Abstract
Background:
In the INRG dataset, the hypothesis that any segmental chromosomal alteration might be of prognostic impact in neuroblastoma without MYCN amplification (MNA) was tested.
Methods:
The presence of any segmental chromosomal alteration (chromosome 1p deletion, 11q deletion and/or chromosome 17q gain) defined a segmental genomic profile. Only tumours with a confirmed unaltered status for all three chromosome arms were considered as having no segmental chromosomal alterations.
Results:
Among the 8800 patients in the INRG database, a genomic type could be attributed for 505 patients without MNA: 397 cases had a segmental genomic type, whereas 108 cases had an absence of any segmental alteration. A segmental genomic type was more frequent in patients >18 months and in stage 4 disease (P<0.0001). In univariate analysis, 11q deletion, 17q gain and a segmental genomic type were associated with a poorer event-free survival (EFS) (P<0.0001, P=0.0002 and P<0.0001, respectively). In multivariate analysis modelling EFS, the parameters age, stage and a segmental genomic type were retained in the model, whereas the individual genetic markers were not (P<0.0001 and RR=2.56; P=0.0002 and RR=1.8; P=0.01 and RR=1.7, respectively).
Conclusion:
A segmental genomic profile, rather than the single genetic markers, adds prognostic information to the clinical markers age and stage in neuroblastoma patients without MNA, underlining the importance of pangenomic studies.
Keywords: neuroblastoma, genetic alterations, genomic profile, prognosis, INRG
Neuroblastoma (NB), the most frequent extracranial solid tumour of childhood, is characterised by a wide variability of its clinical course. Clinical markers, such as stage and age at diagnosis, are insufficient to precisely predict outcome in all patients. This has led to the search for additional markers that might enable a more robust prognostic classification of NB (Maris, 2010). A large number of recurrent genetic alterations have been identified in NB. Amplification of the MYCN oncogene (MNA) is observed in approximately 20% of cases and is clearly associated with a poor outcome (Seeger et al, 1985). Variations of ploidy have also been described in NB, with near-triploidy associated with an excellent outcome, and diploidy/tetraploidy correlating with a poorer outcome (Look et al, 1991). Finally, a number of segmental chromosomal alterations have been reported, including deletions of chromosomes 1p, 3p, 4p and 11q, thought to harbour tumour suppressor genes, and gains of chromosomes 1q, 2p and 17q, harbouring putative oncogenes. A large number of these segmental alterations have been shown to be of prognostic impact in univariate analyses (Maris et al, 1995; Caron et al, 1996; Schleiermacher et al, 1996; Bown et al, 1999; Spitz et al, 2003, 2004; Attiyeh et al, 2005; Pezzolo et al, 2009; Jeison et al, 2010).
The development of high-throughput techniques such as chromosomal or array CGH has enabled the analysis of these parameters in a multivariate setting. It has been shown that the different genetic alterations combine to define distinct genetic subtypes of NB associated with specific clinical and prognostic characteristics (Lastowska et al, 2001; Vandesompele et al, 2005; Maris et al, 2007). Indeed, a genomic profile characterised by the presence of any of the segmental alterations typically observed in NB is strongly associated with a higher risk of relapse as well as a poorer outcome as compared with patients having numerical alterations only (Schleiermacher et al, 2007). Such a genomic profile is also associated with a higher risk of relapse, and a higher risk of metastatic relapse, not only in the overall population but also among patients with localised disease. Furthermore, patients whose tumours have genomic profiles harbouring both numerical and segmental alterations share the poor outcome of those with segmental alterations only (Janoueix-Lerosey et al, 2009). In multivariate analysis, taking into account clinical markers, single genetic markers as well as the genomic profile, the latter is the strongest independent prognostic factor. Segmental chromosomal changes are associated with a higher risk of relapse in infants with localised unresectable or metastatic NB without MNA (Schleiermacher et al, 2011). Recent studies also indicate that NB progression is associated with an accumulation of segmental chromosomal changes, and that a higher number of segmental changes is observed in tumours from children with advanced age (Caren et al, 2010; Schleiermacher et al, 2010).
