Abstract
To confer abscisic acid (ABA) and/or stress-inducible gene expression, an ABA-response complex (ABRC1) from the barley (Hordeum vulgare L.) HVA22 gene was fused to four different lengths of the 5′ region from the rice (Oryza sativa L.) Act1 gene. Transient assay of β-glucuronidase (GUS) activity in barley aleurone cells shows that, coupled with ABRC1, the shortest minimal promoter (Act1–100P) gives both the greatest induction and the highest level of absolute activity following ABA treatment. Two plasmids with one or four copies of ABRC1 combined with the same Act1–100P and HVA22(I) of barley HVA22 were constructed and used for stable expression of uidA in transgenic rice plants. Three Southern blot-positive lines with the correct hybridization pattern for each construct were obtained. Northern analysis indicated that uidA expression is induced by ABA, water-deficit, and NaCl treatments. GUS activity assays in the transgenic plants confirmed that the induction of GUS activity varies from 3- to 8-fold with different treatments or in different rice tissues, and that transgenic rice plants harboring four copies of ABRC1 show 50% to 200% higher absolute GUS activity both before and after treatments than those with one copy of ABRC1.
Drought and high salinity are the most important environmental factors that cause osmotic stress and dramatically limit plant growth and crop productivity (Boyer, 1982). Therefore, production of drought- and NaCl-tolerant transgenic plants is very important for agriculture. In recent years different stress-tolerant transgenic plants have been obtained (Tarczynski et al., 1993; Kishor et al., 1995; Pilon-Smits et al., 1995; Holmström et al., 1996; Xu et al., 1996; Hayashi et al., 1997) by producing either a low-Mr osmoprotectant (such as Gly betaine, mannitol, inositol, Pro, fructan, or trehalose) or a late-embryogenesis-abundant protein. However, under normal environmental conditions, overproduction of these compounds or proteins need extra energy and building blocks and may hamper the normal growth of plants. Thus, it is desirable to generate transgenic plants that synthesize a high level of an osmoprotectant or a protein only under stress conditions.
The phytohormone ABA is thought to mediate physiological processes in response to osmotic stress in plants (King, 1976; Jones et al., 1987). Water stress by NaCl or dehydration can cause endogenous ABA levels to increase in plant tissues (Henson, 1984; Jones et al., 1987). Mundy and Chua (1988) found that ABA controls the accumulation of specific mRNAs and proteins, both from developmental studies with seeds and physiological studies with water-stressed tissues. Specific genes are expressed under stress conditions and can also be induced in unstressed tissues by the application of exogenous ABA (Singh et al., 1987; Gomez et al., 1988; Mundy and Chua, 1988; Chandler and Robertson, 1994; Ingram and Bartels, 1996).
In addition to the studies on the physiological roles of ABA, efforts are being made to investigate the molecular mechanism of ABA action, including the definition of ABREs, and the trans-acting factors that interact with ABREs. It was reported that a 75-bp fragment of the ABA-inducible wheat Em gene, when fused to a truncated CaMV 35S promoter, conferred a more than 10-fold ABA induction of GUS activity in rice (Oryza sativa L.) protoplasts (Guiltinan et al., 1990). They also found a Leu-zipper DNA-binding protein, EmBP-1, which binds the ABRE sequence (CACGTGGC) in this 75-bp region. Transient assays in rice protoplasts revealed a 40-bp ABA-responsive fragment in the rice rab 16B promoter (Ono et al., 1996). Two separate ABREs, motif I (AGTACGTGGC) and motif III (GCCGCGTGGC), are required for ABA induction; however, each can substitute for the other. The 40-bp fragment-containing motif I fused to a truncated CaMV 35S promoter showed an approximately 4- to 5-fold induction by ABA (Ono et al., 1996). The ABREs are very similar to the G-box, which, as has been pointed out by Guiltinan et al. (1990), is present in some genes that are responsive to other environmental and physiological stimuli such as light (Giuliano et al., 1988) and auxin (Liu et al., 1994).
Studies on the promoter of the barley ABA-responsive HVA22 gene indicate that G-box sequences are necessary but not sufficient for an ABA response (Shen and Ho, 1995). Instead, an ABA-responsive complex consisting of a G-box, namely, ABRE3 (GCCACGTACA), and a novel coupling element, CE1 (TGCCACCGG), is sufficient for high-level ABA induction. The results of linker-scan analyses and gain-of-function studies showed that the 49-bp ABRC1 is the minimal sequence governing high-level ABA induction. In addition, the HVA22(I) of the HVA22 gene is also required for high-level ABA induction of HVA22 expression. The results of transient assay of GUS activity in barley (Hordeum vulgare L.) aleurone cells showed that four copies of this 49-bp ABRC1, linked to a truncated (−60 to +57) barley α-amylase promoter (Amy64) and coupled with the HVA22(I), can cause more than 120-fold ABA-inducible uidA expression, whereas approximately 30-fold induction of uidA expression can be detected with one copy of ABRC1. A similar investigation on ABA induction of a barley late-embryogenesis-abundant gene, HVA 1 (Shen et al., 1996), was conducted, and it was found that the ABRC3 of this gene consists of a 10-bp element (CCTACGTGGC) with an ACGT core (A2) and a sequence directly upstream, named CE3 (ACGCGTGTCCTC). Only one copy of this ABRC3 is sufficient to confer ABA induction when fused to a minimal promoter (Amy64). Thus, two types of ABRCs were reported by Shen and Ho (1995) and Shen et al. (1996), namely, ABRC1 (used in this study), consisting of ABRE3 and CE1 from the HVA22 gene, and ABRC3, composed of CE3 and A2 from the HVA1 gene.
As mentioned above, most of the data regarding ABA-responsive expression were obtained from transient assays of GUS activity in suspension cells, protoplasts, or barley aleurone cells. In the present study we used one or four copies of ABRC1 from the barley HVA22 gene to confer ABA and/or stress-inducible uidA expression in transgenic rice plants. The ABA action conferred by the ABRC1-containing transgene in transgenic rice plants may be different from that of individual cells, such as barley aleurone cells, in a transient assay. Another goal for this research was to construct stress-inducible expression plasmids that can be used for subsequent production of stress-tolerant transgenic rice.
