Abstract
The immunoregulatory cytokine interleukin 10 (IL-10) is expressed mainly by T helper type 2 (TH2) cells but also by TH1 cells during chronic infection. Here we observed plasticity in the expression of IL-10 and IL-13 after chronic TH1 stimulation; furthermore, the expression of Il10 and Il13 was regulated by the transcription factor E4BP4. Chronically stimulated E4BP4-deficient (Nfil3−/−; called ‘E4bp4−/−’ here) TH1 cells, regulatory T cells (Treg cells) and natural killer T cells (NKT cells) had attenuated expression of IL-10 and IL-13. Enforced expression of E4bp4 initiated the production of IL-10 and IL-13 by conventional TH1 cells. E4bp4−/− TH2 cells showed impairment of IL-10 production with no effect on IL-13. Our results indicate that E4BP4 has multiple functions in controlling the plasticity of IL-13 in TH1 cells and IL-10 in TH1 cells, TH2 cells, Treg cells and NKT cells.
After being activated by antigen-presenting cells (APC), naive CD4+ T cells differentiate into helper T cells that acquire the ability to secrete effector cytokines. Helper T cells can be categorized into at least three subsets. T helper type 1 (TH1) cells, which secrete interleukin 2 (IL-2) and the proinflammatory cytokines interferon (IFN-γ) and tumor necrosis factor (TNF), promote cellular immune responses mainly to intracellular pathogens and viruses. TH2 cells secrete IL-4, IL-5, IL-6, IL-10 and IL-13 and promote humoral immune responses mainly to extracellular pathogens1. Cells of the TH17 helper T cell subset, which secrete IL-17A, IL-17F and IL-22, mediate immune responses mainly to extracellular pathogenic bacteria and fungi2 or have been linked to autoimmune disease3.
IL-13 was originally identified as a TH2 cytokine4; however, it is now known that IL-13 is produced by a variety of cell types, such as natural killer T cells (NKT cells), natural killer cells (NK cells), eosinophils and basophils5. As all of these cells also produce IL-4, it has been speculated that both IL-13 expression and IL-4 expression are controlled by common transcriptional mechanisms6,7. However, TH1 cells produce IL-13 in response to a combination of antigenic stimulation and IL-18 (ref. 8). Human patients with atopic or nonatopic asthma who have high titers of allergen-specific immunoglobulin E (IgE) can have more production of the TH1 cytokine IFN-γ, along with substantially more IL-13 production9,10. Therefore, TH1 cells could also be a source of IL-13 because of their plasticity, which is controlled by chronic antigen stimulation. However, the molecular mechanisms that regulate the plasticity of IL-13 production in TH1 cells is unclear.
IL-10 is also secreted by a wide variety of cells, such as TH2 cells, NKT cells, NK cells, natural regulatory T cells (Treg cells), regulatory B cells, macrophages, dendritic cells (DCs) and monocytes11. TH17 cells also express IL-10 in the presence of IL-27 (ref. 12). The mechanism by which IL-10 production is abrogated in TH1 cells has been thought to be stable and immutable. However, IL-10 production by TH1 cells has been reported in mice chronically infected with parasites such as Toxoplasma gondii13,14 or Leishmania major15. TH1 cells also produce IL-10 in response to high-dose antigenic stimulation and hyperactivation of T cell antigen receptor (TCR)-mediated signaling leading to continuous phosphorylation of the mitogen-activated protein kinases Erk1 and Erk2. Mechanistically, activation of the transcription factor STAT4 seems to elicit this plasticity16. IL-10 is an immunoregulatory cytokine that can downregulate active immune responses and acts mainly on T cells or macrophages to attenuate inflammatory cytokine production and antigen presentation. IL-10-deficient mice die with spontaneous colitis17, and this phenotype is partly retained even in mice lacking IL-10 only in Treg cells18. IL-10 therefore has an important role regulating overactive responses that would otherwise result in autoinflammatory disease. The plasticity of IL-10 production by TH1 cells in response to excess antigen stimuli could be an effective failsafe mechanism by which immune homeostasis is maintained. The differentiation of TH1, TH2 and TH17 cells is regulated by distinct signaling pathways19. Despite differences in their developmental origin, these T cell subsets share the ability to make IL-10 (ref. 11), although the molecular mechanisms that control this remain unclear.
The expression of Il10 and Il13 is regulated by GATA-3, the master transcriptional regulator for TH2 differentiation. GATA-3 coordinately regulates a group of genes encoding TH2 cytokines. Indeed, GATA-3 acts directly on the Il13 promoter20 and initiates chromatin remodeling at the Il10 locus21. Activated T cell–specific conditional deletion of Gata3 markedly attenuates IL-13 expression in committed TH2 cells22. However, IL-10 is also expressed by TH1 cells and NK cells, which normally do not express GATA-3, and the mechanism by which IL-10 is expressed in the absence of GATA-3 remains unclear16. Therefore, an unknown regulator of plasticity may substitute for the function of GATA-3, particularly in these cell types.
E4BP4 (E4 promoter–binding protein 4) was originally identified as a basic leucine zipper transcription factor. E4BP4 regulates circadian rhythm by competing for DNA binding with a member of the related PAR family of basic leucine zipper transcription factors that has a DNA-binding domain similar to that of E4BP4. PAR factors have been characterized as transcriptional activators, whereas E4BP4 is a repressor. E4BP4 and the PAR proteins may switch back and forth between the on state and off state on target genes to control the central circadian clock23. In the immune system, E4BP4 is also known as NFIL-3, as it regulates the Il3 promoter and promotes IL-3-mediated cell survival24,25. E4BP4 is a critical regulator of NK cell development through its induction of the transcriptional inhibitor Id2 (refs. 26,27) and also regulates IgE class switching in B cells28. Expression of the gene encoding E4BP4 (Nfil3; called ‘E4bp4’ here) is specific to TH2 cells in both mice and humans29,30. However, the functional importance of TH2 cell–specific expression of E4BP4 is unclear.
In this study, we demonstrate that E4BP4 is the unknown factor important for the plasticity in the production of IL-10 and IL-13 by TH1 cells after chronic antigen stimulation and that E4BP4 induced IL-10 production in the absence of GATA-3. Furthermore, E4BP4 regulated the transcription of Il10 and Il13 in natural Treg cells and NKT cells. However, IL-13 expression by TH2 cells was independent of E4BP4, because TH2 cells lacking E4bp4 did not produce IL-10 but IL-13 production was intact. Therefore, our data indicate that E4BP4 is a critical transcriptional regulator for IL-10, especially in CD4+ T cells. In addition, our data provide evidence of the functional importance of the expression of E4BP4 by TH2 cells, in which it is an essential regulator of IL-10 production.
RESULTS
Plasticity in IL-13 production by TH1 cells
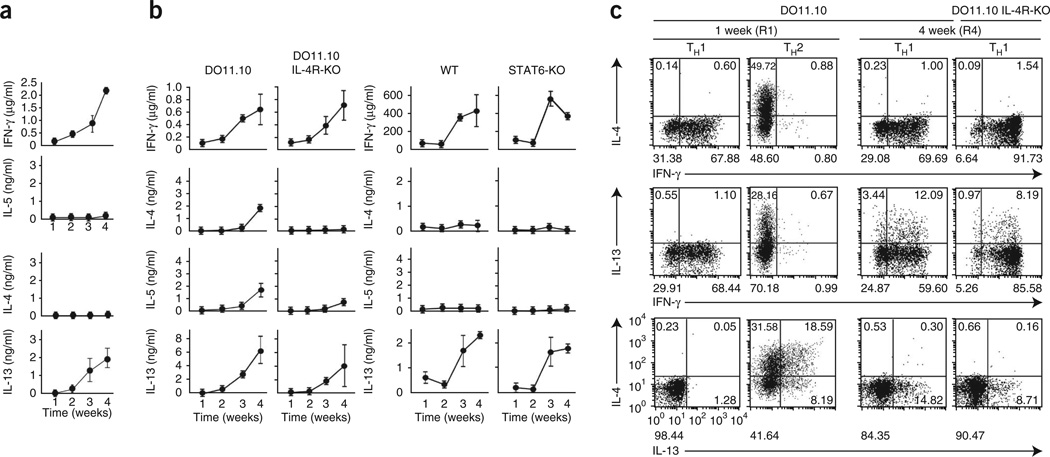
In an airway hyper-responsiveness model, Il4−/− and Il4r−/− mice lacking TH2 cells still had substantial airway hyper-responsiveness, similar to that of control C57BL/6 mice. To confirm the involvement of IL-13 in this response, we isolated CD4+ T cells from bronchoalveolar lavage fluid and mesenteric lymph nodes of Il4−/− mice. These T cells produced IL-13 (Supplementary Fig. 1) despite their lack of detectable expression of GATA-3 protein. These results demonstrate that noncanonical IL-13-producing CD4+ T cells can be generated under TH1-priming conditions. We speculated that repetitive TH1 priming would engender plasticity in TH1 cells, enabling them to express IL-13. To test our hypothesis, we obtained naive CD4+ T cells from mice transgenic for the ovalbumin (OVA)-specific TCR (DO11.10 mice), stimulated the cells weekly for 1–4 weeks (rounds 1–4 (R1–R4)) with OVA in the presence of APCs under TH1-skewing conditions and analyzed cytokine production every week up to 4 weeks. As expected, abundant IFN-γ was induced after the initial antigen priming, and this gradually increased with serial stimulation. Although the cells never made IL-4 or IL-5, IL-13 was detectable after the third round of stimulation (R3), and this increased with further stimulation (Fig. 1a). We observed similar IL-13-producing TH1 cells derived from Il4r−/− and Stat6−/− mice (Fig. 1b), both of which have very few TH2 cells. Intracellular cytokine staining (ICS) analysis showed that 8–12% of the IFN-γ-producing Il4r−/− TH1 cells also expressed IL-13 after four rounds of stimulation (R4), although no cells produced IL-13 after the first round (R1; Fig. 1c). Repetitive stimulation therefore induces IL-13 production by TH1-skewed CD4+ T cells.
Figure 1.
