Abstract
As part of an ongoing investigation of filamentous fungi for anticancer leads, an active culture was identified from the Mycosynthetix library (MSX 70741, of the order Hypocreales, Ascomycota). The fungal extract exhibited cytotoxic activity against the H460 (human non-small cell lung carcinoma) cell line, and bioactivity-directed fractionation yielded peptaibols 1–12 and harzianums A (13) and B (14). Structure elucidation of 1–12 was facilitated by high-resolution MS/MS obtained on a Thermo LTQ Orbitrap XL using Higher-Energy Collisional Dissociation (HCD) and by high field NMR (950 MHz). The absolute configuration was determined by Marfey’s analysis of the individual amino acids; the time required for such analysis was decreased via the development of a 10 min UPLC method. The isolated peptaibols (1–12), along with three other peptaibols isolated and elucidated from a different fungus (MSX 57715) of the same Order (15–17), were examined for activity in a suite of biological assays, including those for cytotoxic, antibacterial, and anthelmintic activities.
Keywords: peptaibols, cytotoxicity, anthelmintic, Hypocreales, Higher-Energy Collisional Dissociation (HCD)
Introduction
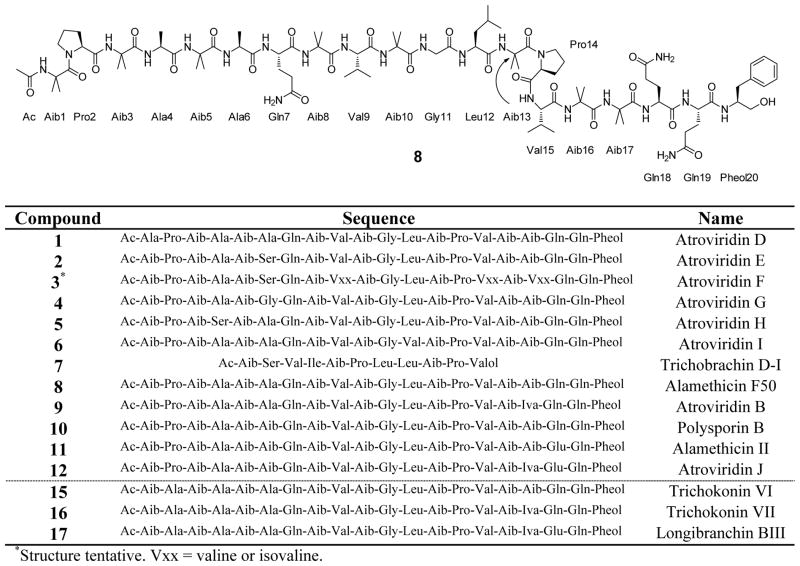
By most measures of scientific progress, peptaibols have been investigated rather extensively. An entire book [1] and an issue of a two separate journals [2, 3] have been devoted to the subject, and various aspects have been reviewed extensively, especially when dealing with a common source (Trichoderma sp.) [4] or the most well studied class of peptaibols (the alamethicins) [5]. However, in the course of a collaborative project to identify anticancer leads from diverse natural product sources [6, 7], extracts of filamentous fungi from the Mycosynthetix library, representing over 55,000 accessions, have not yielded peptaibols to date. In fact, most of the compounds discovered in this program, which is driven by bioactivity-directed fractionation guided by a suite of cytotoxicity and mechanism of action based assays, have been of a molecular weight well under 1000 a.m.u. [8–11]. Hence, uncovering a series of both new and known compounds of significantly greater molecular weight was of interest, both from the standpoint of evaluating their biological activity in assays that pertain to anticancer activity and from examining their chemical diversity relative to the library of fungal isolates. In the course of this research, state-of-the-art technologies were applied to the structure elucidation processes, thereby developing tools that could be applied to research on peptaibols or related compounds. For example, determining the sequence of residues was facilitated via the use of UPLC coupled to high-resolution MS/MS using Higher-Energy Collisional Dissociation (HCD) on a Thermo LTQ Orbitrap XL. This was complemented by the resolution enhancements observed when analyzing the TOCSY and NOESY spectra on a 950 MHz NMR spectrometer. Moreover, the time required to determine the absolute configuration of the residues using Marfey’s analysis was decreased by the development of a 10 min UPLC procedure, compared to 30 to 40 min run times for similar analyses of peptaibols by HPLC [12, 13]. In short, the bioactivity-directed fractionation study of fungus MSX 70741 resulted in the isolation and characterization of a series of peptaibols (1–12; compounds 1–7 and 12 being new) and two known trichothecene analogues [harzianum A (13) and harzianum B (14)]. The isolated peptaibols (1–12), along with three other known peptaibols (15–17) isolated from a different fungus of the same order (MSX 57715) were examined for cytotoxicity, antibacterial and anthelmintic activities, and activity in a mitochondrial transmembrane potential assay. Figure 1 illustrates the structures/sequences for the isolated peptaibols (1–12 and 15–17).
Figure 1.
Structure of Alamethicin F50 (8) and sequences of the other isolated peptaibols.
Materials and Methods
General Experimental Procedures
NMR experiments were conducted in CD3OH with presaturation of the OH peak at δH 4.9 ppm. NMR instrumentation was a Bruker Ultrashield Plus with Avance III console, Topspin software version 2.1, and a QNP style Cryoprobe (operating at 950.30 MHz for 1H). For comparison of these NMR data to that of a 500 MHz spectrometer a JEOL ECA-500 was utilized. HRESIMS was performed on a Thermo LTQ Orbitrap XL system equipped with HCD cell. UPLC was carried out on a Waters Acquity system with data collected and analyzed using Empower software (build 2154). Preparative HPLC was performed on Varian Prostar HPLC systems equipped with Prostar 210 pumps and a Prostar 335 photodiode array detector (PDA), with data collected and analyzed using Galaxie Chromatography Workstation software (version 1.9.3.2). For preparative HPLC, a Phenomenex Synergi Max-RP 80 (4 μm; 250 × 21.2 mm) column was used at a 15 mL/min flow rate, while for UPLC, a BEH C18 (1.7 μm; 50 × 2.1 mm) column was used with a 0.61 mL/min flow rate (0.5 mL/min for Marfey’s analysis), both monitored at 205 nm (340 nm for Marfey’s analysis). Flash chromatography was performed on a Teledyne ISCO CombiFlash Rf using a 40 g Silica Gold column and monitored by UV and evaporative light-scattering detectors. Reference standards of amino acids and Marfey’s reagent were obtained from Sigma-Aldrich. All other reagents and solvents were obtained from Fisher Scientific and were used without further purification.
Producing Organisms and Fermentations
Mycosynthetix fungal strain MSX 70741 was isolated in April 1993 from wood collected in a humid mountain forest and strain MSX 57715 was isolated in October 1991 from leaf litter from a predominately oak, humid forest, both by Dr. Barry Katz of MYCOsearch and later acquired by Mycosynthetix. DNA analyses were performed by MIDI Labs, Inc. (Newark, DE), and the D2 variable region of the large subunit (LSU) rRNA was sequenced and compared to their database; in both cases, the closest match could only determine that these fungi were of the order Hypocreales, Ascomycota; these data were deposited in Genbank (accession Nos. JN377382 and JN377381, respectively). The cultures were stored on malt extract slants and were transferred periodically. Fresh cultures were grown on a similar slant, and a piece was transferred to a medium containing 2% soy peptone, 2% dextrose, and 1% yeast extract (YESD media). Following incubation (7 d) at 22 °C with agitation, the cultures were used to inoculate 50 mL of a rice medium, prepared using rice to which was added a vitamin solution and twice the volume of rice with H2O, in a 250 mL Erlenmeyer flask. This was incubated at 22 °C until the cultures showed good growth (approximately 14 d) to generate the screening cultures. The scale-up cultures, used for isolation of the peptaibols, were grown in a 2.8 L Fernbach flask containing 150 g rice and 300 mL H2O and were inoculated using a seed culture grown in YESD medium. These were incubated at 22 °C for 14 d.