To develop an improved pre-treatment risk assessment for NB patients, the INRG (International Neuroblastoma Risk Group) task force created a database uniting clinical, histological, radiological and biological data sets from 8800 international NB patients. Analysis of these data led to the development of a pre-treatment INRG risk classification that includes the genetic parameters MYCN status, 11q status and ploidy (Ambros et al, 2009; Cohn et al, 2009; Monclair et al, 2009). However, the prognostic value of individual segmental chromosomal alterations was evaluated in this analysis, rather than genomic types. We have now analysed the INRG dataset to determine if the analysis of recurrent genetic alterations observed in NB allows the classification of NB into distinct groups according to the genomic profile in this large retrospective cohort (INRG database). We furthermore sought to determine if a segmental genomic profile is of prognostic impact in this cohort, especially in patients without MNA.
Materials and Methods
The INRG dataset comprises data for 8800 international patients (Cohn et al, 2009). The following data were retrieved from the INRG database: clinical data (date of birth, sex; date of diagnosis, tumour stage, relapse or progression (if yes: date); date of last follow-up and status at last follow-up); biological data (LDH, urinary catecholamines at diagnosis); and tumour characteristics (MYCN status, ploidy, chromosome 1p status, chromosome 11q status, chromosome 17q status). These genetic markers were determined using different techniques, including fluorescent in situ hybridisation (FISH) and loss of heterozygosity (LOH) analysis (Ambros et al, 2009; Cohn et al, 2009). Subsequently, unbalanced 11q LOH and 11q aberrations data were combined into a single variable: ‘11q aberration’. Similarly, 1p LOH and 1p aberrations were combined into the variable ‘1p aberration’.
A segmental chromosomal alteration was defined as either the presence of an aberration of the respective chromosome arm, as determined by FISH, or in the case of deletion or imbalance, as determined by LOH. The presence of any segmental chromosomal alteration in a tumour (chromosome 1p deletion and/or 11q deletion and/or chromosome 17q gain) was taken into account to define a segmental genomic profile, whereas those tumours with a confirmed unaltered status for all three chromosome arms were considered as having no segmental changes. Tumours with a DNA index of ⩽1 were termed diploid, whereas tumours with a DNA index of >1 were termed hyperdiploid.
Correlation between clinical and molecular data was assessed by using the chi-square test, or Fisher’s exact test when indicated. Event-free survival (EFS) and overall survival (OS) were estimated with the Kaplan–Meier method and compared by the log-rank test with a P-value of less than 0.05 considered to be significant. Event-free survival was calculated from diagnosis until the date of last follow-up or event (first occurrence of relapse, progression, secondary malignancy, or death from any cause). Overall survival was calculated from diagnosis to the last follow-up or death from any cause. Multivariate analysis was conducted on EFS and OS, using a Cox regression model, with a backward procedure.
Results
Among the 8800 patients in the INRG database, MYCN status was known in 7102 cases, with MNA present in 1155 cases. Chromosome 1p status was known in 2152 patients with 493 presenting with 1p deletion, chromosome 11q status was known in 1064 patients with 220 having 11q deletion and chromosome 17q status was known in 362 cases with 175 having 17q gain. For 342 cases, the status at all three chromosome arms 1p, 11q and 17q was known.
A genomic type could be attributed to 814 patients (257 with MNA, 505 without MNA and 52 with unknown MYCN status). In 119 cases, all three chromosome arms 1p, 11q and 17q had a normal status, and these cases were considered as having no segmental chromosome alterations. In a further 695 cases, a segmental alteration of either chromosome 1p and/or 11q and/or 17q was observed, and these cases were termed segmental genomic type. A strong association between a segmental genomic type and MNA was observed (P<0.0001). Indeed, in only six tumours with MNA a normal status for chromosome 1p, 11q and 17q was observed, four of whom have relapsed.