MATERIALS AND METHODS
Construction of Plasmids Containing (ABRC1)4 Sequences, Different Lengths of Truncated Act1 Promoters, HVA22(I), and uidA for Transient Assay of ABA-Induced GUS Activity in Barley Aleurone Cells
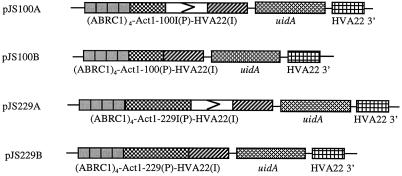
For ABA-inducible uidA expression, a minimal promoter is required in addition to ABRC1, HVA22(I) of the barley (Hordeum vulgare L.) HVA22 gene (Shen and Ho, 1995). To elucidate the relationship between ABA-inducible uidA expression and different lengths of minimal promoters, four fragments of the rice (Oryza sativa L.) Act1 promoter were isolated and tested as potential “minimal” promoters for transient assay of ABA-induced GUS activity in barley aleurone cells. A 789-bp Act1–229I fragment with the Act1 intron was isolated by HphI-EcoRI digestion from the plasmid pBY505 (Wang and Wu, 1995). The other three fragments (Act1–229, Act1–100I, and Act1–100) were isolated from the Act1–229I-derived intermediate plasmids (data not shown) by cutting the NruI and BstEII sites present in the Act1–229I fragment (McElroy et al., 1990) in combination with other restriction sites located in the intermediate plasmids. These four fragments of truncated Act1 promoters were used to replace the Amy64 promoter in the pQS120 plasmid (Shen and Ho, 1995), which also contained four copies of ABRC1 elements and one copy each of HVA22(I) of HVA 22, uidA, and the HVA22 3′ region, to create plasmids pJS229A, pJS229B, pJS100A, and pJS100B (Fig. 1). The four truncated Act1 promoters and all of the border regions between different functional elements were confirmed by sequence analysis. These four plasmids were used for transient assays of GUS activity in barley aleurone cells.
Figure 1.
Schematic diagram of plasmids used for transient assay of ABA-induced GUS activity in barley aleurone cells. For the construction of plasmid pJS100A, Act1–100I(P) was inserted into pQS120 by replacing the Amy64 promoter. Act1–100I(P) contains a truncated Act1 promoter (−100 to +560 including Act1 intron). Similarly, the construction of pJS100B started with Act1–100(P), which includes a truncated Act1 promoter (−100 to +80 without Act1 intron). The construction of pJS229A started with Act1–229I(P), which includes a truncated Act1 promoter (−229 to +560 including Act1 intron). The construction of pJS229B started with Act1–229(P), which includes a truncated Act1 promoter (−229 to +80 without Act1 intron).
Transient Assay of GUS Activity in Barley Aleurone Cells
Seeds of barley cv Himalaya (1988 harvest; Department of Agronomy and Soils, Washington State University, Pullman) were used. Preparations of embryoless half-seeds and aleurone cells, particle bombardment, homogenization of the bombarded seed, and GUS and luciferase assays were conducted essentially as described previously (Lanahan et al., 1992).
Test for Tissue Specificity and Histochemical Analysis
Leaves and roots from 10-d-old rice (cv Kenfong) seedlings grown in solid MS (Murashige and Skoog, 1962) medium were used as transformation materials and bombarded with tungsten particles coated with the pJS100B plasmid, essentially as described by Cao et al. (1992). The bombarded leaves and roots were transferred to fresh solid MS medium and cultured in a growth room (27°C with photoperiod of 12 h) for 2 d. Then the transformed leaves and roots were induced in liquid MS medium in the presence of 20 μm ABA for 20 h and subjected to histochemical staining with a solution containing 1 mm X-gluc and 50 mm sodium phosphate buffer (pH 7.0) as described by Jefferson et al. (1987).
Construction of Plasmids for Analyzing ABA- and/or Stress-Inducible uidA Expression in Transgenic Rice Plants
A previous report (Shen and Ho, 1995) indicated that four copies of ABRC1 confer ABA-responsive induction of uidA expression in barley aleurone cells four times higher than that with one copy. To compare the functional difference between one and four copies of ABRC1 in transgenic rice plants, we constructed two plasmids harboring either one or four copies of ABRC1. For construction of a plasmid containing one copy of ABRC1, the ABRC1 fragment from plasmid pJS115 (Shen and Ho, 1995) was isolated by EcoRI-XbaI digestion and subcloned into EcoRI-XbaI-digested pBluescript-KS(±). An Act1–100 promoter joined to the HVA22(I), which is abbreviated as Act1–100P-HVA22(I), was excised from pJS100B (see Table I) by BamHI digestion and subcloned at the BamHI site downstream of ABRC1 in pBluescript-KS(±) to produce the ABRC1-Act1–100P-HVA22(I) fragment.
Table I.
ABA-induced GUS activity in barley aleurone cells
| Constructs | Normalized Relative GUS
Activity
|
Induction | |
|---|---|---|---|
| −ABA | +ABA | ||
| -fold | |||
| pJS100A | 3101 ± 452 | 14829 ± 3229 | 5 |
| pJS100B | 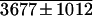
|
77685 ± 3320 | 21 |
| pJS229A | 24571 ± 1963 | 45023 ± 4680 | 2 |
| pJS229B | 5627 ± 423 | 37454 ± 3465 | 7 |
Normalized relative GUS activity was calculated based on luciferase activity (Lanahan et al., 1992). Each value represents the average of four independent analyses ± se. The maximum induction value is underlined.
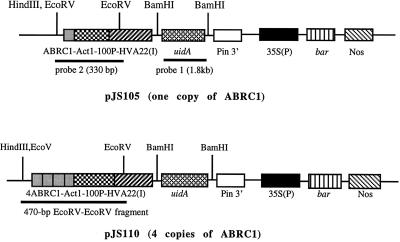
We chose Act1–100 as the minimal promoter (Act1–100P) because it was the best among the four fragments listed in Figure 1, as determined by transient assay of GUS activity in barley aleurone cells (see Results). The fragment ABRC1-Act1–100P-HVA22(I) was further cloned into the Act1 5′ region-deleted pBY505 to create the pJS104 plasmid, which contains ABRC1-Act1–100P-HVA22(I)/polylinker/Pin2 3′//CaMV 35S(P)/bar/Nos. The bar cassette, 35S(P)/bar/Nos, was used for the selection of rice transformants. By using the same procedure (except that four tandem copies of ABRC1 were isolated from pQS120 [Shen and Ho, 1995]), pJS109, which contained 4ABRC1-Act1–100P-HVA22(I)/polylinker/Pin2 3′//35S(P)/bar/Nos, was also constructed. Both plasmids pJS104 and pJS109 may serve as expression vectors for construction of the plasmids containing stress-tolerant genes. The GUS coding sequence (uidA) was cloned into the SmaI site of pJS104 and pJS109 to create pJS105 and pJS110, respectively (the components of the latter two plasmids are shown in Fig. 2). The plasmids pJS105 and pJS110 were used for transformation of rice and for testing ABA and/or stress-inducible uidA expression in the transgenic rice plants.
Figure 2.