Extraction and Isolation
To the large scale solid fermentation of MSX 70741 was added 500 mL of 1:1 MeOH:CHCl3. The mixture was shaken for 16 h then filtered, and the solvent was evaporated. The material (2.07 g) was then dissolved in 1:1 CHCl3:MeOH and adsorbed onto Celite 545 and fractionated by flash silica-gel chromatography. The solvent conditions were 100% hexane to 100% CHCl3 over 10 column volumes (CV), then 100% CHCl3 for 7 CV, followed by increasing amounts of MeOH in CHCl3 from 0–10% over 20 CV, 10–20% over 5 CV, 20–100% over 2 CV, and 100% MeOH for the remaining 8 CV, all at 40 mL/min. The peptaibol-enriched material (100% MeOH; 600 mg) was purified via six separate injections on preparative HPLC using a gradient that initiated with 40:60 CH3CN:H2O and increased linearly to 100:0 CH3CN:H2O over 30 min.
UPLC-HRMS of 1–12 and 15–17
High resolution UPLC-MS was carried out on a Waters Acquity UPLC system coupled to Thermo LTQ Orbitrap XL. Sample injections of 3 μL of 0.5 mg/mL were separated via UPLC at 0.61 mL/min; an in-line column flow tee was used to divert the mobile phase 1:1 between the electrospray source and waste. UPLC conditions were 20% CH3CN in 0.1% aqueous formic acid for 0.5 min., then 20–100% CH3CN from 0.5–3.0 min. Peptaibols have a labile bond between the central Aib and Pro residues that is cleaved under conventional electrospray ionization conditions [14]. As a result, two fragments (referred to as in-source fragments hence forth) are commonly observed at higher abundance than that of the monoisotopic parent ion. Mass spectra were collected in positive mode ESI with the following source parameters: capillary temp. = 275°C, sheath gas = 15, auxiliary gas = 5, sweep gas = 2, source voltage = 4.50 kV, capillary voltage = 46 V, tube lens = 115 V. The MS/MS fragmentation was performed using an instrument method comprised of four scan events. During scan event 1, a full scan mass spectrum of mass range 75–2000 a.m.u. was collected; scan event 2 was used to conduct HCD fragmentation of the low molecular weight (MW) fragment (in-source) at an optimized collision energy of 22 (Table 1); scan event 3 was used to perform HCD fragmentation of the high MW fragment (in-source) at a collision energy of 15; scan event 4 was for HCD fragmentation of the high MW fragment (in-source) at a collision energy of 35.
Table 1.
High-Resolution Mass Data for Compounds 1–12
| Compound | [M + H]+ | Theoretical [M + H]+ | Mass Error (ppm) | Formula | Low MW Fragment | High MW Fragment |
|---|---|---|---|---|---|---|
| 1 | 1949.1166 | 1949.1219 | −2.7 | C91H149N23O24 | 774.4487 | 1175.6748 |
| 2 | 1979.1276 | 1979.1324 | −2.4 | C92H151N23O25 | 774.4483 | 1205.6868 |
| 3 | 1993.1443 | 1993.1480 | −1.9 | C93H153N23O25 | 788.4641 | 1205.6873 |
| 4 | 1949.1136 | 1949.1219 | −4.2 | C91H149N23O24 | 774.4479 | 1175.6745 |
| 5 | 1979.1300 | 1979.1324 | −1.2 | C92H151N23O25 | 774.4482 | 1205.6870 |
| 6 | 1949.1145 | 1949.1218 | −3.8 | C91H149N23O24 | 774.4482 | 1175.6758 |
| 7 | 1120.7307 | 1120.7340 | −3.0 | C55H97N11O13 | 512.3060/609.4315/920.5790* | |
| 8 | 1963.1321 | 1963.1375 | −2.8 | C92H151N23O24 | 774.4478 | 1189.6898 |
| 9 | 1977.1481 | 1977.1532 | −2.6 | C93H153N23O24 | 788.4638 | 1189.6911 |
| 10 | 1977.1468 | 1977.1532 | −3.2 | C93H153N23O24 | 774.4478 | 1203.7051 |
| 11 | 1978.1309 | 1978.1371 | −3.2 | C93H152N22O25 | 775.4318 | 1203.7056 |
| 12 | 1992.1476 | 1992.1528 | −2.6 | C94H154N22O25 | 789.4475 | 1203.7075 |
Three major in-source fragments were present with compound 7
Collision energies selected in this study were optimized using the LTQ Tune software’s automatic tuning feature. To do so, a pure solution of a selected peptaibol was infused directly under UPLC flow conditions. In-source fragments were then subjected to a range of collision energies (10–35 NCE; normalized collision energy is a resonance excitation process used for inducing fragmentation while compensating for mass dependence). Fragmentation energies used in this study were selected to generate a broad molecular weight range of fragments of the highest signal possible. MSn of the large (in-source) fragment was collected at two different fragmentation energies (15 and 35 NCE) during two different scan events because we were not able to produce all of the observed fragments at a single NCE value.
Marfey’s Analysis of 1–12
Our method was based on Kjer et al. [15] with the following modifications. Approximately 0.2 mg of each amino acid standard was weighed into separate glass 2 mL reaction vials. To each standard was added 50 μL of H2O, 20 μL of 1 M NaHCO3, and 100 μL 1% Marfey’s reagent in acetone. The reaction mixtures were agitated at 40°C for 1 h. The reactions were halted by the 7addition of 10 μL of 2N HCl. The product of the reactions was dried under a stream of air and dissolved in ~1.7 mL of MeOH. Each derivatized standard was injected individually (0.7 μL) onto the UPLC. Also, aliquots of all of the derivatized standards were combined to give a mixed standard, which was injected just prior to the digested and derivatized peptaibols (see below). UPLC conditions were 10–70% MeOH in H2O over 10 min on the aforementioned BEH column and eluent monitored at 340 nm.
To generate the digested and derivatized peptaibols, approximately 0.2–0.3 mg of compounds 1–12 were weighed separately into 2 mL reaction vials, to which was added 0.5 mL of 6N HCl. The compounds were hydrolyzed at 110°C for 24 h, at which time they were evaporated under a stream of air. To each hydrolysis product was then added 25 μL H2O, 10 μL 1 M NaHCO3, and 50 μL of 1% Marfey’s reagent in acetone. The reaction mixtures were agitated at 40°C for 1 h. The reactions were halted by the addition of 5 μL of 2N HCl. The mixtures were dried under a stream of air and brought up in ~200 μL of MeOH and injected onto the UPLC using the same conditions as for the standards.
Cytotoxicity Assays
The cytotoxicity measurements against the MCF-7 [16] human breast carcinoma (Barbara A. Karmanos Cancer Center), NCI-H460 [17] human large cell lung carcinoma (HTB-177, American Type Culture Collection (ATCC), and SF-268 [18] human astrocytoma (NCI Developmental Therapeutics Program) cell lines were performed as described previously [19, 20]. Moreover, a second cytotoxicity assay was performed on only the isolated compounds using the MDA-MB-435 [21] human melanoma (HTB-129, ATCC) cell line as described previously [8] and with the following modifications. After treating the MDA-MB-435 cells with test substances and 96 h incubation at 37°C, the cells were evaluated for viability with a commercial absorbance assay (CellTiter 96 AQueous One Solution Cell Proliferation Assay, Promega Corp, Madison, WI). The compounds were also tested in the IMR90 fibroblast cell line (ATCC CCL-186) [22], a normal diploid cell line that proliferates in culture for approximately 58 generations prior to senescence. Positive control data for all cell lines are provided in the legend of Table 2.
