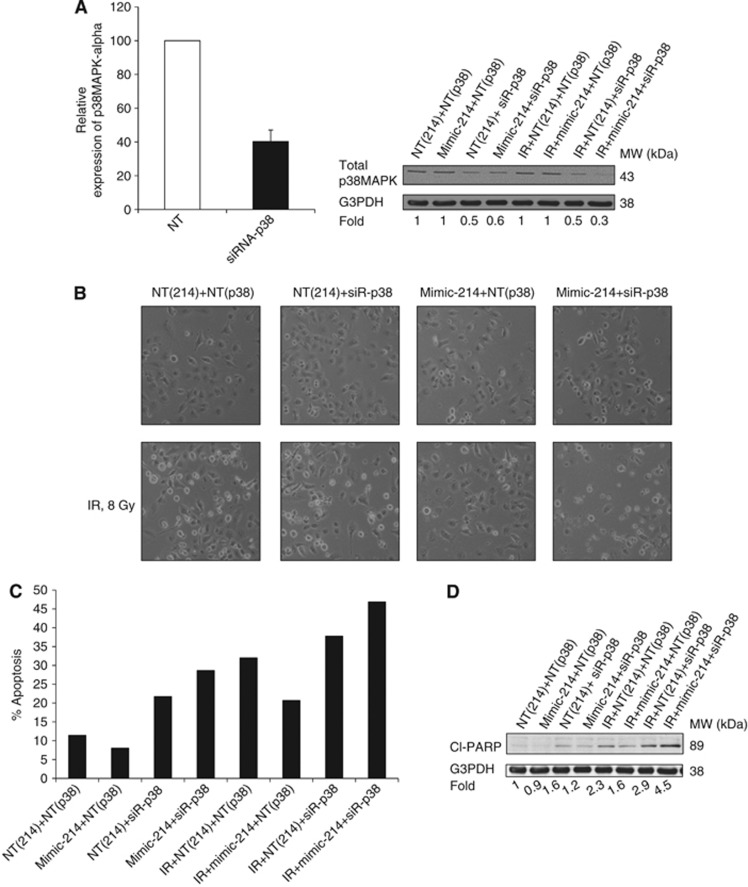
We previously documented that IR resistance of NSCLC cells is at least in part attributed to IGF-1R-p38MAPK pathway, which is activated in response to radiation (Cosaceanu et al, 2007). As shown in Figures 4B and 5E overexpression of miRNA-214 rendered the H23 cells more resistant to IR, which was associated with increased phosphorylation of p38MAPK. This suggested that p38MAPK might be one of the downstream kinases involved in the observed miRNA-214-mediated protection against IR-induced apoptosis. To reveal whether this was the case, we used siRNA approach to knockdown p38MAPK-α alone or in combination with miRNA-214 overexpression with or without IR (Figure 6). First, using q-RT–PCR and western blot, we were able to confirm that efficient knockdown of p38MAPK-α was achieved after siRNA transfection of H23 cells (Figure 6A). Evidently p38MAPK-α expression was reduced by about 60% at the mRNA level after knockdown with siRNA (Figure 6A). Similarly, a 50% reduction of total p38MAPK was observed at the protein level (Figure 6A). To further prove the inhibition of p38MAPK pathway after the siRNA transfection, phosphorylation of HSP27 at Ser82, a downstream target of p38MAPK (Hedges et al, 1999), was analysed using western blot (Supplementary Figure S6). Phosphorylation of HSP27 was reduced after silencing of the p38MAPK-α, which was even more significant after combined treatment with IR (Supplementary Figure S6). Next, we examined the effect of p38MAPK downregulation on cell viability (Figure 6B). Analysis of cellular morphology revealed that while IR enhanced cell death markedly (Figure 6B), upregulation of miRNA-214 rendered H23 cells refractory to IR (Figure 6B). Interestingly, knockdown of p38MAPK-α alleviated the protective effect of miRNA-214 overexpression on IR-induced cell death as there were lower amounts of surviving cells (Figure 6B). Then, the effect of p38MAPK siRNA on apoptosis induction in H23 cells with miRNA-214 overexpression and IR was analysed (Figure 6C). Ionising radiation-induced apoptosis in ∼30% of the cells transfected with NT mimic miRNA and control siRNA (Figure 6C). After overexpression of miRNA-214 the percentage of apoptosis was reduced to 20%, showing the protective effect of miRNA-214 against IR-induced apoptosis (Figure 6C). Importantly, p38MAPK-α downregulation clearly alleviated miRNA-214-mediated radioprotective effect and increased apoptosis two-fold (to 40%) (Figure 6C). To further document the induction of apoptosis, cleavage of the caspase substrate, PARP, was assessed (Figure 6D). In the non-irradiated cells, as a result of downregulation of p38MAPK-α, there was a slight increase in the amount of cleaved PARP (Figure 6D). However, in the irradiated cells, a two-fold increase in cleaved PARP was noted, which was reduced to almost baseline level after upregulation of the miRNA-214 (Figure 6D). Knockdown of the p38MAPK-α significantly (four-fold) increased the amount of cleaved PARP compared with control (Figure 6E). Altogether, these data demonstrate that p38MAPK-α is involved in the miRNA-214-regulated protection against IR-induced cell death in NSCLC cells.
