Abstract
Background:
We previously reported that bone marrow (BM) was a homing site for gastric cancer (GC) cells leading to haematogenous metastases. There has been little study that microRNAs regulated pathways in malignant cells or host cells in BM, and thereby regulated the progression of GC.
Methods:
Both microRNA microarray and gene expression microarray analyses of total RNA from BM were conducted, comparing five early and five advanced GC patients. We focused on miR-144-ZFX axis as a candidate BM regulator of GC progression and validated the origin of the microRNA expression in diverse cell fractions (EpCAM+CD45−, EpCAM−CD45+, and CD14+) by magnetic-activated cell sorting (MACS).
Results:
Quantitative reverse-transcriptase (RT)–PCR analysis validated diminished miR-144 expression in stage IV GC patients with respect to stage I GC patients (t-test, P=0.02), with an inverse correlation to ZFX (ANOVA, P<0.01). Luciferase reporter assays in five GC cell lines indicated their direct binding and validated by western blotting. Pre-miR144 treatment and the resultant repression of ZFX in GC cell lines moderately upregulated their susceptibility to 5-fluorouracil chemotherapy. In MACS-purified BM fractions, the level of miR-144 expression was significantly diminished in disseminated tumour cell fraction (P=0.0005). Diminished miR-144 expression in 93 cases of primary GC indicated poor prognosis.
Conclusion:
We speculate that disseminated cancer cells could survive in BM when low expression of miR-144 permits upregulation of ZFX. The regulation of the miR-144-ZFX axis in cancer cells has a key role in the indicator of the progression of GC cases.
Keywords: cell fraction, microRNA, disseminated tumour cell, host cells
Cancer initiation and progression may involve microRNAs, which are small non-coding RNAs that can regulate gene expression (Calin and Croce, 2006b). Recent studies of microRNA expression profiles of primary human tumours have identified expression signatures associated with classification, diagnosis, prognosis, and even outcome after treatment (Calin et al, 2004; Iorio et al, 2005; Yanaihara et al, 2006; Bottoni et al, 2007; Calin and Croce, 2007; Gottardo et al, 2007; Kulshreshtha et al, 2007; Visone et al, 2007; Garzon et al, 2008; Liu et al, 2008a, 2008b; Lujambio et al, 2008; Marcucci et al, 2008; Schetter et al, 2008; Zhang et al, 2008). Profiling has also been used to identify microRNAs that might represent downstream targets of activated oncogenic pathways in primary cancers (Calin and Croce, 2006a). However, there have been few efforts in gastric cancer (GC) to identify bone marrow (BM) microRNAs and the target genes involved in the progression of disease. The primary endpoints of such studies are the establishment of significant microRNA/target gene profiles in BM and a determination of how they contribute to the progression of GC. This information could clarify how a given microRNA gene axis in BM cells contributes to the progression of GC.
Primary cancer cells are released from their initial sites and spread via the peripheral circulation to the BM. However, both circulating tumour cells and host factors in BM can determine whether metastasis and/or recurrence is likely after curative surgery. It is believed that few cancer stem cells (CSC) initially have the capacity for tumorigenesis and cancer metastasis (Alix-Panabieres et al, 2008; Dirks, 2010). Kang (2009) disclosed the metastatic behaviour of CSCs derived from tumours. As a result, we have focused our studies on CSCs in BM. Kaplan et al (2005) reported the important role of BM progenitor cells as niches for tumour cells (Lyden et al, 2001). After receiving signals from primary GC cells, haematopoietic progenitor cells from BM or endothelial progenitor cells in peripheral blood are recruited to pre-metastatic niches (Mimori et al, 2008). Thus, primary circulating tumour cells actively move to pre-metastatic niches, leading to the development of metastasis. It is postulated that both host BM cells and circulating tumour cells have important roles in the progression of malignant disease. However, it is not known which cells in BM are responsible for the progression of GC in a supportive niche. To approach this problem, we identified the microRNAs expressed among diverse BM cell populations, including the disseminated tumour cell fraction.
Using two microarray platforms, we initially focused on candidate microRNAs regulating GC progression. We postulated that the use of two independent microarray analyses in a double-blind manner would ensure reliability of the data. Then, candidate target genes were subjected to further analysis using several mammalian microRNA target databases, such as miRanda, TargetScan, and PICTAR. We performed gene expression microarray analysis of BM from GC cases that were applied to the microRNA microarray study. We found genes overexpressed in BM in stage IV GC patients. On the basis of in silico analyses, we then compared the genes expressed in BM in stage IV GC patients to determine their contribution to progression. Quantitative RT–PCR assays and assessment of direct binding by luciferase assays were conducted to validate the clinical significance of microRNA-target gene expression.
Patients and methods
Patients
We performed a microRNA microarray comparison between microRNA isolated from the BM of five GC cases with peritoneal dissemination and five cancer cases in whom invasion did not reach the sub-mucosal layer, and lymph node metastasis was absent in our hospital (Supplementary Table S1).
Then, physicians collected BM samples from 205 Japanese GC patients who underwent surgery from 2001 to 2004 at the Central Hospital, National Cancer Center, Tokyo, Japan. Documented informed consent was obtained from all patients, and the local ethics committee approved the study protocol. The average age of the 131 male and 74 female patients was 61.5 years, with a range of 27–86 years. On the basis of the Treaty for Japanese Gastric Cancer Association (Maruyama et al, 2006), 52 cases were classified as stage I, 49 cases as stage II, 51 cases as stage III, and 53 cases as stage IV. Normal negative controls consisted of BM samples collected from 20 patients with no malignancies (e.g., gallstone and hernia cases) from April 2000 to March 2003. Among the 205 cases, we randomly selected 10 representative GC cases, 5 in stage IV and 5 in stage I for the two microarray studies.
Total RNA from 93 cases of primary GCs in our hospital were collected and examined for further analysis.
BM collection
Aspiration of BM was conducted under general anaesthesia immediately before surgery as previously described (Mimori et al, 2008). The BM aspirate was obtained from the sternum using a BM aspiration needle. A volume of 3 ml of BM was added to 4.0 ml of Isogen-LS (Nippon Gene, Toyama, Japan), which was shaken vigorously and stored at −80 °C until RNA extraction.
Gene expression profiles in stage IV and stage I GC cases: comparison with database of predicted target genes
We performed microarray analyses of BM-extracted RNA in five cases of stage IV and those of stage I using Toray’s platform (Nagino et al, 2006; Ito et al, 2007; Iwahashi et al, 2007). It is possible to monitor gene expression profiles even with very small amounts of RNA (0.1–0.01 μg of total RNA) without amplification. Using the 3D-gene platform technology, RNA samples can be detected down to the attomole level.