Table 2.
Biological Activities of Peptaibols 1–12 and 15–17.
| Compound | Cytotoxicity IC50 values (in μM)a | MICb (μg/mL) | IC50c (μg/mL) | |||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| MCF-7 | H460 | SF268 | IMR90 | MDA- MB-435 | S. aureus | MRSA | H. contortus | |
|
| ||||||||
| Atroviridin D (1) | >10 | >10 | >10 | >10 | NTd | NT | NT | NT |
| Atroviridin E (2) | 3.3 | 3.0 | 6.8 | 7.7 | NT | NT | NT | NT |
| Atroviridin F (3) | 3.8 | 3.2 | 5.3 | >10 | NT | NT | NT | NT |
| Atroviridin G (4) | 2.7 | 1.3 | 1.9 | 5.9 | NT | NT | NT | NT |
| Atroviridin H (5) | 1.8 | 2.5 | 4.4 | 6.1 | NT | NT | NT | NT |
| Atroviridin I (6) | 2.1 | 2.3 | 5.1 | 9.8 | NT | 44 | 44 | NT |
| Trichobrachin D-I (7) | 3.4 | 4.1 | 7.0 | >10 | 7.3 | 113 | 113 | NT |
| Alamethicin F50 (8) | 2.2 | 3.4 | 2.3 | 4.8 | 8.9 | 35 | 140 | 0.2 |
| Atroviridin B (9) | 1.3 | 2.5 | 1.9 | 3.6 | 4.2 | 6 | 13 | 0.4 |
| Polysporin B (10) | 1.3 | 2.0 | 1.5 | 2.9 | 3.2 | 102 | 102 | NT |
| Alamethicin II (11) | 1.1 | 1.6 | 1.7 | 4.0 | 6.2 | 12 | 23 | NT |
| Atroviridin J (12) | 1.0 | 1.0 | 2.6 | 5.2 | NT | 9 | 18 | NT |
| Trichokonin VI (15) | 2.3 | 2.7 | 2.0 | 4.5 | 3.8 | 21 | 43 | NT |
| Trichokonin VII (16) | 1.3 | 2.2 | 1.5 | 5.1 | 3.0 | 8 | 17 | >8.3 |
| Longibranchin BIII (17) | 0.8 | 0.8 | 1.4 | 4.6 | NT | 4 | 8 | 3.0 |
| Camptothecine | 0.05 | 0.008 | 0.03 | 0.18 | NT | NT | NT | NT |
| Vinblastinee | NT | NT | NT | NT | f | NT | NT | NT |
| Vancomycine | NT | NT | NT | NT | NT | 1.5 | 0.8 | NT |
| Ivermectine | NT | NT | NT | NT | NT | NT | NT | 0.006 |
IC50 values were determined as the concentration required to reduce cellular proliferation by 50% relative to untreated controls following 72 h of continuous exposure (96 h for MDA-MB-435).
Minimal inhibitory concentration (MIC) is the lowest concentration of compound completely inhibiting growth as expressed in μg/mL [19].
IC50 values were determined as the concentration required to inhibit larval motility by 50% relative to untreated controls following 72 h of continuous exposure as expressed in μg/mL.
Indicates ‘not tested’.
Positive controls.
The positive control for MDA-MB-435 was vinblastine tested at 2.0 nM and 1.0 nM, which resulted in 41% and 68% viable cells after treatment, respectively.
Antimicrobial Assay
The compounds were screened initially for antimicrobial activity using an agar plate diffusion assay. Overnight cultures of Escherichia coli, Pseudomonas aeruginosa, Staphylococcus aureus, Mycobacterium smegmatis, Candida albicans, and Bacillus subtilis were used to inoculate molten LB media or Middlebrook 7H9 media (Difco) with 1% glycerol, containing 1.5% agar and kept at 50°C; these were then used to prepare assay plates. Samples (dissolved in 10 μL MeOH) were applied to the surface of the assay dish, and positive controls were treated in a similar manner (penicillin G, gentamicin, novobiocin, and streptomycin; all from Sigma). The bioassay plates were incubated overnight at 37°C. Biological activity of the standards could be detected to 1 μg/mL (except that penicillin G was active against E. coli at 100 μg/mL only).
Measurement of Antibiotic Activity against Methicillin-resistant S. aureus (MRSA)
The samples were tested against a suite of methicillin-resistant S. aureus (MRSA) isolates (data not shown); only representative data are shown in Table 2 against S. aureus (ATCC 6538) and an unrelated MRSA strain (ATCC 43300), which were both acquired from Danville Community Hospital (Danville, VA, USA). Minimal inhibitory concentration (MIC) measurements were performed as described previously [23, 24]. All measurements were made in duplicate and susceptibilities of the strains to vancomycin as a positive control were measured in parallel.
Anthelmintic Assay
The compounds were screened against Haemonchus contortus infective larvae (HcL3) in a L3 motility assay; this assay evaluates the effect of the compounds on the body wall musculature since this is not a feeding stage. Third stage larvae of an isolate of H. contortus maintained in goats were obtained from the laboratory of Dr. Raymond Kaplan (University of Georgia, Athens, GA). The nematode L3 bioassay was as described [25] with slight modifications. Briefly, in a 15 mL centrifugation tube, HcL3s were incubated at rt in a 0.15% sodium hypochlorite solution for 25 min, centrifuged at 100 rcf for 5 min, and then the supernatant was removed and rinsed with distilled H2O. HcL3s were concentrated in phosphate buffered saline to contain approximately 800 HcL3s/mL. The L3 motility assay was conducted in triplicate in 96-well plates; each well had a total volume of 75 μL and contained roughly 50 exsheathed HcL3s. Plates were incubated in a humidified chamber at 25–30°C for 72 h and percent immobility was calculated. Ivermectin (positive control) and DMSO only (negative control) wells were included on every plate, and percent immobility was calculated using the mean counts from three replicates. The peptaibols were dissolved in DMSO and evaluated at 20 μg/mL in triplicate; compounds were considered active if they had any immobilizing effects on HcL3s. Active compounds were evaluated further in dose response, and IC50 values were calculated using a log probit regression analysis.
Mitochondria Transmembrane Potential Assay
The mitochondrial transmembrane potential assay was performed as described previously [8].
Results
The crude 1:1 CHCl3:MeOH extract of fungus MSX 70741 was partitioned with 4:1:5 CHCl3:MeOH:H2O. The organic soluble material was active (>95% growth inhibition of H460 cells at 20 μg/mL), and was fractionated initially by flash silica-gel chromatography. Harzianum A (13, 6.8 mg) [26] and harzianum B (14, 0.9 mg) [27] eluted in the 4–5% MeOH in CHCl3 fraction (>96% growth inhibition of H460 at 2 μg/mL); these were purified by HPLC and their NMR data were in excellent agreement with the literature (see Figure S1 for structures of 13 and 14) [27]. The peptaibols eluted in the flash chromatography system in the 100% MeOH fraction (>98% growth inhibition of H460 at 20 μg/mL). Final purification of the peptaibols was accomplished by preparative scale RP-HPLC (see Figure S2). Compound 1 eluted at 12.0 min (1.1 mg), 2 at 13.1 min (3.3 mg), 3 at 14.0 min (0.7 mg), 4 at 14.3 min (2.0 mg), 5 at 15.7 min (2.6 mg), 6 at 17.7 min (6.7 mg), 7 at 18.7 min (12.4 mg), 8 between 19–21 min (242.5 mg), 9 between 21.3–22.3 min (77.6 mg), 10 between 22.3–23.5 min (52.6 mg), 11 between 23.5–24.5 min (28.8 mg), and 12 at 25.3 min (4.1 mg). Using the exact same conditions as above, fungus MSX 57715 was extracted and fractionated to isolate peptaibols 15 (205.6 mg), 16 (93.6 mg), and 17 (7.5 mg) with HPLC elution times of 17–19 min, 19–20.5 min, and 21.1 min, respectively. Compounds 8–11 were the major isolates and were known peptaibols, while 1–7 and 12 were new. The numbering of these compounds corresponds to their elution order on RP-HPLC: 1 eluted earliest and 12 eluted latest.