Schematic diagram of plasmids pJS105 and pJS110. Each plasmid consists of two gene expression cassettes, the uidA cassette, in which uidA expression is regulated by the ABRC1-Act1–100P-HVA22(I) promoter complex and the potato Pin 2 3′ region, and the bar cassette, in which the bar gene is controlled by the CaMV 35S promoter and the Nos 3′ region, and serves as the selectable marker for transformation of rice. Only those restriction sites used for DNA digestion in DNA-blot hybridization are indicated. HindIII is a unique site in these two plasmids.
Production of Transgenic Rice Plants
Calli were induced in Linsmaier and Skoog medium (Linsmaier and Skoog, 1965) from mature rice cv Kenfong embryos. Suspension cultures were initiated from embryogenic calli in liquid AA medium (Cao et al., 1991). Fine suspension cells (subcultured for 3 d prior to bombardment) were bombarded with tungsten particles coated with either the pJS105 or the pJS110 plasmid, according to the procedure described by Cao et al. (1992). Resistant calli were selected in KPR medium (Zhang and Wu, 1988), supplemented with 8 mg L−1 Bialaphos as a selective agent, for 6 weeks (subcultured every 2 weeks). The resistant calli were transferred to MS regeneration medium containing 3 mg L−1 Bialaphos to regenerate into plants. Regenerated plants were transplanted into sterilized soil and grown in the greenhouse (32°C day/22°C night with a supplemental photoperiod of 10 h).
The presence of the transgenes in regenerated rice plants was first indicated by the herbicide resistance of the plants. To test herbicide resistance, leaves on 3-month-old transgenic rice plants were painted on both sides with 0.25% (v/v) of the herbicide Basta (containing 162 g L−1 glufosinate ammonium; Hoechst-Roussel Agri-Vet Co., Somerville, NJ) and 0.05% (v/v) Tween 20. One week later, the resistant or sensitive phenotypes were scored.
DNA-Blot Hybridization Analysis of Transgenic Rice Plants
Genomic DNA from transgenic rice plants was prepared as described by Zhao et al. (1989). Eight micrograms of genomic DNA was digested with restriction enzymes, electrophoresed through 0.8% (for uidA probe) and 1.2% (for probe 2 shown in Fig. 2) agarose gels, and transferred to nylon membranes (Nytran, Schleicher & Schuell). Probe preparation and hybridization were performed by following the manufacturer's instructions for nonradioactive DIG labeling and for the detection kit (Boehringer Mannheim).
RNA-Blot-Hybridization Analysis of Transgenic Rice Plants
Total RNA from leaves of R1 transgenic rice plants was isolated as described by Hihara et al. (1996). Five micrograms of total RNA from the transgenic rice was subjected to electrophoresis in a 1.0% formaldehyde agarose gel. After electrophoresis, RNA was transferred to a nylon membrane (Boehringer Mannheim). The 1.8-kb GUS coding region was used as a probe and labeled with [α-32P]dCTP using a random primer DNA-labeling kit (GIBCO-BRL). Gel preparation, hybridization, and washing were carried out as described by Sambrook et al. (1989).
ABA, Water Deficit, and NaCl Treatments of Transgenic Rice
For ABA treatment, seedlings of R1 plants were used. Rice embryos of R1 mature seeds both from transgenic and nontransgenic plants were germinated in solid one-half-strength MS medium and cultured in a growth room for 5 weeks for RNA-blot hybridization or for 2 weeks for assaying ABA-induced GUS activity. Then the 5- or 2-week-old R1 plants were transferred to liquid one-half-strength MS medium containing 50 μm ABA for 20 h in the growth room. For stress treatments (water deficit and NaCl), R1 plants grown in soil were used. R1 seeds were first germinated in one-half-strength MS medium for 7 d, and were then transplanted into soil in pots (8 × 8 in) with holes in the bottom. The pots were kept in flat-bottomed trays containing water. The seedlings were grown for an additional 7 weeks before they were exposed to stress conditions. To induce water deficit, water was withheld from the trays for up to 8 d. The absolute water content of the soil during the stress period and before treatment was determined. Nonstressed plants were supplied with water continuously from the trays. For NaCl treatment, water containing 150 mm NaCl solution was used to water 8-week-old plants, including nontransgenic plants. Leaves and roots were collected from the same plant after different periods of stress treatments and used for assaying stress-induced GUS activity.
Quantitative Assay of GUS Activity in Transgenic Rice Plants
To detect the GUS activity in R0 plants before treatment and in R1 transgenic rice plants after treatment with ABA, water deficit, and NaCl, a quantitative assay of GUS activity was carried out as described by Jefferson et al. (1987). Different leaves (adjacent) or roots from the same R1 plant of each line were collected before treatment or at the different stages of treatments: 20 h for ABA treatment; 4, 6, and 8 d for water stress; and 48, 72, and 96 h for NaCl treatment. Control experiments in parallel to ABA and NaCl treatments in the absence of ABA or NaCl were also performed to test for possible injury effect on GUS activity. Collected leaves or roots were frozen immediately in liquid N2 and homogenized in extraction buffer (50 mm phosphate buffer, pH 7.0, 10 mm EDTA, 0.1% Triton X-100, 0.1% Sarkosyl, 10 mm β-mercaptoethanol, and 25 μg mL−1 PMSF). After centrifugation (12,000 rpm for 15 min at 4°C) the crude extract, containing 20 μg of protein from leaves or roots, was directly used for spectrofluorometric assay. Protein concentration of the crude extract was determined by the dye-binding method of Bradford (1976) with a protein assay reagent (Bio-Rad).
RESULTS
The Shortest Truncated Act1 Promoter (Act1–100P) Confers the Highest ABA Induction in Barley Aleurone Cells
To get ABA- and stress-inducible gene expression in transgenic rice plants, a truncated promoter (termed the “minimal promoter”) is required in addition to ABRC1 and HVA22(I) of the HVA 22 gene. Before stable transformation of rice, transient expression assay of ABA-induced GUS activity was first performed in barley aleurone cells by using four different lengths of truncated Act1 promoters as the minimal promoters. The results (Table I) indicated that the plasmid with the shortest promoter (Act1–100P) shows not only the highest induction (21-fold), but also the highest GUS activity after exogenous ABA application. The Act1 intron is not necessary for ABA-inducible uidA expression. In fact, it inhibits uidA expression when the HVA22 intron is also present in the plasmid (see Table I).
Tissue specificity of uidA expression driven by the ABA-responsive promoter complex (4ABRC1-Act1–100P-HVA22(I)) was also tested in this study. After histochemical analysis following ABA induction, blue spots were observed in the detached leaves and roots bombarded with plasmid pJS100B (data not shown). This result indicated a lack of tissue specificity for ABA-inducible uidA expression driven by the ABA-responsive promoter complex. According to the results mentioned above, Act1–100P was used as a minimal promoter for plasmid constructs suitable for stable transformation of rice plants.