IL-13 production induced by repetitive antigen stimulation in TH1 cells. (a) Enzyme-linked immunosorbent assay (ELISA) of IFN-γ, IL-4, IL-5 and IL-13 in cells generated from naive CD4+ T cells (from DO11.10 mice on a recombination-activating gene 2–deficient background) repeatedly stimulated with OVA peptide in the presence of BALB/c APCs every week under TH1-skewing conditions, then restimulated with mAb to TCRβ at 1–4 weeks after initial stimulation. (b) ELISA of cytokines in cells generated from naive CD4+ T cells obtained from Il4r+/+ (DO11.10) or Il4r−/− (DO11.10 IL-4R-KO) DO11.10 mice (on the BALB/c background) and stimulated with OVA peptide in the context of APCs (far left and middle left), or generated from naive CD4+ T cells obtained from wild-type (WT) or Stat6−/− (STAT6-KO) mice and stimulated by mAb to TCRβ and mAb to CD28 under TH1-skewing conditions (far right and middle right). (c) ICS detection of cells producing IFN-γ, IL-13 and IL-4 among cells generated from CD4+ T cells (from DO11.10 mice on a recombination-activating gene 2–deficient or Il4r−/− background) stimulated with OVA peptide in the context of APCs every week under TH1-skewing conditions, followed by restimulation with mAb to TCRβ. Numbers in or below quadrants indicate percent cells in each throughout. Data are from three independent experiments (a,b; mean ± s.e.m.) or are representative of three experiments with similar results (c).
E4BP4 regulates IL-13 expression by TH1 cells
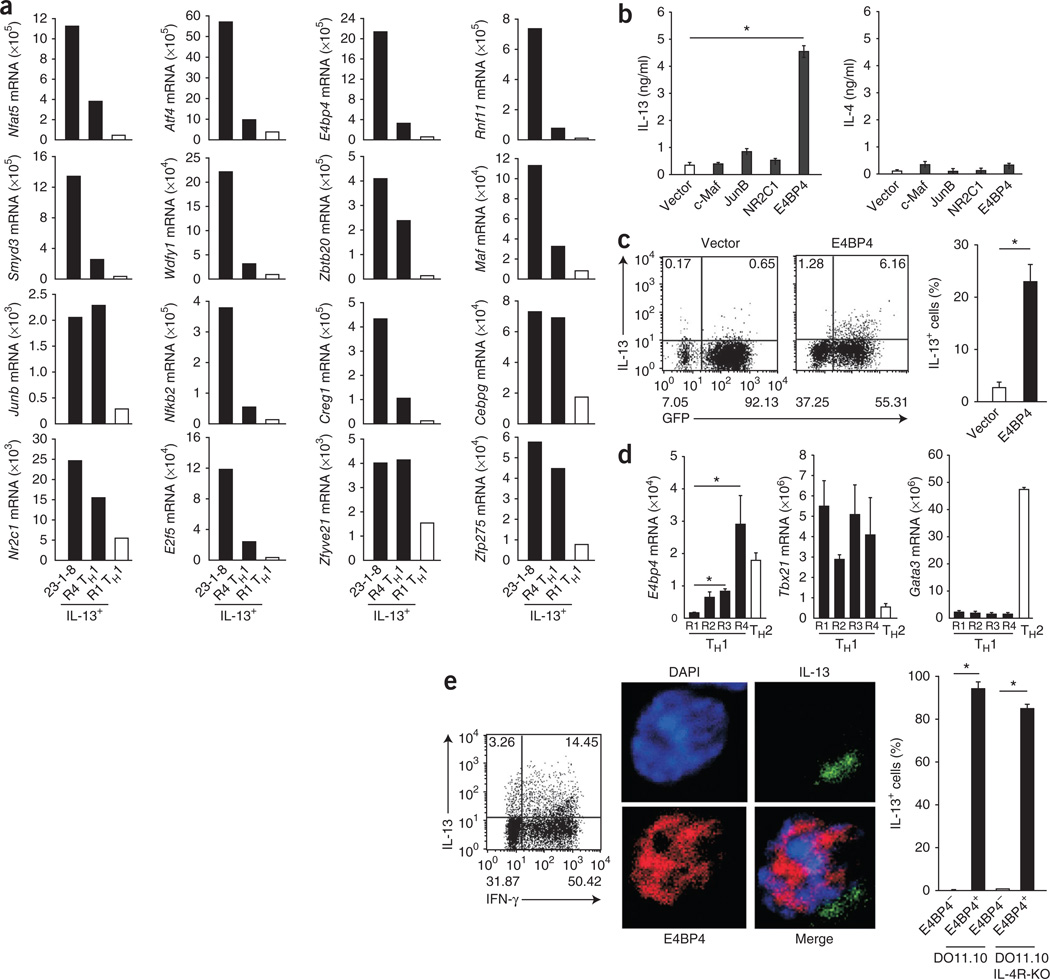
Although Il13 expression by TH2 cells is regulated mainly by GATA-3 (refs. 20,22,31), our data suggested that Il13 expression by TH1 cells is regulated by unknown mechanisms that are independent of this transcription factor. Activation of these mechanisms correlated with repetitive T cell activation; thus, we sought to identify the IL-13 regulators specifically expressed in R4 TH1 cells but not in conventional TH1 cells. Microarray analysis identified more than 50 genes with fivefold higher mRNA expression in R4 TH1 cells (Supplementary Fig. 2). Among these genes, 16 encoded molecules with features characteristic of transcription factors, and we confirmed their enhanced expression in R4 TH1 cells by quantitative RT-PCR (Fig. 2a). To determine which of the candidate genes encoded a molecule that could induce IL-13 expression, we ectopically expressed each in conventional TH1 cells by retroviral transduction. Only overexpression of E4bp4 induced readily detectable IL-13 protein (Supplementary Fig. 3). In contrast, overexpression of Maf, Junb or Nr2c1, which all encode molecules thought to be regulators of TH2 differentiation, did not induce IL-13 production by TH1 cells (Fig. 2b). However, IL-4 was nearly below the level of detection in the E4bp4-transduced cells (Fig. 2b). The tempo of IL-13 production was accelerated in the transduced cells; single-cell analysis after intracellular cytokine staining demonstrated that more than 20% of the E4BP4-expressing cells produced IL-13 after only a single round of TH1 stimulation (Fig. 2c).
Figure 2.
E4BP4 regulates IL-13 expression in TH1 cells. (a) Quantitative RT-PCR analysis of the expression of 16 candidate genes identified by comparative transcriptome analysis in TH1 cells given one round (R1) or four rounds (R4) of simulation and then restimulated with mAb to TCRβ, and also in 23-1-8 cells, a TH1 cell clone corresponding to R4 TH1 cells (primer sequences, Supplementary Table 1). (b) ELISA of IL-4 and IL-13 in supernatants of cultures of BALB/c CD4+ T cells given initial stimulation with mAb to TCRβ and mAb to CD28, then transduced for 7 d with retroviral plasmid encoding green fluorescent protein (GFP) alone (Vector) or plasmid encoding GFP plus E4BP4, c-Maf, JunB or NR2C1, followed by restimulation of GFP+ cells for 24 h with mAb to TCRβ. (c) ICS detection (left) of IL-13-producing cells among BALB/c CD4+ T cells given initial stimulation with mAb to TCRβ and mAb to CD28, then transduced for 7 d with retroviral plasmid alone or plasmid encoding E4BP4, followed by restimulation of GFP+ cells for 6 h with mAb to TCRβ; right, frequency of IL-13-producing cells among all CD4+ T cells. (d) Quantitative RT-PCR analysis of the expression of E4bp4, Tbx21 and Gata3 in R1–R4 TH1 cells and in TH2 cells (primers, Supplementary Table 1). (e) ICS detection (left) of IL-13- and IFN-γ-producing cells among R4 TH1 cells from DO11.10 mice; immunocytochemistry analysis (middle) of E4BP4 and IL-13 in DO11.10 T cells; and frequency of IL-13+ cells among E4BP4+ or E4BP4− cells (right). Original magnification (middle), ×640. DAPI, DNA-intercalating dye. *P < 0.01 (Student’s t-test). Data are representative of two experiments with 100 cells expressing both IFN-γ and IL-13 (a) or are from three independent experiments (b–e; mean and s.e.m.).
Published studies based on microarray data have shown E4bp4 expression mainly in TH2 cells29,30. We confirmed that finding but also found that E4bp4 mRNA was markedly enhanced in TH1 cells after chronic TH1 stimulation (Fig. 2d). This promiscuous expression in both subsets was unique to E4bp4, as expression of mRNA for the transcription factor T-bet (Tbx21 mRNA) and Gata3 mRNA was limited to TH1 and TH2 cells, respectively. Next, to determine whether E4bp4 expression directly correlated with IL-13 expression at the single-cell level, we did immunofluorescence staining of R4 TH1 cells generated from DO11.10 mice that lacked TH2 cells because of Il4r deletion (Il4r−/− DO11.10 mice). E4BP4 protein was located mainly in the nucleus, and 90% of IL-13-producing cells had readily detectable E4BP4 expression (Fig. 2e). As expected, IL-13 was coexpressed with IFN-γ in the stimulated cells (Fig. 2e).
To test the generality of our observations in mice, we examined naive (CD45RAhi) CD4+ T cells from human peripheral blood (Supplementary Fig. 4a). Three rounds of stimulation with monoclonal antibody (mAb) to CD3 under TH1 conditions induced IL-13 production in ~9% of cells, of which ~97% were double producers of IL-13 and IFN-γ (Supplementary Fig. 4b). These cells also had nuclear expression of E4BP4 protein, and about 90% of E4BP4-expressing cells CD4+ T cells produced IL-13 (Supplementary Fig. 4c). Therefore, there was a correlation between E4BP4 protein and IL-13 expression in human TH1 cells after chronic TCR stimulation.
Expression of E4bp4, as well as of Gata3, was inducible in TH2 cells29,30 (Fig. 2d). To begin to assess the mechanisms of regulation of E4bp4 expression in TH2 cells, we first tested the importance of the IL-4–STAT6 axis and confirmed that both neutralization of IL-4 and deficiency in Stat6 attenuated E4bp4 expression in TH2-skewed cells (Supplementary Fig. 5). However, ectopic expression of Gata3 failed to induce E4bp4 expression (Supplementary Fig. 5), which suggested that E4bp4 expression in TH2 cells is regulated by the IL-4–STAT6 pathway and not by GATA-3.