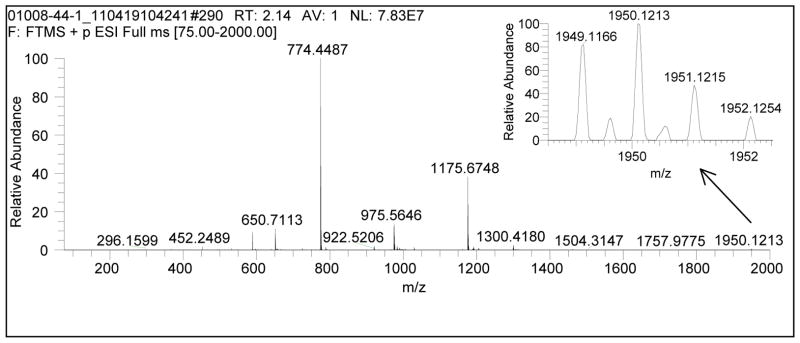
The high-resolution protonated monoisotopic precursor ion data, [M + H]+, and the resultant molecular formulae for compounds 1–12, are listed in Table 1; the structure for 8 and the sequence for the other peptaibols are displayed in Figure 1. An important characteristic of the MS spectra for typical 20-residue peptaibols was in-source fragmentation between the Aib13 and Pro14 residues, leaving two major fragments (Figure 2) [14]. Other diagnostic peaks were the doubly-and triply-charged species, as well as the triply-charged dimer (Figure 2). The MS data for the two major fragments (termed here, “low MW fragment” and “high MW fragment”) of 1–12 are listed in Table 1; note that 7 had three major in-source fragments. These major fragments were then subjected to MS/MS to elucidate the sequence.
Figure 2.
Full-scan HRMS (single scan) of atroviridin D (1). The two diagnostic in-source fragments are apparent at 774.4487 (low MW fragment) and 1175.6748 (high MW fragment). The multiply charged species are apparent at 650.7113 [M + 3H]3+, 975.5646 [M + 2H]2+, and 1300.4180 [2M + 3H]3+.
The MS/MS data, in conjunction with NMR data, were used to sequence the major metabolite (8), identified as the known peptaibol alamethicin F50 (or atroviridin A) [28]. The absolute configuration of 8 was confirmed by Marfey’s analysis. As with all of the peptaibols isolated in this study, the individual amino acids had the L- configuration except for isovaline, which was D-configuration. Compounds 9–11 were likewise found to be the known compounds atroviridin B (9) [29], polysporin B (10) [30], and alamethicin II (11) [31].
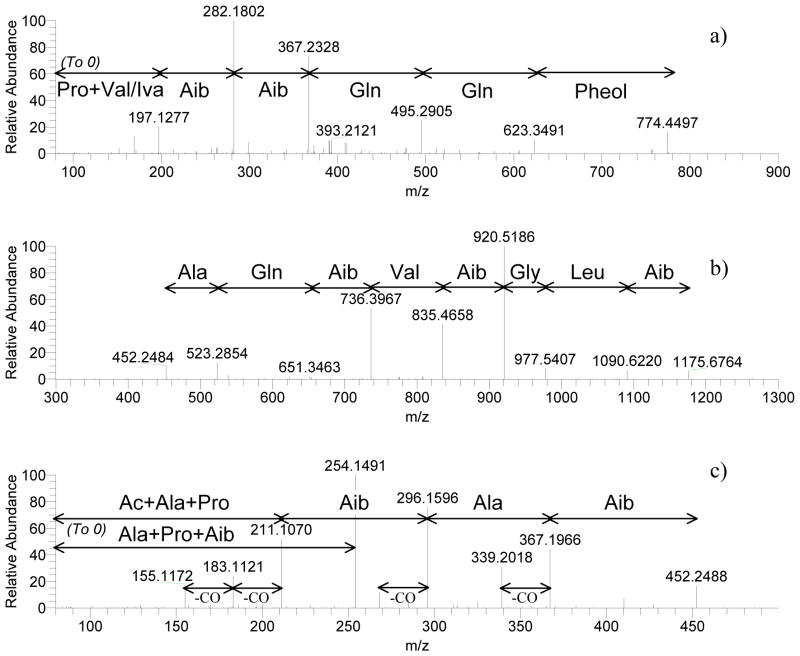
The sequences and structures of compounds 1–7 and 12 were elucidated in a similar fashion by MS/MS, NMR, and Marfey’s analysis. Examples of MS/MS analysis (Figure 3), of 1H NMR and NOESY spectra (Figures 4 and 5, respectively), and of Marfey’s analysis (Figure 6) are provided. Due to paucity of material, quality NMR data for compounds 1 and 3 could not be obtained; however, MS/MS and Marfey’s analysis was sufficient for the complete structure elucidation for 1. Alternatively, structure 3 should be considered tentative because the locations of valine vs isovaline could not be determined unequivocally, since both were present in the Marfey’s analysis. It is highly likely that residues 9 and 15 were Val and residue 17 was Iva, due to similarity with known peptaibols; however, this could not be proven.
Figure 3.
HR-MS/MS by HCD of Atroviridin D (1). a) HCD of 774 peak, CE = 22; b) HCD of 1175 peak, CE = 15; c) HCD of 1175 peak, CE = 35.
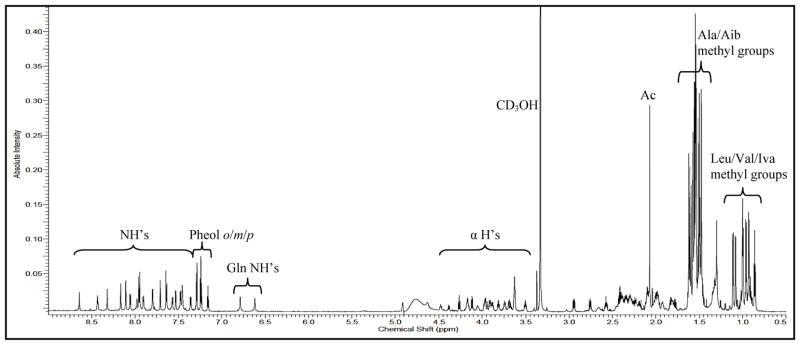
Figure 4.
1H NMR Spectrum of Atroviridin J (12), (CD3OH, 950 MHz). Spectrum was obtained with presaturation of the -OH peak at δH 4.9.
Figure 5.
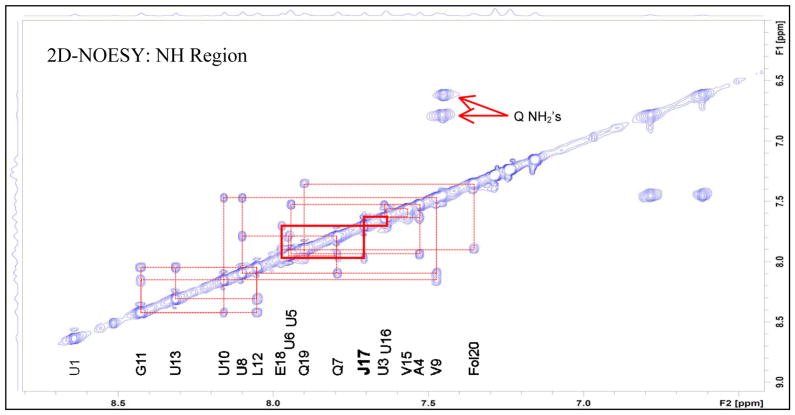
NOESY spectrum of Atroviridin J (12), highlighting the correlations proving the position of the Iva (J) residue (CD3OH, 950 MHz).