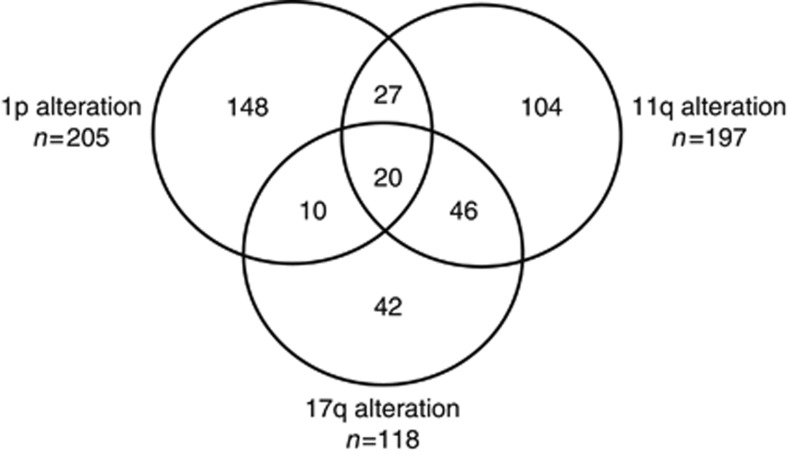
As the prognostic impact of MNA has been widely documented, we then focused our analysis on patients without MNA. For 505 patients without MNA for whom a genomic type could be determined, the medium follow-up was 63 months (range 0–167 months), with 207 events and 129 deaths having occurred. Among these 505 patients, 108 cases had no segmental chromosomal alterations, whereas in 397 other cases a segmental genomic type was observed, with the chromosome status known at all three chromosomal loci in 221 cases (Figure 1). A segmental genomic type was observed more frequently in stage 4 disease and in children aged >18 months (Table 1).
Figure 1.
Frequency of detection the genetic alterations chromsome 1p deletion, chromosome 11q deletion and chromosome 17q gain among 397 patients without MYCN amplification harbouring at least one genetic alteration in the INRG database.
Table 1. Frequency of a segmental genomic type in neuroblastoma without MYCN amplification.
| No segmental alterations a | ‘Segmental’ type b | Chi-square | |
|---|---|---|---|
| Stage | |||
| Localised (n=236) | 67 | 169 | |
| Stage 4 (n=234) | 27 | 207 | P<0.0001 |
| Stage 4s (n=35) | 14 | 21 | |
| Age | |||
| <18 months (n=240) | 73 | 167 | P<0.0001 |
| >18 months (n=265) | 35 | 230 | |
| Ploidy | |||
| Diploid (n=76) | 1 | 75 | |
| Hyperdiploid (n=43) | 1 | 42 | NS (Fischer test) |
| Missing data (n=386) | |||
Abbreviation: NS=not significant.
For the definition of no segmental alterations, all three markers have to have a normal status, leading to the determination of an altogether lower number of ‘no segmental alteration’ cases (see text).
Defined as a normal status at all three chromosome arms 1p, 11q and 17q.
Defined as the presence of a segmental chromosomal alteration of either chromosome 1p and/or 11q and/or 17q.
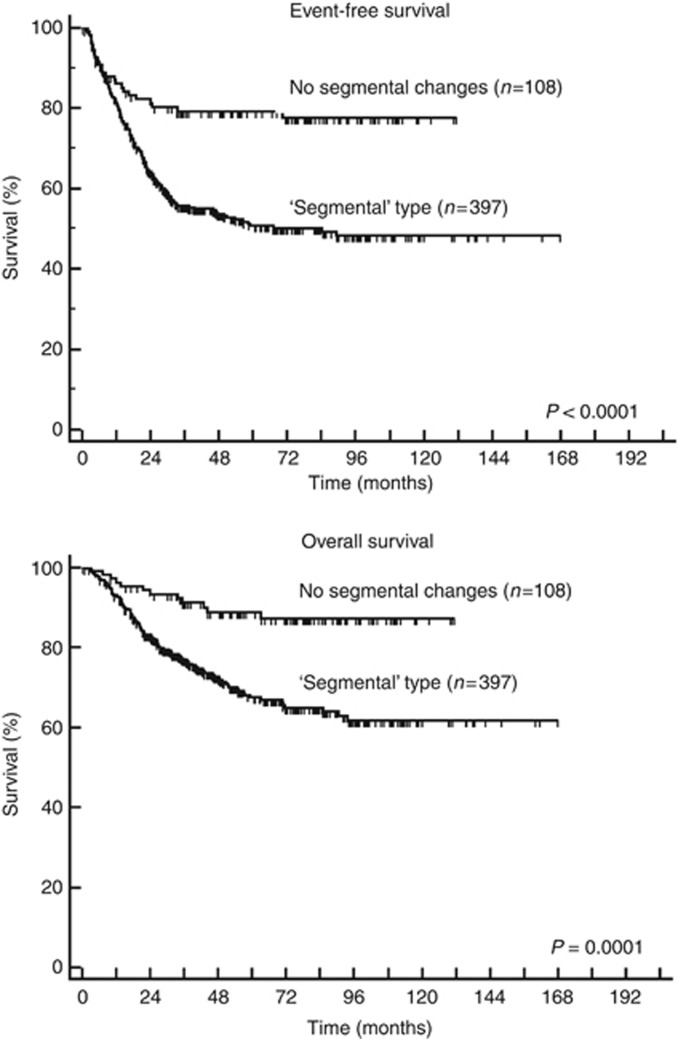
