Abstract
The tissue microenvironment directs stem/progenitor cell behavior. Cancer cells are also influenced by the microenvironment. It has been shown that, when placed into blastocysts, cancer cells respond to embryonic cues and differentiate according to the tissue type encountered during ontological development. Previously, we showed that the mouse mammary gland was capable of redirecting adult mouse testicular and neural stem/progenitor cells toward a mammary epithelial cell fate during gland regeneration. Here, we report that human embryonal carcinoma cells proliferate and produce differentiated mammary epithelial cell progeny when mixed with mouse mammary epithelial cells and inoculated into the epithelium-free mammary fat pads of athymic nude mice. Fluorescence in situ hybridization confirmed the presence of human cell progeny in the mammary outgrowths for human centromeric DNA, as well as immunochemistry for human-specific breast epithelial cytokeratins and human-specific milk proteins in impregnated transplant hosts. It was found that the number of human cells increased by 66- to 660-fold during mammary epithelial growth and expansion as determined by human cytokeratin expression. All features found in primary outgrowths were recapitulated in the secondary outgrowths from chimeric implants. These results show that human embryonal carcinoma–derived progeny interact with mouse mammary cells during mammary gland regeneration and are directed to differentiate into cells that exhibit diverse mammary epithelial cell phenotypes. This is the first demonstration that human cells are capable of recognizing the signals generated by the mouse mammary gland microenvironment present during gland regeneration in vivo.
Introduction
The responsiveness of cancer cells to developmental signals is a seldom-utilized facet of cancer biology. Research has shown that cancer cells may lose their tumorigenic potential and display embryonic cellular behavior when placed into ontogenic environments (1–4). Specifically, Mintz and Illmensee (3) reported in 1975 that embryonal carcinoma cells from transplantable mouse teratomas were incorporated into normal adult tissues in genetically mosaic mice produced from blastocysts of fertilized mouse zygotes injected with those cancer cells. One year later, these authors showed that single teratocarcinoma cells could produce a clone capable of populating multiple tissue types with normally functioning progeny when injected into genetically marked mouse blastocysts (1). Further, malignant melanoma of the mouse reverted to a normal phenotype when transplanted to embryonic skin during a narrow developmental window. In 1998, McCullough and colleagues (4) showed that tumorigenic liver cells, which consistently form tumors at ectopic sites, differentiate to form normal hepatocytes in young rat livers but not in aged rat livers. This evidence suggests that the cancer phenotype may be controlled by signals generated in normally developing tissues. Previously, we showed that the mouse mammary microenvironment could redirect adult mouse cells of nonmammary origins to expand and differentiate to mammary epithelial cell fates during glandular regeneration in vivo (5, 6). Although it is clear that the mouse mammary gland possesses the ability to generate signals that redirect nonnative adult progenitor cells, it is unknown if similar signals can be generated in situ to reprogram cancer cells. Therefore, we hypothesized that embryonal carcinoma cells would respond to signals produced by the regenerating mouse mammary gland and contribute to gland regeneration in vivo. To test this hypothesis, NTERA-2cl (NT2) tumor cells were mixed with normal mouse mammary epithelial cells and placed into epithelium-divested mammary fat pads of pubertal mice. Here, we show that embryonal carcinoma cells from a human testicular tumor are subject to redirection from their tumorigenic phenotype to differentiation into human-specific mammary epithelial cells through interaction with the mouse mammary microenvironment in vivo. The detection of human mammary–specific keratin expression and the secretion of human-specific milk protein in lactating hosts showed the conversion of human carcinoma cells to bona fide human mammary epithelial cells. This shows that human embryonal cancer cells are subject to the normal tissue–specific developmental signals present during the developmental regeneration of mouse mammary gland. The responsiveness of human cancer cells to differentiate and lose their tumorigenic capacity when exposed to the signals present in developing mouse mammary gland suggests the that human cancer may be susceptible to control in situ by signals generated within developing mammalian tissues.
Materials and Methods
Mice
Three-week-old female athymic nude mice were used as hosts for transplantation studies. All mice were housed in Association for Assessment and Accreditation of Laboratory Animal Care–accredited facilities in accordance with the NIH Guide for the Care and Use of Laboratory Animals. The National Cancer Institute (NCI) Animal Care and Use Committee approved all experimental procedures.
Cells
NTERA-2c1 (NT2) cells [NTERA-2 cl.D1 (NT2/D1), American Type Culture Collection (ATCC) CRL-1973], derived from a human male embryonic carcinoma and characterized by a pseudotriploid chromosomal content (7), were purchased from ATCC. The cell line was authenticated by ATCC using tests recommended in their technical bulletin (8). Cells were grown in McCoy's media supplemented with 15% fetal bovine serum (FBS) and 1% penicillin-streptomycin at 37°C with 5% CO2. NT2 cells were grown on glass chamber slides and were fixed and stained for CD133, estrogen receptor α (ERα), and human keratins 8, 5, and 14. No staining for the human keratins or ERα was observed. The cells were essentially 100% positive for CD133. Mammary epithelial cells from 10- to 12-week-old FVB/n or Balb/C females were collected from primary mammary cultures after 4 to 7 days on plastic culture flasks in DMEM supplemented with 10% FBS, insulin (1.0 µg/mL), and epidermal growth factor (10 ng/mL). Fibroblasts were reduced before collection of the epithelial cells by differential trypsinization.