Figure 6.
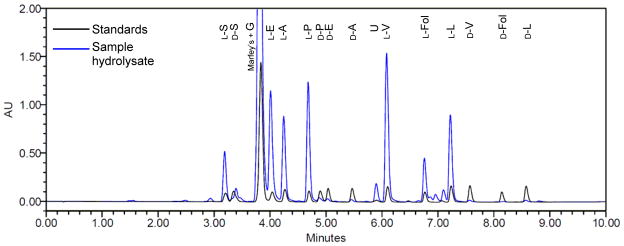
Marfey’s Analysis of Atroviridin E (2). Note that the peak in the sample hydrolysate at 3.4 min that appears to indicate the presence of D-serine is an artifact of the derivatization reaction that was present in all hydrolysate chromatograms. The retention times, while close, do not match. For procedure and chromatographic conditions, see Section 2.5.
In work of a similar vein from another fungus from the Mycosynthetix library (specifically, MSX 57715), other known peptaibols were isolated and elucidated, along with trichodermin, which is a simple trichothecene. These peptaibols were identified as trichokonin VI (15) [32], trichokonin VII (16) [32], and longibranchin BIII (17) [33, 34]. These peptaibols were evaluated in the same bioassays as 1–12 to determine the significance of Pro2 vs. Ala2.
The isolated peptaibols were evaluated in a series of biological assays. With respect to anticancer activity, they were tested for cytotoxicity against a panel of human tumor cell lines and a human fibroblast cell line (Table 2). They were also examined for activity in a mitochondria transmembrane potential assay but found inactive (data not shown). Based on literature precedent for antimicrobial activity for some peptaibols [35], they were tested against a battery of assays, including those for Gram positive and Gram negative bacteria, and C. albicans. The most promising antimicrobial activity was with respect to Staphylococcus aureus, which spawned further examination against methicillin-resistant S. aureus (Table 2). Finally, a few peptaibols have been reported to have anthelmintic activity [36, 37], and thus the compounds were examined in an assay for larval motility against Haemonchus contortus infective stages. Larval motility was inhibited completely by four of the peptaibols (8, 9, 16 and 17) when tested at 20 μg/ml; these were evaluated further for dose response with a top concentration of 8.3 μg/ml (Table 2). In summary, the biological potential of these compounds were examined extensively.
Discussion
In this work, a single injection on UPLC-MS/MS using HCD on a Thermo LTQ Orbitrap XL gave high-resolution fragmentation and nearly complete sequence data in a three minute run. HCD fragmentation is becoming a routine tool in proteomics research, especially for quantitation studies that utilize various isotopic-labeling methods and for peptide sequencing studies [38]; to the best of our knowledge, this technique has not been applied previously to research peptaibols. The advantages of the HCD collision cell in the LTQ Orbitrap XL system include its ability to generate “rich” fragmentation spectra that include low m/z values and Orbitrap (high resolution/high mass accuracy) detection of mass fragments, allowing for molecular formula assignment of a given m/z value [39].
Compound 1 was identical to the major metabolite 8 except for the substitution of Ala1 for Aib1. To our knowledge, the only other peptaibol that has been reported to have Ala1 is trichokonin IIb [40], which was identical to 1 except for the replacement of Pro2 with Ala2. The result of substituting Aib1 with Ala1 on the cytotoxic activity was interesting, as 1 was inactive compared to 8 (Table 2).
Compounds 2–6 were also structurally related to alamethicin F50 (8; Fig. 1). Compounds 2, 4, 5, and 6 differed from 8 by a single amino acid residue substitution. Residue 6 appeared to be the most variable site, with substitution of Ser, Gly, or Aib for Ala. Compound 2 was identical to 8 except for substitution of Ser6 for Ala6. Compound 3 likely included Iva17 instead of Aib17 in addition to Ser6 versus Ala6, though the structure elucidation of 3 was incomplete due to paucity of material. Compound 5 was identical to 8 except for substitution of Ser4 for Ala4. Compound 6 was identical to 8 except for substitution of Val12 for Leu12. Compound 12 was more closely related to alamethicin II (11), and differed from 11 only by substitution of Iva17 for Aib17. Due to their similarity to the known atroviridins, compounds 1–6 and 12 were ascribed the trivial names of atroviridin D through atroviridin J (Fig. 1), respectively.
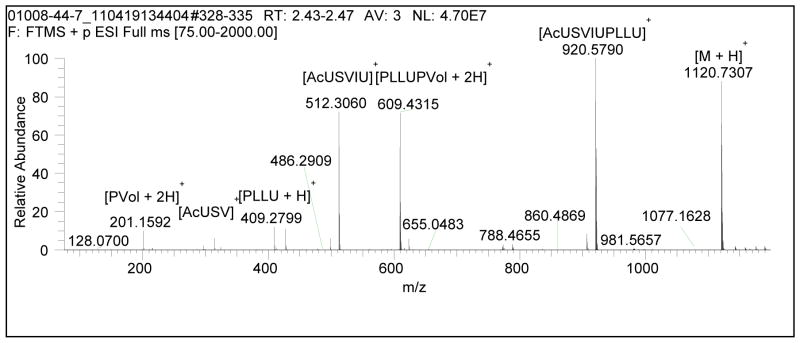
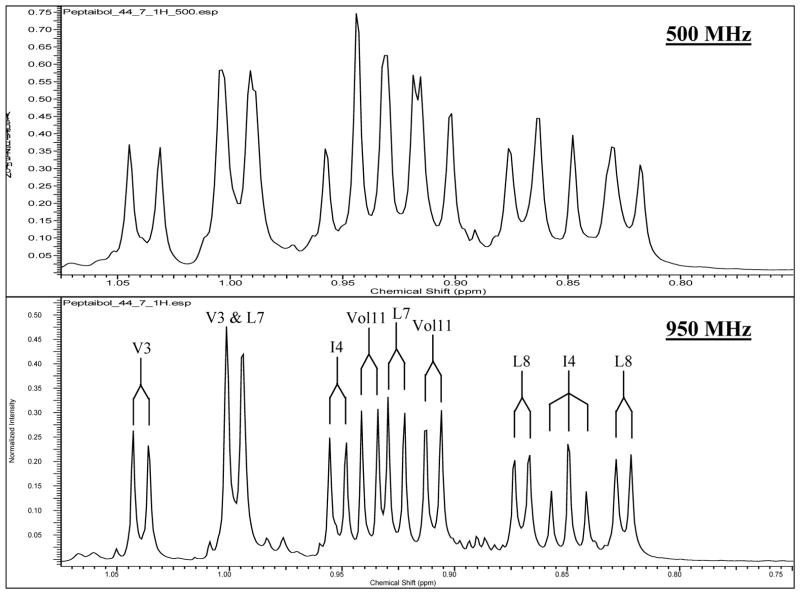
Compound 7 was unique with respect to the other peptaibols isolated from MSX 70741 (Fig. 1), in that 7 contained 11 amino acid residues whereas the others were 20-mers. Also, the HRMS of 7 showed three major in-source fragments instead of two, as with the other peptaibols (see Fig. 7). The 1H NMR also showed a methyl triplet at δH 0.85 (see Fig. 8), and from 2D TOCSY and NOESY spectra this triplet was due to isoleucine (Ile) at residue 4. Isoleucine was not present in any of the other peptaibols in this study. Compound 7 was most closely related to the trichobrachin [41, 42], hypomurocin [43], and trichobrevin [44, 45] subclasses of 11-mer peptaibols. The effect of a shorter chain on cytotoxic activity was negligible (Table 2), although it seemed to diminish antibacterial activity (Table 2) and was inactive in the HcL3 motility assay at 20 μg/mL. The NMR data for 7 illustrated the benefit of higher field on resolution (see Fig. 8). The Ile4 methyl triplet for 7 was fully resolved at 950 MHz, whereas at 500 MHz, the triplet could not be identified due to overlap with the adjacent methyl doublets from the Leu8 residue. Four methyl doublets from δH 0.90–0.95 were also completely resolved at 950 MHz, whereas at 500 MHz there was too much overlap to distinguish the signals. The 950 MHz NMR data was not only well-resolved for the methyl peaks of compound 7, but it also resulted in greatly improved resolution of NH-NH correlations, which were essential for sequencing. While it is well appreciated in the natural products community that higher field provides better resolution, the example of 500 vs 950 MHz NMR spectra of compound 7 provides a cogent example of how this can be applied to structure elucidation of peptaibols. Compound 7 most closely resembled trichobrevin B–IIb [45], having the same “rough” amino acid sequence; meaning that the leucine/isoleucine and valine/isovaline residues were not unambiguously determined for trichobrevin B-IIb. As compound 7 also closely resembled the trichobrachin peptaibol series, it was ascribed the trivial name trichobrachin D–I, to differentiate it from the trichobrachin A series (which all have Asp2, Aib9, and Pro10), the trichobrachin B series (which contain Asp2, Val9, and Pro10), and the trichobrachin C series (which have Gln2, Aib9, and Pro10). All trichobrachins, including 7, are also characterized by Aib5 and Pro6 [41, 42].