Among these 505 patients without MNA, for whom a genomic type could be attributed, diploidy, chromosome 11q deletion and chromosome 17q gain were significantly associated with poorer EFS and OS in univariate survival analysis, as was a segmental genomic type (Table 2, Figure 2). In a multivariate analysis modelling for EFS, the parameters age >18 months, stage 4 disease and a segmental genomic type were associated with a higher risk of relapse, whereas the individual genetic markers and ploidy were not (P<0.0001 and RR=2.6; P=0.0002 and RR=1.8; P=0.01 and RR=1.7, respectively, Table 3). Likewise, in a multivariate analysis modelling for OS, the same parameters age, stage and a segmental genomic type were retained in the model (P<0.0001 and RR=4.6; P<0.0001 and RR=3.1; P=0.05 and RR=1.8, respectively). In particular, a segmental genomic type was associated with a significantly poorer EFS in the 86 patients older than 18 months with non-stage 4 disease (P<0.02).
Table 2. Univariate analysis of EFS and OS according to genetic markers in 505 patients without MYCN amplification for whom a genomic type could be attributed.
| Parameter | 4-year EFS (%±s.d.) | Log rank ( P ) | 4-year OS (%±s.d.) | Log rank ( P ) |
|---|---|---|---|---|
| Ploidy | ||||
| Hyperdiploid (n=43) | 65±6.1 | 0.05 | 74±6.6 | 0.0025 |
| Diploid (n=76) | 45±8.9 | 44±12 | ||
| 1p status | ||||
| Normal (n=290) | 63±2.9 | 0.06 | 79±2.6 | 0.11 |
| Deletion (n=205) | 55±3.7 | 72±3.3 | ||
| 11q status | ||||
| Normal (n=207) | 75±3 | <0.0001 | 88±2.4 | <0.0001 |
| Deletion (n=197) | 42±3.8 | 65±3.9 | ||
| 17q status | ||||
| Normal (n=146) | 75±3.6 | 0.0002 | 86±2.9 | 0.0001 |
| Gain (n=118) | 49±4.7 | 72±4.3 | ||
| No segmental alterations (n=108) | 79±3.9 | <0.0001 | 88±3.2 | 0.0001 |
| ‘Segmental’ type (n=397) | 53±2.7 | 71±2.5 | ||
Abbreviations: EFS=event-free survival; OS=overall survival.
Figure 2.
Event-free and overall survival in 505 patients without MYCN amplification for whom a genomic type could be attributed. ‘Segmental’ genomic type: presence of either chromosome 1p deletion and/or chromosome 11q deletion and/or chromosome 17q gain. No segmental changes: no alteration for all three genetic markers.
Table 3. Multivariate analysis modelling EFS in a backward model.
| RR | CI | P -value | |
|---|---|---|---|
| Age >18 months | 2.6 | 1.8–3.6 | <0.0001 |
| Stage 4 | 1.8 | 1.3–2.5 | 0.0002 |
| ‘Segmental’ type | 1.7 | 1.1–2.7 | 0.01 |
The individual genetic markers 1p deletion, 11q deletion and 17q gain were not retained in the model.