E4bp4 regulates the expression of IL-13 and IL-10 in CD4+ T cells
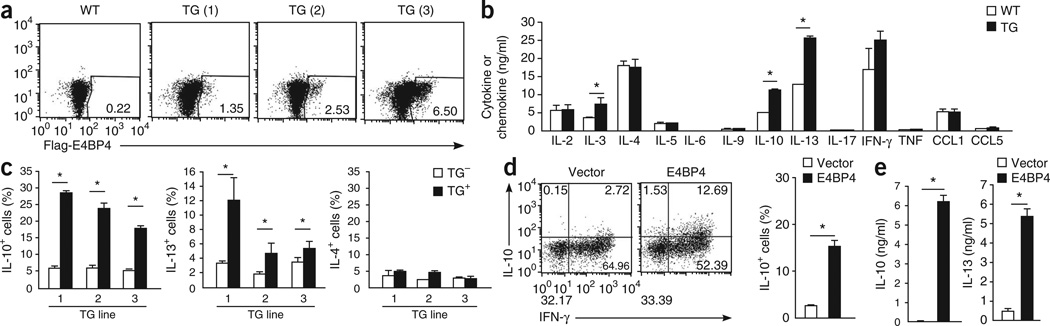
To study the role of E4BP4 in vivo, we established mice expressing a transgene encoding Flag-tagged E4BP4. Three independent lines had expression of Flag-tagged E4BP4 in 1–6% of CD4+ T cells (Fig. 3a). Enforced expression of E4BP4 in CD4+ T cells resulted in more IL-13 production at 48 h after initial activation without affecting the production of IL-4 or IL-5 (Fig. 3b). Furthermore, this E4bp4 transgene also induced the expression of IL-3 as well as of IL-10 (Fig. 3b). The effect on IL-3 production was expected, as E4BP4 is a known inducer of this cytokine24; however, the effect on IL-10 expression was completely unexpected. To further examine the possible regulation of both Il13 and Il10 by E4BP4, we assessed cytokine production and transgene expression in single cells. Production of IL-10 and IL-13 was augmented in cells expressing the E4bp4 transgene compared with that in transgene-negative CD4+ T cells, whereas IL-4 production was the same in both cell types (Fig. 3c). Moreover, retroviral overexpression of E4bp4 induced the production of IL-10 as well as of IL-13 in TH1 cells (Fig. 3d,e). These results suggested that E4BP4 ‘preferentially’ induces the expression of both Il10 and Il13.
Figure 3.
E4bp4 overexpression induces the expression of IL-10 and IL-13 by CD4+ T cells. (a) Intracellular staining to detect transgene expression by CD4+ T cells purified from the spleens of a wild-type mouse or C57BL/6 mice expressing a transgene (TG) encoding Flag-tagged E4BP4 (three independent lines, TG (1)–TG (3)), assessed with anti-Flag. (b) Concentration of cytokines and chemokines in wild-type and E4bp4-transgenic CD4+ T cells stimulated for 7 d with mAb to TCRβ and mAb to CD28, then restimulated with mAb to TCRβ. *P < 0.05 (Student’s t-test). (c) Intracellular staining to detect T cells producing IL-4, IL-10 and IL-13 among Flag-positive cells (TG+) or Flag-negative cells (TG−) generated from E4bp4-transgenic CD4+ T cells stimulated for 7 d as in b, then restimulated with mAb to TCRβ. *P < 0.05 (Student’s t-test). (d) Intracellular staining to detect T cells producing IL-10 and IFN-γ (left) among GFP+ T cells generated from C57BL/6 CD4+ T cells transduced with plasmid encoding GFP alone or GFP plus E4BP4 and activated under TH1 conditions; right, frequency of IL-10+ cells. (e) Production of IL-10 and IL-13 in the GFP+ T cells isolated in d. *P < 0.01 (Student’s t-test). Data are representative of two experiments (a) or are from three independent experiments (b–d; mean and s.e.m.).
E4BP4 is essential for the expression of Il10 and Il13
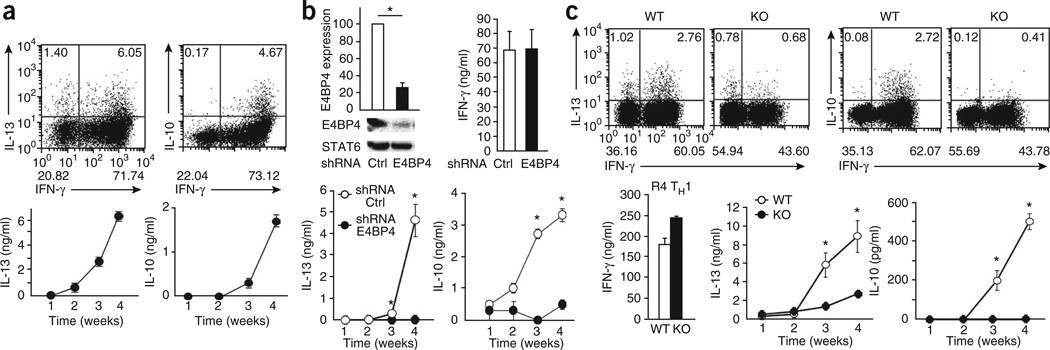
Given their apparent coregulation by E4BP4, we next examined the production of both IL-10 and IL-13 in R4 TH1 cells. Repetitive TH1 priming induced both cytokines (Fig. 4a). To assess whether the expression of Il10 and Il13 required E4BP4 expression in TH1 cells, we used a retrovirus vector encoding short hairpin RNA (shRNA) specific for E4bp4 to suppress its expression. Knockdown of E4bp4 under TH1-skewing conditions markedly inhibited the production of IL-10 and IL-13 normally observed after the third or fourth round of stimulation but had no effect on IFN-γ production during the first round of stimulation (Fig. 4b), which indicated that E4BP4 is the critical factor that regulates the production of both IL-10 and IL-13 in chronically activated TH1 cells.
Figure 4.
E4BP4 regulates the expression of Il10 and Il13 in TH1 cells. (a) ICS analysis (top) and ELISA (bottom) of IL-13, IL-10 and IFN-γ in cells generated from C57BL/6 CD4+ T cells stimulated four times with mAb to TCRβ and mAb to CD28 under TH1 conditions. Data are representative of three independent experiments (mean ± s.e.m.). (b) Immunoblot analysis (top left) of E4BP4 and STAT6 in CD4+ T cells stimulated for 48 h under TH2 conditions and transduced with retrovirus encoding control shRNA (Ctrl) or E4bp4-specific shRNA (E4BP4), assessed 1 week later by probing with anti-E4BP4, followed by densitometry analysis (above; normalized to STAT6 values and presented relative to E4BP4 in cells transduced with the control shRNA, set as 100); and ELISA of IFN-γ in R4 TH1 cells (top right) or of IL-10 and IL-13 in TH1 cells stimulated 1–4 weeks (R1–R4; bottom). *P < 0.01 (Student’s t-test). Data are from three independent experiments (mean ± s.e.m.). (c) ICS analysis (top) and ELISA (bottom) of IL-13, IL-10 and IFN-γ in CD4+ T cells derived from E4bp4−/− mice (KO) and their wild-type littermates (WT), then stimulated for 4 weeks under TH1-skewing conditions. *P < 0.01 (Student’s t-test). Data are representative of three independent experiments (ICS) or are from three independent experiments (mean ± s.e.m.).
To better understand the mechanistic effects of E4BP4 on T cell function, especially in terms of regulation of cytokine production, we generated mice lacking E4bp4 (Supplementary Fig. 6a). The development of thymic T cells and splenic T cells and B cells in E4bp4-deficient mice was similar to that of their littermates (Supplementary Fig. 6b). Moreover, T cell proliferation after TCR engagement and the expression profiles of the T cell–activation markers CD25 and CD69 were similar for both wild-type and E4bp4-deficient splenic T cells (Supplementary Fig. 6c,d). In contrast, there was significantly less production of IL-10 and IL-13 by R4 TH1 cells derived from the E4bp4−/− mice than by those from their littermates (Fig. 4c). We next examined the effect of E4bp4 deficiency on the differentiation of TH1, TH2, TH17 and induced Treg cells. E4bp4−/− T cells showed normal production of IFN-γ, IL-4 and IL-1,7 as well as normal induction of Treg cells by transforming growth factor-β (Supplementary Fig. 6e and data not shown).
Mouse memory TH1 cells and human TH1 cells produce IL-13 when they are generated in the presence of IL-18 (refs. 32,33). However, E4BP4 is dispensable for IL-18-induced IL-13 production, because E4bp4−/− TH1 cells expressed IL-13 after stimulation with IL-18 (Supplementary Fig. 7a). IL-10 production by TH1 cells after stimulation with IL-27 has also been reported34. Indeed, control TH1 cells expressed IL-10 in the presence of IL-27, whereas E4bp4−/− TH1 cells did not (Supplementary Fig. 7b). Moreover, although IL-10 production has been reported in TH17-polarizing conditions35, we were unable to confirm that result (Supplementary Fig. 7c). Therefore, all these data support the hypothesis that E4BP4 is required for IL-13 expression by TH1 cells in which plasticity has been induced by chronic stimulation and for IL-10 expression when plasticity is induced by IL-27 stimulation.
E4BP4 regulates Il10 expression by TH2 cells
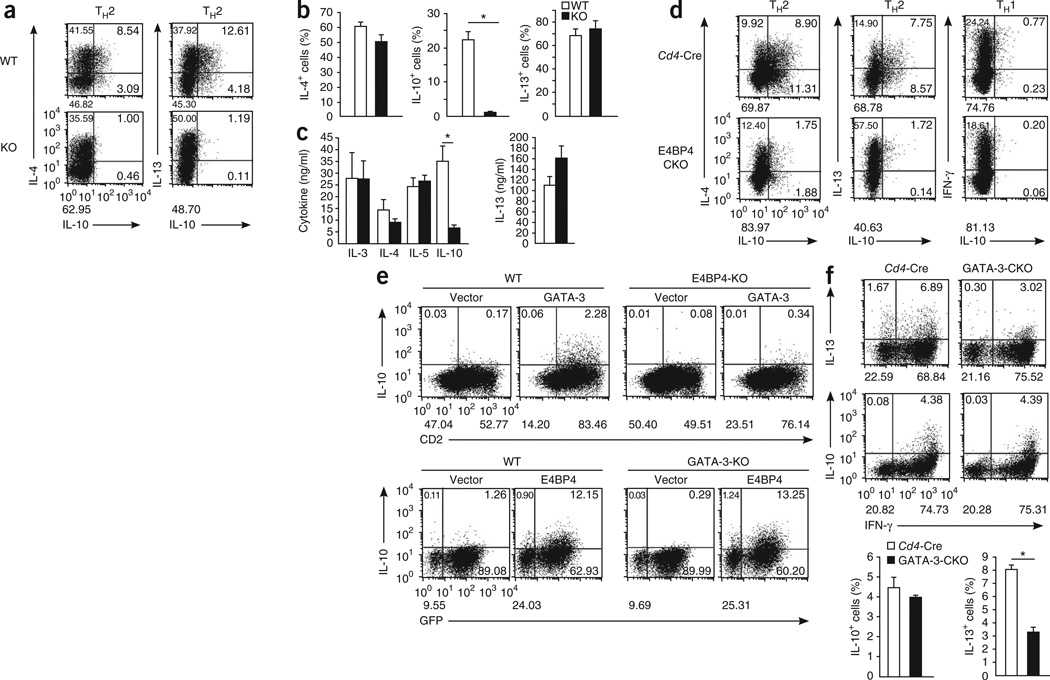
Next we analyzed the role of E4BP4 in the production of IL-10 and IL-13 by TH2 cells. We obtained naive T cells from E4bp4−/− or wild-type mice and stimulated the cells under TH2-polarizing conditions. About 50% of the E4bp4+/+ T cells generated under TH2 conditions produced IL-13 or IL-10, ~20% of the IL-4+ cells were also IL-10+, and ~25% of the IL-13+ cells also produced IL-10. In contrast, the population of IL-10+ cells among the E4bp4−/− TH2 cells, was much smaller than that of cells from the wild-type littermates (Fig. 5a), although the production of IL-4, IL-5 and IL-13 by these cells was similar to that of wild-type T cells (Fig. 5b,c).