Total RNA extraction and first-strand complementary DNA synthesis
Samples transferred from Tokyo to Beppu remained frozen while in transit. Total RNA was extracted from BM according to the manufacturer’s protocol as described elsewhere (Liu et al, 2008a). The RT reaction was performed as previously described (Mori et al, 1995). The first complementary DNA (cDNA) strand was synthesised from 2.7 μg of total RNA in 30 μl reaction mixtures containing 5 μl 5 × RT buffer (Gibco BRL, Gaithersburg, MD, USA), 200 μℳ deoxynucleotide triphosphates (dNTPs), 100 μℳ of a random hexadeoxynucleotide mixture, 50 units of RNasin (Promega, Madison, WI, USA), 2 μl of 0.1 ℳ dithiothreitol, and 100 units of Maloney leukaemia virus RT (BRL, Tokyo, Japan). The mixture was incubated at 37 °C for 60 min, heated to 95 °C for 10 min, and then chilled on ice.
microRNA expression profiling by two independent experiments: the first assay
Extracted total RNA was labelled with Hy5 using the miRCURY LNA Array miR labelling kit (Exiqon, Vedbaek, Denmark). Labelled RNAs were hybridised onto 3D-Gene Human microRNA Oligo chips containing 837 anti-sense probes printed in duplicate spots (Toray, Kamakura, Japan). The annotation and oligonucleotide sequences of the probes conformed to the miRBase microRNA database (http://microRNA.sanger.ac.uk/sequences/). After stringent washes, fluorescent signals were scanned with the ScanArray Express Scanner (PerkinElmer, Waltham, MA, USA) and analysed using GenePix Pro version 5.0 (Molecular Devices, Sunnyvale, CA, USA). The raw data for each spot was normalised by substitution with a mean intensity of the background signal determined from all blank spots’ signal intensities with 95% confidence intervals. Measurements of duplicate spots with signal intensities greater than two s.d. of the background signal intensity were considered to be valid. The relative expression level of a given microRNA was calculated by comparing the signal intensities of the averaged valid spots with their mean value throughout the microarray experiments after normalisation by their median values. MicroRNAs differentially expressed between GC stage I and IV were statistically identified using the Welch t-test.
microRNA microarray analysis: the second assay
The concentrations and purities of total RNAs were assessed spectrophotometrically, and RNA integrity was verified using an Agilent 2100 Bioanalyzer (Agilent Technologies, Tokyo, Japan). The RNA labelling and hybridisation on microRNA microarray chips were performed as described elsewhere (Liu et al, 2008a). Briefly, 5.0 μg of total RNA from each sample were reverse transcribed using a biotin end-labelled random-octamer oligonucleotide primer. Hybridisation of biotin-labelled cDNA was performed on Ohio State University’s microRNA microarray chip (version 4.0, microRNACHIPv4), which contains 4104 microRNA probes, including 474 human and 373 mouse microRNA genes, spotted in duplicate. The hybridised chips were washed and processed to detect biotin-containing transcripts by streptavidin–Alexa647 conjugate and scanned on an Axon 4000B microarray scanner (Molecular Devices).
Quantitative real-time RT–PCR for microRNA
We performed quantitative analysis of miR-144. We synthesised miR-144- and RNU6B (internal control)-specific cDNAs from total RNA using gene-specific primers according to the TaqMan MicroRNA Assays Protocol (Roche Applied Science, Indianapolis, IN, USA). Reverse transcriptase reactions contained 10 ng total RNAs, 50 nmol l−1 stem-loop RT primer, 1 × RT buffer, 0.25 mmol l−1 each dNTP, 3.33 units per μl MultiScribe RT, and 0.25 units per μl RNase inhibitor. Reactions were incubated in a BIO-RAD i-Cycler (Bio-Rad Laboratories, Tokyo, Japan) in 96-well plates for 30 min at 16 °C, 30 min at 42 °C, 5 min at 85 °C, and then maintained at 4 °C. Real-time PCR was done using an Applied Biosystems 7500 real-time PCR system. The 10 μl PCR mixture included 0.67 μl RT products, 1 × TaqMan Universal PCR master mix, and 1 μl primers and TaqMan microRNA Assays probe mix. Reactions were incubated in 96-well optical plates at 95 °C for 10 min. Relative quantification of microRNA expression was calculated as follows: raw data were presented as the relative quantity of target microRNA normalised with respect to RNU6B and relative to a calibrated sample.
Quantitative RT–PCR for candidate genes
The following primer sequences were used to amplify ZFX mRNA (Sakhinia et al, 2007); ZFX forward: 5′-ATA GCA CTA CAG ATG CCT CAG-3′, and ZFX reverse: 5′-GAA TGA GAC TGA TTG GCT TTA-3′. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an internal control, with the following primers: sense, 5′-TGA ACG GGA AGC TCA CTG G-3′ antisense, 5′-TCC ACC ACC CTG TTG CTG TA-3′. Real-time monitoring of PCR reactions was performed using the LightCycler System (Roche Applied Science) and SYBR-Green I dye (Roche Diagnostics, Tokyo, Japan). Monitoring was performed according to the manufacturer’s instructions, as previously described (Mimori et al, 2008). In brief, reactions were prepared on ice, containing 1 μl of cDNA, 1 × DNA Master SYBR-Green I, 50 ng of primers, and 3 mℳ MgCl2. The final volume was adjusted to 20 μl with water. After the reaction mixture was loaded into glass capillary tubes, quantitative RT–PCR was performed with the following cycling conditions: initial denaturation at 95 °C for 10 min, followed by 40 cycles of 95 °C for 10 s, annealing at 62 °C for 10 s, and extension at 72 °C for 10 s. After amplification, the amplicons were subjected to a temperature gradient from 67 °C to 95 °C at 0.2 °C s−1 under continuous fluorescence monitoring, to produce a melting curve of the products.
Data analysis for RT–PCR
After proportional baseline adjustment, the fit point method was employed to determine the cycle in which the log-linear signal was first distinguishable from the baseline. This cycle number was used as the crossing point value. Measuring the crossing point of each standard value and plotting it against the logarithmic concentration value produced a standard curve. Concentrations of unknown samples were calculated by plotting their crossing points against the standard curve and dividing by the GAPDH content.
After raw data were measured, we subtracted the background value, which yielded the background value (average)+2 s.d. (BG substitution). The value was converted into log2 data (BG substitution (LOG2)). Then, we found the z-transform value as the standard normal distribution value adjusted by the normal distribution.
Western blot analysis
Total protein was extracted from cell lines using protein extraction solution (PRO-PREP, iNtRON Biotechnology, Seongnam Si, Korea). Total protein (40 μg) was electrophoresed in 10% concentration READY GELS J (Bio-Rad Laboratories) and electroblotted onto pure nitrocellulose membranes (Trans-Blot Transfer Medium; Bio-Rad Laboratories) at 0.2 A for 120 min. The ZFX protein was detected using rabbit polyclonal antibodies (Santa Cruz Biotechnology, Santa Cruz, CA, USA) diluted 1 : 200. The ZFX protein level was normalised to the level of β-actin protein (Cytoskeleton, Denver, CO, USA) diluted 1 : 1000. Blots were developed with horseradish-peroxidase-linked anti- rabbit immunoglobulin (Promega) diluted 1 : 2000. The ECL Detection Reagents (Amersham Biosciences, Piscataway, NJ, USA) were used to detect antigen–antibody reactions.
Cell culture and luciferase assay
Note also that the detection and characterisation of micrometastatic cells or host cells in the BM of patients with cancer are of prognostic and therapeutic importance (Braun et al, 2000, 2005; Choesmel et al, 2004). Therefore, we examined and compared levels of microRNA expression in cellular fractions, such as epithelial cancer cells (EpCAM+CD45−), haematopoietic cells (EpCAM−CD45+), macrophages (CD14+), and other fractions separated by magnetic-activated cell sorting (MACS; Schmitz et al, 1994; Engel et al, 1999). On the basis of the analysis of cell populations, we identified the origin of cells expressing specific microRNA-target gene pathways in the BM.
The 3′-UTR and ORF of ZFX was amplified by RT–PCR. The amplified product was subcloned and ligated into the pmirGLO Dual-Luciferase microRNA Target Expression Vector (Promega). The resultant reporter vector position was confirmed by sequencing and termed pmirGLO-ZFX. Luciferase assays were conducted using 1 × 104 MKN45 cells, MKN7 cells, AZ521 cells, MKN74 cells, and NUGC4 cells plated in a 96-well plate. Transfections were performed using Lipofectamine 2000 (Invitrogen, Tokyo, Japan) in OptiMEM-reduced serum media (GIBCO, Tokyo, Japan). Cells were transfected with 30 ng of pmirGLO-ZFX or pmirGLO empty vector, and either 100 nℳ of pre-miR-negative control or pre-miR-144. Twenty-four hours following transfection, cells were assayed for both firefly and Renilla luciferase using Dual-Glo Luciferase Assay System (Promega). All transfection experiments were conducted in triplicate.