Discussion
In NB, a number of genetic aberrations have been identified that are strongly associated with outcome, including deletions of chromosomes 1p, 3p, 4p and 11q, and gains of chromosomes 1q and 17q (Maris et al, 1995; Caron et al, 1996; Schleiermacher et al, 1996; Bown et al, 1999; Spitz et al, 2003; Attiyeh et al, 2005; Pezzolo et al, 2009). Gains of chromosome 2p, harbouring the MYCN gene, have also been shown to be associated with a poor outcome (Spitz et al, 2004; Jeison et al, 2010). The extensive INRG dataset includes information on ploidy, chromosome 1p, 11q and 17q status, as well as on MYCN status, distinguishing MNA from absence of amplification, but no information on other copy number variations including 2p gain is given. The major unfavourable prognostic impact of MNA has led to the inclusion of the MYCN status in the INRG pre-treatment risk classification system. The other genetic parameters retained in the INRG pre-treatment classification scheme are 11q status and ploidy, with 11q aberration distinguishing stage L2 or MS patients with a worse outcome and diploidy identifying stage M infants with a poorer outcome (Bagatell et al, 2009; Cohn et al, 2009; Monclair et al, 2009).
Several different techniques, including FISH and LOH studies, were used for the identification of structural aberrations. As the INRG retrospective data does not include higher throughput data such as pangenomic data obtained by multiplex ligation-dependent probe amplification (MLPA), array-CGH or SNP-arrays, a complete genetic dataset was available for only a few patients. However, the information available in the INRG database can be used to confirm the absence of segmental chromosome alterations at chromosomes 1p, 11q and 17q, thus identifying a non-segmental genomic type. Recent analyses show that 17q gain, 1p deletion and 11q deletion are the most frequent segmental chromosome alterations in tumours without MNA (Tomioka et al, 2008; Janoueix-Lerosey et al, 2009; Coco et al, 2012). Taking into account segmental alterations of any of the three chromosome arms 1p/11q/17q will thus identify the majority of all tumours with a segmental genomic type. Indeed, in previous studies, 155 out of 175 cases with a segmental genomic type without MNA, and 43 out of 48 infants with a segmental genomic type without MNA, had alterations of either chromosome 1p, and/or 11q, and/or 17q (Janoueix-Lerosey et al, 2009; Schleiermacher et al, 2011). We now show that in the INRG database, a segmental genomic type defined by the presence of any segmental chromosome alteration, at the three evaluated chromosome arms, is associated with clinical parameters predicting poor outcome, and in patients without MNA, a segmental genomic type is associated with a higher risk of relapse and a poorer outcome than in patients with a non-segmental genomic type. Indeed, a genomic profile characterised by any of the three segmental alterations adds prognostic information to the parameters ‘age’ and ‘stage’ rather than the individual genetic markers.
The more stringent definition of the absence of segmental chromosome alterations, for which the status at all three chromosome arms had to be known, when compared with the definition of a segmental type, for which an alteration of any of the chromosome arms was sufficient, might lead to an underrepresentation of non-segmental cases. However, survival analyses revealed comparable results when analysing the population of patients with all genetic markers known.
Several recent studies indicate that the presence of segmental chromosomal alterations, rather than individual genetic markers, may provide prognostic information in patients with MYCN non-amplified tumours (Tomioka et al, 2008; Janoueix-Lerosey et al, 2009; Caren et al, 2010; Schleiermacher et al, 2011). According to one hypothesis, the genomic type is thought to reflect an underlying genomic abnormality. In tumours with numerical aberrations, an abnormality in the mitotic segregation of the chromosomes may exist. On the other hand, the segmental chromosomal alterations in high- and intermediate-risk tumours are most frequently due to unbalanced chromosomal translocations, which in turn are thought to arise from improperly repaired DNA double-strand breaks, suggesting that a DNA maintenance or repair pathway is most likely impaired. Thus, the specific genetic markers analysed here could serve as surrogate markers for an underlying abnormality that will confer additional selective advantage (Schleiermacher et al, 2010).