Figure 6.
Knockdown of p38MAPK resensitises H23 cells to IR after upregulation of miRNA-214. (A) Left panel: the efficient knockdown of p38MAPK-α was confirmed by q-RT–PCR at 6 h post-transfection. Data shown are the relative expression of p38MAPK-α after transfection with p38MAPK-α siRNA as compared with an NT-siRNA in which p38MAPK-α Ct value was subtracted with internal control 18s rRNA, as described in the Materials and Methods. Data represent one out of three separate experiments. Right panel: knockdown of the p38MAPK-α was analysed at the protein level at 48 h post IR. (B) Morphology of H23 cells transfected with mimic miRNA-214 or NT-mimic or with siRNA-p38MAPK-α was analysed at 48 h post IR by light microscopy. Representative images from three biological replicates are shown. (C) H23 cells were treated as in B. Cell nuclei were stained with DAPI and nuclear morphology of H23 cells was examined at 48 h post IR. The percentage of apoptotic cells among at least 200 cells was calculated. (D) PARP cleavage with or without mimic-214, siRNA-p38MAPK-α and IR was examined in H23 cells treated as in B and C. The fold of PARP cleavage was normalised to the NT-siRNA-treated samples. G3PDH was used as a loading control.
Discussion
Deregulation of miRNA expression has recently been shown in various cancers including LC, where certain miRNAs have been suggested to work as oncogenes and others as tumour suppressor genes. Moreover, certain miRNAs have been linked to LC prognosis, for example, high miRNA-155 and low let-7a-2 expression are both associated with poor LC patient survival (Yanaihara et al, 2006). The altered miRNA expression was not only associated with LC prognosis, but also influenced response to different treatment modalities (Chen et al, 2010; Chun-Zhi et al, 2010; Galluzzi et al, 2010; Jiang et al, 2010; Oh et al, 2010; Arora et al, 2011). Radiotherapy is one of the common treatment regimes in NSCLC but, as mentioned above, resistance remains a major problem. There are several reports emphasising a role of miRNAs in RT response of different cancer types (Chen et al, 2010; Chun-Zhi et al, 2010; Jiang et al, 2010; Oh et al, 2010; Yan et al, 2010). However, there are no studies on global miRNA expression profiling in LC cells in relation to RT responsiveness. Therefore, we performed miRNA profiling in a panel of SCLC and NSCLC cell lines with different RT sensitivity. Our analysis revealed that a number of miRNAs, that is, miRNA-214, miRNA-1827, miRNA-21, miRNA-625, miRNA-339-5p, miRNA-768-3p, miRNA-532-3p, miRNA-1301, miRNA-324-5p, miRNA-1227, miRNA-423-3p, miRNA-1249, miRNA-2160 and miRNA-558, can potentially be linked to the RR or RS phenotypes of NSCLC and SCLC.