Tissue/cell transplantation
DeOme and colleagues (9) first described the technique of transplanting tissue fragments into mammary fat pads cleared of endogenous mammary epithelium. Following these methods, NT2 cells were mixed with mouse mammary epithelial cells at ratios of 1:5 (6 glands) and 1:50 (12 glands) in PBS (5, 6). Ten microliters were immediately injected into the epithelium-divested inguinal mammary fat pads of 3-week-old Nu/Nu female mice. NT2 cells alone (10 K; 6 glands) and mouse mammary epithelial cells alone (50 K; 12 glands) were inoculated into epithelium-free fat pads as controls. Eight to 10 weeks later, mice were mated or maintained as virgins, then subsequently euthanized. The resulting fat pads were removed and either fragments taken for reimplantation as secondary outgrowths, dissociated in collagenase and prepared as primary tissue cultures, or examined after sectioning paraffin embedded whole mounts through immunostaining.
Whole-mount analysis
The number 4 mammary inguinal mammary gland was excised and spread on a glass slide with overnight fixation at 4°C in Carnoy's fixative. Glands were stained in carmine alum at room temperature for 2 to 4 hours and dehydrated through a series of graded ethanols and xylenes to remove fatty stroma.
Immunochemistry
Immunohistochemistry was performed on 6-µm-thick paraffin sections cut from the whole mounts described above after removal of the coverslips in xylene and rehydration through a graded series of alcohol. Primary antibodies included anti-mouse total caseins (1:500, rabbit polyclonal) (10), anti-human lysozyme (1:100; NeoMarkers), antihuman α-lactalbumin (1:100; Santa Cruz), anti-human prolactin (15 µg/mL; R & D Systems), anti-human CD133 (1:100; Miltenyi Biotec), anti-human Keratin 8 (1:100; Thermo Scientific), anti-human Keratin 5 (1:25; Thermo Scientific), anti-human Keratin 14 (1:100; Thermo Scientific), anti-mouse Keratin 14 (1:100, a gift from D. Roop; Baylor College of Medicine, Houston, Texas) (11), anti-ERα (1:300; Santa Cruz), and anti–progesterone receptor (PR; 1:100; DAKO Cytomation). Immunohistochemistry was performed for anti-ERα, anti-PR, anti–human Keratin 5, anti–human Keratin 8, and anti–human Keratin 14 following antigen retrieval by boiling slides for 10 minutes (ERα, PR) or 3.5 minutes (K8, K5, K14) in 10 mmol/L citrate buffer (pH 6.0). Slides were incubated with primary antibodies at 4°C overnight. Secondary antibodies were conjugated to Alexa 568 (1:200; Molecular Probes), Alexa 488 (1:200; Molecular Probes), FITC (1:3,500; Abcam), or rhodamine (1:3,000; Abcam), and were incubated on slides for 1 hour at room temperature. Slides were mounted using ProLong Gold anti-fade reagent with diamno-2-phenylindole (DAPI; Invitrogen).
Fluorescent in situ hybridization
Slides were deparaffinized by three treatments in xylene and then dehydrated in 100% ethanol. They were washed in 2× SSC at 70°C for 10 minutes followed by treatment in 4 mg/mL pepsin in 0.2 mol/L HCL at 37°C for 15 minutes in a humidified chamber. The slides were washed in 2× SSC, denatured in 70% formamide 2× SSC, and dehydrated in ethanol. Dual-color fluorescent in situ hybridization (FISH) was performed on serial sections using human probe PAHC0001-G in FITC with mouse bac clone chromosome 6 in digoxigenin-dUTP in 10 µL at 37°C in a humidified chamber overnight, and washed. Slides were stained with immunophenotype antibody CD133, washed, and counterstained with DAPI. Analyses were performed using an Axioplan 2 plus (Zeiss) fluorescence microscope coupled with a CCD camera (Photometrics), and images were captured with FISHview 4.5 software (Applied Spectral Imaging, Inc.).
Magnetic cell sorting
Cells were enumerated, then incubated for 30 minutes at 4°C with mouse anti-human CD133/2 primary antibody. Biotin-conjugated goat anti-mouse IgG secondary antibody was applied for 15 minutes at 4°C. Cells were incubated with Streptavidin Microbeads for 15 minutes at 4°C, transferred to a prewetted LD MACS separation column (Miltenyi Biotec), and allowed to elute. Following two subsequent washes and elution of the negative fraction, the column was removed from the magnet, and human CD133 positive fraction was collected. A second elution was carried out to increase positive fraction recovery.