Cell sorting with antibodies by MACS
We performed immuno-magnetic cell sorting of 30 ml of BM from nine GC patients (there were six cases of stage I GC and three cases of stage IV GC). The mononuclear cell fraction was isolated by carefully loading 30 ml of whole blood onto 10 ml of Ficoll density medium (GE Healthcare Bio-Sciences, Tokyo, Japan) in 50 ml polypropylene tubes. The gradients were centrifuged for 30 min at room temperature at 450 g, and the interface collected after aspirating and discarding the supernatant. The cells were washed with 20 ml PBS and centrifuged at 150 g for 5 min at room temperature. The supernatant was aspirated and the cells were washed with PBS a second time.
CD326 (EpCAM)-PE, human MicroBeads was used for the positive selection of viable epithelial tumour cells from peripheral blood. To detect human leukocytes, we used human CD45-FITC. Then, CD14-APC was applied for detection of human monocytes and macrophages (Miltenyi Biotec, Bergisch Gladbach, Germany).
We measured miR-144 expression level per one cell to compare the average between stage I and stage IV in each fraction.
Cell culture assays for altered sensitivity to chemotherapy by anti-microRNA
To determine the role of ZFX as a regulator of chemotherapy sensitivity in cancer cells, we introduced pre-miR-144, control anti-miR, anti-miR-144 (100 pmol), and anti-miR-144 (200 pmol) into MKN45 parental cells. The cells were treated with 5-fluorouracil (5-FU; 0, 1, 5 and 10 μg ml−1) to compare levels of toxicity.
Statistical analysis
For continuous variables, data were expressed as the means±s.d. The relationship between ZFX mRNA expression and clinicopathological factors was analysed using a χ2-test and Student’s t-test. Findings were considered significant when the P-value was <0.05. All tests were performed using JMP software (SAS Institute Inc., Cary, NC, USA).
The best false discovery rate (FDR) value: (predictive pseudo positive number)/(number of genes satisfied with the threshold)=(number of genes to calculate) × (P-value of the gene)/number of genes below P-value.
Results
Expression profile of microRNA microarray
We performed microRNA microarray analyses to establish microRNA expression profiles for BM cells obtained from GC patients with and without metastasis. To maximise reliability, two different platforms were applied independently (Supplementary Figure S1). We observed downregulated and upregulated microRNAs in BMs of stage IV vs stage I GC cases. The number of significantly downregulated microRNAs in stage IV than stage I in the Japanese platform was 29 microRNAs (FDR<0.05; Supplementary Table S2), whereas the platform from Ohio State University revealed 13 microRNA (FDR<0.25; Supplementary Table S3). From these results, we selected three downregulated microRNAs identified by both platforms: hsa-miR-144, hsa-miR-19a, and hsa-miR-370.
We also performed gene expression microarray analysis of total BM RNAs from GC patients with and without metastases. We identified 180 BM probes with more abundant expression in stage IV than stage I GC (Supplementary Table S4). Using the miRanda, TargetScan, and PICTAR databases, we identified the ZFX gene as a target molecule, which has a high homology to SEED sequences in the 3′-UTR region of 7mer-m8, that is, miR-144.
Expression profiles of genes and identification of an miR gene axis by real microarray analysis and three databases
We compared total RNAs from the BMs of 10 GC patients, 5 cases in stage I and 5 cases in stage IV. Supplementary Table S4 shows the 180 upregulated genes in the BMs of stage IV GC cases vs stage I. The ZFX gene was significantly overexpressed in BM from stage IV GC cases with respect to stage I (P=0.00006; Supplementary Table S4).
Candidate target genes for miR-144 were predicted by three databases (miRands, Target Scan, and Pictar). We also undertook quantitative comparisons of gene expression in patients with stage IV and stage I GC. Five genes (ZFX, FOSB, SUCLA2, LSM14A, and HDHD2) were expressed at significantly higher levels in stage IV than stage I (P<0.05). These genes are candidate miR-144 target genes according to the three databases (Supplementary Table S5 and Supplementary Figure S1). Among the five genes, we focused on ZFX as a possible candidate target gene for miR-144.
ZFX is an X-linked, zinc finger protein. Galan-Caridad et al (2007) reported that the deletion of Zfx impaired the self-renewal of murine embryonic stem cells. Furthermore, Zfx deletion abolished the maintenance of hematopoietic stem cells (Galan-Caridad et al, 2007). Huang et al (2009) reported that ZFX was upregulated in side-population (SP) cells in oesophageal cancer, and that SP cells possessed CSC characteristics of oesophageal carcinoma. Therefore, we expected that the abundant expression of ZFX in cancer cells or in BM host cells might have crucial roles in the maintenance of human CSCs or niche cells, respectively, and their development of the metastatic phenotype.
Direct binding between miR-144 and ZFX by luciferase assay and by western blotting
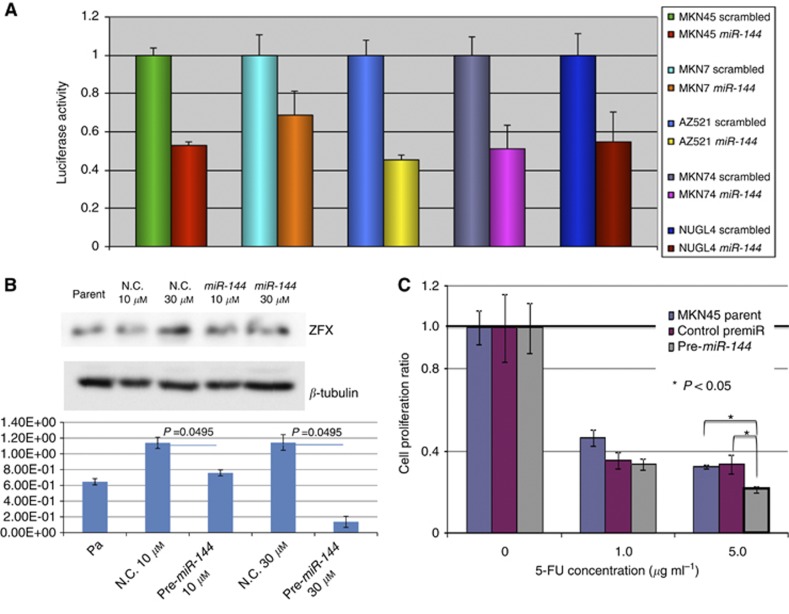
We investigated direct binding of miR-144 and the ZFX gene by luciferase assay. In five GC cell lines (MKN45, MKN7, AZ521, MKN74, and NUGC4), we observed reduction of the luciferase activity in the pre-miR-144-treated lines in comparison with lines treated with the scrambled control (0.53-, 0.69-, 0.45-, 0.51-, and 0.55-fold, respectively) with statistical significance in each experiment (Figure 1A).
Figure 1.
(A) Luciferase assay for interaction of miR-144 and ZFX gene in five GC cell lines, MKN45, MKN7, AZ521, MKN74, and NUGC4. Every cell line attenuated with miR-144 showed reduced luciferase activity in comparison with control cells. (B) Western blotting assay for disclosing the inhibitory effect of miR-144 on ZFX protein expression. The average ZFX expression in parent cells, normal control cells with 10 μℳ of control vector, pre-miR144 10 μℳ, normal control cells with 30 μℳ of control vector, and pre-miR144 30 μℳ were 6.46E-01, 1.14E+00, 7.58E-01, 1.14E+00, and 1.37E-01, respectively. There were significant differences between pre-miR and control vector in each concentration. (C) Pre-miR144 administration acquired the capability of resistance to 5-FU treatment. Anti-tumour effects of 5-FU (0, 1.0, and 10.0 μg ml−1) treatment evaluated by cell proliferation (MTT) assay were reduced 0.94, 0.85, and 0.76 times in the group of pre-miR144 administration in comparison with control cell lines with statistical significance by ANOVA analysis (P<0.05).