The wide availability and cost reduction of multi-locus techniques such as MLPA and pangenomic assays such as array-CGH or SNP-arrays strongly favour the prospective analysis of all neuroblastic tumours (Ambros et al, 2009). Further, our data strongly support the idea that pangenomic analysis should be performed for all NBs, and that a genomic profile defined by the presence or absence of specific segmental chromosome alterations will be clinically useful for risk stratification particularly in NBs without MNA.
Acknowledgments
GS is supported by the Annenberg Foundation. The INRG database harbours biology data from different national groups and laboratories responsible for neuroblastoma genetics. For SIOPEN, the following members of the SIOPEN Biology group contributed data: Clare Bedwell, Newcastle, UK; Klaus Beiske, Oslo, Norway; Jean Bénard, Villejuif, France; Nick Bown, Newcastle, UK; Valérie Combaret, Lyon, France; Jerome Couturier, Paris, France; Raffaella Defferrari, Genoa, Italy; Nicole Gross, Lausanne, Switzerland; Marta Jeison, Petah Tikva, Israel: John Lunec, Newcastle, UK; Tommy Martinsson, Goteborg, Sweden; Barbara Marques, Lisbon, Portugal; Katia Mazzocco, Genoa, Italy; Rosa Noguera, Valencia, Spain; Gianpaolo Tonini, Genoa, Italy; Deborah Tweddle, Newcastle, UK; Alexander Valent, Villejuif, France; Nadine Van Roy, Ghent, Belgium; Luigi Varesio, Genoa, Italy; Alex Vicha, Prague, Czech Republic; Eva Villamon, Valencia, Spain. For GPOH, the following biology group members contributed data: Franck Berthold, Cologne, Germany; Jessica Theissen, Cologne, Germany; Manfred Schwab, Heidelberg, Germany; Frank Westermann, Heidelberg, Germany; Freimut Schilling, Stuttgart, Germany; Sabine Stegmaier, Stuttgart, Germany; Felix Niggli, Zurich, Switzerland. For COG, the following biology group members contributed data: Michael Hogarty, Philadelphia, USA; Rochelle Bagatell, Philadelphia, USA. For Japan, the following biology group members contributed data: Miki Ohira, Chiba, Japan; Yasuhiko Kaneko, Saitama, Japan; Junko Takita, Tokyo, Japan; Hajime Ohkita, Tokyo, Japan (Ambros et al, 2009).
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
10/04/2012
This paper has been modified since advance online publication, an acknowledgement has been added
References
- Ambros PF, Ambros IM, Brodeur GM, Haber M, Khan J, Nakagawara A, Schleiermacher G, Speleman F, Spitz R, London WB, Cohn SL, Pearson AD, Maris JM (2009) International consensus for neuroblastoma molecular diagnostics: report from the International Neuroblastoma Risk Group (INRG) Biology Committee. Br J Cancer 100: 1471–1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attiyeh EF, London WB, Mosse YP, Wang Q, Winter C, Khazi D, McGrady PW, Seeger RC, Look AT, Shimada H, Brodeur GM, Cohn SL, Matthay KK, Maris JM (2005) Chromosome 1p and 11q deletions and outcome in neuroblastoma. N Engl J Med 353: 2243–2253 [DOI] [PubMed] [Google Scholar]
- Bagatell R, Beck-Popovic M, London WB, Zhang Y, Pearson AD, Matthay KK, Monclair T, Ambros PF, Cohn SL (2009) Significance of MYCN amplification in international neuroblastoma staging system stage 1 and 2 neuroblastoma: a report from the International Neuroblastoma Risk Group database. J Clin Oncol 27: 365–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bown N, Cotterill S, Lastowska M, O’Neill S, Pearson AD, Plantaz D, Meddeb M, Danglot G, Brinkschmidt C, Christiansen H, Laureys G, Speleman F, Nicholson J, Bernheim A, Betts DR, Vandesompele J, Van Roy N (1999) Gain of chromosome arm 17q and adverse outcome in patients with neuroblastoma. N Engl J Med 340: 1954–1961 [DOI] [PubMed] [Google Scholar]