DNA isolation and PCR
DNA was made from human CD133-positive cells using the DNeasy Blood and Tissue kit (Qiagen) according to the manufacturer's instructions. A reaction mixture containing 200 µg DNA, 45 µL PCR SuperMix, and 1 µL each of 10 µmol/L primers was assayed in a GeneAmp PCR System 9700 thermalcycler. The 50 µL reaction mixture was heated to 94°C for 5 minutes, and 35 PCR cycles were carried out as follows: denaturation at 94°C for 1 minute, annealing at 62°C for 2 minutes, and extension at 72°C for 2 minutes. The reaction was heated at 72°C for 10 minutes and subsequently cooled to 4°C indefinitely. Electrophoresis was performed on a 2% agarose gel. Primer sequences for the human SRY sex determining Y gene were as described in Nagafuchi and colleagues (12): forward, 5′-CAG TGT GAA ACG GGA GAA AAC AGT-3′; reverse, 5′-CTT CCG ACG AGG TCG ATA CTT ATA-3′, yielding a 270-bp band for the human SRY gene. The DNA was further purified using a PureLink Quick Gel Extraction kit (Invitrogen), amplified using the PCR conditions previously stated, and final electrophoresis carried out on a 2% agarose gel. DNA positive for the human SRY gene was visualized using an UVP BioImaging System EpiChem 3 Darkroom (UVP, LLC).
Propidium iodine staining and flow cytometry
Cells were washed, fixed with 70% ethanol for at least 24 hours, and stained with propidium iodine (0.1% Triton X-100, 50 µg/mL RNase A, and 20 µg/mL propidiumiodine in PBS) for 30 minutes at room temperature. DNA content was determined using a FACSCanto flow cytometer (Becton Dickinson), and data were analyzed with the Flowjo Software (Tree Star).
Metaphase spreads
Cells were treated with 0.1 µg/mL colcemid for 6 to 24 hours. Cells in metaphase were removed by repeated tapping of the flask and resuspended in 0.075M KCl at 37°C for 20 minutes. Cells were fixed in methanol and acetic acid (3:1) and dropped from ~50 cm onto glass slides. Spreads were stained with DAPI (0.2 ng/µL).
Results
Human NT2 cells are incorporated into the mammary gland epithelium and differentiate
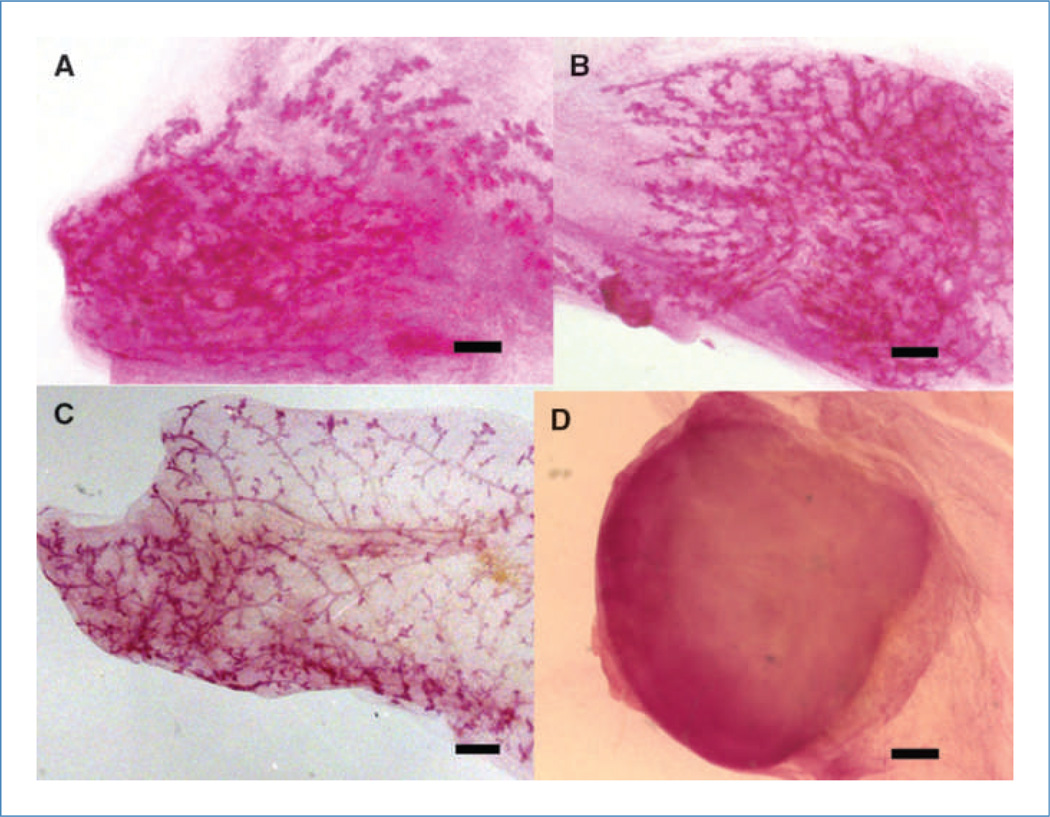
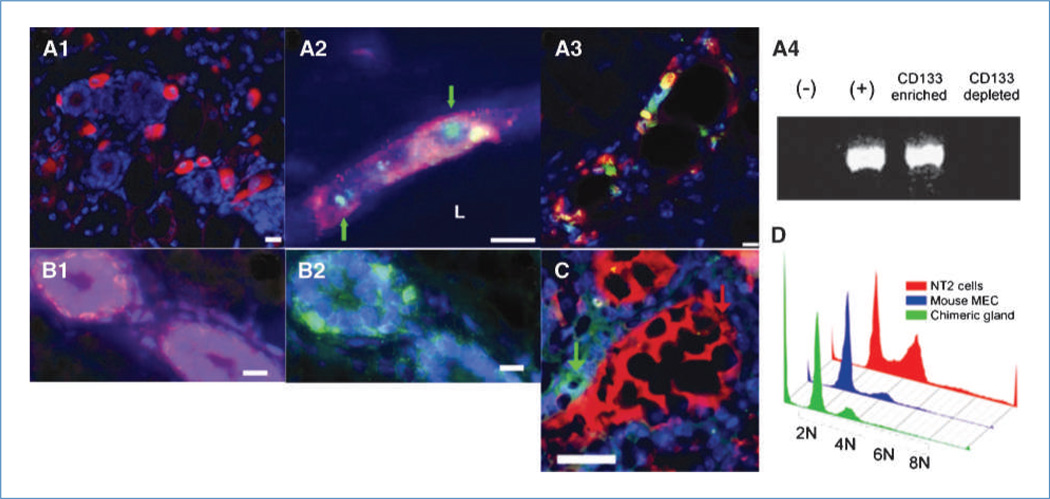
NT2 human embryonal carcinoma cells incorporate into chimeric mouse/human mammary outgrowths when transplanted in concert with wild-type mouse mammary epithelial cells in the epithelium-divested inguinal mammary fat pads of juvenile Nu/Nu female mice. NT2 cell progeny were detected in mammary outgrowths developed in first and secondary transplant generations (Supplementary Table S1). NT2 cells alone did not produce outgrowths, although occasional tumors were formed (2 of 6) in the inoculated fat pads (Supplementary Table S1). Some of the observed human/mouse mammary chimeric outgrowths displayed a morphology (Fig. 1A and B) distinct from that observed when mouse mammary epithelial cells (50 K) were inoculated alone (Fig. 1C). However, when 10 K NT2 cells were inoculated alone (without mammary epithelial cells), tumors formed in two of the six fat pads examined (Supplementary Table S1; Fig. 1D). Second-generation outgrowths resulted in fragments taken from both 1 K (10 of 10) and 10 K (8 of 8) chimeric NT2/mouse mammary glands (Supplementary Table S1). No tumors were seen for 1 K NT2/mammary epithelial cell or 10 K NT2/mammary epithelial cell in either first- or second-generation transplants, respectively (Supplementary Table S1). Some NT2 cells maintained the expression of human CD133 in the implanted mammary fat pads (Fig. 2A1 and A2). Further characterization of the human/mouse chimeras by FISH combined with immunologic staining for human CD133 showed that