Production of Transgenic Rice Plants and Southern-Blot-Hybridization Analyses
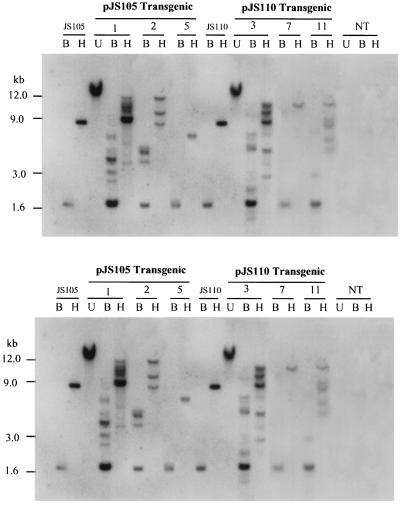
Two plasmids, pJS105 (containing one copy of ABRC1) and pJS110 (containing four copies of ABRC1), were constructed for expression of uidA in transgenic rice plants. The structures of these two plasmids are shown in Figure 2. After particle bombardment of suspension cells by using the two plasmids, eight Basta-resistant and Southern-blot-positive lines were regenerated, of which six (three lines for each plasmid) showed the correct hybridization pattern. The other two lines had rearranged bands and, therefore, were not further studied. The six desired transgenic lines were all fertile and their R1 generations were used for further analyses. The results of Southern-blot hybridization with the 1.8-kb uidA coding region as the probe (Fig. 2, probe 1) are shown in Figure 3. Both rice genomic and plasmid DNA were digested by BamHI or HindIII. BamHI digestion released a 1.8-kb hybridizing band corresponding to the size of uidA. HindIII is a unique site in the plasmids pJS105 and pJS110, thus each hybridization band created by HindIII digestion represents one copy of the transgene uidA, except in cases when HindIII fragments cannot be resolved. Each line has its own specific hybridization pattern except the expected 1.8-kb band, indicating that these six transgenic lines were derived from independent transformation events.
Figure 3.
Southern-hybridization analysis of gusA-transgenic rice plants. Eight micrograms of rice genomic DNA was digested by BamHI (two sites in the plasmids) or HindIII (a unique site in the plasmids) and separated in a 0.8% agarose gel. A DIG-labeled, 1.8-kb GUS coding region (probe 1; see Fig. 2) was used as the probe. Molecular sizes (kb) of the 1-kb DNA ladder are indicated on the left side. B, BamHI; H, HindIII; U, undigested; and NT, DNA from nontransgenic plants.
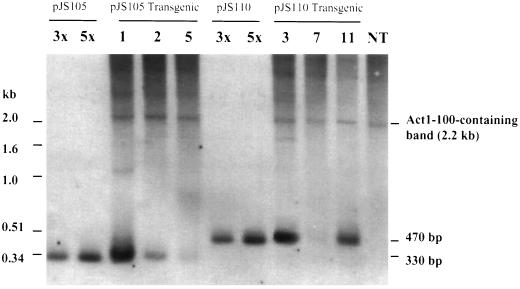
To verify that one copy of ABRC1 and four copies of ABRC1 were also integrated into the genome of transgenic rice plants, an additional Southern-blot hybridization was conducted by using the 330-bp probe 2 (see Fig. 2). The results (Fig. 4) indicated that transgenic lines 1, 2, and 5 contained one copy of ABRC1 corresponding to the size (330 bp) of the expected band of pJS105, whereas lines 3, 7, and 11 contained four copies of ABRC1 corresponding to the size (470 bp) of the expected band of pJS110. This result also showed that the one copy of ABRC1 or four copies of ABRC1, fused to the Act1–100P with HVA22(I), were integrated into the rice genome. The copy number of the transgenes was estimated both by HindIII digestion, which has only one restriction site in the plasmids (Fig. 3), and by using the Act1–100P-containing band as an internal standard. Previous work (McElroy et al., 1990) indicated the presence of only one copy of the Act1 gene in the rice genome. Since there is also one copy of the Act1–100P in plasmids pJS105 and pJS110, the ratio of the intensity of hybridization bands (the 330-bp band for pJS105 transgenic lines, and 470-bp band for pJS110 transgenic lines) to the band (2.2 kb) corresponding to that of nontransgenic plants should give the copy number of the transgene in a given transgenic plant (Table II).
Figure 4.
Southern-hybridization analysis of gusA-transgenic rice plants. Eight micrograms of genomic DNA was digested by EcoRV (see EcoRV sites in Fig. 2) and the digested DNA was separated in a 1.2% agarose gel. A DIG-labeled, 330-bp of probe 2 (indicated in Fig. 2) was used. Molecular sizes of the 1-kb DNA ladder are indicated on the left side. 3x and 5x plasmid DNA represent 3 and 5 genome equivalents of DNA relative to 8 μg of rice genomic DNA, respectively. NT, DNA from nontransgenic plants.
Table II.
Approximate copy number of transgenes in pJS105- and pJS110-transgenic lines
| pJS105 Transgenic | pJS110 Transgenic | |||||
|---|---|---|---|---|---|---|
| Lines | 1 | 2 | 5 | 3 | 7 | 11 |
| Transgene copy no. | 9 | 3 | 1 | 7 | 1 | 5 |
GUS Activity in R0 Transgenic Rice Plants
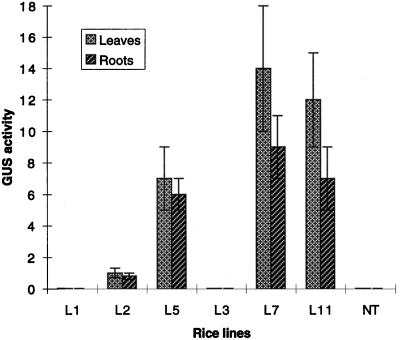
The promoter complex in plasmids pJS105 and pJS110 is composed of ABRC1, the Act1–100P minimal promoter, and HVA22(I), in which the Act1–100P promoter plays an important role in conferring the basal level of uidA expression. Before starting to test for ABA- and stress-induced GUS activity, we first examined the basal level of GUS activity in 4-month-old R0 transgenic plants. The results are shown in Figure 5. Of the six transgenic lines, L5, L7, and L11 showed high levels of GUS activity and L2 showed low activity (≤ 1 nmol h−1 mg−1 protein). No GUS activity was detected in either leaves or roots of L1 and L3. L2 and L5 (pJS105 transformants) and L7 and L11 (pJS110 transformants) were used for assaying ABA- and stress-inducible uidA expression.