We present clear evidence that miRNA-214 can be directly linked to the radiosensitivity of NSCLC. We showed that overexpression of miRNA-214 in sensitive NSCLC cells leads to their radioresistance in part as a consequence of reduced apoptotic propensity upon radiation treatment. Along the same line, downregulation of miRNA-214 was found to sensitise RR NSCLC cells to IR. In this case, the mechanism did not involve increased apoptotic signalling, but was instead mediated by increased senescence. One may ask why silencing of miRNA-214 did not alter apoptotic signalling in the RR cells, whereas the overexpression of miRNA-214 in RS cells did. In the search for the possible signalling mechanisms involved, we first examined the p38MAPK, as its role in regulation of survival in response to DNA damage has been demonstrated previously (Choi et al, 2006; Cosaceanu et al, 2007; Thornton and Rincon, 2009). We found that overexpression of miRNA-214 in H23 cells increased total p38MAPK expression and its phosphorylation at Thr180/Tyr182 site. Moreover, whereas in H23 cells transfected with NT mimic IR blocked p38MAPK phosphorylation, a further increase in p38MAPK phosphorylation was observed upon combined miRNA-214 overexpression and treatment with IR. This observed increase in p38MAPK phosphorylation is responsible for the increased RT resistance; this finding is in accordance with our previous results in which p38MAPK activity conferred RT resistance (Cosaceanu et al, 2007). In contrast to the observed alteration in p38MAPK in H23 cells upon miRNA-214 overexpression, a non-specific increase in both total p38MAPK and phosphorylation was found in U-1810 cells after miRNA-214 ablation. The fact that miRNA-214 downregulation was not associated with p38MAPK deactivation in U-1810 cells may in part explain the lack of IR-induced apoptosis as well. Moreover, it has been shown in certain LC cell lines, such as A549 and H1299, that in response to radiation the level of anti-apoptotic proteins Bcl-2 and Bcl-XL is increased, which can also prevent IR-induced apoptosis (Ho et al, 2010). If miRNA-214 interferes with apoptotic signalling pathway downstream of Bcl-2 family proteins or in a parallel independent way, this might explain why antagomir for miRNA-214 did not sensitise U-1810 cells to IR-induced apoptosis.
A global strategy was applied to look for putative miRNA-214 targets using several target prediction tools and, as in the case of most other miRNAs, a large number of targets (around 400) for miRNA-214 were suggested. We decided to focus on targets that could be linked to signalling of apoptosis or senescence (Table 2), as these two modes of cell death were demonstrated to be influenced by miRNA-214 in the NSCLC cell lines. This work was performed to elucidate possible mechanisms of miRNA-214-mediated resistance to RT. One predicted target of miRNA-214 was PTEN, a major negative regulator of the cell survival PI3K-Akt pathway. It has been proved experimentally that in ovarian carcinoma cells miRNA-214 directly binds to the 3′end of PTEN and inhibits its expression (Yang et al, 2008). In line with this we found that downregulation of miRNA-214 in U-1810 cells caused upregulation of PTEN to some extent at the protein level which in turn reduced the phosphorylation of Akt independently of IR, whereas a non-significant increase of PTEN at the mRNA level could be due to post-transcriptional modification of PTEN by the miRNA-214. Overexpression of miRNA-214 in H23 cells neither affected PTEN at protein nor at mRNA level, forcing us to conclude that miRNA-214 may not be involved in PTEN regulation in NSCLC cells. As downregulation of miRNA-214 sensitised the NSCLC U-1810 cells to IR by driving them into senescence, the expression level of different predicted targets of miRNA-214 involved in senescence, such as CDK6 and FoxO4, was also examined. Our results indicate that miRNA-214 does not regulate RT sensitivity through targeting CDK6 or FoxO4 in NSCLC. Two well-known regulators of cell cycle and senescence, p21WAF1/Cip1 and p27Kip1, which also often show deregulation in tumours, were also analysed (Alexander and Hinds, 2001; Hershko, 2010). No alteration in p21WAF1/Cip1level upon miRNA-214 ablation was observed. This is not surprising, as U-1810 cells are p53-mutant and activation of p21WAF1/Cip1 is p53-dependent upon DNA damage, suggesting the minor importance of p21WAF1/Cip1 in the