cells carrying human chromosomes were integrated into mammary structures (Supplementary Fig. S1; Fig. 2A2). CD133+ NT2 cells also differentiated into ERα- and PR-positive luminal cells that were found in mammary ducts (Fig. 2A3). Figure 2B1 and 2B2 show human keratin 14 (hK14) and mouse keratin 14 location in consecutive sections from the same mammary duct, showing that both human and mouse cells lie juxtaposed in basal positions within the chimeric outgrowth. In early studies, secondary outgrowths were harvested at 10 weeks and placed into culture conditions favorable to the growth and expansion of NT2 cells. The cultures were subsequently dissociated and cells were magnetically sorted for the expression of human-specific CD133. Approximately 20% of the total cells harvested were found in the CD133enriched fraction. DNA isolated from the CD133enriched and CD133depleted cell fractions was subjected to PCR analysis for the presence of the human male Y–specific chromosomal marker SRY (13). Only the CD133enriched fraction contained the SRY gene (Fig. 2A4), providing evidence that the human NT2 cells proliferated and contributed to progeny in the secondary outgrowths.
Figure 1.
NT2 cells contribute to the formation of the mouse mammary gland. Seven to 12 weeks post-transplantation, mammary gland outgrowths were harvested, fixed in Carnoy's fixative, and stained overnight with carmine alum. A and B, representative gland whole mounts of mouse mammary glands that were inoculated with both NT2 and mammary epithelial cells. C, representative mouse mammary gland outgrowth of a gland inoculated with 50 K mammary epithelial cells. D, a tumor (dark indistinct mass) that appeared when the fat pad was inoculated with 10 K NT2 cells alone. Scale bars, 2 mm.
Figure 2.
NT2-derived cells are incorporated into the mammary epithelium and differentiate but do not show increased ploidy. A1, CD133-positive NT2 cells (red) are present within the confines of the fat pad containing regenerated mammary ducts. A2, human NT2 cells are present within the mammary outgrowths as determined by human-specific FISH (green, nuclear; identified with green arrows) and human-specific immunocytochemical staining for CD133 (red). A3, NT2 cells in regenerated mammary epithelium, as determined by expression of human CD133 (red), differentiate into luminal epithelial cells and express ER-α (green). A4, PCR results following magnetic separation based on human CD133 expression show that the NT2 cells before transplantation (+) as well as the CD133enriched fraction contain human Y-chromosome–specific markers, whereas normal mouse mammary epithelial cells (−) and the CD133depleted fraction do not. B1 and B2, the location of basal cells (B1, red) expressing hK14 and mouse K14 (B2, green) in consecutive sections showing the same chimeric duct. C, human and mouse proteins are secreted into the same lumen. Simultaneous staining of chimeric mammary outgrowth for human α lactalbumin (green) and mouse caseins (red) shows that production and secretion of both mouse and human milk proteins are present in the same chimeric duct. D, NT2 cells, mouse mammary epithelial cells, and cells isolated from second-generation chimeric outgrowths were fixed and stained with propidium iodide, and DNA content was determined by flow cytometry. The chimeric gland did not have a greater proportion of cells with abnormal ploidy compared with cultures of mouse mammary epithelial cells (MEC) or NT2 cells. All fluorescent sections are counterstained with DAPI (blue). Scale bars, 20 µm (A1), 15 µm (A2 and A3), 10 µm (B1 and B2), and 25 µm (C).