Figure 5.
GUS activity in the R0 transgenic plants without any treatment. L1, L2, and L5 represent pJS105-transgenic lines 1, 2, 5; and L3, L7, and L11 represent pJS110-transgenic lines 3, 7, and 11. Mean ± se values of GUS activity (4-methylumbelliferone, nmol h−1 mg−1 protein) are: Leaves: L1, 0.02 ± 0.01; L2, 1 ± 0.3; L5, 7 ± 2; L3, 0.02 ± 0.01; L7, 14 ± 4; L11, 12 ± 3; and NT, 0.02 ± 0.01. NT, DNA from nontransgenic plants. Roots: L1, 0.01 ± 0.01; L2, 0.8 ± 0.2; L5, 6 ± 1; L3, 0.01 ± 0.01; L7, 9 ± 2; L11, 7 ± 2; and NT, 0.01 ± 0.01. Data represent the average results of four experiments by using different tillers of the same R0 line. Bar represents the se.
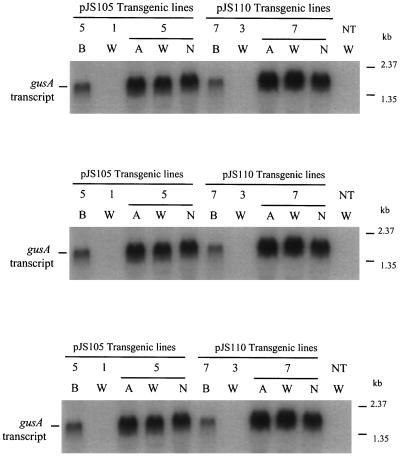
ABA-, Water Deficit-, and NaCl-Induced uidA mRNA Level in Transgenic Rice Plants
To test the ABA- or stress-inducible uidA expression, we first examined the transcript level of the uidA transgene in R1 leaves before and after water-deficit treatment for 6 d in the greenhouse (see Methods). Three transgenic lines (L5, L7, and L11) were found to express uidA (data not shown). L5 from the pJS105 construct and L7 from the pJS110 construct were selected for further treatments and analyses. ABA and NaCl were also found to induce uidA expression (Fig. 6). By densitometry tracing, the induction level varied from 6- to 8-fold. No uidA transcripts were detected in R1 leaf RNA from the other two Southern-blot-positive lines (L1 and L3) or from nontransgenic plants even after water-deficit treatment.
Figure 6.
ABA-, water-deficit-, and NaCl-induced gusA expression confirmed by northern-hybridization analysis. Five micrograms of total RNA was fractionated in a 1% formaldehyde agarose gel and blotted onto a nylon membrane hybridized with [α-32P]dCTP-labeled gusA coding sequence. Equal loading of the RNA samples was confirmed by ethidium bromide staining of rRNA in a parallel-running gel. Molecular sizes (kb) of two fragments from the RNA ladder are indicated on the right side. A, ABA: 50 μm for 20 h; B, basal level without any treatment; W, water deficit: water withheld for 6 d; and N, NaCl: 150 mm NaCl, for 72 h. (For detailed procedure, see Methods.)
ABA-Induced GUS Activity in Transgenic Rice Plants
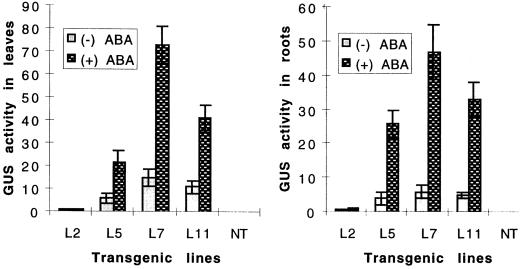
A previous report (Shen and Ho, 1995) indicated that ABRC1 confers a high degree of ABA induction for gene expression by a transient assay in barley aleurone cells. To examine the ABA-induction level of uidA expression conferred by the ABA-responsive promoter complex, ABRC1-Act1–100P-HVA22(I), in transgenic rice leaves and roots, a quantitative assay of GUS activity before and after ABA treatment of 2-week-old seedlings was carried out. At the 2-week stage, most R1 seedlings had two normal-sized leaves. Of 10 plants tested, 8 showed GUS activity and ABA inducibility. A lower leaf of an R1 seedling was cut off and used for GUS activity assay before applying exogenous ABA. An upper leaf of the same seedling was collected for assaying ABA-inducible uidA expression after supplying 50 μm ABA for 20 h. The results of this analysis are given in Figure 7. It is shown that the absolute level of GUS activity in pJS110-transformed plants is higher than that of pJS105-transformed plants both before and after ABA induction. A control experiment using upper leaves of L7 (with the highest GUS activity) after collection of the lower leaf was carried out in the absence of ABA and no increase of GUS activity was found. Thus, removing leaf tissues from plants did not show any adverse effect on GUS activity.
Figure 7.
ABA-induced GUS activity (4-methylumbelliferone, nmol h−1 mg−1 protein) in 2-week-old R1 seedlings of transgenic plants. All data were derived from the results of eight seedlings. pJS105 (one copy of ABRC1), L2 and L5; pJS110 (four copies of ABRC1), L7 and L11. NT, Nontransgenic plants. x indicates the -fold induction. Bars represents the se. Left panel, Leaves. Mean ± se values of ABA-induced GUS activity are: L2, 1 ± 0.2 (−ABA), 1.2 ± 0.3 (+ABA), 1.2x; L5, 6 ± 2 (−ABA), 22 ± 5 (+ABA), 4x; L7, 15 ± 4 (−ABA), 73 ± 8 (+ABA), 5x; L11, 11 ± 3 (−ABA), 41 ± 6 (+ABA), 4x; and NT, 0.02 ± 0.01 (−ABA), 0.02 ± 0.01 (+ABA), 1x. Right panel, Roots. Mean ± se values of ABA-induced GUS activity are: L2, 0.8 ± 0.2 (−ABA), 0.9 ± 0.2 (+ABA), 1x; L5, 4 ± 2 (−ABA), 26 ± 5 (+ABA), 7x; L7, 6 ± 2 (−ABA), 48 ± 10 (+ABA), 8x; L11, 5 ± 1 (−ABA), 33 ± 5 (+ABA), 7x; and NT, 0.01 ± 0.01 (−ABA), 0.01 ± 0.01 (+ABA), 1x.