- Caren H, Kryh H, Nethander M, Sjoberg RM, Trager C, Nilsson S, Abrahamsson J, Kogner P, Martinsson T (2010) High-risk neuroblastoma tumors with 11q-deletion display a poor prognostic, chromosome instability phenotype with later onset. Proc Natl Acad Sci USA 107: 4323–4328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caron H, van Sluis P, de Kraker J, Bokkerink J, Egeler M, Laureys G, Slater R, Westerveld A, Voute PA, Versteeg R (1996) Allelic loss of chromosome 1p as a predictor of unfavorable outcome in patients with neuroblastoma. N Engl J Med 334: 225–230 [DOI] [PubMed] [Google Scholar]
- Cohn SL, Pearson AD, London WB, Monclair T, Ambros PF, Brodeur GM, Faldum A, Hero B, Iehara T, Machin D, Mosseri V, Simon T, Garaventa A, Castel V, Matthay KK (2009) The International Neuroblastoma Risk Group (INRG) classification system: an INRG Task Force report. J Clin Oncol 27: 289–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coco S, Theissen J, Scaruffi P, Stigliani S, Moretti S, Oberthuer A, Valdora F, Fischer M, Gallo F, Hero B, Bonassi S, Berthold F, Tonini GP (2012) Age-dependent accumulation of genomic aberrations and deregulation of cell cycle and telomerase genes in metastatic neuroblastoma. Int J Cancer 131: 1591–1600 [DOI] [PubMed] [Google Scholar]
- Janoueix-Lerosey I, Schleiermacher G, Michels E, Mosseri V, Ribeiro A, Lequin D, Vermeulen J, Couturier J, Peuchmaur M, Valent A, Plantaz D, Rubie H, Valteau-Couanet D, Thomas C, Combaret V, Rousseau R, Eggert A, Michon J, Speleman F, Delattre O (2009) Overall genomic pattern is a predictor of outcome in neuroblastoma. J Clin Oncol 27: 1026–1033 [DOI] [PubMed] [Google Scholar]
- Jeison M, Ash S, Halevy-Berko G, Mardoukh J, Luria D, Avigad S, Feinberg-Gorenshtein G, Goshen Y, Hertzel G, Kapelushnik J, Ben Barak A, Attias D, Steinberg R, Stein J, Stark B, Yaniv I (2010) 2p24 Gain region harboring MYCN gene compared with MYCN amplified and nonamplified neuroblastoma: biological and clinical characteristics. Am J Pathol 176: 2616–2625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lastowska M, Cullinane C, Variend S, Cotterill S, Bown N, O’Neill S, Mazzocco K, Roberts P, Nicholson J, Ellershaw C, Pearson AD, Jackson MS (2001) Comprehensive genetic and histopathologic study reveals three types of neuroblastoma tumors. J Clin Oncol 19: 3080–3090 [DOI] [PubMed] [Google Scholar]
- Look AT, Hayes FA, Shuster JJ, Douglass EC, Castleberry RP, Bowman LC, Smith EI, Brodeur GM (1991) Clinical relevance of tumor cell ploidy and N-myc gene amplification in childhood neuroblastoma: a Pediatric Oncology Group study. J Clin Oncol 9: 581–591 [DOI] [PubMed] [Google Scholar]
- Maris JM (2010) Recent advances in neuroblastoma. N Engl J Med 362: 2202–2211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maris JM, Hogarty MD, Bagatell R, Cohn SL (2007) Neuroblastoma. Lancet 369: 2106–2120 [DOI] [PubMed] [Google Scholar]
- Maris JM, White PS, Beltinger CP, Sulman EP, Castleberry RP, Shuster JJ, Look AT, Brodeur GM (1995) Significance of chromosome 1p loss of heterozygosity in neuroblastoma. Cancer Res 55: 4664–4669 [PubMed] [Google Scholar]