Figure 7.
Full-scan HRMS (sum of 8 mass spectra) of Trichobrachin D–I (7).
Figure 8.
Comparison of the Upfield Methyl Region of the 500 MHz vs 950 MHz 1H NMR Data of Trichobrachin D–I (7).
Compounds 2, 3, and 5 were novel in that they contain Ser at positions 4 or 6, which, to the best of our knowledge, has not been reported previously in 20-mer peptaibols. While Ser2 has been reported in a number of classes of 18-mer peptaibols, such as the hypomurocin B series [43], the trichokindins [46], the trichorzin MA series [47], and the trichorzin PA series [33, 34], the presence of Ser2 is rare for 11-mer peptaibols (as in compound 7), having only been reported in the trichobrevin B class [44, 45]. The presence of Ser in 2, 3, 5, and 7, according to the MS/MS fragmentation work, was confirmed by the TOCSY NMR spectra (except for 3) as well as the Marfey’s analyses, which also confirmed the stereochemistry as L-Ser (see Figure 6).
Three known peptaibols were isolated from a second Mycosynthetix fungus (MSX 57715; Fig. 1). Trichokonin VI (15) was the major peptaibol isolate, and was identical to alamethicin F50 (8), except for replacement of Pro2 with Ala2; compounds 8 and 15 had nearly the same cytotoxic activity profile (Table 2). Trichokonin VII (16) was analogous to atroviridin B (9) with the same substitution of Ala2 for Pro2; these two compounds were essentially equipotent in the cytotoxicity and antibacterial assays (Table 2). Longibranchin BIII (17) substituted Glu18 for Gln18 of trichokonin VII (16); the former was slightly more potent in both cytotoxicity and antibacterial assays (Table 2), although possibly within experimental error of the assays.
A range of bioactivities were observed for the isolated peptaibols (Table 2). Several of the peptaibols exhibited not only cytotoxicity, but cancer cell selectivity (Table 2). Selectivity was assessed via the IMR90 cell lines, and they are non-transformed but proliferating cells. For example, the positive control camptothecin exhibited 3.6-fold selectivity in MCF-7 cells to 23-fold selectivity in H460 cells relative to IMR90 cells. Alamethicin II (11), atroviridin J (12), trichokonin VII (16), and longibranchin BIII (17) were most noteworthy in exhibiting 3.6-fold, 5.2-fold, 3.9-fold, and 5.8-fold selectivity, respectively, in MCF-7 vs. IMR90 cells. These data were consistent with a previous report of the growth inhibitory activity of trichokonin VI (15) against hepatocellular carcinoma cells, in which 15 did not obviously affect normal liver cells at lower concentrations [48]. Three of these four compounds contained Iva17 and were the more hydrophobic peptaibols. A general trend of higher activity with higher hydrophobicity has been reported previously [42], although the effect was much less pronounced in our work. With respect to antibacterial activity, the compounds were essentially equipotent to both S. aureus and MRSA (or within experimental error), except compound 8, which was approximately four times more potent against S. aureus than MRSA. Compound 8 was also the most potent in the assay for larval motility against Haemonchus contortus infective stages, although still two orders of magnitude less potent than the positive control.
In summary, a series of twelve structurally-related peptaibols (1–12) were isolated from MSX 70741, including eight new peptaibols, four of which featured the incorporation of serine. These peptaibols were sequenced by MS/MS using HCD. The method was designed to enable full scan intact mass analysis followed by sequential fragmentation of the low and high MW fragments from a single UPLC injection. This streamlined approach enabled nearly complete amino acid sequencing capabilities of peptaibols from a small volume of sample, over a 10-fold decrease in separation time, and accurate mass measurements to confirm amino acid identities. Fragmentation of peptaibols by high-resolution was important due to the fact that some peptaibols contain both leucine (and/or isoleucine) as well as hydroxyproline residues, an example of which is clonostachin [49]. Leucine, isoleucine, and hydroxyproline fragments have the same nominal mass (113); however, hydroxyproline has a different formula, and therefore can only be distinguished in MS by high-resolution measurements. Leucine and isoleucine have the same formula and therefore have to be distinguished by NMR. The NMR experiments confirmed the sequencing of the peptaibols and were critical for distinguishing the locations of constitutional isomers that were present in the same compound, such as leucine and isoleucine in compound 7 or valine and isovaline in compound 12. The Marfey’s analysis was also a good demonstration of the power of UPLC compared to HPLC, where analyses were completed in 10 min with excellent resolution of the amino acid-Marfey derivative standards. Finally, from a mycological perspective, both organisms were prolific producer of peptaibols, generating >400 mg (MSX 70741) and >300 mg (MSX 57715) of peptaibols per solid phase culture in a 2.8 L Fernbach flask, using standard techniques with no growth optimization studies.
Supplementary Material
Acknowledgments
This research was supported by P01 CA125066 from the National Cancer Institute/National Institutes of Health, Bethesda, MD, USA. The anthelmintic studies were supported in part by a Collaborative Funding Grant (2011-CFG-8008) from the North Carolina Biotechnology Center and the Kenan Institute for Engineering, Technology & Science. Mycology technical support was provided by Blaise Darveaux and Maurica Lawrence. The authors acknowledge the technical assistance of Ms. Myra D. Williams (Virginia Tech) in measuring MICs. The authors also thank Dr. Kevin Knagge of the David H. Murdock Research Institute, Kannapolis, NC, for 950 MHz NMR data and Ms. Tamam El-Elimat for helpful discussions.