Water-Deficit-Induced GUS Activity in Transgenic Rice Plants
As mentioned previously, ABA mediates gene expression involved in plant physiological responses to stress such as drought and salinity. The ABA-induced uidA expression in this report encouraged us to explore the water-deficit-induced GUS activity in the transgenic rice. Before water-deficit treatment, the third leaf from the bottom and about one-tenth the amount of roots of 8-week-old R1 plants with four to five leaves were collected, frozen in liquid N2, and used for assaying the basal level of GUS activity. These plants were subjected to water-deficit treatment for 4, 6, and 8 d. The other three leaves from the same plant used for assaying basal level were collected at 4, 6, and 8 d, respectively, after the treatment and used for testing the induced activity. At the same time, one-tenth the amount of roots was also collected at each time point following the leaf collection. Three plants for each line were used for each experiment to calculate the degree of induction of uidA expression by water-deficit treatment. The results from three independent experiments are listed in Table III. After 4 d of treatment GUS activity in rice leaves increased only slightly. With an increase of treatment days, GUS activity increased rapidly and reached a peak at 8 d, resulting in 5- to 6-fold induction. Beyond 8 d (data not shown), the treated leaves started to wilt. In rice roots the GUS activity reached a peak after 6 d. A longer treatment (e.g. 8 d) gave a slightly reduced level of uidA expression in the roots of transgenic rice by withholding water.
Table III.
Water-deficit-induced GUS activity in R1 leaves and roots of transgenic rice plants
| Days of Treatment | Water Content of Soil | GUS Activity
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| pJS105
Transgenic
|
pJS110 Transgenic
|
||||||||||
| L2
|
L5
|
L7
|
L11
|
NT
|
|||||||
| Leaves | Roots | Leaves | Roots | Leaves | Roots | Leaves | Roots | Leaves | Roots | ||
| % | 4-MU nmol h−1 mg−1 protein | ||||||||||
| 0 | 37 | 1 ± 0.2 | 0.9 ± 0.2 | 7 ± 3 | 6 ± 2 | 14 ± 4 | 13 ± 3 | 10 ± 3 | 7 ± 2 | 0.02 ± 0.01 | 0.01 ± 0.01 |
| 4 | 24 | 1 ± 0.2 | 0.9 ± 0.2 | 10 ± 3 | 11 ± 3 | 18 ± 4 | 28 ± 3 | 14 ± 4 | 16 ± 3 | 0.02 ± 0.01 | 0.01 ± 0.01 |
| 6 | 14 | 1 ± 0.2 | 0.9 ± 0.2 | 18 ± 4 | 34 ± 5 | 35 ± 6 | 88 ± 6 | 27 ± 6 | 41 ± 5 | 0.02 ± 0.01 | 0.01 ± 0.01 |
| 8 | 9 | 1 ± 0.2 | 0.9 ± 0.2 | 40 ± 6 | 31 ± 5 | 81 ± 7 | 80 ± 6 | 47 ± 7 | 38 ± 5 | 0.02 ± 0.01 | 0.01 ± 0.01 |
| 8 d/0 d | 1 | 6 | 6 | 5 | 1 | ||||||
| 6 d/0 d | 1 | 6 | 7 | 6 | 1 | ||||||
Mean ± se values of the GUS activity were calculated from the results of three independent experiments and three plants were used for each experiment. R1 plants were grown in a greenhouse and treated without water for 4, 6, and 8 d. 0 d represents the basal level before water-deficit treatment. 8 d/0 d indicates the induction of GUS activity in rice leaves by withholding water for 8 d, and 6 d/0 d indicates the induction in rice roots by withholding water for 6 d. NT, Nontransgenic; 4-MU, 4-methylumbelliferone. Maximum induction values are underlined.
NaCl-Induced GUS Activity in Transgenic Rice Plants
To test the extent of induction of uidA expression by NaCl treatment, a 150 mm NaCl solution was used to create a salinity-stress condition. Water was withheld for 24 h from 8-week-old plants with four to five leaves grown in the greenhouse, and then a 150 mm NaCl solution was added to the plant-containing pots and the tray. The NaCl solution was changed every 24 h. Samples were collected in the same way as in the water-deficit treatment except that the third leaf of each plant used for assaying the basal level of GUS activity was collected after 24 h of withholding water (0 h treatment by NaCl). Table IV indicates the results of this analysis in the leaves and roots of transgenic rice. As compared with the results of ABA and water-deficit treatments, the GUS activity and induction levels were both lower. Similar to the water-deficit treatment, the NaCl-induced GUS activity in the roots of transgenic rice plants reached its peak at 72 h of treatment. A longer treatment (such as 96 h) showed a slightly reduced level of uidA expression. A control experiment using leaves collected at 48, 72, and 96 h after cutting the first leaves was also performed in the absence of NaCl and no increase of GUS activity was observed. Thus, removing leaf tissues from plants did not show any adverse effect on GUS activity.
Table IV.
NaCl-induced GUS activity in R1 leaves and roots of transgenic rice plants
| NaCl Treatment | GUS Activity
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| pJS105 Transgenic
|
pJS110
Transgenic
|
|||||||||
| L2
|
L5
|
L7
|
L11
|
NT
|
||||||
| Leaves | Roots | Leaves | Roots | Leaves | Roots | Leaves | Roots | Leaves | Roots | |
| h | 4-MU nmol h−1 mg−1 protein | |||||||||
| 0 | 0.9 ± 0.2 | 0.8 ± 0.2 | 6 ± 2 | 5 ± 1 | 13 ± 3 | 12 ± 2 | 10 ± 2 | 9 ± 2 | 0.02 ± 0.01 | 0.01 ± 0.01 |
| 48 | 0.9 ± 0.2 | 0.8 ± 0.2 | 10 ± 3 | 11 ± 3 | 20 ± 4 | 25 ± 4 | 16 ± 4 | 20 ± 4 | 0.02 ± 0.01 | 0.01 ± 0.01 |
| 72 | 0.9 ± 0.2 | 0.8 ± 0.2 | 14 ± 3 | 20 ± 5 | 28 ± 6 | 46 ± 6 | 21 ± 4 | 25 ± 5 | 0.02 ± 0.01 | 0.01 ± 0.01 |
| 96 | 0.9 ± 0.2 | 0.8 ± 0.2 | 17 ± 4 | 16 ± 3 | 59 ± 7 | 40 ± 4 | 28 ± 5 | 22 ± 3 | 0.02 ± 0.01 | 0.01 ± 0.01 |
| 96 h/0 h | 1 | 3 | 4 | 3 | 1 | |||||
| 72 h/0 h | 1 | 4 | 4 | 3 | 1 | |||||
Mean ± se values of NaCl-induced GUS activity were calculated from the results of three independent experiments and three plants were used for each experiment. Eight-week-old R1 plants were grown in the greenhouse. After withholding water for 24 h, the third leaf or one-tenth the amount of roots was collected and used for a basal level test of GUS activity (0 h). Then, the plants were supplied with 150 mm NaCl solution. At 48, 72, and 96 h, the other three leaves or one-tenth the amount of roots were collected, respectively, and used for assaying NaCl-induced GUS activity. Maximum induction values are underlined. 96 h/0 h indicates the -fold induction of GUS activity in rice tissues after 96 h of treatment with 150 mm NaCl. 72 h/0 h indicates the induction. NT, Nontransgenic; 4-MU, 4-methylumbelliferone.