Figure 5.
Impaired production of IL-10 and IL-13 in E4bp4-deficient T cells. (a–c) Intracellular staining analysis (a,b) and ELISA (c) of IL-4, IL-10 and IL-13 in CD4+ T cells obtained from E4bp4−/− mice (KO) and their E4bp4+/+ littermates (WT) and stimulated under TH2 conditions. *P < 0.01 (Student’s t-test). Data are representative of (a) or from (b,c) three independent experiments (mean and s.e.m. in b,c). (d) Intracellular staining analysis of IL-4, IL-10 and IL-13 in cells generated from naive CD4+ T cells obtained from Cd4-Cre mice or E4bp4f/fCd4-Cre mice (E4BP4-CKO), then stimulated with mAb to TCRβ and mAb to CD28 under TH2 conditions or TH1 conditions. Data are representative of three independent experiments. (e) Intracellular staining analysis of IL-10 in cells generated from activated CD4+ T cells derived from E4bp4−/− mice (E4BP4-KO) and their E4bp4+/+ littermates (WT) and transduced with empty plasmid or plasmid encoding GATA-3 (top), or derived from Gata3f/f (GATA-3-KO) mice or C57BL/6 mice (WT) and transduced with plasmid encoding GFP and E4BP4 (E4BP4) or plasmid encoding human CD8 (Vector; bottom), followed by restimulation with mAb to TCRβ. GATA-3-deficient T cells (bottom) were isolated by gating on cells positive for human CD8. Data are representative of three independent experiments. (f) ICS analysis (top) of IL-10 and IL-13 in CD4+ T cells prepared from Cd4-CreGata3f/f mice (GATA-3-CKO) and Cd4-Cre mice, then stimulated for 4 weeks under TH1-skewing conditions; and frequency of IL-10+ and IL-13+ cells (bottom). *P < 0.01 (Student’s t-test). Data are representative of three independent experiments (ICS) or are from three independent experiments (mean and s.e.m.).
To further confirm that the effect of E4BP4 on CD4+ T cells was intrinsic, we crossed mice transgenic for Cd4-driven expression of Cre recombinase (Cd4-Cre) with our mouse line with conditional deficiency in E4bp4 to generate mice with CD4+ T cell–specific deletion of E4bp4 (E4bp4f/fCd4-Cre mice); although the deletion of E4bp4 in CD4+ T cells was partial, it was still greater than 80% (Supplementary Fig. 6a). Again, IFN-γ production by E4bp4f/fCd4-Cre TH1 cells and the expression of IL-4 and IL-13 by E4bp4f/fCd4-Cre TH2 cells were similar to that of the Cd4-Cre control mice. However, IL-10 expression in TH2 cells was much lower than that of the Cd4-Cre control cells (Fig. 5d). As expected, neither wild-type nor E4bp4f/f Cd4-Cre TH1 cells had detectable production of IL-10.
We next determined whether the production of IL-10 or IL-13 by T cells was also dependent on GATA-3. To determine if E4BP4 requires GATA-3 for IL-10 production, we transduced E4bp4+/+ and E4bp4−/− T cells with a Gata3-expressing retroviral vector. Exogenous Gata3 induced the generation of IL-10-producing E4bp4+/+ T cells slightly, but the response was completely abolished in E4bp4−/− T cells (Fig. 5e, top row). In addition, we transduced a retroviral vector expressing Cre into T cells with loxP-flanked Gata3 (Gata3f/f) and into wild-type C57BL/6 T cells. We further transduced cells with a retroviral vector expressing E4bp4 and found that ectopic expression of E4BP4 protein significantly induced the development of IL-10+ T cells with or without Gata3 expression (Fig. 5e, bottom). We further examined the production of IL-10 and IL-13 by TH1 cells using CD4+ T cells derived from Gata3f/fCd4-Cre mice. IL-13 production in R4 TH1 cells lacking Gata3 was ~50% lower, but there was no effect on IL-10 production (Fig. 5f). These results indicate that E4BP4 is a critical factor for Il10 expression in CD4+T cells independently of Gata3 expression and that Il13 expression in TH1 cells requires low Gata3 expression.
E4BP4 regulates the expression of Il13 and Il10 in NKT cells
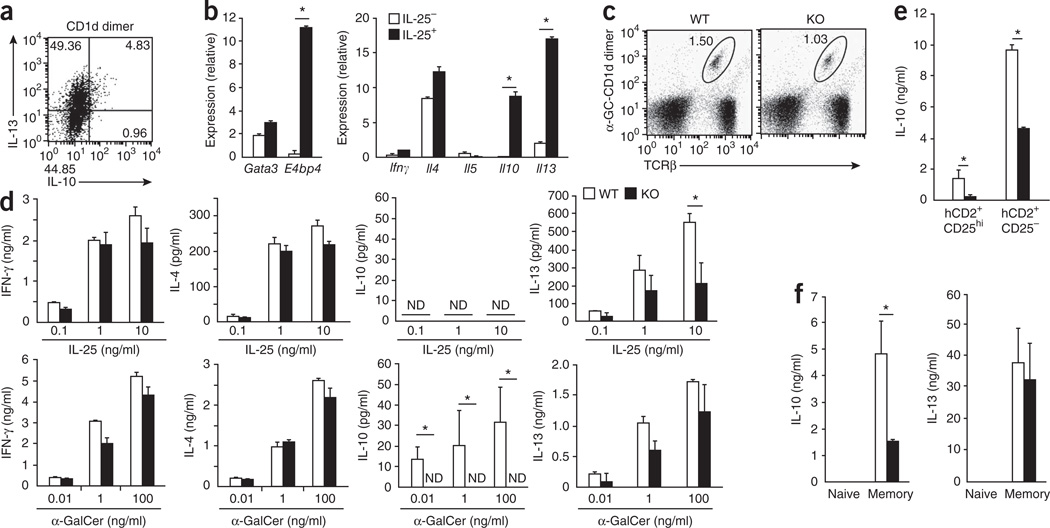
Various T cell subsets, including NKT cells and Treg cells, produce IL-10 and IL-13, and we hypothesized that E4BP4 is a regulator of the expression of Il10 and Il13 in these cells as well. To test our hypothesis, we first determined if there was a correlation between the expression E4bp4 and the expression of both Il10 and Il13 by NKT cells. We stimulated whole splenocytes with the NKT cell ligand α-galactosyl ceramide (α-GalCer) and analyzed cytokine production cells by ICS after gating on cells that recognized a dimer of α-GalCer and the antigen-presenting molecule CD1d. About 55% of NKT cells produced IL-13, and all of the IL-10-producing cells also produced IL-13 (Fig. 6a). We also observed that E4bp4 expression was induced by stimulation with α-GalCer, which suggested a possible role for E4BP4 in the expression of Il10 and Il13 by NKT cells. It has been reported that the treatment of IL-17RB+ NKT cells with IL-25 induces high Il13 expression36; therefore, we focused on a possible role for E4BP4 in inducing the expression of Il10 and Il13 in this NKT cell subset. We isolated IL-17RB+ NKT cells from BALB/c mice and stimulated the cells with IL-25 in the presence of DCs. E4bp4 expression was induced considerably in the IL-25-treated cells, and its expression correlated well with the expression of Il10 and Il13 (Fig. 6b). The frequency of splenic NKT cells was nearly identical in E4bp4−/− and wild-type mice (Fig. 6c); however, in response to stimulation with α-GalCer, E4bp4−/− NKT cells had no detectable IL-10 expression and lower IL-13 expression, and in response to IL-25, E4bp4−/− NKT cells had lower IL-13 expression and both wild-type and E4bp4−/− NKT cells had no detectable IL-10, whereas the expression of IFN-γ and IL-4 was unaffected (Fig. 6d).
Figure 6.
Impaired production of IL-10 and IL-13 by CD4+ T cell subsets lacking E4BP4. (a) ICS detection of IL-13- and IL-10-producing cells among whole BALB/c splenocytes stimulated for 48 h with the α-GalCer–CD1d dimer (CD1d dimer). (b) Quantitative RT-PCR analysis of gene expression among total RNA from IL-17RB+ NKT cells generated from IL-17RB+α-GalCer–CD1d+TCRβ+ cells sorted from spleen cells and cultured for 24 h in the presence (IL-25+) or absence (IL-25−) of IL-25 together with bone marrow–derived DCs induced by granulocyte-macrophage colony-stimulating factor. (c) Flow cytometry analysis of NKT cells from the spleens of E4bp4−/− mice and their E4bp4+/+ littermates. Numbers adjacent to outlined areas indicate percent α-GalCer–CD1d+TCRβ+ cells. (d) ELISA of cytokine production in IL-17RB+ NKT cells derived from E4bp4−/− mice and their E4bp4+/+ littermates (C57BL/6 background) and stimulated for 24 h with various concentrations of IL-25 (top) or α-GalCer (bottom) in the presence of bone marrow–derived DCs induced by granulocyte-macrophage colony-stimulating factor. ND, not detected. (e,f) ELISA of cytokines in culture supernatants of Foxp3+CD4+ (CD25− or CD25hi) T cells (e) and naive (Foxp3−CD62LhiCD44lo) and memory (Foxp3−CD62LloCD44hi) CD4+ T cells (f) purified from reporter mice expressing Foxp3 from human CD2 (hCD2), on an E4bp4+/+ or E4bp4−/− background, then stimulated for 48 h with mAb to TCRβ and mAb to CD28. *P < 0.01 (Student’s t-test). Data are representative of two experiments (a) or three experiments (b–f; mean and s.e.m.).
To study the involvement of E4BP4 in IL-10 production by Treg cells, we isolated naive CD4+ T cells (Foxp3−CD44loCD62Lhi), memory CD4+ T cells (Foxp3−CD44hiCD62Llo) and natural Treg cells (Foxp3+CD25− or Foxp3+CD25hi) from mice expressing the transcription factor Foxp3 from a human CD2 reporter (Foxp3hCD2 mice)37, which allowed better separation of Foxp3+ cells from other live cells on the basis of the expression of human CD2, and stimulated the cells through the TCR. Natural Treg cells expressed IL-10 but not IL-13 after stimulation of the TCR (Supplementary Fig. 8a). We found high expression of E4bp4 in Il10-expressing natural Treg cells, particularly in the CD25− subset, and we also detected this in memory T cells, which also expressed Il10. In contrast, naive T cells had low expression of both E4bp4 mRNA and Il10 mRNA (Supplementary Fig. 8a). IL-10 production was impaired in natural Treg cells and memory T cells derived from E4bp4−/− mice, but IL-13 production was not (Fig. 6e,f and Supplementary Fig. 8b,c). These results indicate that E4bp4 expression is correlated with the expression of Il10 and Il13 by NKT cells, memory T cells and IL-10-expressing natural Treg cells, which suggests that E4BP4 regulates Il10 expression in all T cell subsets and Il13 expression in NKT cells.