experimental system analysed. In contrast, we found increased p27Kip1 expression in miRNA-214 knocked down cells, which also show G2 accumulation after IR and go later on to senescence. With respect to G2 cell cycle accumulation and p27Kip expression, our results are in accordance with a previous study showing that upregulation of p27Kip1 can arrest MEFs in the G2M phase of the cell cycle (Payne et al, 2008; Hsu et al, 2011). At present, however, it is hard to conclude whether the effect of miRNA-214 on p27Kip1 expression is direct or indirect, as p27Kip1 is not a predicted target of the miRNA-214. Nevertheless, there are results supporting a role for p27Kip1 in senescence, which further corroborate our findings (Alexander and Hinds, 2001). Intriguingly, although downregulation of miRNA-214 induced senescence in the U-1810 cells, there was no specific change in the different phases of the cell cycle distribution which one would expect if cells undergo senescence. It is known that IR induces cell cycle arrest at G2/M, which is mediated by p53. However, as U-1810 cells express mutant p53, upon silencing of miRNA-214 level of p27Kip1 increases, leading to a further prolongation of the G2 arrest and subsequently to senescence.
In H23 cells with overexpressed miRNA-214, we observed increased phosphorylation of both p38MAPK and FoxO4. Regulation of FoxO transcription factors by survival signals such as IGF-1/IGF-1R and regulation of FoxO4 by Akt has been demonstrated (Rena et al, 2001; Van Der Heide et al, 2004; Liu et al, 2005; Lam et al, 2006; Huang and Tindall, 2007; Lupertz et al, 2008; Yamagata et al, 2008; Essaghir et al, 2009). Urbich et al showed that increased FoxO4 phosphorylation by statins decreased pro-apoptotic protein Bim expression and resulted in inhibition of apoptosis in endothelial progenitor cells (Urbich et al, 2005). However, the major regulators of increased phosphorylation of FoxO4 in NSCLC cells and its role in regulation of cell resistance to radiation are still unknown. It has been documented previously that increased FoxO4 expression confers resistance to oxidative stress, an effect influenced by p38MAPK signalling (Srisuttee et al, 2011). Here we also show that miRNA-214 overexpression in NSCLC cells is associated with an increased p38MAPK phosphorylation and expression. Therefore, it is tempting to speculate that p38MAPK signalling is involved in the observed FoxO4 phosphorylation. We observed that miRNA-214 overexpression, which rescued H23 cells from IR-induced apoptosis, indeed caused an upregulation of p38MAPK phosphorylation. Furthermore, by knockdown of p38MAPK-α, we demonstrated that this kinase is an important factor in the protective activity of miRNA-214 against IR-induced cell death. The higher level of IR-induced apoptosis after miRNA-214 overexpression and knockdown of p38MAPK was further demonstrated through detection of enhanced PARP cleavage. As previously reported, p38MAPK exerts a protective effect on IR-induced cell death (Cosaceanu et al, 2007), and IR-induced activation of IGF-1R resulted in increased p38MAPK phosphorylation. Blocking of either IGF-1R itself or p38MAPK-sensitised NSCLC cells to IR-induced cell death. Given our finding that the protective function of miRNA-214 overexpression on IR-induced apoptosis could be augmented by inhibition of p38MAPK-α expression further suggest a role of p38MAPK in protection of LC cells to IR-induced cell death.
In summary, analysis of the miRNA profiling in NSCLC and SCLC revealed that miRNAs may inflict RT responsiveness. We demonstrated a functional role for miRNA-214 in this respect and showed that by manipulating miRNA-214 expression, the RT response of NSCLC cells can be significantly altered. Downregulation of miRNA-214 sensitised NSCLC cells to IR by inducing senescence mechanisms. Consistently, overexpression of miRNA-214 protected the RS NSCLC cells against IR-induced cell death. We also established that concomitantly with impaired RT-induced apoptotic signalling, miRNA-214 overexpression increased p38MAPK and FoxO4 phosphorylation. Inhibition of p38MAPK-α resulted in resensitisation of the NSCLC cells to IR upon miRNA-214 overexpression, supporting a protective role for p38MAPK-α in IR-induced apoptotic signalling. Further analysis of miRNA-214 and its targets are still of considerable interest, as they may reveal novel RT sensitise strategies for this type of tumour.