References
- 1.Toniolo C, Bruckner H. Peptaibiotics: Fungal peptides containing α-dialkyl α-amino acids. Zurich: Vrlag Helvetica Chimica Acta; 2009. p. 702. [Google Scholar]
- 2.Toniolo C, Bruckner H. Peptaibiotics. Chem Biodivers. 2007;4:1021–2. doi: 10.1002/cbdv.201300139. [DOI] [PubMed] [Google Scholar]
- 3.Bruckner H. Peptaibols/Peptaibiotics - Editorial. J Pept Sci. 2003;9:659. [Google Scholar]
- 4.Daniel JF, Filho ER. Peptaibols of Trichoderma. Nat Prod Rep. 2007;24:1128–41. doi: 10.1039/b618086h. [DOI] [PubMed] [Google Scholar]
- 5.Leitgeb B, Szekeres A, Manczinger L, Vagvolgyi C, Kredics L. The history of alamethicin: a review of the most extensively studied peptaibol. Chem Biodivers. 2007;4:1027–51. doi: 10.1002/cbdv.200790095. [DOI] [PubMed] [Google Scholar]
- 6.Kinghorn AD, Carache de Blanco EJ, Chai HB, Orjala J, Farnsworth NR, Soejarto DD, Oberlies NH, Wani MC, Kroll DJ, Pearce CJ, et al. Discovery of anticancer agents of diverse natural origin. Pure Appl Chem. 2009;81:1051–63. doi: 10.1351/PAC-CON-08-10-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Orjala J, Oberlies NH, Pearce CJ, Swanson SM, Kinghorn AD. Discovery of potential anticancer agents from aquatic cyanobacteria, filamentous fungi, and tropical plants. In: Tringali C, editor. Bioactive Compounds from Natural Sources. Natural Products as Lead Compounds in Drug Discovery. London, UK: Taylor & Francis; 2012. pp. 37–63. [Google Scholar]
- 8.Ayers S, Graf TN, Adcock AF, Kroll DJ, Matthew S, Carache de Blanco EJ, Shen Q, Swanson SM, Wani MC, Pearce CJ, et al. Resorcylic acid lactones with cytotoxic and NF-κB inhibitory activities and their structure-activity relationships. J Nat Prod. 2011;74:1126–31. doi: 10.1021/np200062x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ayers S, Graf TN, Adcock AF, Kroll DJ, Shen Q, Swanson SM, Wani MC, Darveaux BA, Pearce CJ, Oberlies NH. Obionin B: An o-pyranonaphthoquinone decaketide from an unidentified fungus (MSX 63619) from the Order Pleosporales. Tetrahedron Lett. 2011;52:5128–230. doi: 10.1016/j.tetlet.2011.07.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sy-Cordero AA, Graf TN, Adcock AF, Kroll DJ, Shen Q, Swanson SM, Wani MC, Pearce CJ, Oberlies NH. Cyclodepsipeptides, sesquiterpenoids, and other cytotoxic metabolites from the filamentous fungus Trichothecium sp. (MSX 51320) J Nat Prod. 2011;74:2137–42. doi: 10.1021/np2004243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ayers S, Ehrmann BM, Adcock AF, Kroll DJ, Wani MC, Pearce CJ, Oberlies NH. Thielavin B methyl ester: A cytotoxic benzoate trimer from an unidentified fungus (MSX 55526) from the Order Sordariales. Tetrahedron Lett. 2011;52:5733–5. doi: 10.1016/j.tetlet.2011.08.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Summers MY, Kong F, Feng X, Siegel MM, Janso JE, Graziani EI, Carter GT. Septocylindrins A and B: peptaibols produced by the terrestrial fungus Septocylindrium sp. LL- Z1518. J Nat Prod. 2007;70:391–6. doi: 10.1021/np060571q. [DOI] [PubMed] [Google Scholar]
- 13.Mitova MI, Murphy AC, Lang G, Blunt JW, Cole AL, Ellis G, Munro MH. Evolving trends in the dereplication of natural product extracts. 2. The isolation of chrysaibol, an antibiotic peptaibol from a New Zealand sample of the mycoparasitic fungus Sepedonium chrysospermum. J Nat Prod. 2008;71:1600–3. doi: 10.1021/np800221b. [DOI] [PubMed] [Google Scholar]
- 14.el Hajji M, Rebuffat S, Lecommandeur D, Bodo B. Isolation and sequence determination of trichorzianines A antifungal peptides from Trichoderma harzianum. Int J Pept Protein Res. 1987;29:207–15. doi: 10.1111/j.1399-3011.1987.tb02247.x. [DOI] [PubMed] [Google Scholar]
- 15.Kjer J, Debbab A, Aly AH, Proksch P. Methods for isolation of marine-derived endophytic fungi and their bioactive secondary products. Nat Protoc. 2010;5:479–90. doi: 10.1038/nprot.2009.233. [DOI] [PubMed] [Google Scholar]
- 16.Soule HD, Vazguez J, Long A, Albert S, Brennan M. A human cell line from a pleural effusion derived from a breast carcinoma. J Natl Cancer Inst. 1973;51:1409–16. doi: 10.1093/jnci/51.5.1409. [DOI] [PubMed] [Google Scholar]
- 17.Carney DN, Gazdar AF, Bunn PA, Jr, Guccion JG. Demonstration of the stem cell nature of clonogenic tumor cells from lung cancer patients. Stem Cells. 1982;1:149–64. [PubMed] [Google Scholar]
- 18.Rosenblum ML, Gerosa MA, Wilson CB, Barger GR, Pertuiset BF, de Tribolet N, Dougherty DV. Stem cell studies of human malignant brain tumors. Part 1: Development of the stem cell assay and its potential. J Neurosurg. 1983;58:170–6. doi: 10.3171/jns.1983.58.2.0170. [DOI] [PubMed] [Google Scholar]
- 19.Alali FQ, El-Elimat T, Li C, Qandil A, Alkofahi A, Tawaha K, Burgess JP, Nakanishi Y, Kroll DJ, Navarro HA, et al. New colchicinoids from a native Jordanian meadow saffron, Colchicum brachyphyllum: Isolation of the first naturally occurring dextrorotary colchicinoid. J Nat Prod. 2005;68:173–8. doi: 10.1021/np0496587. [DOI] [PubMed] [Google Scholar]
- 20.Li C, Lee D, Graf TN, Phifer SS, Nakanishi Y, Riswan S, Setyowati FM, Saribi AM, Soejarto DD, Farnsworth NR, et al. Bioactive constituents of the stem bark of Mitrephora glabra. J Nat Prod. 2009;72:1949–53. doi: 10.1021/np900572g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rae JM, Creighton CJ, Meck JM, Haddad BR, Johnson MD. MDA-MB-435 cells are derived from M14 melanoma cells--a loss for breast cancer, but a boon for melanoma research. Breast Cancer Res Treat. 2007;104:13–9. doi: 10.1007/s10549-006-9392-8. [DOI] [PubMed] [Google Scholar]
- 22.Nichols WW, Murphy DG, Cristofalo VJ, Toji LH, Greene AE, Dwight SA. Characterization of a new human diploid cell strain, IMR-90. Science. 1977;196:60–3. doi: 10.1126/science.841339. [DOI] [PubMed] [Google Scholar]
- 23.Williams AA, Sugandhi EW, Macri RV, Falkinham JO, 3rd, Gandour RD. Antimicrobial activity of long-chain, water-soluble, dendritic tricarboxylato amphiphiles. J Antimicrob Chemother. 2007;59:451–8. doi: 10.1093/jac/dkl503. [DOI] [PubMed] [Google Scholar]
- 24.Maisuria BB, Actis ML, Hardrict SN, Falkinham JO, 3rd, Cole MF, Cihlar RL, Peters SM, Macri RV, Sugandhi EW, Williams AA, et al. Comparing micellar, hemolytic, and antibacterial properties of di- and tricarboxyl dendritic amphiphiles. Bioorg Med Chem. 2011;19:2918–26. doi: 10.1016/j.bmc.2011.03.036. [DOI] [PubMed] [Google Scholar]