E4BP4 binds specifically to the Il13 and Il10 loci
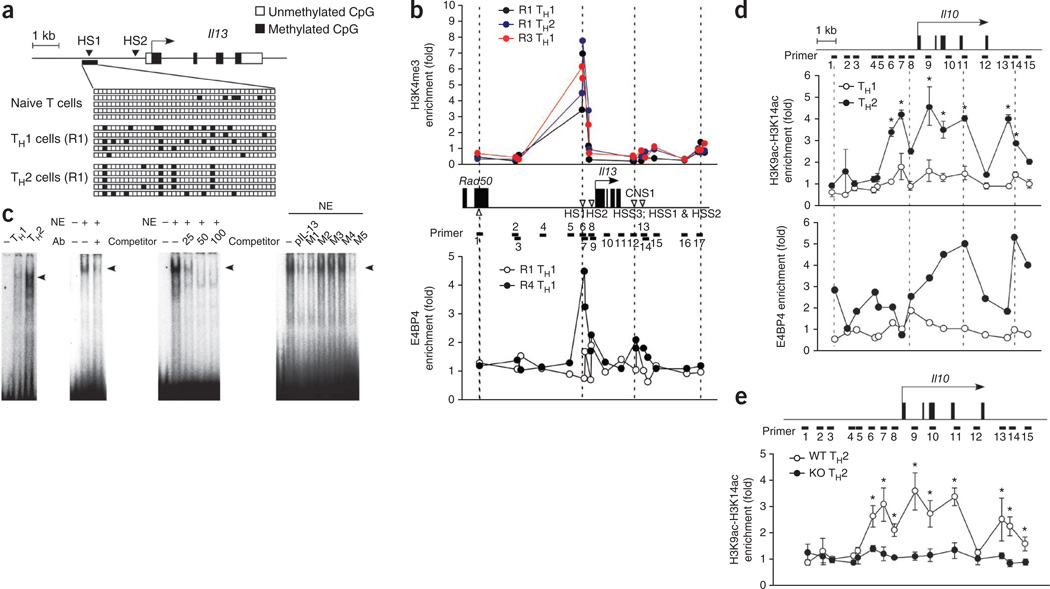
The Il4 locus in TH1 cells is thought to be maintained in a repressed state by epigenetic modifications38. However, as shown above, chronic TH1 stimulation promoted IL-13 production, which suggests that the Il13 and Il4 loci may have distinct modes of regulation. Therefore, we studied methylation and histone modifications of the Il13 locus in TH1 cells. The Il13 distal promoter contains two well characterized DNase I–hypersensitive sites, HS1 and HS2, which are highly conserved in mammals. We used a bisulfite-sequencing PCR assay to investigate the DNA-methylation status, especially of the HS1 region, in naive T cells and in developing TH1 and TH2 cells (Fig. 7a). Most CpG dinucleotides were equally unmethylated in naive CD4+ cells; however, we observed a few methylated sites in both developing TH1 cells and TH2 cells (Fig. 7a). We also investigated histone modification of the Il13 and Il4 loci in various TH1 cell developmental stages by chromatin immunoprecipitation (ChIP) and found considerable enrichment of histone H3 trimethylated at Lys4 in the HS1 region of the Il13 promoter, even in TH1 cells stimulated for 1 week (R1; Fig. 7b, top). We then used ChIP analysis with nuclear extracts prepared from conventional TH1 and R4 TH1 cells to determine whether E4BP4 binds to the Il13 promoter. There was considerable enrichment of specific binding of E4BP4 to the HS1 site in R4 TH1 cells but not in conventional TH1 cells (Fig. 7b, bottom).
Figure 7.
E4BP4 protein binds specifically to the Il13 promoter and in the Il10 locus. (a) Methylation of genomic DNA (bottom) isolated from naive CD4+ T cells and from TH1 cells and TH2 cells given 1 week of stimulation (R1), analyzed by sequencing with primers specific for the Il13 promoter (top) after bisulfite treatment. (b) ChIP analysis of the enrichment of histone H3 trimethylated at Lys4 (H3K4me3) in R1 TH1 cells, R1 TH2 cells or R3 TH1 cells (top) and of E4BP4 in R1 TH1 and R4 TH1 cells (bottom), both at the Il13 locus (middle). CNS1, conserved noncoding sequence 1; HSS, hypersensitive site; Rad50, gene encoding a protein involved in the repair of DNA double-strand breaks. (c) Electrophoretic mobility-shift assay of nuclear extracts (NE) of TH1 and TH2 cells, with an oligonucleotide from a specific region of the Il13 promoter as the probe (far left); supershift analysis of nuclear extracts from TH2 cells with anti-E4BP4 (Ab; middle left); and competition analysis with 25-, 50- or 100-fold excess competitor (middle right) or mutant oligonucleotides (M1–M5) and the Il13 promoter (pIL-13; far right). (d) ChIP analysis of the enrichment of histone H3 acetylated at Lys9 and Lys14 (H3K9ac–H3K14ac; top) or of E4BP4 (bottom) at the Il10 locus (above) in chromatin fractions extracted from TH1 or TH2 cells; results are presented relative to those obtained with input DNA prepared from untreated chromatin. (e) ChIP analysis of the enrichment of histone H3 acetylated at Lys9 and Lys14 at the Il10 locus (above) in TH2 cells from E4bp4−/− and wild-type mice; results presented as in d. *P < 0.01 (Student’s t-test). Data are representative of two experiments (a–c) or are from three independent experiment (d,e; mean and s.e.m.).
We then used electrophoretic mobility-shift assay to identify the putative E4BP4-binding site on the Il13 promoter. We prepared nuclear extracts of R4 TH1 cells and TH2 cells and tested for binding of E4BP4 to oligonucleotides corresponding to the putative binding sites in the adenovirus E4 promoter and the mouse Il13 promoter. We found a distinctly shifted band in TH2 cells and a slight sift even in TH1 cells (Fig. 7c). This binding was blocked by treatment with either antibody to E4BP4 (anti-E4BP4) or an excess of unlabeled competitor oligonucleotide corresponding to the HS1 site of the Il13 promoter (Fig. 7c). The HS1 site corresponds to a conserved GATA-3-response element (CGRE) identified as a distal promoter for Il13, and CGRE contains a consensus E4BP4-binding sequence as well as a GATA-3-binding site39. We found that 2-kilobase fragments including the CGRE had strong promoter activity in a TCR-stimulation assay, and deletion of the CGRE from the promoter region attenuated this activity even when E4bp4 was overexpressed (Supplementary Fig. 9a). Thus, binding of E4BP4 to the CGRE regulates Il13 promoter activity. Together these results demonstrate that binding of E4BP4 to the CGRE site of the Il13 promoter promotes the expression of IL-13 in T cells. The HS1 site in the Il13 promoter is transcriptionally permissive even in TH1 cells, which suggests that E4BP4 expression may directly govern the plasticity of Il13 in TH1 cells.
We next did a similar study of the Il10 locus, in which we identified 15 possible binding sites on the basis of the predicted E4BP4-binding sequence (Fig. 7d). ChIP analysis showed enrichment for histone H3 acetylated at Lys9 or Lys14 in some of the binding sites. We observed such enrichment in these regions mainly in TH2 cells but not in conventional TH1 cells. Further ChIP analysis showed that E4BP4 bound to intron 4 and the 3′ noncoding region of the Il10 locus (Fig. 7d). Two 5′ upstream fragments of Il10 400 and 1,000 in length that do not contain consensus binding sequences for E4BP4 had no promoter activity in the T cell–activation assay (Supplementary Fig. 9a), which indicated that E4BP4 does not regulate the activation-induced promoter activity. To unequivocally demonstrate a role for E4BP4 in the epigenetic modification of the Il10 locus, we analyzed E4bp4−/− cells. The strong enrichment for acetylated histone H3 in the Il10 locus noted above was completely absent from E4bp4−/− TH2 cells (Fig. 7e). From these results, we conclude that E4BP4 directly regulates the permissive status of the Il10 locus at the chromatin level.
E4BP4 in the control for autoimmune responses
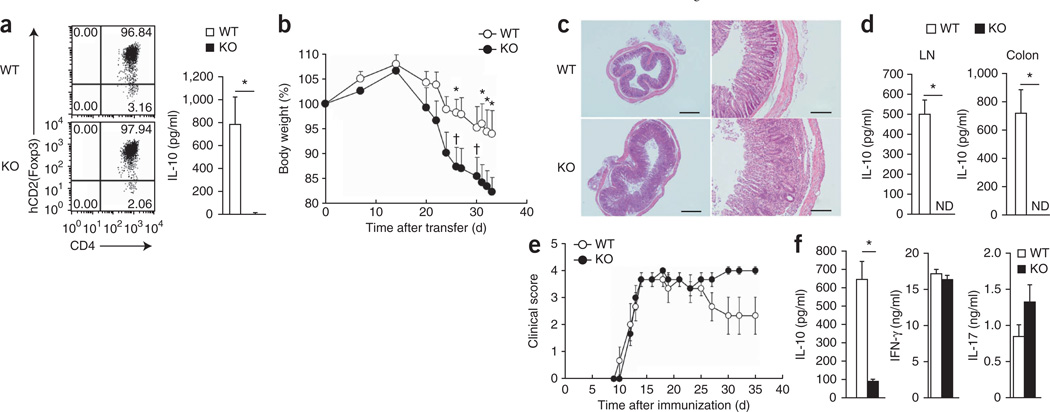
Mice with complete ablation of Il10 or T cell–specific ablation of Il10 (Il10f/fCd4-Cre mice) develop spontaneous intestinal inflammation40, and Treg cell–specific ablation of Il10 also results in spontaneous colitis, albeit a milder version18. Such results demonstrate that IL-10 produced by Treg cells has an important role in suppressing immune inflammation in the intestine. Indeed, about half of the E4bp4−/− mice spontaneously developed mild diarrhea (data not shown), and induced Treg cells generated from E4bp4−/− mice had impaired IL-10 production after TCR stimulation even though control induced Treg cells had detectable, but low, expression of IL-10 (Fig. 8a). Given those findings, we examined the involvement of E4BP4 in T cells in a T cell–transfer colitis model in which we reconstituted lymphopenic mice deficient in recombination-activating gene 1 with CD4+CD44hiCD62LloCD25− naive T cells from control littermate mice or E4bp4−/− mice and monitored the recipient mice for the development of inflammatory disease. Mice reconstituted with control T cells gained weight for the first 2 weeks but then began to gradually lose weight (Fig. 8b). In contrast, mice reconstituted with E4bp4−/− T cells gained less weight and had more severe weight loss and a worse clinical score, including death (two of ten mice; Fig. 8b). Five of the ten mice reconstituted with E4bp4−/− T cells began to have diarrhea after 4 weeks (data not shown). We confirmed the clinical symptoms by evaluating colonic inflammation morphologically and by histology (Fig. 8c) and also found that E4bp4−/− T cells derived from intestinal lymph nodes and colon did not express IL-10 (Fig. 8d). Thus, regulation of IL-10 in T cells by E4BP4 is important for tolerance induction to prevent the development of chronic inflammatory disease in the colon.