Acknowledgments
This study was supported by grants from the Swedish Cancer Society (RL,BZ), the Stockholm Cancer Society (RL, KV, BZ), the Stockholm County Council (to Karolinska Biomics Centre (RL)), the Swedish Research Foundation (RL, BZ), the Swedish Childhood Cancer Foundation (BZ) and the European Union (Chemores and Apo-Sys projects) (RL, BZ)]. Financial support was received from the Higher Education Commission in Pakistan (NA), from Mr Ahmad Ismael via the Ministry of Higher Education and Scientific Research in Iraqi-Kurdistan Regional Government (HS) and from the Wenner-Gren Foundation (AHV). We are grateful to Mrs Birgitta Mörk for technical assistance.
Supplementary Information accompanies the paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
The authors declare no conflict of interest.
10/04/2012
This paper has been modified since advance online publication, an acknowledgement has been added
Supplementary Material
References
- Alexander K, Hinds PW (2001) Requirement for p27(KIP1) in retinoblastoma protein-mediated senescence. Mol Cell Biol 21: 3616–3631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora H, Qureshi R, Jin S, Park AK, Park WY (2011) miR-9 and let-7g enhance the sensitivity to ionizing radiation by suppression of NFkappaB1. Exp Mol Med 43: 298–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arteaga CL (2006) EGF receptor mutations in lung cancer: from humans to mice and maybe back to humans. Cancer Cell 9: 421–423 [DOI] [PubMed] [Google Scholar]
- Bergh J, Larsson E, Zech L, Nilsson K (1982) Establishment and characterization of two neoplastic cell lines (U-1285 and U-1568) derived from small cell carcinoma of the lung. Acta Pathol Microbiol Immunol Scand A 90: 149–158 [DOI] [PubMed] [Google Scholar]
- Bergh J, Nilsson K, Ekman R, Giovanella B (1985) Establishment and characterization of cell lines from human small cell and large cell carcinomas of the lung. Acta Pathol Microbiol Immunol Scand A 93: 133–147 [DOI] [PubMed] [Google Scholar]
- Bonci D, Coppola V, Musumeci M, Addario A, Giuffrida R, Memeo L, D’Urso L, Pagliuca A, Biffoni M, Labbaye C, Bartucci M, Muto G, Peschle C, De Maria R (2008) The miR-15a-miR-16-1 cluster controls prostate cancer by targeting multiple oncogenic activities. Nat Med 14: 1271–1277 [DOI] [PubMed] [Google Scholar]
- Brunet A, Bonni A, Zigmond MJ, Lin MZ, Juo P, Hu LS, Anderson MJ, Arden KC, Blenis J, Greenberg ME (1999) Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell 96: 857–868 [DOI] [PubMed] [Google Scholar]
- Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M, Croce CM (2004) Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA 101: 2999–3004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmichael J, Degraff WG, Gamson J, Russo D, Gazdar AF, Levitt ML, Minna JD, Mitchell JB (1989) Radiation sensitivity of human lung cancer cell lines. Eur J Cancer Clin Oncol 25: 527–534 [DOI] [PubMed] [Google Scholar]
- Carney DN, Mitchell JB, Kinsella TJ (1983) In vitro radiation and chemotherapy sensitivity of established cell lines of human small cell lung cancer and its large cell morphological variants. Cancer Res 43: 2806–2811 [PubMed] [Google Scholar]
- Chen G, Zhu W, Shi D, Lv L, Zhang C, Liu P, Hu W (2010) MicroRNA-181a sensitizes human malignant glioma U87MG cells to radiation by targeting Bcl-2. Oncol Rep 23: 997–1003 [DOI] [PubMed] [Google Scholar]