NT2 cells contribute to the formation of both luminal and basal cellular structures in the mouse mammary gland
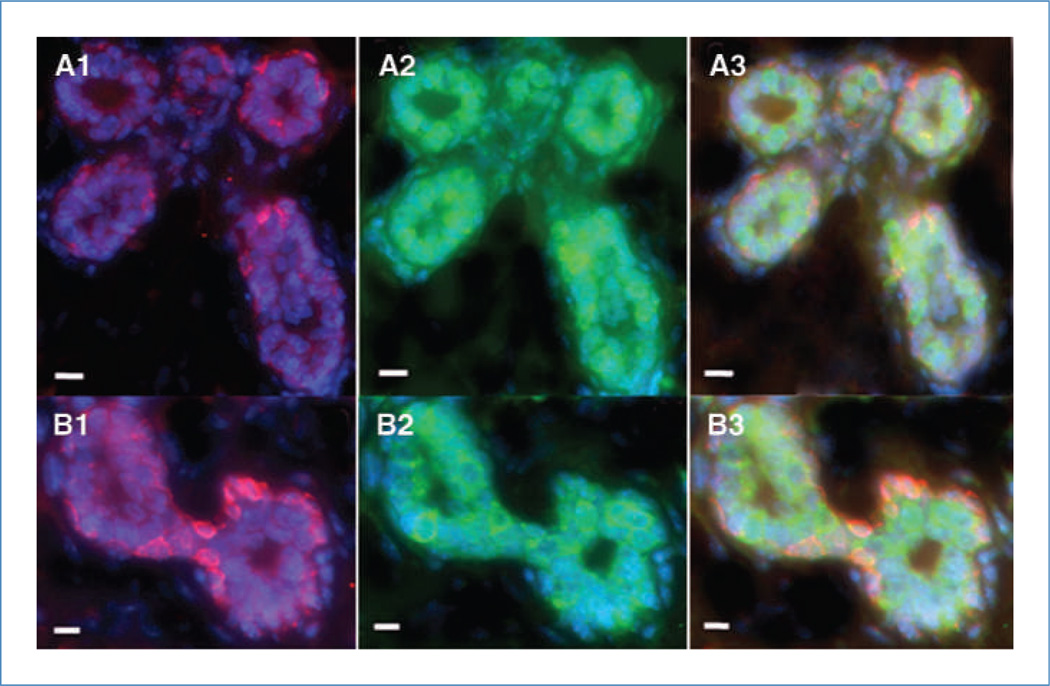
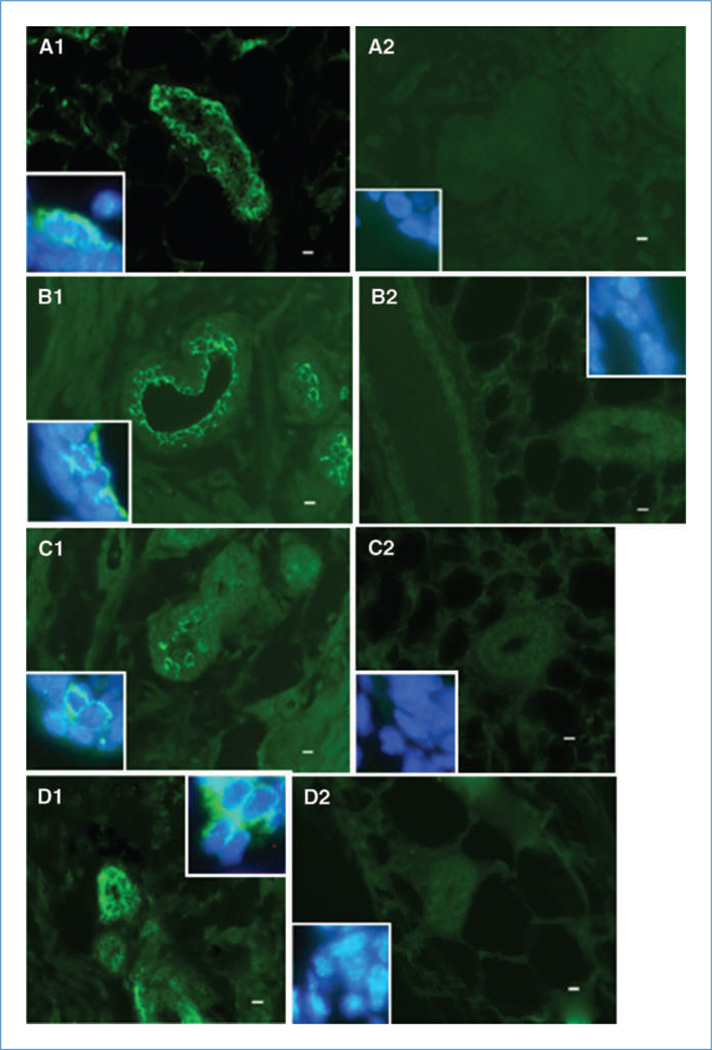
NT2 cells in the chimeric mammary glands also expressed the human mammary myoepithelial marker human keratin 5 (hK5) in basal cells (Fig. 3A1) and the luminal marker human K8 (hK8) in luminal epithelial cells (Fig. 3A2). The myoepithelial marker K14 was shown in human and mouse cells detected in basal positions throughout the first-generation chimeric mammary outgrowths (merge, Fig. 3A3). Similar results were seen in second-generation chimeric mammary outgrowths (Fig. 3B1–B3). Neither the human- or mouse-specific cytokeratin antibodies were cross-reactive with mouse or human epithelium, respectively (Fig. 4A1–D2). Additionally, of the 100 cells counted in five separate primary outgrowths, approximately one in three cells in the chimeric gland was positive for hK5 and hK8 keratins, whereas hK14 was expressed in one in four cells. These results indicate that the NT2 cancer cells contribute to the formation of both luminal and basal mammary structures in the chimeric gland. Furthermore, it was found that cells derived from the original NT2 population continue to proliferate and contribute to mammary epithelial cell progeny in secondary outgrowths upon transplantation. In particular, it was estimated by human-specific keratin expression that the number of human cells present in the regenerated chimeric gland was 66- to 660-fold greater than the quantity of human cancer cells that were initially inoculated, revealing a significant expansion of human cell progeny during chimera generation. This preceding statement is meant only to highlight the increase in the number of demonstrably human cells (through human keratin expression) that has occurred following implantation. These data do not indicate that all inoculated NT2 cells contribute progeny toward the human/mouse mammary chimera, only that the number of human-derived cells significantly increased during organogenesis.
Figure 3.
Human cancer cells contribute to the formation of both luminal (hK8 positive) and basal (hK5 positive) epithelial cells in the chimeric mammary gland. NT2 cells incorporated into the mammary gland express (A1 and B1) human-specific myoepithelial markers keratin 5 (red; rhodamine) and (A2 and B2) luminal marker keratin 8 (green; Alexa Fluor 488) in the primary (A1–A3) and secondary (B1–B3) generation transplants. A3 and B3, merge. All fluorescent sections are counterstained with DAPI (blue). Scale bars, 10 µm.