Figure 8.
Exacerbated colitis and EAE in E4bp4−/− mice. (a) Flow cytometry analysis of the expression of human CD2 (as a marker of Foxp3) and CD4 (left), and ELISA of IL-10 (right) in Foxp3+ Treg cells induced from naive T cells from Foxp3hCD2 reporter mice on the E4bp4−/− or wild-type background by culture with transforming growth factor-β. (b) Body weight of mice deficient in recombination-activating gene 1 (n = 10 per group) given intravenous transfer of naive CD4+ T cells (4 × 105) from the spleens of E4bp4−/− mice (n = 10) or wild-type mice (n = 10), presented relative to initial body weight on day 0. † indicates death (two of ten mice died). (c) Hematoxylin and eosin staining of colon tissue sections at day 34 in the recipient mice in b. Scale bars, 200 µm (left) or 50 µm (right). (d) ELISA of IL-10 in CD4+ T cells isolated at day 34 from the lymph nodes (LN) and colons of the recipient mice in b, then stimulated for 48 h with mAb to TCRβ and mAb to CD28. (e) EAE clinical scores of E4bp4−/− mice (n = 5) and wild-type mice (n = 5) immunized at day 0 with a peptide of myelin oligodendrocyte glycoprotein (amino acids 35–55) in complete Freund’s adjuvant. (f) ELISA of cytokines in CD4+ T cells isolated at day 35 from the lymph nodes of mice immunized as in e, then stimulated for 48 h with mAb to TCRβ and mAb to CD28. *P < 0.01 (Student’s t-test). Data are from three (a,c,d,f) or ten (b) independent experiments (mean and s.e.m. in a,b,d–f).
We further examined the importance of the regulation of IL-10 by E4BP4 in an experimental autoimmune encephalomyelitis (EAE) model. We immunized E4bp4−/− mice and their control littermates with a peptide of myelin oligodendrocyte glycoprotein (amino acids 35–55) emulsified in complete Freund’s adjuvant. Control and E4bp4−/− mice showed notable EAE development that reached a maximum at 15 d (Fig. 8e). Control mice began to recover, showing improvement in their clinical scores after 25 d. In contrast, E4bp4−/− mice showed no evidence of recovery and had worse EAE scores after 30 d (Fig. 8e). E4bp4−/− T cells from the regional lymph node expressed no IL-10 and had a slightly more IL-17 production (Fig. 8f). We further monitored Il10 transcription in mice with ongoing disease using a reporter system in which sequence encoding the yellow fluorescent protein Venus was knocked into the 3′ untranslated region of the Il10 locus41. Fewer E4bp4−/− T cells from the spleen, among intestinal intraepithelial lymphocytes or from the lamina propria or bone marrow produced IL-10 than did those from control mice (Supplementary Fig. 10). Therefore, E4BP4 induction of IL-10 is also important for EAE remission.
DISCUSSION
We have demonstrated here that chronic antigen stimulation resulted in plasticity of TH1 cells, which then secreted certain TH2 cytokines, including IL-10 and IL-13, but not IL-4 or IL-5. Acquisition by TH1 cells of the ability to express IL-10 and IL-13 was regulated by E4BP4, whose own expression was upregulated by chronic antigenic stimulation. E4BP4 also regulated the production of IL-10 and IL-13 in NKT cells, memory T cells and Treg cells. In contrast, Il13 regulation in TH2 cells was totally independent of E4BP4, which indicated that E4BP4 regulates Il13 differently in distinct CD4+ T cell subsets but commonly regulates Il10 in all CD4+ T cells. Therefore, we propose that E4BP4 has multiple functions in the regulation of genes encoding cytokines and is a transcriptional factor essential to the regulation of IL-10 production. Our results provide the new insight that different TH2 cytokines are regulated independently by a distinct combination of transcription factors in different T cell subsets.
During the past two decades, it has become widely accepted that Il10 and Il13 are controlled together with Il4 by common transcriptional mechanisms. This idea has been supported by many observations; for example, that upregulation of Gata3 expression is essential for the expression of IL-4 and IL-13 in TH2 cells42, and that conditional deletion of Gata3 in activated CD4+ T cells attenuates the production of both cytokines22. However, we found no defect in IL-13-induced airway hyper-responsiveness in Il4−/− mice, which completely lack TH2 cells, which indicated that TH2 cells are not the source of IL-13 under certain conditions43,44. The source of IL-13 in Il4−/− mice has thus far remained unknown, but our study has shown that because of their plasticity, TH1 cells are an alternative source of this cytokine. In terms of IL-10, its production by TH1 cells has been reported after chronic infection with T. gondii or L. major13,15. IL-10 production is thus induced by relatively strong chronic stimulation, as in parasite infection, and this is consistent with our data showing that chronic antigen stimulation induced the expression of E4BP4, which controls the production of IL-10 and IL-13 by TH1 cells.
E4bp4 is expressed mainly by TH2 cells29,30, and we have demonstrated that E4bp4 expression was controlled by the IL-4–STAT6 pathway. However, the question of how chronic TH1 stimulation induces E4bp4 expression remains. A published report has indicated that calcium signaling–mediated activation of the transcription factor NFAT induces E4bp4 expression45, and Nfat5 was one of the genes we found was induced by repetitive stimulation, although its overexpression did not induce E4bp4 expression. None of the 15 other candidate genes that showed differences in upregulation in R4 TH1 cells affected E4bp4 expression; therefore, further investigation is need to clarify how chronic TH1 stimulation induces E4bp4 expression.
E4BP4 was originally identified as a negative regulator of the mammalian circadian oscillatory system23. E4bp4−/− mice were born at normal Mendelian ratios and grew normally, which indicated that deficiency in E4BP4 has no severe phenotypic consequences except the spontaneous development of mild diarrhea. In the absence of E4BP4, the phenotype of CD4+ T cells, including NKT cells, was fairly normal, consistent with the published observation that deletion of E4bp4 affects mainly NK cell development with no change in the proportion of other T cell lineages26,27. T cells lacking E4bp4 had normal TH1, TH2, induced Treg and TH17 differentiation, although the proportion of TH1 cells and TH2 cells was slightly lower than that in wild-type mice. E4bp4-deficient TH2 cells were considerably impaired in IL-10 production but not in GATA-3 expression or the production of other TH2 cytokines such as IL-4 and IL-5, and IL-13 production was even enhanced in E4bp4−/− TH2 cells.
GATA-3 directly regulates the Il13 promoter20 and, furthermore, conditional deletion of Gata3 in TH2-committed T cells completely attenuates Il13 expression22,31, which suggests that GATA-3 is indispensable for Il13 transcription in TH2 cells but E4BP4 is not. In contrast, E4BP4 was absolutely required for IL-13 expression in TH1 cells. The distal Il13 promoter, CGRE, 1.4–1.8 kilobases upstream of exon 1, contains 42 CpG dinucleotides46, and analysis of DNA dimethylation and methylation of histone H3 at Lys4 indicated that the CGRE is transcriptionally accessible in naive T cells, TH1 cells and TH2 cells. Reporter assays indicated the activity of this promoter is regulated by E4BP4. Therefore, IL-13 production by TH1 cells seems to be determined by the regulation of the promoter at the level of E4BP4 expression rather than by epigenetic regulation.
We have demonstrated that E4BP4 was also essential for IL-10 expression in most of the CD4+ subsets we examined. The precise mechanism by which IL-10 expression is regulated in T cells has remained unclear, although a contribution by GATA-3 has been demonstrated by retroviral overexpression experiments21. Our data indicated that E4BP4 induced IL-10 production in the absence of GATA-3, which suggests that GATA-3 is dispensable for IL-10 production. Therefore, E4BP4 seems to be an essential regulator of IL-10 expression not only in TH2 cells but also in TH1 cells, which typically have very low expression of GATA-3. However, unlike its binding to Il13, E4BP4 did not bind to the Il10 promoter region, binding instead to an intronic region and a 3′ noncoding sequence, both of which are well conserved between humans and mice. TH2 cells lacking E4BP4 had no permissive chromatin structure in these regions, which suggests that Il10 is epigenetically regulated by E4BP4 binding. However, T cells are not the only source of IL-10, and IL-10 is reported to be secreted by NK cells, regulatory B cells and regulatory DCs. Indeed, regulatory DCs from the lamina propria of E4bp4−/− mice had lower expression of IL-10 (data not shown), which suggests that E4BP4 may function as a common transcriptional regulator for IL-10 production in a variety of cell types.
ONLINE METHODS
Mice
DO11.10 mice on the BALB/c background were provided by K. Murphy; and Stat6−/− mice, Il4r−/− mice and Cd4-Cre mice were gifts from S. Akira47, F. Brombacher48 and C. Wilson, respectively. Foxp3hCD2 reporter mice37 and Venus reporter mice41 were maintained on the C57BL/6 background. Mice were kept in specific pathogen–free conditions in accordance with the guidelines of the Institutional Animal Care Committee of the RIKEN Institute.
For the generation of E4bp4-transgenic mice, cDNA encoding a Flag-tagged E4BP4 fusion protein was inserted downstream of the Eµ enhancer of the immunoglobulin heavy-chain locus and was injected into fertilized mouse eggs. Three independent transgenic mouse lines (1, 2 and 3) were selected for study on the basis of intracellular staining with anti-Flag (M-2; Sigma). The mice were backcrossed to C57BL/6 mice for more than ten generations.
E4bp4-deficient mice were generated by homologous recombination and replacement of E4bp4, in 129/C57BL/6 hybrid embryonic stem cells, with a loxP-flanked cassette (RIKEN Center for Developmental Biology) containing the coding sequence of mouse E4bp4 (Supplementary Fig. 6). A neomycin-resistance expression cassette flanked by flippase recognition target sites was located 3′ of the targeting site. After germline deletion of those elements by crossing of those mice with mice expressing Cre recombinase (CAGcre-transgenic mice)49, the resulting mice were backcrossed with C57BL/6 mice for five generations.