- Monclair T, Brodeur GM, Ambros PF, Brisse HJ, Cecchetto G, Holmes K, Kaneko M, London WB, Matthay KK, Nuchtern JG, von Schweinitz D, Simon T, Cohn SL, Pearson AD (2009) The International Neuroblastoma Risk Group (INRG) staging system: an INRG Task Force report. J Clin Oncol 27: 298–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pezzolo A, Rossi E, Gimelli S, Parodi F, Negri F, Conte M, Pistorio A, Sementa A, Pistoia V, Zuffardi O, Gambini C (2009) Presence of 1q gain and absence of 7p gain are new predictors of local or metastatic relapse in localized resectable neuroblastoma. Neuro Oncol 11: 192–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleiermacher G, Delattre O, Peter M, Mosseri V, Delonlay P, Vielh P, Thomas G, Zucker JM, Magdelenat H, Michon J (1996) Clinical relevance of loss heterozygosity of the short arm of chromosome 1 in neuroblastoma: a single-institution study. Int J Cancer 69: 73–78 [DOI] [PubMed] [Google Scholar]
- Schleiermacher G, Michon J, Huon I, d’Enghien CD, Klijanienko J, Brisse H, Ribeiro A, Mosseri V, Rubie H, Munzer C, Thomas C, Valteau-Couanet D, Auvrignon A, Plantaz D, Delattre O, Couturier J (2007) Chromosomal CGH identifies patients with a higher risk of relapse in neuroblastoma without MYCN amplification. Br J Cancer 97: 238–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleiermacher G, Janoueix-Lerosey I, Ribeiro A, Klijanienko J, Couturier J, Pierron G, Mosseri V, Valent A, Auger N, Plantaz D, Rubie H, Valteau-Couanet D, Bourdeaut F, Combaret V, Bergeron C, Michon J, Delattre O (2010) Accumulation of segmental alterations determines progression in neuroblastoma. J Clin Oncol 28: 3122–3130 [DOI] [PubMed] [Google Scholar]
- Schleiermacher G, Michon J, Ribeiro A, Pierron G, Mosseri V, Rubie H, Munzer C, Benard J, Auger N, Combaret V, Janoueix-Lerosey I, Pearson A, Tweddle DA, Bown N, Gerrard M, Wheeler K, Noguera R, Villamon E, Canete A, Castel V, Marques B, de Lacerda A, Tonini GP, Mazzocco K, Defferrari R, de Bernardi B, di Cataldo A, van Roy N, Brichard B, Ladenstein R, Ambros I, Ambros P, Beiske K, Delattre O, Couturier J (2011) Segmental chromosomal alterations lead to a higher risk of relapse in infants with MYCN-non-amplified localised unresectable/disseminated neuroblastoma (a SIOPEN collaborative study). Br J Cancer 105: 1940–1948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeger RC, Brodeur GM, Sather H, Dalton A, Siegel SE, Wong KY, Hammond D (1985) Association of multiple copies of the N-myc oncogene with rapid progression of neuroblastomas. N Engl J Med 313: 1111–1116 [DOI] [PubMed] [Google Scholar]
- Spitz R, Hero B, Ernestus K, Berthold F (2003) Deletions in chromosome arms 3p and 11q are new prognostic markers in localized and 4s neuroblastoma. Clin Cancer Res 9: 52–58 [PubMed] [Google Scholar]
- Spitz R, Hero B, Skowron M, Ernestus K, Berthold F (2004) MYCN-status in neuroblastoma: characteristics of tumours showing amplification, gain, and non-amplification. Eur J Cancer 40: 2753–2759 [DOI] [PubMed] [Google Scholar]
- Tomioka N, Oba S, Ohira M, Misra A, Fridlyand J, Ishii S, Nakamura Y, Isogai E, Hirata T, Yoshida Y, Todo S, Kaneko Y, Albertson DG, Pinkel D, Feuerstein BG, Nakagawara A (2008) Novel risk stratification of patients with neuroblastoma by genomic signature, which is independent of molecular signature. Oncogene 27: 441–449 [DOI] [PubMed] [Google Scholar]
- Vandesompele J, Baudis M, De Preter K, Van Roy N, Ambros P, Bown N, Brinkschmidt C, Christiansen H, Combaret V, Lastowska M, Nicholson J, O’Meara A, Plantaz D, Stallings R, Brichard B, Van den Broecke C, De Bie S, De Paepe A, Laureys G, Speleman F (2005) Unequivocal delineation of clinicogenetic subgroups and development of a new model for improved outcome prediction in neuroblastoma. J Clin Oncol 23: 2280–2299 [DOI] [PubMed] [Google Scholar]