In addition, we performed western blotting assay for disclosing the inhibitory effect of miR-144 on ZFX protein expression (Figure 1B). The average ZFX expression in parent cells, normal control cells with 10 μℳ of control vector, pre-miR144 10 μℳ, normal control cells with 30 μℳ of control vector, and pre-miR144 30 μℳ were 6.46E-01, 1.14E+00, 7.58E-01, 1.14E+00, and 1.37E-01, respectively. There were significant difference between pre-miR and control vector in each concentration.
Clinicopathological significance of miR-144 and the target protein ZFX
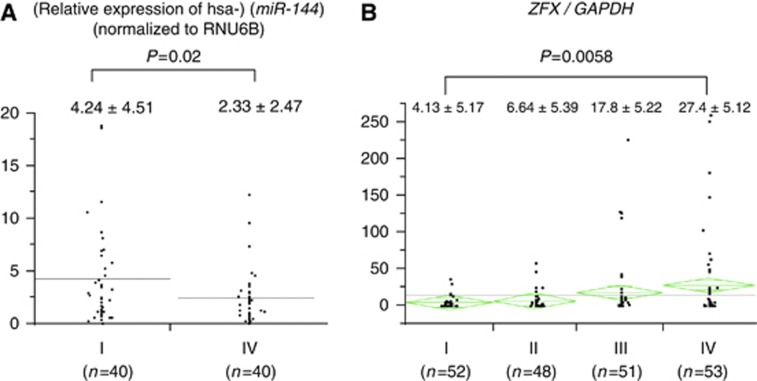
In clinicopathological analysis, there was no significant association between miR-144 expression and any clinicopathological variable. The expression ratios comparing values to the universal internal control (miR-144 RNU6B) were 4.24±4.51 for stage I and 2.33±2.47 for stage IV (Figure 2A and Table 1A). These results indicated diminished miR-144 expression during progression of disease from stage I to stage IV in GC (P=0.02). On the other hand, the expression of ZFX gene increased with progression of the disease (P=0.0058). The expression ratios for ZFX/GAPDH were as follows: 4.13±5.17 (stage I), 6.64±5.39 (stage II), 17.8±5.22 (stage III), and 27.4±5.12 (stage IV; Figure 2B and Table 1B).
Figure 2.
Verification of the expression of miR-144 and ZFX in BM from 80 and 214 GC patients, respectively. (A) miR-144 expression was diminished in BM from stage IV GC patients (2.33±2.47) compared with those of stage I (4.24±4.51; P<0.02). (B) As expected, ZFX expression increased with Dukes stage: 4.13±5.17 (stage I), 6.64±5.39 (stage II), 17.8±5.22 (stage III), and 27.4±5.12 (stage IV) (P<0.0058).
Table 1A. Clinicopathological significance of miR-144 expression in BM from GC cases.
|
Low expression group (
n
=99)
|
High expression group (
n
=99)
|
||||
|---|---|---|---|---|---|
| Factors | Number | % | Number | % | P -value |
| Age (mean±s.d.) | 62.1±1.22 | 61.6±1.22 | 0.77 | ||
| Sex | 0.21 | ||||
| Male | 65 | 65.7 | 73 | 73.7 | |
| Female | 34 | 34.3 | 26 | 26.3 | |
| Tumour stage | 0.054 | ||||
| m, sm | 29 | 29.3 | 42 | 42.4 | |
| mp, ss, se, si | 70 | 70.7 | 57 | 57.6 | |
| Lymph node metastasis | 0.56 | ||||
| Absent | 37 | 37.4 | 41 | 41.4 | |
| Present | 62 | 62.6 | 58 | 58.6 | |
| Lymphatic invasion | 0.78 | ||||
| Absent | 44 | 44.4 | 46 | 46.5 | |
| Present | 55 | 55.6 | 53 | 53.5 | |
| Venous invasion | 0.32 | ||||
| Absent | 72 | 72.7 | 78 | 78.8 | |
| Present | 27 | 27.3 | 21 | 21.2 | |
| Peritoneal dissemination | 1 | ||||
| Absent | 95 | 96 | 95 | 96 | |
| Present | 4 | 4 | 4 | 4 | |
| Liver metastasis | 1 | ||||
| Absent | 98 | 98.9 | 98 | 98.9 | |
| Present | 1 | 1.1 | 1 | 1.1 | |
| Distant metastasis | 0.3 | ||||
| Absent | 98 | 98.9 | 96 | 97 | |
| Present | 1 | 1.1 | 3 | 3 | |
| TNM stage | 0.049* | ||||
| I | 26 | 26.3 | 39 | 39.4 | |
| II, III, IV | 73 | 73.7 | 60 | 60.6 | |
Abbreviations: BM=bone marrow; GC=gastric cancer; m=mucosa; mp=musclaris propria; se=serosa exposed; si=serosa infiltrating; sm=submucosa; ss=subserosa; TNM=tumour, node, metastasis.
*P<0.05, there is a statistical significance by Student’s t-test.
Table 1B. Clinicopathological significance of ZFX expression in BM from GC cases.
| Expression ratio | ||||
|---|---|---|---|---|
| Clinicopathological factors | Status | n | ZFX/GAPDH | P -value |
| Tumour size | 0.0499* | |||
| <5 cm | 92 | 8.80±4.00 | ||
| ⩾5 cm | 113 | 19.4±3.61 | ||
| Depth of tumour invasion | 0.0107* | |||
| m, sm | 60 | 3.99±4.92 | ||
| mp, ss, se, si | 145 | 19.1±3.17 | ||
| Lymph node metastasis | 0.0499* | |||
| Absent | 67 | 7.06±4.70 | ||
| Present | 138 | 18.3±3.27 | ||
| Lymphatic permeation | 0.871 | |||
| Absent | 80 | 13.4±4.34 | ||
| Present | 124 | 15.6±3.49 | ||
| Venous permeation | 0.93 | |||
| Absent | 138 | 14.6±3.31 | ||
| Present | 66 | 15.0±4.78 | ||
| Peritoneal dissemination | 0.0102* | |||
| Absent | 162 | 11.1±3.00 | ||
| Present | 43 | 28.1±5.82 | ||
| Clinical stage | 0.0101* | |||
| I | 52 | 4.13±5.25 | ||
| II | 49 | 8.70±5.41 | ||
| III | 51 | 17.8±5.30 | ||
| IV | 53 | 27.4±5.20 |
Abbreviations: BM=bone marrow; GAPDH=glyceraldehyde-3-phosphate dehydrogenase; GC=gastric cancer; m=mucosa; mp=musclaris propria; se=serosa exposed; si=serosa infiltrating; sm=submucosa; ss=subserosa. *P<0.05, there is a statistical significance by Student’s t-test.