Cell preparation and reagents
CD4+ T cells were isolated from spleen with the IMag magnetic bead system (BD Biosciences) and naive (CD44lo CD62Lhi) cells were isolated by cell sorting with a FACSVantage (BD Biosciences). T cells (1 × 106 cells per ml) were stimulated with plate-bound mAb to TCRβ (H57-597) and mAb to CD28 (PV-1). For DO11.10 CD4+ T cells, cells were stimulated with the antigenic peptide Loh15 (1 µg/ml) in the presence of irradiated spleen cells (1 × 107 cells per ml). For the induction of TH2 cells, cells were stimulated in the presence of mouse IL-4 (100 U/ml) and mAb to IFN-γ (R46A2). For TH1 polarization, cells were cultured in the presence of mouse IL-12 and mAb to IL-4 (11B11). For TH17 and Treg induction, cells were cultured with recombinant human transforming growth factor-β (10 ng/ml) with or without recombinant mouse IL-6 (4 ng/ml) in the presence of mAb to IL-4 (11B11) and mAb to IFN-γ (R46A2). For the preparation of NKT cells and Treg cells, cells were sorted with biotinylated mAb to IL-17RB (3H8)36, phycoerythrin-conjugated mAb to CD25 (PC61.5; 12-0251; eBioscience), biotinylated mAb to human CD2 (RPA2.10; 300203; BioLegend) and allophycocyanin-conjugated mAb to CD44 (1M7; 559250; BD Biosciences).
Human CD4+ T cells were isolated from peripheral blood mononuclear cells with IMag beads, and cells were stimulated for 7 d with mAb to CD3 (OKT3; 14-0037-82; eBioscience) and mAb to CD28 (CD28.2; 555726; BD Biosciences) in the presence of human IL-12 (5 ng/ml) and mAb to IL-4 (8D48; 556917; BD Biosciences) for the induction of TH1 cells.
Measurement of cytokine production
T cells were stimulated for 24 h with plate-bound mAb to TCR plus mAb to CD28. The concentrations of IL-4, IL-5, IL-13 and IFN-γ in culture supernatants were measured by ELISA50; mAb to IFN-γ (R4-6A2; 551216; BD Biosciences), mAb to IL-4 mAb(BVD4-1D11; 554387; BD Biosciences), mAb to IL-5 (JES6-1A12; 554393; BD Biosciences) and mAb to IL-13 (DY413; 38213; R&D Systems) were used for captured, and mAb to IFN-γ (XMG1.2; 554410; BD Biosciences), mAb to IL-4 (BVD6-24G2; 13–7042; eBioscience), mAb to IL-5 (JES6-5H4; 554397; BD Biosciences) and mAb to IL-13 (DY413) were used for detection. Cytokines were measured by a suspension array system (Luminex 200; Bio-Rad Laboratories).
Intracellular cytokine staining and confocal microscopy
CD4+ T cells were restimulated for 6 h with mAb to TCRβ in the presence of 2 µM monensin. Cells were fixed with 4% (vol/vol) paraformaldehyde and were made permeable with 0.5% (vol/vol) Triton X-100. After blocking, cells were stained with the following: fluorescein isothiocyanate–conjugated mAb to IFN-γ (XMG1.2; 554410; BD Biosciences), allophycocyanin-conjugated mAb to IL-4 (11B11; 554436; BD Biosciences) and Alexa Fluor 647–conjugated mAb to IL-13 (eBio-13A; 51–7133; eBioscience). GATA-3 was detected with mAb to GATA-3 (L50-823; 560163; BD Biosciences). A FACSCalibur and CellQuest software (BD Biosciences) were used for flow cytometry. For confocal microscopy, cells were made permeable as described above and then stained with rabbit anti-E4BP4 (H-300; sc-28203; Santa Cruz) and fluorescein isothiocyanate–conjugated rat antibody to rabbit IgG (111-095-003; Jackson Laboratories). Fixed cells were imaged with an LSM 510 confocal microscopy system (Leica Microsystems).
Microarray analysis and quantitative RT-PCR
Total RNA was isolated with TRIzol (Invitrogen). An Affymetrix microarray chip (Mouse Genome 430 2.0 array) was used for microarray analysis, and results were normalized with Genespring software (Silicon Genetics). A 7500 real-time PCR system (Applied Biosystems) was used for quantitative real-time PCR (primers, Supplementary Table 1).
Retroviral transduction
The retroviral vector pMX-IRES-GFP was transfected into PLAT-E packaging cells (a gift from T. Kitamura) with FuGENE 6 (Roche), and culture supernatants were used as a source of viral particles. Naive CD4+ T cells were stimulated with mAb to TCR and mAb to CD28. Viral particles were transduced into the purified CD4+ T cells, and GFP+ cells were sorted after 2 d.
Generation of shRNA for knockdown for E4bp4
For generation of the E4bp4-specific shRNA plasmid construct, the targeting oligonucleotide 5′-GTTGCATCTCAGTCATCAAGC-3′ was cloned into the pSIREN-RetroQ vector (Clontech); shRNA specific for luciferase was used as control. Each shRNA was introduced into activated T cells by retroviral transduction, and transduced cells were selected with puromycin (2 ng/ml).
Bisulfite sequencing PCR assay
DNA methylation was analyzed with a bisulfite-sequencing PCR assay kit (Chemicon International). Genomic DNA was chemically modified by treatment with sodium bisulfite, then modified DNA was amplified with specific primers by PCR and DNA methylation was detected by sequencing.
ChIP assay
Nuclear fractions were extracted as chromatin from sonicated cells. After preclearance with protein G agarose (Amersham), chromatin fractions were incubated with the appropriate antibody. Input samples were defined as DNA extracts prepared from untreated chromatin. DNA was extracted from immunoprecipitated chromatin, and concentrations were quantified with a PicoGreen fluorescence assay (Molecular Probes). Equivalent masses of immunoprecipitated and input DNA were compared by real-time PCR (primers, Supplementary Table 1), with results presented as the ratio of the change in threshold value after immunoprecipitation to value to the change in threshold value of input DNA.
Electrophoretic mobility-shift assay
Nuclear proteins were incubated for 30 min with 32P-labeled oligonucleotide probes. Samples were separated by 4% PAGE in a buffer of low ionic strength. Shifts in mobility were visualized with a BAS-2500 system (oligonucleotide probes, Supplementary Table 1).
Colitis model
CD4+ T cells were prepared from the spleen with IMag magnetic beads, and CD44loCD25− naive cells were isolated by cell sorting. Cells (4 × 105) were transferred intravenously into mice deficient in recombination-activating gene 1, and body weight and symptoms of clinical disease were monitored. Mice were killed after 34 d. Colons were fixed with 4% (vol/vol) paraformaldehyde and sections were stained with hematoxylin and eosin.
EAE induction
Mice were immunized subcutaneously on day 0 with a peptide of myelin oligodendrocyte glycoprotein amino acids 35–55 emulsified in complete Freund’s adjuvant (100 µg per mouse), and were given intravenous injection of pertussis toxin (200 ng per mouse; List Biological Laboratories) on days 0 and 2. EAE was assessed as follows: 0, no disease; 1, limp tail; 2, weak hindlimbs; 3, partially paralyzed hindlimbs; 4, complete hindlimb paralysis; and 5, death.
Supplementary Material
Acknowledgments
We thank K. Murphy (Washington University) for DO11.10 mice on the BALB/c background; S. Akira (Osaka University) for Stat6−/− mice; F. Brombacher (University of Cape Town) for Il4r−/− mice; C. Wilson (Washington University) for Cd4-Cre mice; R. Abe (Tokyo University of Science) for the PV-1 mAb to CD28; T. Kitamura (Tokyo University) for Platinum-E packaging cells; H. Fujimoto, Y. Suzuki, K. Ikari and E. Hayashi for technical assistance; and P. Burrows for comments on the manuscript. Supported by RIKEN (Y. Motomura), the Ministry of Education, Culture, Sports, Science and Technology of Japan, and the Program for Promotion of Fundamental Studies in Health Sciences of the National Institute of Biomedical Innovation.
Footnotes
Note: Supplementary information is available on the Nature Immunology website.