- Choi SY, Kim MJ, Kang CM, Bae S, Cho CK, Soh JW, Kim JH, Kang S, Chung HY, Lee YS, Lee SJ (2006) Activation of Bak and Bax through c-abl-protein kinase Cdelta-p38 MAPK signaling in response to ionizing radiation in human non-small cell lung cancer cells. J Biol Chem 281: 7049–7059 [DOI] [PubMed] [Google Scholar]
- Chun-Zhi Z, Lei H, An-Ling Z, Yan-Chao F, Xiao Y, Guang-Xiu W, Zhi-Fan J, Pei-Yu P, Qing-Yu Z, Chun-Sheng K (2010) MicroRNA-221 and microRNA-222 regulate gastric carcinoma cell proliferation and radioresistance by targeting PTEN. BMC Cancer 10: 367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosaceanu D, Budiu RA, Carapancea M, Castro J, Lewensohn R, Dricu A (2007) Ionizing radiation activates IGF-1R triggering a cytoprotective signaling by interfering with Ku-DNA binding and by modulating Ku86 expression via a p38 kinase-dependent mechanism. Oncogene 26: 2423–2434 [DOI] [PubMed] [Google Scholar]
- Dijkers PF, Birkenkamp KU, Lam EW, Thomas NS, Lammers JW, Koenderman L, Coffer PJ (2002) FKHR-L1 can act as a critical effector of cell death induced by cytokine withdrawal: protein kinase B-enhanced cell survival through maintenance of mitochondrial integrity. J Cell Biol 156: 531–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dijkers PF, Medema RH, Pals C, Banerji L, Thomas NS, Lam EW, Burgering BM, Raaijmakers JA, Lammers JW, Koenderman L, Coffer PJ (2000) Forkhead transcription factor FKHR-L1 modulates cytokine-dependent transcriptional regulation of p27(KIP1). Mol Cell Biol 20: 9138–9148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y, Watanabe H, Shibuya H, Miura M (2002) The phosphatidylinositol-3 kinase pathway is not essential for insulin-like growth factor I receptor-mediated clonogenic radioresistance. J Radiat Res (Tokyo) 43: 325–329 [DOI] [PubMed] [Google Scholar]
- Edelman MJ (2004) Second-line chemotherapy and beyond for non-small-cell lung cancer. Clin Adv Hematol Oncol 2: 373–378 [PubMed] [Google Scholar]
- Essaghir A, Dif N, Marbehant CY, Coffer PJ, Demoulin J-B (2009) The transcription of FOXO genes is stimulated by FOXO3 and repressed by growth factors. J Biol Chem 284: 10334–10342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franken NA, Rodermond HM, Stap J, Haveman J, van Bree C (2006) Clonogenic assay of cells in vitro. Nat Protoc 1: 2315–2319 [DOI] [PubMed] [Google Scholar]
- Galluzzi L, Morselli E, Vitale I, Kepp O, Senovilla L, Criollo A, Servant N, Paccard C, Hupe P, Robert T, Ripoche H, Lazar V, Harel-Bellan A, Dessen P, Barillot E, Kroemer G (2010) miR-181a and miR-630 regulate cisplatin-induced cancer cell death. Cancer Res 70: 1793–1803 [DOI] [PubMed] [Google Scholar]
- Hammerschmidt S, Wirtz H (2009) Lung cancer: current diagnosis and treatment. Deutsches Arzteblatt International 106: 809–818, ; quiz 819–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashita Y, Osada H, Tatematsu Y, Yamada H, Yanagisawa K, Tomida S, Yatabe Y, Kawahara K, Sekido Y, Takahashi T (2005) A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res 65: 9628–9632 [DOI] [PubMed] [Google Scholar]
- He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, Hammond SM (2005) A microRNA polycistron as a potential human oncogene. Nature 435: 828–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges JC, Dechert MA, Yamboliev IA, Martin JL, Hickey E, Weber LA, Gerthoffer WT (1999) A role for p38(MAPK)/HSP27 pathway in smooth muscle cell migration. J Biol Chem 274: 24211–24219 [DOI] [PubMed] [Google Scholar]