Figure 4.
Neither human- or mouse-specific cytokeratin antibodies were cross-reactive with murine or human epithelium, respectively. A1, mouse-specific keratin 14 reacts positively with mouse tissue (A1, green, FITC). A2, mouse-specific keratin 14 does not react with human tissue (A2, green, FITC). B1, human-specific keratin 8 reacts positively with human tissue (B1, green, FITC). B2, human-specific keratin 8 does not react with mouse tissue (B2, green, FITC). C1, human-specific keratin 5 reacts positively with human tissue (C1, green, FITC). C2, human-specific keratin 5 does not react with mouse tissue (C2, green, FITC). D1, human-specific keratin 14 reacts positively with human tissue (D1, green, FITC). D2, human-specific keratin 14 does not react with mouse tissue (D2, green, FITC). All insets, magnification of representative areas with merge of FITC (green) plus DAPI (blue). Scale bars, 10 µm.
Both mouse and human milk proteins were present in the ductal lumen of lactating outgrowths of human/mouse mammary tissue
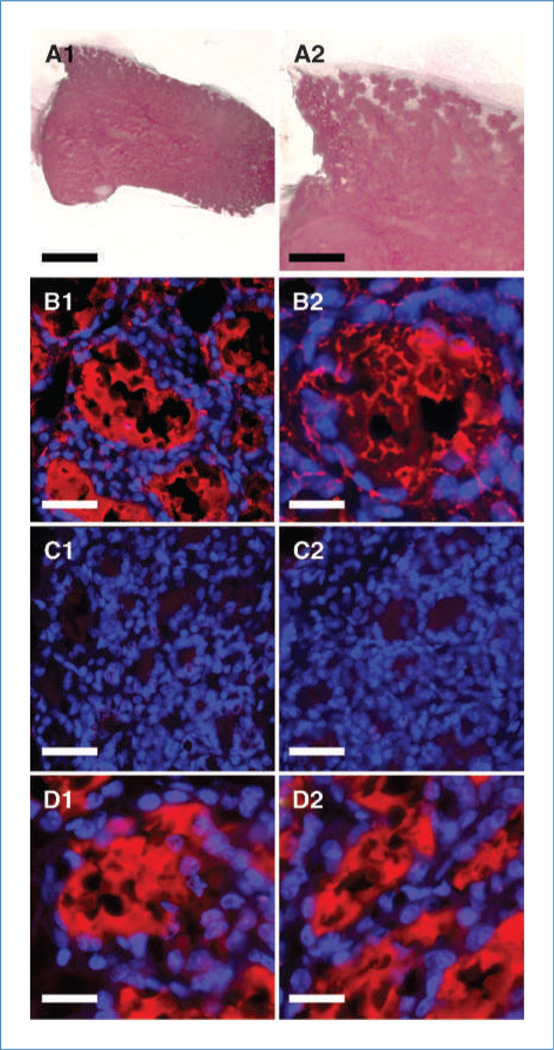
During pregnancy, the mammary gland undergoes alveolar proliferation and differentiation, resulting in the formation of a functional secretory system comprised of luminal epithelial cells that produce and secrete milk proteins, in the intact mammary gland and the implanted mammary outgrowths (14). To determine if NT2 cell progeny formed secretory mammary epithelial cells, chimeric mammary outgrowths were removed following a full-term pregnancy at day 2 of lactation. Outgrowths were found to completely fill the mammary fat pad and exhibited extensive development of secretory acini (Fig. 5A1 and A2). In addition, the human milk proteins α-lactalbumin and lysozyme were detected (Fig. 5B1 and B2). Human α-lactalbumin and lysozyme were not detected in sections from the intact lactating mammary glands of the recipient host (Fig. 5C1 and C2). Antibodies directed toward total mouse caseins produced positive staining in the secretory lumen in chimeric mammary outgrowths (Fig. 5D1) and the intact mammary glands of the host recipients (Fig. 5D2). More importantly, simultaneous immunochemical staining for both human and mouse milk proteins showed that individual cells within the same acinus were positive for either human or mouse milk proteins (Fig. 2C). Furthermore, it was found that NT2 cellular progeny, as determined by the expression of human K8, in secondary chimeric outgrowths in lactating hosts contributed to the production of extracellular prolactin (Supplementary Fig. S2A–D).