In conclusion, ABA, water deficit, and 150 mm NaCl induced uidA expression both at the RNA and protein levels (GUS activity) conferred by the ABA-induced promoter in transgenic rice plants. Transgenic rice plants harboring the plasmid with four copies of ABRC1 exhibited 50% to 200% higher GUS activity than those with one copy of ABRC1 among the tested transgenic rice lines. The Act1–100P minimal promoter coupled with ABRC1 and HVA22(I) of the barley HVA22 gene conferred ABA- and stress-inducible uidA expression in transgenic rice. These results suggest that the expression vectors pJS104 (containing one copy of ABRC1) and pJS109 (four copies of ABRC1) can be used for other plasmid constructions to produce stress-induced osmotolerant transgenic rice plants.
DISCUSSION
Different stress treatments and exogenous ABA application caused different extents of induction of uidA expression in both transgenic rice leaves and roots. In this study water-deficit treatment caused the highest induction of GUS activity, about 5- to 6-fold in rice leaves, followed by ABA application with a 4- to 5-fold increase, and NaCl treatment with 3- to 4-fold increase of GUS activity. In roots ABA treatment resulted in the highest induction of GUS activity, with a 7- to 8-fold increase, followed by water-deficit treatment with a 6- to 7-fold induction, and NaCl treatment with a 3- to 4-fold increase.
Strong and constitutive promoters are beneficial for high-level expression of selectable marker genes, which is necessary for efficient selection and generation of transgenic plants. However, constitutively active promoters are not always desirable for plant genetic engineering because constitutive overexpression of a transgene may compete for energy and building blocks for synthesis of proteins, RNA, etc., which are also required for plant growth under normal conditions. Either one copy of ABRC1 or four tandem copies of ABRC1 coupled with Act1–100P and HVA22(I) of the HVA22 gene confer ABA- and stress-induced uidA expression in transgenic rice.
Transgene expression in transgenic plants is often correlated with copy number (Hobbs et al., 1993; Matzke et al., 1994) and integration position of transgenes (position effect) in the genome (Peach and Velten, 1991; Bhattacharyya et al., 1994). Thus, it is difficult to conclude which type of promoter complex (either one copy of ABRC1 or four copies of ABRC1) would be better for generation of stress-tolerant transgenic rice plants. According to the results in this study, we prefer to use four copies of the ABRC1-containing promoter complex because it can give approximately 50% to 200% higher GUS activity (e.g. plant L7) than one copy of the ABRC1-containing promoter complex (plant L5). We are currently using the stress-induced expression vectors to construct plasmids containing other potentially useful genes for transformation of rice. We believe that transgenic rice plants with foreign genes driven by a stress-induced promoter are expected to develop and grow better than those with genes driven by a constitutive promoter because the transgenes would be highly expressed only under stress conditions.
The primary goal of this study was to test recombinant gene constructs, the expression of which is induced by ABA and stress conditions in transgenic rice plants. The information obtained in this study will be valuable in future work attempting to express useful genes in transgenic plants under stress. Since it is well established that environmental stresses such as water deficit and salinity usually lead to enhanced levels of endogenous ABA (Zeevaart and Creelman, 1988), we reason that an ABA-responsive promoter could also be induced by stress conditions. Indeed, the ABA-responsive gene constructs tested in this study are all induced by water-deficit and NaCl treatment. For an ABA-/stress-responsive promoter to be useful in driving the expression of useful genes, it is better to be highly sensitive and respond quickly to ABA/stress. Indeed, in this work we have shown that the ABRC1/actin minimal promoter responds to mild water stress and salinity within a couple of days (Tables III and IV). Although not determined in this work, we believe that this construct is even more sensitive, because Shen et al. (1993) have shown that HVA22, the promoter of which contains ABRC1, is responsive to ABA concentrations as low as 10−8 m, and that this gene is induced by 10−6 m ABA within 40 min. Therefore, ABRC1 appears to have the desirable features in regulating transgenes encoding useful traits for protecting plants against stress conditions.
Among the ABA-responsive promoter sequences, ABRC, as defined by Shen and Ho (1995) and Shen et al. (1996), appears to be necessary and sufficient for a high level of ABA induction. However, their work was essentially carried out in a highly specialized tissue, the aleurone layers of germination barley seeds. By linking ABRC to a minimal promoter derived from the actin gene, which is constitutively expressed in many cell types, we have shown that our gene constructs can be expressed in at least two major vegetative tissues, leaves and roots, in addition to the aleurone layers. Although the transgenic approach as described in this work has proven to be an efficient means to analyze promoters, ectopic functions of promoters in transgenic plants have also been observed. For example, Sieburth and Meyerowitz (1997) have recently reported that the cis elements for spatial regulation of the Arabidopsis AGAMOUS gene are located intragenically. Thus, it is conceivable that the promoter of a gene does not always contain all of the elements regulating its expression. However, it is clear from our work and from the work of Shen and Ho (1996) that ABRC1 alone is sufficient to confer a high level of ABA inducibility. It is equally significant that the gene constructs tested in this study function well in both rice and barley. Since the ABRC we used was derived from a barley gene with homologs present in many cereal grains (Q. Shen and D. Ho, unpublished data), it is conceivable that our gene constructs could work in other cereals as well.
ACKNOWLEDGMENTS
We thank Dr. Xiongfong Chen for help with preparation of the photos, Cathy Herlache for reading the manuscript, and Miguel Munoz for help with drawing figures in this manuscript.
Abbreviations:
- ABRC
ABA-response complex
- ABREs
ABA-response elements
- CaMV
cauliflower mosaic virus
- DIG
digoxigenin
- HVA22(I)
intron1-exon2-intron2 of barley HVA22 gene
- MS
Murashige and Skoog
- Nos
nopaline synthetase
Footnotes
This work was supported by the Rockefeller Foundation (research grant no. RF93001, allocation no. 194) to R.W. J.S. was supported by a postdoctoral fellowship of the Rockefeller Foundation.