Use of MACS fractionation to analyse BM cells with abundant miR-144 expression in GC patients
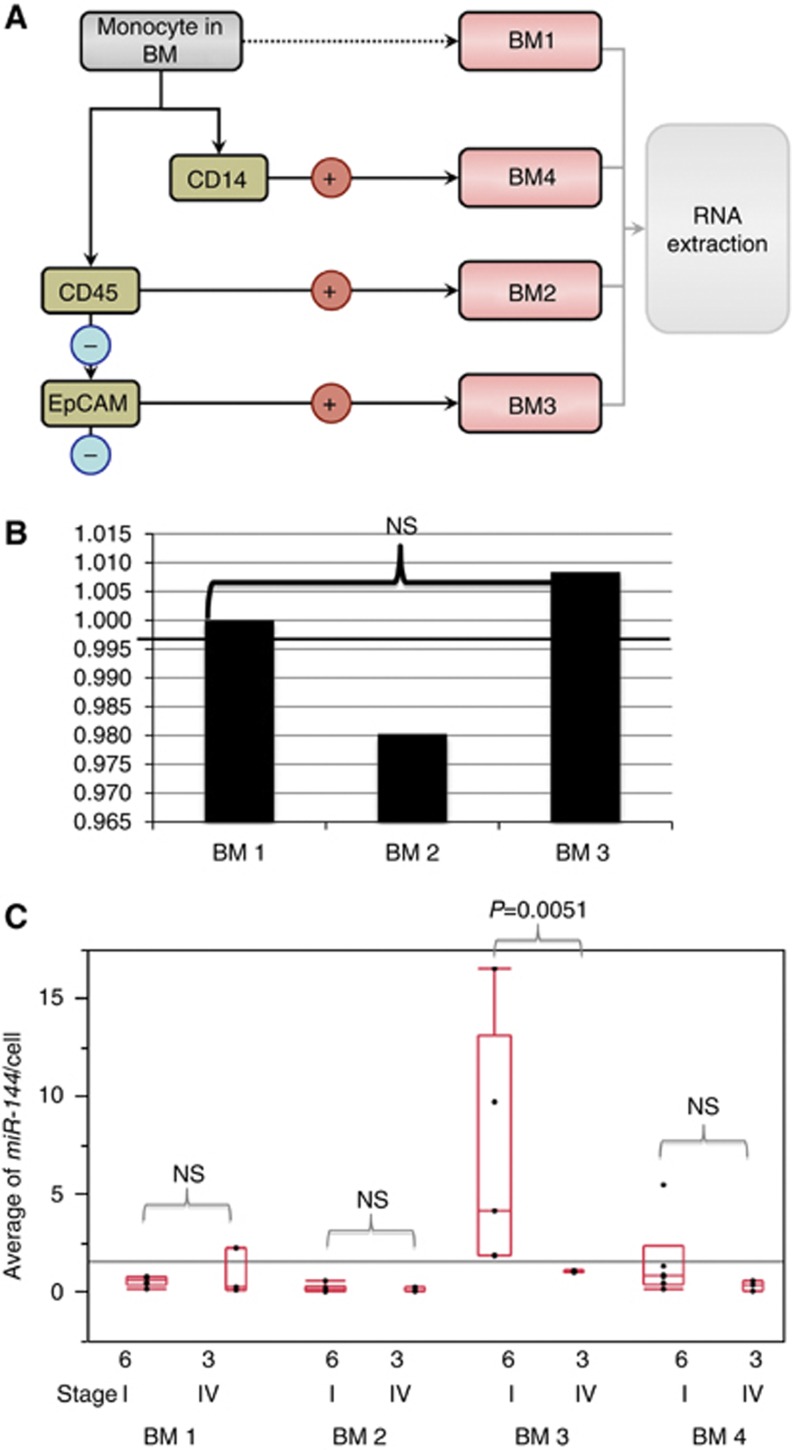
To determine whether miR-144 was expressed in cancer cells or host BM-derived cells, we performed cell sorting of BM by MACS (Figure 3A). Quantitative RT–PCR analysis showed the average miR-144 expression per one cell in six GC cases of stage I and three cases of stage IV (Figure 3C). As we showed in this figure, the expression of miR-144 per cell was significantly higher in BM3 fraction of GC cases of stage I than that of GC cases of stage IV (P=0.0051). Therefore, diminished expression of miR-144 was supposed to be originated from disseminated tumour cells.
Figure 3.
Determining the origin of miR-144-expressing cells from nine representative GC patients with distant metastasis. (A) We sorted BM-derived monocytes. The fraction was enriched for monocytes (6 × 107) from 15 ml of BM for BM1, BM2, and BM3 fractions. The BM2 consisted primarily of lymphocytes (2.8 × 107), and fraction BM3 was the EpCAM+ and CD45− fraction, circulating epithelial cells (1.9 × 106). Another BM-derived monocytes (6 × 107) from 15 ml was stored for BM1 and sorted with antibodies CD14+. The BM4 contained macrophages and monocytes (7.5 × 105). (B) miR-144 expression in BM fractions in normal healthy donor sample. miR-144 expression was 1.1 times higher in BM3 fraction than other cellular fractions, such as BM1 and BM2. However, the difference was really faint among them. (C) Quantitative RT–PCR analysis of the average miR-144 expression per one cell in six GC cases of stage I and three cases of stage IV. As we showed in this figure, the expression of miR-144 per cell was higher in BM3 fraction of GC cases of stage I than that of GC cases of stage IV significantly (P=0.0051).
In normal healthy sample, ZFX expression was 1.1 times higher in BM3 fraction than other cellular fractions, such as BM1 and BM2 (Figure 3B). However, the difference was really small among them. Therefore, the difference of miR-144 expression might be provoked by the presence of disseminated tumour cells in BM in GC cases.
Clinicopathological significance of miR-144 expression in primary GC
Diminished miR-144 expression was observed in primary GC with severe tumour invasion, whereas higher miR-144 expression was observed in GC patients with less pronounced invasion (Table 2A). There was a significant association between them (Fisher’s exact test, P=0.037). In fact, higher miR-144 expression was observed in stage I and II GC patients, whereas lower miR-144 expression was seen in stages III and IV (Fisher’s exact test, P=0.016).
Table 2A. Clinicopathological significance of microRNA144 expression in primary gastric cancer cases.
|
miR-144
primary gastric cancer |
|||
|---|---|---|---|
| High expression | Low expression | ||
| n =47 | n =46 | P -value | |
| Age | 67.2±10.8 | 62.9±12.9 | 0.08 |
| Gender | 0.2 | ||
| Male | 29 | 34 | |
| Female | 18 | 12 | |
| Tumour size | 0.08 | ||
| <5 cm | 24 | 15 | |
| 5 cm⩽ | 22 | 29 | |
| Depth of tumour invasion | 0.037a | ||
| m, sm | 13 | 5 | |
| mp, ss, se, si | 34 | 41 | |
| Lymph node metastasis | 0.21 | ||
| Absent | 18 | 12 | |
| Present | 29 | 34 | |
| Lymphatic permeation | 0.11 | ||
| Absent | 16 | 9 | |
| Present | 31 | 37 | |
| Venous permeation | 0.09 | ||
| Absent | 37 | 29 | |
| Present | 10 | 17 | |
| Peritoneal dissemination | 0.39 | ||
| Absent | 40 | 36 | |
| Present | 7 | 10 | |
| Clinical stage | 0.016a | ||
| I,II | 29 | 17 | |
| III,IV | 18 | 29 | |
There is a statistical significance (Fisher’s exact test, P=0.037). Gastric cancer cases with advanced stage (III and IV) indicated much lower mi-R144 expression than stage I and II cases with statistical significance (Fisher’s exact test, P=0.016).
Gastric cancer with lower invasion showed significantly higher miR-144 expression.