- Herbst RS, Heymach JV, Lippman SM (2008) Lung cancer. N Engl J Med 359: 1367–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko DD (2010) Cyclin-dependent kinase inhibitor p27 as a prognostic biomarker and potential cancer therapeutic target. Future Oncol 6: 1837–1847 [DOI] [PubMed] [Google Scholar]
- Ho JN, Kang GY, Lee SS, Kim J, Bae IH, Hwang SG, Um HD (2010) Bcl-XL and STAT3 mediate malignant actions of gamma-irradiation in lung cancer cells. Cancer Sci 101: 1417–1423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu JD, Kao SH, Ou TT, Chen YJ, Li YJ, Wang CJ (2011) Gallic acid induces G2/M phase arrest of breast cancer cell MCF-7 through stabilization of p27(Kip1) attributed to disruption of p27(Kip1)/Skp2 complex. J Agric Food Chem 59: 1996–2003 [DOI] [PubMed] [Google Scholar]
- Huang H, Tindall DJ (2007) Dynamic FoxO transcription factors. J Cell Sci 120: 2479–2487 [DOI] [PubMed] [Google Scholar]
- Hui RC, Gomes AR, Constantinidou D, Costa JR, Karadedou CT, Fernandez de Mattos S, Wymann MP, Brosens JJ, Schulze A, Lam EW (2008) The forkhead transcription factor FOXO3a increases phosphoinositide-3 kinase/Akt activity in drug-resistant leukemic cells through induction of PIK3CA expression. Mol Cell Biol 28: 5886–5898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvagner G, Zamore PD (2002) RNAi: nature abhors a double-strand. Curr Opin Genet Dev 12: 225–232 [DOI] [PubMed] [Google Scholar]
- Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T, Thun MJ (2008) Cancer statistics, 2008. CA Cancer J Clin 58: 71–96 [DOI] [PubMed] [Google Scholar]
- Jiang P, Rao EY, Meng N, Zhao Y, Wang JJ (2010) MicroRNA-17-92 significantly enhances radioresistance in human mantle cell lymphoma cells. Radiat Oncol 5: 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ (2005) RAS is regulated by the let-7 microRNA family. Cell 120: 635–647 [DOI] [PubMed] [Google Scholar]
- Ketting RF, Fischer SE, Bernstein E, Sijen T, Hannon GJ, Plasterk RH (2001) Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev 15: 2654–2659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam EW, Francis RE, Petkovic M (2006) FOXO transcription factors: key regulators of cell fate. Biochem Soc Trans 34: 722–726 [DOI] [PubMed] [Google Scholar]
- Liu W, Wang S, Wei S, Sun L, Feng X (2005) Receptor activator of NF-kappaB (RANK) cytoplasmic motif, 369PFQEP373, plays a predominant role in osteoclast survival in part by activating Akt/PKB and its downstream effector AFX/FOXO4. J Biol Chem 280: 43064–43072 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR (2005) MicroRNA expression profiles classify human cancers. Nature 435: 834–838 [DOI] [PubMed] [Google Scholar]
- Lupertz R, Chovolou Y, Unfried K, Kampkotter A, Watjen W, Kahl R (2008) The forkhead transcription factor FOXO4 sensitizes cancer cells to doxorubicin-mediated cytotoxicity. Carcinogenesis 29: 2045–2052 [DOI] [PubMed] [Google Scholar]
- Oh JS, Kim JJ, Byun JY, Kim IA (2010) Lin28-let7 modulates radiosensitivity of human cancer cells with activation of K-Ras. Int J Radiat Oncol Biol Phys 76: 5–8 [DOI] [PubMed] [Google Scholar]