AUTHOR CONTRIBUTIONS
Y. Motomura built the initial constructs, generated mouse lines and confirmed them in vivo, and did most of the experiments; H.K. and A.H. did microarray and bioinformatics analysis; Y. Matsunaga, K.M. and H.I. did airway hyper-responsiveness experiments; K.A. provided Il10 reporter mice; S.H. analyzed Treg cells; H.W. and M.T. analyzed NKT cells; J.Z. provided materials; and M.K. designed and coordinated experiments, supervised the project and wrote the paper.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Murphy KM, Reiner SL. The lineage decisions of helper T cells. Nat. Rev. Immunol. 2002;2:933–944. doi: 10.1038/nri954. [DOI] [PubMed] [Google Scholar]
- 2.Dong C. TH17 cells in development: an updated view of their molecular identity and genetic programming. Nat. Rev. Immunol. 2008;8:337–348. doi: 10.1038/nri2295. [DOI] [PubMed] [Google Scholar]
- 3.Iwakura Y, Nakae S, Saijo S, Ishigame H. The roles of IL-17A in inflammatory immune responses and host defense against pathogens. Immunol. Rev. 2008;226:57–79. doi: 10.1111/j.1600-065X.2008.00699.x. [DOI] [PubMed] [Google Scholar]
- 4.McKenzie AN, et al. Interleukin 13, a T-cell-derived cytokine that regulates human monocyte and B-cell function. Proc. Natl. Acad. Sci. USA. 1993;90:3735–3739. doi: 10.1073/pnas.90.8.3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wynn TA. IL-13 effector functions. Annu. Rev. Immunol. 2003;21:425–456. doi: 10.1146/annurev.immunol.21.120601.141142. [DOI] [PubMed] [Google Scholar]
- 6.Lee GR, Fields PE, Griffin TJ, Flavell RA. Regulation of the Th2 cytokine locus by a locus control region. Immunity. 2003;19:145–153. doi: 10.1016/s1074-7613(03)00179-1. [DOI] [PubMed] [Google Scholar]
- 7.Wilson CB, Rowell E, Sekimata M. Epigenetic control of T-helper-cell differentiation. Nat. Rev. Immunol. 2009;9:91–105. doi: 10.1038/nri2487. [DOI] [PubMed] [Google Scholar]
- 8.Hayashi N, et al. T helper 1 cells stimulated with ovalbumin and IL-18 induce airway hyperresponsiveness and lung fibrosis by IFN-γ and IL-13 production. Proc. Natl. Acad. Sci. USA. 2007;104:14765–14770. doi: 10.1073/pnas.0706378104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li L, Xia Y, Nguyen A, Feng L, Lo D. Th2-induced eotaxin expression and eosinophilia coexist with Th1 responses at the effector stage of lung inflammation. J. Immunol. 1998;161:3128–3135. [PubMed] [Google Scholar]
- 10.Smart JM, Kemp AS. Increased Th1 and Th2 allergen-induced cytokine responses in children with atopic disease. Clin. Exp. Allergy. 2002;32:796–802. doi: 10.1046/j.1365-2222.2002.01391.x. [DOI] [PubMed] [Google Scholar]
- 11.Saraiva M, O’Garra A. The regulation of IL-10 production by immune cells. Nat. Rev. Immunol. 2010;10:170–181. doi: 10.1038/nri2711. [DOI] [PubMed] [Google Scholar]
- 12.Batten M, et al. Cutting edge: IL-27 is a potent inducer of IL-10 but not FoxP3 in murine T cells. J. Immunol. 2008;180:2752–2756. doi: 10.4049/jimmunol.180.5.2752. [DOI] [PubMed] [Google Scholar]
- 13.Jankovic D, et al. Conventional T-bet+Foxp3− Th1 cells are the major source of host-protective regulatory IL-10 during intracellular protozoan infection. J. Exp. Med. 2007;204:273–283. doi: 10.1084/jem.20062175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shaw MH, et al. Tyk2 negatively regulates adaptive Th1 immunity by mediating IL-10 signaling and promoting IFN-γ-dependent IL-10 reactivation. J. Immunol. 2006;176:7263–7271. doi: 10.4049/jimmunol.176.12.7263. [DOI] [PubMed] [Google Scholar]
- 15.Anderson CF, Oukka M, Kuchroo VJ, Sacks D. CD4+CD25−Foxp3− Th1 cells are the source of IL-10-mediated immune suppression in chronic cutaneous leishmaniasis. J. Exp. Med. 2007;204:285–297. doi: 10.1084/jem.20061886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saraiva M, et al. Interleukin-10 production by Th1 cells requires interleukin-12-induced STAT4 transcription factor and ERK MAP kinase activation by high antigen dose. Immunity. 2009;31:209–219. doi: 10.1016/j.immuni.2009.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuhn R, Lohler J, Rennick D, Rajewsky K, Muller W. Interleukin-10-deficient mice develop chronic enterocolitis. Cell. 1993;75:263–274. doi: 10.1016/0092-8674(93)80068-p. [DOI] [PubMed] [Google Scholar]
- 18.Rubtsov YP, et al. Regulatory T cell-derived interleukin-10 limits inflammation at environmental interfaces. Immunity. 2008;28:546–558. doi: 10.1016/j.immuni.2008.02.017. [DOI] [PubMed] [Google Scholar]
- 19.O’Shea JJ, Paul WE. Mechanisms underlying lineage commitment and plasticity of helper CD4+ T cells. Science. 2010;327:1098–1102. doi: 10.1126/science.1178334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kishikawa H, Sun J, Choi A, Miaw SC, Ho IC. The cell type-specific expression of the murine IL-13 gene is regulated by GATA-3. J. Immunol. 2001;167:4414–4420. doi: 10.4049/jimmunol.167.8.4414. [DOI] [PubMed] [Google Scholar]
- 21.Shoemaker J, Saraiva M, O’Garra A. GATA-3 directly remodels the IL-10 locus independently of IL-4 in CD4+ T cells. J. Immunol. 2006;176:3470–3479. doi: 10.4049/jimmunol.176.6.3470. [DOI] [PubMed] [Google Scholar]
- 22.Zhu J, et al. Conditional deletion of GATA-3 shows its essential function in TH1-TH2 responses. Nat. Immunol. 2004;5:1157–1165. doi: 10.1038/ni1128. [DOI] [PubMed] [Google Scholar]
- 23.Cowell IG. E4BP4/NFIL3, a PAR-related bZIP factor with many roles. Bioessays. 2002;24:1023–1029. doi: 10.1002/bies.10176. [DOI] [PubMed] [Google Scholar]
- 24.Zhang W, et al. Molecular cloning and characterization of NF-IL3A, a transcriptional activator of the human interleukin-3 promoter. Mol. Cell. Biol. 1995;15:6055–6063. doi: 10.1128/mcb.15.11.6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ikushima S, et al. Pivotal role for the NFIL3/E4BP4 transcription factor in interleukin 3-mediated survival of pro-B lymphocytes. Proc. Natl. Acad. Sci. USA. 1997;94:2609–2614. doi: 10.1073/pnas.94.6.2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gascoyne DM, et al. The basic leucine zipper transcription factor E4BP4 is essential for natural killer cell development. Nat. Immunol. 2009;10:1118–1124. doi: 10.1038/ni.1787. [DOI] [PubMed] [Google Scholar]
- 27.Kamizono S, et al. Nfil3/E4bp4 is required for the development and maturation of NK cells in vivo. J. Exp. Med. 2009;206:2977–2986. doi: 10.1084/jem.20092176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kashiwada M, et al. IL-4-induced transcription factor NFIL3/E4BP4 controls IgE class switching. Proc. Natl. Acad. Sci. USA. 2010;107:821–826. doi: 10.1073/pnas.0909235107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lund R, Aittokallio T, Nevalainen O, Lahesmaa R. Identification of novel genes regulated by IL-12, IL-4, or TGF-β during the early polarization of CD4+ lymphocytes. J. Immunol. 2003;171:5328–5336. doi: 10.4049/jimmunol.171.10.5328. [DOI] [PubMed] [Google Scholar]
- 30.Lund R, et al. Identification of genes involved in the initiation of human Th1 or Th2 cell commitment. Eur. J. Immunol. 2005;35:3307–3319. doi: 10.1002/eji.200526079. [DOI] [PubMed] [Google Scholar]
- 31.Pai SY, Truitt ML, Ho IC. GATA-3 deficiency abrogates the development and maintenance of T helper type 2 cells. Proc. Natl. Acad. Sci. USA. 2004;101:1993–1998. doi: 10.1073/pnas.0308697100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hata H, Yoshimoto T, Hayashi N, Hada T, Nakanishi K. IL-18 together with anti-CD3 antibody induces human Th1 cells to produce Th1- and Th2-cytokines and IL-8. Int. Immunol. 2004;16:1733–1739. doi: 10.1093/intimm/dxh174. [DOI] [PubMed] [Google Scholar]
- 33.Sugimoto T, et al. Interleukin 18 acts on memory T helper cells type 1 to induce airway inflammation and hyperresponsiveness in a naive host mouse. J. Exp. Med. 2004;199:535–545. doi: 10.1084/jem.20031368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stumhofer JS, et al. Interleukins 27 and 6 induce STAT3-mediated T cell production of interleukin 10. Nat. Immunol. 2007;8:1363–1371. doi: 10.1038/ni1537. [DOI] [PubMed] [Google Scholar]
- 35.McGeachy MJ, et al. TGF-β and IL-6 drive the production of IL-17 and IL-10 by T cells and restrain TH-17 cell-mediated pathology. Nat. Immunol. 2007;8:1390–1397. doi: 10.1038/ni1539. [DOI] [PubMed] [Google Scholar]
- 36.Terashima A, et al. A novel subset of mouse NKT cells bearing the IL-17 receptor B responds to IL-25 and contributes to airway hyperreactivity. J. Exp. Med. 2008;205:2727–2733. doi: 10.1084/jem.20080698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Komatsu N, et al. Heterogeneity of natural Foxp3+ T cells: a committed regulatory T-cell lineage and an uncommitted minor population retaining plasticity. Proc. Natl. Acad. Sci. USA. 2009;106:1903–1908. doi: 10.1073/pnas.0811556106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koyanagi M, et al. EZH2 and histone 3 trimethyl lysine 27 associated with Il4 and Il13 gene silencing in Th1 cells. J. Biol. Chem. 2005;280:31470–31477. doi: 10.1074/jbc.M504766200. [DOI] [PubMed] [Google Scholar]
- 39.Tanaka S, et al. The enhancer HS2 critically regulates GATA-3-mediated Il4 transcription in TH2 cells. Nat. Immunol. 2011;12:77–85. doi: 10.1038/ni.1966. [DOI] [PubMed] [Google Scholar]
- 40.Roers A, et al. T cell-specific inactivation of the interleukin 10 gene in mice results in enhanced T cell responses but normal innate responses to lipopolysaccharide or skin irritation. J. Exp. Med. 2004;200:1289–1297. doi: 10.1084/jem.20041789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Atarashi K, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zheng W, Flavell RA. The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell. 1997;89:587–596. doi: 10.1016/s0092-8674(00)80240-8. [DOI] [PubMed] [Google Scholar]
- 43.Grunig G, et al. Requirement for IL-13 independently of IL-4 in experimental asthma. Science. 1998;282:2261–2263. doi: 10.1126/science.282.5397.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Walter DM, et al. Critical role for IL-13 in the development of allergen-induced airway hyperreactivity. J. Immunol. 2001;167:4668–4675. doi: 10.4049/jimmunol.167.8.4668. [DOI] [PubMed] [Google Scholar]
- 45.Nishimura Y, Tanaka T. Calcium-dependent activation of nuclear factor regulated by interleukin 3/adenovirus E4 promoter-binding protein gene expression by calcineurin/nuclear factor of activated T cells and calcium/calmodulin-dependent protein kinase signaling. J. Biol. Chem. 2001;276:19921–19928. doi: 10.1074/jbc.M010332200. [DOI] [PubMed] [Google Scholar]
- 46.Smirnov DV, Smirnova MG, Korobko VG, Frolova EI. Tandem arrangement of human genes for interleukin-4 and interleukin-13: resemblance in their organization. Gene. 1995;155:277–281. doi: 10.1016/0378-1119(94)00720-d. [DOI] [PubMed] [Google Scholar]
- 47.Takeda K, et al. Essential role of Stat6 in IL-4 signalling. Nature. 1996;380:627–630. doi: 10.1038/380627a0. [DOI] [PubMed] [Google Scholar]
- 48.Mohrs M, et al. Differences between IL-4- and IL-4 receptor α-deficient mice in chronic leishmaniasis reveal a protective role for IL-13 receptor signaling. J. Immunol. 1999;162:7302–7308. [PubMed] [Google Scholar]
- 49.Sakai K, Miyazaki J. A transgenic mouse line that retains Cre recombinase activity in mature oocytes irrespective of the cre transgene transmission. Biochem. Biophys. Res. Commun. 1997;237:318–324. doi: 10.1006/bbrc.1997.7111. [DOI] [PubMed] [Google Scholar]
- 50.Yagi R, et al. The IL-4 production capability of different strains of naive CD4+ T cells controls the direction of the TH cell response. Int. Immunol. 2002;14:1–11. doi: 10.1093/intimm/14.1.1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.