Figure 5.
Chimeric mammary outgrowths produce both human and mouse milk proteins. A1, whole mount of chimeric mammary gland at the 2nd day of lactation stained with carmine alum; A2, higher magnification of A1 illustrating the development of secretory acini. Immunofluorescent staining for the human-specific milk proteins α-lactalbumin (red) in B1 and, for lysozyme in B2, of a section of the chimeric NT2/mouse mammary outgrowth. No immunofluorescent staining for the human milk proteins (red) α-lactalbumin (C1) and lysozyme (C2) is present in the intact axillary mammary gland from the same animal. Immunofluorescent staining for total mouse casein (red) in the chimeric mammary outgrowth (D1) and in the intact axillary mammary gland (D2). All fluorescent sections counterstained with DAPI (blue). Scale bars, 1.0 mm (A1), 0.4 mm (A2), 50 µm (B1, C1, and C2), and 25 µm (B2, D1, and D2).
Human NT2 and mouse mammary epithelial cells did not fuse during mammary gland regeneration
To determine if any of the preceding results were attributable to fusion between the NT2 and host mammary cells, cells were isolated from the second-generation chimeric gland and stained with propidium iodide. DNA content was determined by flow cytometry. As shown in Fig. 2D, propidium iodide–stained cells isolated from chimeric glands were compared with similarly stained cells from NT2 and mammary epithelial cell cultures. The proportion of chimera cells with DNA content greater to 4N or intermediate between 2N and 4N did not differ from mouse epithelial cells or NT2 cells alone. This result is inconsistent with the presence of human/mouse cell fusion. For further confirmation of DNA content, metaphase spreads were generated from cells taken from the cocultured human NT2 cells and mouse mammary epithelial cells, as well as from cultures generated from secondary chimeric outgrowths (Supplementary Table S2; Supplementary Fig. S3A–D). As shown in Supplementary Fig. S3A and B, human and mouse chromosomes can be easily distinguished by morphology, and none of the metaphase spreads exhibited evidence of mouse/human cell fusion. Cells isolated from second-generation chimeric outgrowths were also analyzed (n = 30) and were found to contain either all human (n = 5) or mouse (n = 25) chromosomes. Furthermore, of the nuclei containing chromosomes with mouse morphology, all but one had a euploid number (i.e., 40) of chromosomes (Supplementary Fig. S3C), with one cell containing 80 chromosomes (all possessed normal mouse chromosome morphology). Cells with human chromosomes all had 45 to 59 chromosomes (Supplementary Fig. S3D), which is within the chromosome range seen with metaphase spreads from NT2 cell cultures (Supplementary Table S2). These results show that the human gene activities detected in the second-generation chimeric outgrowths are not the result of fusion between human and mouse cells and further show that human cells proliferated independently in the formation of primary and secondary chimeras.
Discussion
To our knowledge, this is the first demonstration that cancer cells from a different mammalian species can be redirected to produce progeny capable of typical mammary epithelial cell function by interaction with a developing adult organ system in vivo. Totipotent human embryonal carcinoma cells readily interpreted the developmental signals generated in the mouse mammary gland during regeneration. The resulting interaction leads to the redirection of human NT2 cancer cell progeny to adopt a human mammary epithelial cell phenotype with an absence of tumorgenic activity. Thus, these results provide direct evidence that the developmental cues in the mouse mammary gland and those present during human breast development may be indistinguishable. Furthermore, the human cancer cell progeny differentiate down two distinctly different mammary epithelial cell pathways, i.e., luminal and basal (myoepithelial), during chimeric mammary regeneration. This indicates a multipotentiality in the cancer cell response to signals from the mouse mammary microenvironment. In the case of mouse stem/progenitor cell activities, evidence for distinct lobule-limited and duct-limited multipotent epithelial cells has been shown (14–16). No such evidence is yet available for human breast stem/progenitor cells. Therefore, isolation and characterization of the human cells present in the mouse/human mammary chimeras will be important in understanding the nature of the reprogrammed human cancer cell progeny. Studies are in progress to label NT2 cells with a fluorescent marker so that these investigations may be possible. The observation that human embryonal carcinoma cells are able to detect and respond to signals generated within the developing mammary tissue suggests that other human cancer cells may also be able to respond in a similar manner. We are currently investigating this hypothesis. In addition, these observations may lead to a new paradigm for control and treatment of cancer in situ. Preliminary studies indicate that tissue extracts may be used in place of live mammary epithelial cells to reprogram adult mouse spermatogenic cells to adopt mammary epithelial cell fate. Likewise, it may be possible to redirect tumorigenic cells to normal cell function by exposure to substances present within normal tissues.