- 25.Lopez-Aroche U, Salinas-Sanchez DO, de Gives PM, Lopez-Arellano ME, Liebano-Hernandez E, Valladares-Cisneros G, Arias-Ataide DM, Hernandez-Velazquez V. In vitro nematicidal effects of medicinal plants from the Sierra de Huautla, Biosphere Reserve, Morelos, Mexico against Haemonchus contortus infective larvae. J Helminthol. 2008;82:25–31. doi: 10.1017/S0022149X07873627. [DOI] [PubMed] [Google Scholar]
- 26.Corley DG, Millerwideman M, Durley RC. Isolation and structure of harziazum A - a new trichothecene from Trichoderma harzianum. J Nat Prod. 1994;57:422–5. doi: 10.1021/np50105a019. [DOI] [PubMed] [Google Scholar]
- 27.Jin HZ, Lee JH, Zhang WD, Lee HB, Hong YS, Kim YH, Lee JJ. Harzianums A and B produced by a fungal strain, Hypocrea sp F000527, and their cytotoxicity against tumor cell lines. J Asian Nat Prod Res. 2007;9:203–7. doi: 10.1080/10286020500531977. [DOI] [PubMed] [Google Scholar]
- 28.Meyer CE, Reusser F. A polypeptide antibacterial agent isolated from Trichoderma viride. Experientia. 1967;23:85–6. doi: 10.1007/BF02135929. [DOI] [PubMed] [Google Scholar]
- 29.Oh SU, Lee SJ, Kim JH, Yoo ID. Structural elucidation of new antibiotic peptides, atroviridins A, B and C from Trichoderma atroviride. Tetrahedron Lett. 2000;41:61–4. [Google Scholar]
- 30.New AP, Eckers C, Haskins NJ, Neville WA, Elson S, HuesoRodriguez JA, RiveraSagredo A. Structures of polysporins A-D, four new peptaibols isolated from Trichoderma polysporum. Tetrahedron Lett. 1996;37:3039–42. [Google Scholar]
- 31.Pandey RC, Cook JC, Rinehart KL. Peptaibophol antibiotics.3. High-resolution and field desorption mass-spectrometry studies and revised structures of alamethicin-I and alamethicin-II. J Am Chem Soc. 1977;99:8469–83. doi: 10.1021/ja00457a063. [DOI] [PubMed] [Google Scholar]
- 32.Huang Q, Tezuka Y, Kikuchi T, Nishi A, Tubaki K, Tanaka K. Studies on metabolites of mycoparasitic fungi .2. metabolites of Trichoderma koningii. Chem Pharm Bull. 1995;43:223–9. doi: 10.1248/cpb.43.223. [DOI] [PubMed] [Google Scholar]
- 33.Leclerc G, Rebuffat S, Bodo B. Directed biosynthesis of peptaibol antibiotics in two Trichoderma strains II. Structure elucidation. J Antibiot. 1998;51:178–83. doi: 10.7164/antibiotics.51.178. [DOI] [PubMed] [Google Scholar]
- 34.Leclerc G, Rebuffat S, Goulard C, Bodo B. Directed biosynthesis of peptaibol antibiotics in two Trichoderma strains I. Fermentation and isolation. J Antibiot. 1998;51:170–7. doi: 10.7164/antibiotics.51.170. [DOI] [PubMed] [Google Scholar]
- 35.Duclohier H. Peptaibiotics and peptaibols: an alternative to classical antibiotics? Chem Biodivers. 2007;4:1023–6. doi: 10.1002/cbdv.200790094. [DOI] [PubMed] [Google Scholar]
- 36.Thirumalachar MJ. Antiamoebin, a new antiprotozoal-anthelmintic antibiotic. I. Production and biological studies. Hindustan Antibiot Bull. 1968;10:287–9. [PubMed] [Google Scholar]
- 37.Schiell M, Hofmann J, Kurz M, Schmidt FR, Vertesy L, Vogel M, Wink J, Seibert G. Cephaibols, new peptaibol antibiotics with anthelmintic properties from Acremonium tubakii DSM 12774. J Antibiot. 2001;54:220–33. doi: 10.7164/antibiotics.54.220. [DOI] [PubMed] [Google Scholar]
- 38.Olsen JV, Macek B, Lange O, Makarov A, Horning S, Mann M. Higher-energy C-trap dissociation for peptide modification analysis. Nat Methods. 2007;4:709–12. doi: 10.1038/nmeth1060. [DOI] [PubMed] [Google Scholar]
- 39.Kim S, Rodgers RP, Marshall AG. Truly “exact” mass: Elemental composition can be determined uniquely from molecular mass measurement at similar to 0.1 mDa accuracy for molecules up to similar to 500 Da. Int J Mass Spectrom. 2006;251:260–5. [Google Scholar]
- 40.Huang Q, Tezuka Y, Hatanaka Y, Kikuchi T, Nishi A, Tubaki K. Studies on metabolites of mycoparasitic fungi .5. Ion-spray ionization mass spectrometric analysis of Trichokonin-II, a peptaibol mixture obtained from the culture broth of Trichoderma koningii. Chem Pharm Bull. 1996;44:590–3. doi: 10.1248/cpb.44.590. [DOI] [PubMed] [Google Scholar]
- 41.Mohamed-Benkada M, Montagu M, Biard JF, Mondeguer F, Verite P, Dalgalarrondo M, Bissett J, Pouchus YF. New short peptaibols from a marine Trichoderma strain. Rapid Commun Mass Spectrom. 2006;20:1176–80. doi: 10.1002/rcm.2430. [DOI] [PubMed] [Google Scholar]
- 42.Ruiz N, Wielgosz-Collin G, Poirier L, Grovel O, Petit KE, Mohamed-Benkada M, du Pont TR, Bissett J, Verite P, Barnathan G, et al. New Trichobrachins, 11-residue peptaibols from a marine strain of Trichoderma longibrachiatum. Peptides. 2007;28:1351–8. doi: 10.1016/j.peptides.2007.05.012. [DOI] [PubMed] [Google Scholar]
- 43.Becker D, Kiess M, Bruckner H. Structures of peptaibol antibiotics hypomurocin A and B from the ascomycetous fungus Hypocrea muroiana Hino et Katsumoto. Liebigs Annalen-Recueil. 1997:767–72. [Google Scholar]
- 44.Degenkolb T, Dieckmann R, Nielsen KF, Grafenhan T, Theis C, Zafari D, Chaverri P, Ismaiel A, Bruckner H, von Dohren H, et al. The Trichoderma brevicompactum clade: a separate lineage with new species, new peptaibiotics, and mycotoxins. Mycol Prog. 2008;7:177–219. [Google Scholar]
- 45.Degenkolb T, Grafenhan T, Nirenberg HI, Gams W, Bruckner H. Trichoderma brevicompactum complex: rich source of novel and recurrent plant-protective polypeptide antibiotics (peptaibiotics) J Agric Food Chem. 2006;54:7047–61. doi: 10.1021/jf060788q. [DOI] [PubMed] [Google Scholar]
- 46.Iida A, Sanekata M, Fujita T, Tanaka H, Enoki A, Fuse G, Kanai M, Rudewicz PJ, Tachikawa E. Fungal metabolites .XVI. Structures of new peptaibols, trichokindins I-VII, from the fungus Trichoderma harzianum. Chem Pharm Bull. 1994;42:1070–5. doi: 10.1248/cpb.42.1070. [DOI] [PubMed] [Google Scholar]
- 47.Goulard C, Hlimi S, Rebuffat S, Bodo B. Trichorzins HA and MA, antibiotic peptides from Trichoderma harzianum . I. Fermentation, isolation and biological properties. J Antibiot. 1995;48:1248–53. doi: 10.7164/antibiotics.48.1248. [DOI] [PubMed] [Google Scholar]
- 48.Shi M, Wang HN, Xie ST, Luo Y, Sun CY, Chen XL, Zhang YZ. Antimicrobial peptaibols, novel suppressors of tumor cells, targeted calcium-mediated apoptosis and autophagy in human hepatocellular carcinoma cells. Mol Cancer. 2010;9:26. doi: 10.1186/1476-4598-9-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chikanishi T, Hasumi K, Harada T, Kawasaki N, Endo A. Clonostachin, a novel peptaibol that inhibits platelet aggregation. J Antibiot. 1997;50:105–10. doi: 10.7164/antibiotics.50.105. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.