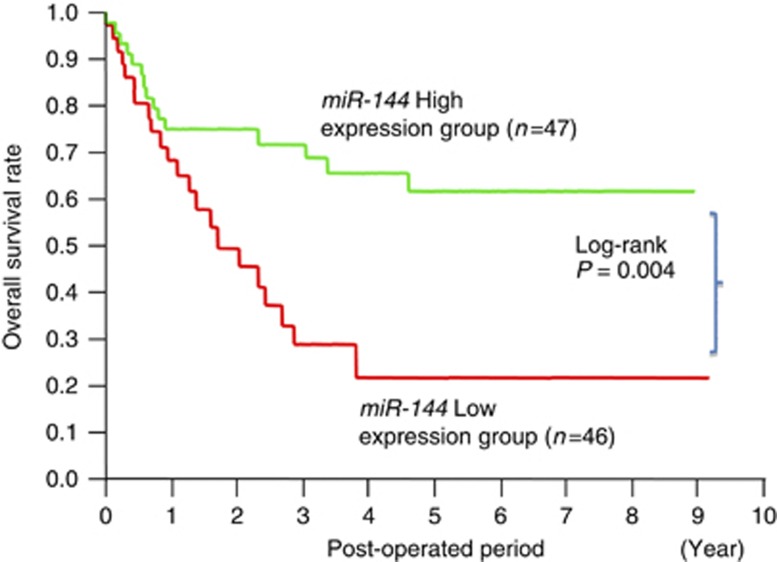
With regard to the prognostic significance of miR-144 (Figure 4), 46 cases of GC with diminished miR-144 in primary tissues had a significantly worse overall survival rate than 47 patients with higher levels of miR-144 expression (by log-rank test, P=0.004). However, the evaluation of miR-144 was not an independent prognostic marker of GC cases with statistical significance by multivariate analysis (Table 2B).
Figure 4.
Comparison of the prognosis between the high and low miR-144 expression groups by Kaplan–Meier method.
Table 2B. Univariate and multivariate analysis of the clinicopathological factors and miR-144 expression in primary gastric cancer cases for overall survival.
| Clinicopathological |
Univariate analysis
|
Multivariate analysis
|
||||
|---|---|---|---|---|---|---|
| variables | RR | 95% CI | P -value | RR | 95% CI | P -value |
| Gender | 0.9 | 0.66–1.2 | 0.5 | — | — | — |
| Depth (T1, T2, T3/T4) | 2.59 | 1.65–4.71 | <0.0001 | 1.03 | 0.99–1.07 | 0.08 |
| Lymph node metastasis | 4.05 | 2.45–8.21 | <0.0001 | 2.97 | 1.58–7.58 | 0.0001 |
| Lymphatic permeation | 3.15 | 1.91–6.38 | <0.0001 | 1.21 | 0.62–3.13 | 0.59 |
| Venous permeation | 1.92 | 1.46–2.53 | <0.0001 | 1.05 | 0.69–1.6 | 0.79 |
| Peritoneal dissemination | 8.3 | 4.49–15.17 | <0.0001 | 3.95 | 1.48–11.3 | 0.005 |
| miR-144 (high/low) | 0.71 | 0.5–0.99 | 0.0431 | 0.96 | 0.65–1.4 | 0.84 |
Abbreviations: RR=relative risk ratio; CI=confidence interval.
In vitro administration of miR-144 inhibits ZFX protein and increases susceptibility to 5-FU
ZFX is believed to maintain the stemness of embryonic stem cells, whereas resistance to chemotherapy is considered to be one of the characteristics of stemness in cancer cells. In this study, we have shown that ZFX is directly targeted by miR-144. Therefore, to confirm the functional relationship between miR-144 and ZFX, we investigated the effect of miR-144 restoration on chemotherapy resistance. Anti-tumour effects of 5-FU (0 μg ml−1, 1.0 μg ml−1, and 10.0 μg ml−1) treatments were evaluated by cell proliferation (MTT) assays. Growth was reduced 0.94-, 0.85-, and 0.76-fold in the group treated with pre-miR-144 in comparison with control cell lines, with statistical significance by ANOVA analysis (Figure 1B). This study indicated that the administration of miR-144 in cancer cells increased responsiveness to chemotherapy.
Discussion
Since Karnoub et al (2007) reported the role of microRNAs in breast cancer metastasis, several studies have investigated whether microRNAs are regulators of the metastatic process. To establish and enhance the reliability of our microRNA microarray data, we utilised two different platforms. We found three preferentially downregulated microRNAs in GC patients’ BM in stage IV compared with stage I. Among these, miR-144 showed the best FDR value. In addition, quantitative RT–PCR assays validated miR-144 as a candidate for further analysis.
For analysis of the gene expression array, we applied 3D gene array (Iwano et al, 2010). This technological innovation enabled us to identify specific molecules even from tiny amounts of materials. Among more than 600 predicted miR-144 target genes, the microarray validated five (ZFX, FOSB, SUCLA2, LSM14A, and HDHD2). Recently, CSC is believed to have an important role in forming cancer metastasis; therefore, we have focused on genes in terms of the word listed among these five genes. In general, considering CSCs, several biological phenotypes should be proven, such as tumorigenesis, expressing specific cell surface antigens, dormant cell cycle, and resistant against treatment. Among those features, chemotherapy resistance is considered to be one of the most consecutive features, and we have focused on it in the current study. The effect of pre-miR144 for recovering susceptibility to 5-FU was observed in MKN45 GC cell (Figure 1C); however, the alteration of susceptibility to chemotherapy rescued by miR-144 was moderate in comparison with control cells and parent cells. The direct inhibition of ZFX by miR-144 in five GC cell lines was proven distinctively by luciferase assay (Figure 1A); however, the actual protein inhibition of ZFX was moderate by miR-144 (Figure 1B). We speculated that ZFX expression was probably rescued by other compensated systems to preserve the role of this critical gene, which disclosed the moderate change in sensitivity to 5-FU in GC cells in the current study (Figure 1C). ZFX was a cancer stem-cell-related gene in terms of chemotherapy resistance; however, this gene was partially inhibited by miR-144 effect. Therefore, we should expect the role of miR-144 expression in BM not as a mimic microRNA to rescue the susceptibility to chemotherapy, but as an indicator of the poorer prognosis of GC cases.
In a previous study, we showed that predicting GC metastasis or recurrence could not be achieved simply by detecting circulating tumour cells. As for the bona fide factors contributing to metastasis, we must consider the presence of circulating (disseminated) tumour cells and BM-derived progenitor cells at premetastatic sites. As a matter of fact, Lyden et al (2001) highlighted the role of host BM-derived cells in the development of metastasis after curative surgery. We previously reported the importance of the simultaneous presence of disseminated tumour cells and host cells expressing VEGFR-1, hematopoietic progenitor cells (Mimori et al, 2008). Therefore, in the current study, we determined in which cells expression of miR-144 was mainly diminished and ZFX upregulated in BM during progression of GC.
Here we showed that miR-144 expression was decreased in stage IV GC, but the expression of miR-144 was retained in stage I GC (Figure 4). Therefore, we disclosed that the loss of miR-144 was specifically observed in epithelial cell fractions in BM. In our previous study, we clarified the existence of cancer cell clusters in BM even in stage I GC cases (Mimori et al, 2008); therefore, we validated that the loss of miR-144 expression in ITC fraction in BM might have a critical role in GC cancer progression. We also found that primary GC tumours with a relatively low level of miR-144 expression showed a poorer prognosis than GC cases with a high level of miR-144 expression. On the other hand, one of the target molecules of miR-144, ZFX, was expressed at higher levels in BM at stage I in comparison with stage IV. Moreover, restoration of miR-144 expression in cancer cell lines restored the cells’ sensitivity to chemotherapy (Figure 1B), suggesting a key role of miR-144 target genes in the development of GC’s resistance to chemotherapy resulting from ZFX upregulation. This in vitro finding supported our current hypothesis that the miR-144-ZFX pathway has an important role primarily in disseminated cancer cells, and not in host BM cells in GC.