Supplementary Material
Acknowledgments
We thank Drs. Paul Goldsmith and Chris Heyer, Antibody and Protein Purification Unit, CCR, NCI for determining by Western blot, which commercially available human milk protein antibodies did not cross-react with mouse milk, and Sandy Burkett, FCRC, for the expert assistance during the FISH analysis of primary and secondary human/mouse chimeric outgrowths.
Grant Support
The intramural research program of the Center for Cancer Research, NCI, NIH supported this work.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Illmensee K, Mintz B. Totipotency and normal differentiation of single teratocarcinoma cells cloned by injection into blastocysts. Proc Natl Acad Sci U S A. 1976;73:549–553. doi: 10.1073/pnas.73.2.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gerschenson M, Graves K, Carson SD, Wells RS, Pierce GB. Regulation of melanoma by the embryonic skin. Proc Natl Acad Sci U S A. 1986;83:7307–7310. doi: 10.1073/pnas.83.19.7307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mintz B, Illmensee K. Normal genetically mosaic mice produced from malignant teratocarcinoma cells. Proc Natl Acad Sci U S A. 1975;72:3585–3589. doi: 10.1073/pnas.72.9.3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McCullough KD, Coleman WB, Ricketts SL, Wilson JW, Smith GJ, Grisham JW. Plasticity of the neoplastic phenotype in vivo is regulated by epigenetic factors. Proc Natl Acad Sci U S A. 1998;95:15333–15338. doi: 10.1073/pnas.95.26.15333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boulanger CA, Mack DL, Booth BW, Smith GH. Interaction with the mammary microenvironment redirects spermatogenic cell fate in vivo. Proc Natl Acad Sci U S A. 2007;104:3871–3876. doi: 10.1073/pnas.0611637104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Booth BW, Mack DL, Androutsellis-Theotokis A, McKay RDG, Boulanger CA, Smith GH. The mammary microenvironment alters the differentation repertoire of neural stem cells. PNAS. 2008;105:14891–14896. doi: 10.1073/pnas.0803214105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andrews PW. Human teratocarcinomas. Biochem Biophys Acta. 1988;948:17–36. doi: 10.1016/0304-419x(88)90003-0. [DOI] [PubMed] [Google Scholar]
- 8.ATCC. Cell line verification test recommendations. ATCC Technical Bulletin. 2007;8:1–3. [Google Scholar]
- 9.DeOme KB, Faulkin LJ, Jr, Bern HA, Blair PB. Development of mammary tumors from hyperplastic alveolar nodules transplanted into gland-free mammary fat pads of female C3H mice. Cancer Res. 1959;19:515–520. [PubMed] [Google Scholar]
- 10.Smith GH, Vonderhaar BK, Graham DE, Medina D. Expression of pregnancy-specific genes in preneoplastic mouse mammary tissues from virgin mice. Cancer Res. 1984;44:3426–3437. [PubMed] [Google Scholar]
- 11.Smith GH, Mehrel T, Roop DR. Differential keratin gene expression in developing, differentiating, preneoplastic, and neoplastic mosue mammary epithelium. Cell Growth Differ. 1990;1:161–170. [PubMed] [Google Scholar]
- 12.Nagafuchi S, Seki S, Nakahori Y, Tamura T, Numabe H, Nakagome Y. PCR detection of structuraly abnormal Y chromosomes. Jpn J Hum Genet. 1992;37:187–193. doi: 10.1007/BF01900712. [DOI] [PubMed] [Google Scholar]
- 13.Sinclair AH, Berta P, Palmer MS, et al. A gene from the human sex-determining region encodes a protein with homology to a conserved DNA-binding motif. Nature. 1990;346:240–244. doi: 10.1038/346240a0. [DOI] [PubMed] [Google Scholar]
- 14.Smith GH. Experimental mammary epithelial morphogenesis in an in vivo model: evidence for distinct cellular progenitors of the ductal and lobular phenotype. Breast Cancer Res Treat. 1996;39:21–31. doi: 10.1007/BF01806075. [DOI] [PubMed] [Google Scholar]
- 15.Boulanger CA, Wagner KU, Smith GH. Parity-induced mouse mammary epithelial cells are pluripotent, self-renewing and sensitive to TGF-#1 expression. Oncogene. 2005;24:552–560. doi: 10.1038/sj.onc.1208185. [DOI] [PubMed] [Google Scholar]
- 16.Wagner KU, Boulanger CA, Henry MD, Sgagias M, Hennighausen L, Smith GH. An adjunct mammary epithelial cell population in parous females: its role in functional adaptation and tissue renewal. Development. 2002;129:1377–1386. doi: 10.1242/dev.129.6.1377. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.