- Payne SR, Zhang S, Tsuchiya K, Moser R, Gurley KE, Longton G, deBoer J, Kemp CJ (2008) p27kip1 deficiency impairs G2/M arrest in response to DNA damage, leading to an increase in genetic instability. Mol Cell Biol 28: 258–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rena G, Prescott AR, Guo S, Cohen P, Unterman TG (2001) Roles of the forkhead in rhabdomyosarcoma (FKHR) phosphorylation sites in regulating 14-3-3 binding, transactivation and nuclear targetting. Biochem J 354: 605–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rio DC, Ares M, Hannon GJ, Nilsen TW (2010) Purification of RNA using TRIzol (TRI reagent). Cold Spring Harb Protoc 2010, pdb prot5439 [DOI] [PubMed] [Google Scholar]
- Saha A, Rudd R (2006) Gemcitabine and carboplatin: is this the best combination for non-small cell lung cancer? Expert Rev Anticancer Ther 6: 165–173 [DOI] [PubMed] [Google Scholar]
- Schmittel A (2011) Second-line therapy for small-cell lung cancer. Expert Rev Anticancer Ther 11: 631–637 [DOI] [PubMed] [Google Scholar]
- Sher T, Dy GK, Adjei AA (2008) Small cell lung cancer. Mayo Clin Proc 83: 355–367 [DOI] [PubMed] [Google Scholar]
- Sirzen F, Zhivotovsky B, Nilsson A, Bergh J, Lewensohn R (1998) Higher spontaneous apoptotic index in small cell compared with non-small cell lung carcinoma cell lines; lack of correlation with Bcl-2/Bax. Lung Cancer 22: 1–13 [DOI] [PubMed] [Google Scholar]
- Srisuttee R, Koh SS, Park EH, Cho IR, Min HJ, Jhun BH, Yu DY, Park S, Park do Y, Lee MO, Castrillon DH, Johnston RN, Chung YH (2011) Up-regulation of Foxo4 mediated by hepatitis B virus X protein confers resistance to oxidative stress-induced cell death. Int J Mol Med 28: 255–260 [DOI] [PubMed] [Google Scholar]
- Thornton TM, Rincon M (2009) Non-classical p38 map kinase functions: cell cycle checkpoints and survival. Int J Biol Sci 5: 44–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbich C, Knau A, Fichtlscherer S, Walter DH, Bruhl T, Potente M, Hofmann WK, de Vos S, Zeiher AM, Dimmeler S (2005) FOXO-dependent expression of the proapoptotic protein Bim: pivotal role for apoptosis signaling in endothelial progenitor cells. FASEB J 19: 974–976 [DOI] [PubMed] [Google Scholar]
- Van Der Heide LP, Hoekman MF, Smidt MP (2004) The ins and outs of FoxO shuttling: mechanisms of FoxO translocation and transcriptional regulation. Biochem J 380: 297–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viktorsson K, Lewensohn R (2007) Apoptotic signaling pathways in lung cancer. J Thorac Oncol 2: 175–179 [DOI] [PubMed] [Google Scholar]
- Wightman B, Ha I, Ruvkun G (1993) Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 75: 855–862 [DOI] [PubMed] [Google Scholar]
- Yamagata K, Daitoku H, Takahashi Y, Namiki K, Hisatake K, Kako K, Mukai H, Kasuya Y, Fukamizu A (2008) Arginine methylation of FOXO transcription factors inhibits their phosphorylation by Akt. Mol Cell 32: 221–231 [DOI] [PubMed] [Google Scholar]
- Yan D, Ng WL, Zhang X, Wang P, Zhang Z, Mo YY, Mao H, Hao C, Olson JJ, Curran WJ, Wang Y (2010) Targeting DNA-PKcs and ATM with miR-101 sensitizes tumors to radiation. PloS One 5: e11397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, Calin GA, Liu CG, Croce CM, Harris CC (2006) Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell 9: 189–198 [DOI] [PubMed] [Google Scholar]
- Yang H, Kong W, He L, Zhao JJ, O’Donnell JD, Wang J, Wenham RM, Coppola D, Kruk PA, Nicosia SV, Cheng JQ (2008) MicroRNA expression profiling in human ovarian cancer: miR-214 induces cell survival and cisplatin resistance by targeting PTEN. Cancer Res 68: 425–433 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.