We speculated that cancer cells’ abundant expression of ZFX in advanced cases means that ZFX might have a crucial role in the maintenance of CSCs in BM. As for the role of ZFX as a regulator of CSCs, recent reports are intriguing. In non-malignant embryonic stem cells, Cellot and Sauvageau (2007) described the transcriptional role of Zfx in the self-renewal/maintenance of both embryonic stem cells and hematopoietic stem cells (Galan-Caridad et al, 2007). Using a new software programme, Ouyang et al (2009) identified ZFX as one of 12 transcription factors upregulated in embryonic stem cells (Ouyang et al, 2009). On the other hand, in malignant cells, Huang et al (2009) reported that ZFX, two ATP-binding cassette (ABC) transporter genes, three Wnt, and two Notch signal pathway-related genes were upregulated in SP cells from two oesophageal cancer cells. In the current study, we demonstrated that in cells with low miR-144 expression, ZFX expression was elevated and the cells were chemotherapy-resistant, presumably because those cancer cells expressed ABC transporter genes at high levels. Our study is the first to demonstrate that the loss of miR-144 expression by disseminated cancer cells and their resultant high level of ZFX expression is critically important in the progression of GC in the marrow. We speculate that only ZFX-expressing cancer cells could survive and prepare for progression of GC.
In conclusion, we demonstrated that low miR-144 expression in cancer cells metastasised to the marrow is associated with GC progression. Decreased expression of miR-144 correlated with progression of the disease. We expected that reestablishing miR-144 might be effectively used in BM to restore GC’s chemosensitivity to 5-FU through its direct targeting of ZFX in the future. The ZFX gene, an X-linked zinc finger protein, has crucial roles in the maintenance of haematopoietic stem cells and the stemness of cancer cells.
Acknowledgments
We thank T Shimooka, K Ogata, M Kasagi, Y Nakagawa, and T Kawano for their technical assistance. We would really appreciate Dr H Iinuma for providing data of cell sorting in bone marrow and Professor Carlo Croce for microarray analysis in Columbus, OH, USA. This work was supported in part by the following grants and foundations: CREST, Japan Science and Technology Agency (JST); Japan Society for the Promotion of Science (JSPS) Grant-in-Aid for Scientific Research, grant numbers 20390360, 20591547, 20790960, 21591644, 21791295, 21791297, 215921014, and 21679006; and the Funding Program for Next Generation World-Leading Researchers (LS094); and NEDO (New Energy and Industrial Technology Development Organization) Technological Development for Chromosome Analysis.
Supplementary Information accompanies the paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
10/04/2012
This paper has been modified since advance online publication, an acknowledgement has been added
Supplementary Material
References
- Alix-Panabieres C, Riethdorf S, Pantel K (2008) Circulating tumor cells and bone marrow micrometastasis. Clin Cancer Res 14: 5013–5021 [DOI] [PubMed] [Google Scholar]
- Bottoni A, Zatelli MC, Ferracin M, Tagliati F, Piccin D, Vignali C, Calin GA, Negrini M, Croce CM, Degli Uberti EC (2007) Identification of differentially expressed microRNAs by microarray: a possible role for microRNA genes in pituitary adenomas. J Cell Physiol 210: 370–377 [DOI] [PubMed] [Google Scholar]
- Braun S, Pantel K, Muller P, Janni W, Hepp F, Kentenich CR, Gastroph S, Wischnik A, Dimpfl T, Kindermann G, Riethmuller G, Schlimok G (2000) Cytokeratin-positive cells in the bone marrow and survival of patients with stage I, II, or III breast cancer. N Engl J Med 342: 525–533 [DOI] [PubMed] [Google Scholar]
- Braun S, Vogl FD, Naume B, Janni W, Osborne MP, Coombes RC, Schlimok G, Diel IJ, Gerber B, Gebauer G, Pierga JY, Marth C, Oruzio D, Wiedswang G, Solomayer EF, Kundt G, Strobl B, Fehm T, Wong GY, Bliss J, Vincent-Salomon A, Pantel K (2005) A pooled analysis of bone marrow micrometastasis in breast cancer. N Engl J Med 353: 793–802 [DOI] [PubMed] [Google Scholar]
- Calin GA, Croce CM (2006a) MicroRNA signatures in human cancers. Nat Rev Cancer 6: 857–866 [DOI] [PubMed] [Google Scholar]
- Calin GA, Croce CM (2006b) MicroRNA-cancer connection: the beginning of a new tale. Cancer Res 66: 7390–7394 [DOI] [PubMed] [Google Scholar]
- Calin GA, Croce CM (2007) Investigation of microRNA alterations in leukemias and lymphomas. Methods Enzymol 427: 193–213 [DOI] [PubMed] [Google Scholar]
- Calin GA, Liu CG, Sevignani C, Ferracin M, Felli N, Dumitru CD, Shimizu M, Cimmino A, Zupo S, Dono M, Dell’Aquila ML, Alder H, Rassenti L, Kipps TJ, Bullrich F, Negrini M, Croce CM (2004) MicroRNA profiling reveals distinct signatures in B cell chronic lymphocytic leukemias. Proc Natl Acad Sci USA 101: 11755–11760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cellot S, Sauvageau G (2007) Zfx: at the crossroads of survival and self-renewal. Cell 129: 239–241 [DOI] [PubMed] [Google Scholar]
- Choesmel V, Anract P, Hoifodt H, Thiery JP, Blin N (2004) A relevant immunomagnetic assay to detect and characterize epithelial cell adhesion molecule-positive cells in bone marrow from patients with breast carcinoma: immunomagnetic purification of micrometastases. Cancer 101: 693–703 [DOI] [PubMed] [Google Scholar]
- Dirks P (2010) Cancer stem cells: invitation to a second round. Nature 466: 40–41 [DOI] [PubMed] [Google Scholar]
- Engel H, Kleespies C, Friedrich J, Breidenbach M, Kallenborn A, Schondorf T, Kolhagen H, Mallmann P (1999) Detection of circulating tumour cells in patients with breast or ovarian cancer by molecular cytogenetics. Br J Cancer 81: 1165–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galan-Caridad JM, Harel S, Arenzana TL, Hou ZE, Doetsch FK, Mirny LA, Reizis B (2007) Zfx controls the self-renewal of embryonic and hematopoietic stem cells. Cell 129: 345–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garzon R, Volinia S, Liu CG, Fernandez-Cymering C, Palumbo T, Pichiorri F, Fabbri M, Coombes K, Alder H, Nakamura T, Flomenberg N, Marcucci G, Calin GA, Kornblau SM, Kantarjian H, Bloomfield CD, Andreeff M, Croce CM (2008) MicroRNA signatures associated with cytogenetics and prognosis in acute myeloid leukemia. Blood 111: 3183–3189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottardo F, Liu CG, Ferracin M, Calin GA, Fassan M, Bassi P, Sevignani C, Byrne D, Negrini M, Pagano F, Gomella LG, Croce CM, Baffa R (2007) Micro-RNA profiling in kidney and bladder cancers. Urol Oncol 25: 387–392 [DOI] [PubMed] [Google Scholar]
- Huang D, Gao Q, Guo L, Zhang C, Jiang W, Li H, Wang J, Han X, Shi Y, Lu SH (2009) Isolation and identification of cancer stem-like cells in esophageal carcinoma cell lines. Stem Cells Dev 18: 465–473 [DOI] [PubMed] [Google Scholar]
- Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M, Menard S, Palazzo JP, Rosenberg A, Musiani P, Volinia S, Nenci I, Calin GA, Querzoli P, Negrini M, Croce CM (2005) MicroRNA gene expression deregulation in human breast cancer. Cancer Res 65: 7065–7070 [DOI] [PubMed] [Google Scholar]
- Ito T, Tanaka E, Kadowaki T, Kan T, Higashiyama M, Shiojima S, Tomoda S, Myoumoto A, Akiyama H, Nobumasa H, Matsumoto S, Miyamoto S, Mitsumori M, Sato F, Watanabe G, Itami A, Meltzer SJ, Tsujimoto G, Shimada Y (2007) An ultrasensitive new DNA microarray chip provides gene expression profiles for preoperative esophageal cancer biopsies without RNA amplification. Oncology 73: 366–375 [DOI] [PubMed] [Google Scholar]
- Iwahashi H, Kitagawa E, Suzuki Y, Ueda Y, Ishizawa YH, Nobumasa H, Kuboki Y, Hosoda H, Iwahashi Y (2007) Evaluation of toxicity of the mycotoxin citrinin using yeast ORF DNA microarray and Oligo DNA microarray. BMC Genomics 8: 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwano S, Ichikawa M, Takizawa S, Hashimoto H, Miyamoto Y (2010) Identification of AhR-regulated genes involved in PAH-induced immunotoxicity using a highly-sensitive DNA chip, 3D-Gene Human Immunity and Metabolic Syndrome 9k. Toxicol In Vitro 24: 85–91 [DOI] [PubMed] [Google Scholar]
- Kang Y (2009) Analysis of cancer stem cell metastasis in xenograft animal models. Methods Mol Biol 568: 7–19 [DOI] [PubMed] [Google Scholar]
- Kaplan RN, Riba RD, Zacharoulis S, Bramley AH, Vincent L, Costa C, MacDonald DD, Jin DK, Shido K, Kerns SA, Zhu Z, Hicklin D, Wu Y, Port JL, Altorki N, Port ER, Ruggero D, Shmelkov SV, Jensen KK, Rafii S, Lyden D (2005) VEGFR1-positive haematopoietic bone marrow progenitors initiate the pre-metastatic niche. Nature 438: 820–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnoub AE, Dash AB, Vo AP, Sullivan A, Brooks MW, Bell GW, Richardson AL, Polyak K, Tubo R, Weinberg RA (2007) Mesenchymal stem cells within tumour stroma promote breast cancer metastasis. Nature 449: 557–563 [DOI] [PubMed] [Google Scholar]
- Kulshreshtha R, Ferracin M, Wojcik SE, Garzon R, Alder H, Agosto-Perez FJ, Davuluri R, Liu CG, Croce CM, Negrini M, Calin GA, Ivan M (2007) A microRNA signature of hypoxia. Mol Cell Biol 27: 1859–1867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CG, Calin GA, Volinia S, Croce CM (2008a) MicroRNA expression profiling using microarrays. Nat Protoc 3: 563–578 [DOI] [PubMed] [Google Scholar]
- Liu CG, Spizzo R, Calin GA, Croce CM (2008b) Expression profiling of microRNA using oligo DNA arrays. Methods 44: 22–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lujambio A, Calin GA, Villanueva A, Ropero S, Sanchez-Cespedes M, Blanco D, Montuenga LM, Rossi S, Nicoloso MS, Faller WJ, Gallagher WM, Eccles SA, Croce CM, Esteller M (2008) A microRNA DNA methylation signature for human cancer metastasis. Proc Natl Acad Sci USA 105: 13556–13561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyden D, Hattori K, Dias S, Costa C, Blaikie P, Butros L, Chadburn A, Heissig B, Marks W, Witte L, Wu Y, Hicklin D, Zhu Z, Hackett NR, Crystal RG, Moore MA, Hajjar KA, Manova K, Benezra R, Rafii S (2001) Impaired recruitment of bone-marrow-derived endothelial and hematopoietic precursor cells blocks tumor angiogenesis and growth. Nat Med 7: 1194–1201 [DOI] [PubMed] [Google Scholar]
- Marcucci G, Radmacher MD, Maharry K, Mrozek K, Ruppert AS, Paschka P, Vukosavljevic T, Whitman SP, Baldus CD, Langer C, Liu CG, Carroll AJ, Powell BL, Garzon R, Croce CM, Kolitz JE, Caligiuri MA, Larson RA, Bloomfield CD (2008) MicroRNA expression in cytogenetically normal acute myeloid leukemia. N Engl J Med 358: 1919–1928 [DOI] [PubMed] [Google Scholar]
- Maruyama K, Kaminishi M, Hayashi K, Isobe Y, Honda I, Katai H, Arai K, Kodera Y, Nashimoto A (2006) Gastric cancer treated in 1991 in Japan: data analysis of nationwide registry. Gastric Cancer 9: 51–66 [DOI] [PubMed] [Google Scholar]
- Mimori K, Fukagawa T, Kosaka Y, Kita Y, Ishikawa K, Etoh T, Iinuma H, Sasako M, Mori M (2008) Hematogenous metastasis in gastric cancer requires isolated tumor cells and expression of vascular endothelial growth factor receptor-1. Clin Cancer Res 14: 2609–2616 [DOI] [PubMed] [Google Scholar]
- Mori M, Mimori K, Inoue H, Barnard GF, Tsuji K, Nanbara S, Ueo H, Akiyoshi T (1995) Detection of cancer micrometastases in lymph nodes by reverse transcriptase-polymerase chain reaction. Cancer Res 55: 3417–3420 [PubMed] [Google Scholar]
- Nagino K, Nomura O, Takii Y, Myomoto A, Ichikawa M, Nakamura F, Higasa M, Akiyama H, Nobumasa H, Shiojima S, Tsujimoto G (2006) Ultrasensitive DNA chip: gene expression profile analysis without RNA amplification. J Biochem 139: 697–703 [DOI] [PubMed] [Google Scholar]
- Ouyang Z, Zhou Q, Wong WH (2009) ChIP-Seq of transcription factors predicts absolute and differential gene expression in embryonic stem cells. Proc Natl Acad Sci USA 106: 21521–21526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakhinia E, Glennie C, Hoyland JA, Menasce LP, Brady G, Miller C, Radford JA, Byers RJ (2007) Clinical quantitation of diagnostic and predictive gene expression levels in follicular and diffuse large B-cell lymphoma by RT-PCR gene expression profiling. Blood 109: 3922–3928 [DOI] [PubMed] [Google Scholar]
- Schetter AJ, Leung SY, Sohn JJ, Zanetti KA, Bowman ED, Yanaihara N, Yuen ST, Chan TL, Kwong DL, Au GK, Liu CG, Calin GA, Croce CM, Harris CC (2008) MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. Jama 299: 425–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz B, Radbruch A, Kummel T, Wickenhauser C, Korb H, Hansmann ML, Thiele J, Fischer R (1994) Magnetic activated cell sorting (MACS)--a new immunomagnetic method for megakaryocytic cell isolation: comparison of different separation techniques. Eur J Haematol 52: 267–275 [DOI] [PubMed] [Google Scholar]
- Visone R, Pallante P, Vecchione A, Cirombella R, Ferracin M, Ferraro A, Volinia S, Coluzzi S, Leone V, Borbone E, Liu CG, Petrocca F, Troncone G, Calin GA, Scarpa A, Colato C, Tallini G, Santoro M, Croce CM, Fusco A (2007) Specific microRNAs are downregulated in human thyroid anaplastic carcinomas. Oncogene 26: 7590–7595 [DOI] [PubMed] [Google Scholar]
- Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, Calin GA, Liu CG, Croce CM, Harris CC (2006) Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell 9: 189–198 [DOI] [PubMed] [Google Scholar]
- Zhang L, Volinia S, Bonome T, Calin GA, Greshock J, Yang N, Liu CG, Giannakakis A, Alexiou P, Hasegawa K, Johnstone CN, Megraw MS, Adams S, Lassus H, Huang J, Kaur S, Liang S, Sethupathy P, Leminen A, Simossis VA, Sandaltzopoulos R, Naomoto Y, Katsaros D, Gimotty PA, DeMichele A, Huang Q, Butzow R, Rustgi AK, Weber BL, Birrer MJ, Hatzigeorgiou AG, Croce CM, Coukos G (2008) Genomic and epigenetic alterations deregulate microRNA expression in human epithelial ovarian cancer. Proc Natl Acad Sci USA 105: 7004–7009 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.