LITERATURE CITED
- Bhattacharyya BA, Stermer BA, Dixon RA. Reduced variation in transgene expression from a binary vector with selectable markers at the right and left T-DNA borders. Plant J. 1994;6:957–968. [Google Scholar]
- Boyer JS. Plant productivity and environment. Science. 1982;218:443. doi: 10.1126/science.218.4571.443. [DOI] [PubMed] [Google Scholar]
- Bradford M. A rapid and sensitive method for quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Bruce WB, Quail PH. cis-Acting elements involved in photoregulation of an oat phytochrome promoter in rice. Plant Cell. 1990;2:1081–1089. doi: 10.1105/tpc.2.11.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J, Duan X, McElory D, Wu R. Regeneration of herbicide resistant transgenic rice plants following microprojectile-mediated transformation of suspension culture cells. Plant Cell Rep. 1992;11:586–591. doi: 10.1007/BF00233098. [DOI] [PubMed] [Google Scholar]
- Cao J, Zhang W, McElory D, Wu R (1991) Assessment of rice genetic transformation techniques. In GH Toenniessen, GS Khush, eds, Rice Biotechnology. C.A.B. International, Oxon, UK, pp 175–198
- Chandler PM, Robertson M. Gene expression regulated by abscisic acid and its regulation to stress tolerance. Annu Rev Plant Physiol Mol Biol. 1994;45:113–114. [Google Scholar]
- Giuliano G, Pichersky E, Malik VS, Timko MP, Scolnik PA, Cashmore AR. An evolutionarily conserved protein binding sequence upstream of a plant light-regulated gene. Proc Natl Acad Sci USA. 1988;85:7089–7093. doi: 10.1073/pnas.85.19.7089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez J, Sanchez-Martinez D, Stiefel V, Rigau J, Puigdomenech P, Pages M. A gene induced by the plant hormone abscisic acid in response to water stress encodes a glycine-rich protein. Nature. 1988;334:262–264. doi: 10.1038/334262a0. [DOI] [PubMed] [Google Scholar]
- Guiltinan MJ, Marcotte WR, Jr, Quatrano RS. A plant leucine zipper protein that recognizes an abscisic acid response element. Science. 1990;250:267–271. doi: 10.1126/science.2145628. [DOI] [PubMed] [Google Scholar]
- Hayashi H. Transformation of Arabidopsis thaliana with codA gene for choline oxidase: accumulation of glycinebetaine and enhanced tolerance to salt and cold stress. Plant J. 1997;12:133–142. doi: 10.1046/j.1365-313x.1997.12010133.x. [DOI] [PubMed] [Google Scholar]
- Henson IE. Effects of atmospheric humidity on abscisic acid accumulation and water in leaves of rice (Oryza sativa L.) Ann Bot. 1984;54:569–582. [Google Scholar]
- Hihara Y, Hara C, Uchimiya H. Isolation and characterization of two cDNA clones for mRNA that are abundantly expressed in immature anthers of rice (Oryza sativa L.) Plant Mol Biol. 1996;30:1181–1193. doi: 10.1007/BF00019551. [DOI] [PubMed] [Google Scholar]
- Hobbs SLA, Warkentin TD, Delong CMO. Transgene copy number can be positively or negatively associated with transgene expression. Plant Mol Biol. 1993;21:17–26. doi: 10.1007/BF00039614. [DOI] [PubMed] [Google Scholar]
- Holmström KO, Mäntyla E, Welin B, Mandal A, Palva ET, Tunnela OE, Londesborough J. Drought tolerance in tobacco. Nature. 1996;379:683–684. [Google Scholar]
- Ingram J, Bartels D. The molecular basis of dehydration tolerance in plants. Annu Rev Plant Physiol Mol Biol. 1996;47:377–404. doi: 10.1146/annurev.arplant.47.1.377. [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. GUS fusion: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones H, Leigh RA, Tomos AD, Jones RGW. The effect of abscisic acid on cell turgor pressures, solute content and growth of wheat roots. Planta. 1987;170:257–262. doi: 10.1007/BF00397896. [DOI] [PubMed] [Google Scholar]
- King RW. Abscisic acid in developing wheat grains and its relationship to grain growth and maturation. Planta. 1976;132:43–51. doi: 10.1007/BF00390329. [DOI] [PubMed] [Google Scholar]
- Kishor PBK, Hong Z, Miao G-H, Hu C-AA, Verma DPS. Overexpression of Δ1-pyrroline-5-carboxylate synthetase increases proline production and confers osmotolerance in transgenic plants. Plant Physiol. 1995;108:1387–1394. doi: 10.1104/pp.108.4.1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanahan MB, Ho T-HD, Rogers SW, Rogers JC. A gibberellin response complex in cereal α-amylase gene promoters. Plant Cell. 1992;4:203–211. doi: 10.1105/tpc.4.2.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linsmaier EM, Skoog F. Organic growth factor requirements of tobacco tissue cultures. Physiol Plant. 1965;18:100–127. [Google Scholar]
- Liu Z-B, Ulmasov T, Shi X, Hagen G, Guilfoyle TJ. Soybean GH3 promoter contains multiple auxin-inducible elements. Plant Cell. 1994;6:645–657. doi: 10.1105/tpc.6.5.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke AJM, Neuhuber F, Park YD, Ambros PF, Matzke MA. Homology-dependent gene silencing in transgenic plants: epistatic silencing loci contain multiple copies of methylated transgenes. Mol Gen Genet. 1994;244:219–229. doi: 10.1007/BF00285449. [DOI] [PubMed] [Google Scholar]
- McElroy D, Zhang W, Cao J, Wu R. Isolation of an efficient actin promoter for use in rice transformation. Plant Cell. 1990;2:163–171. doi: 10.1105/tpc.2.2.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundy J, Chua N-H. Abscisic acid and water-stress induce the expression of a novel rice gene. EMBO J. 1988;7:2279–2286. doi: 10.1002/j.1460-2075.1988.tb03070.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant. 1962;15:473–497. [Google Scholar]
- Ono A, Izawa T, Chua N-H, Shimamoto K. The rab 16B promoter of rice contains two distinct abscisic acid-responsive elements. Plant Physiol. 1996;112:483–491. doi: 10.1104/pp.112.2.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peach C, Velten J. Transgene expression variability (position effect) of CAT and GUS reporter genes driven by linked divergent T-DNA promoters. Plant Mol Biol. 1991;17:49–60. doi: 10.1007/BF00036805. [DOI] [PubMed] [Google Scholar]
- Pilon-Smits EAH, Ebskamp MJM. Improved performance of transgenic fructan-accumulating tobacco under drought stress. Plant Physiol. 1995;107:125–130. doi: 10.1104/pp.107.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Shen Q, Ho T-HD. Functional dissection of an abscisic acid (ABA)-inducible gene reveals two independent ABA-responsive complexes each containing a G-box and a novel cis-acting element. The Plant Cell. 1995;7:295–307. doi: 10.1105/tpc.7.3.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Q, Uknes SJ, Ho T-HD. Hormone response complex of a novel abscisic acid and cycloheximide inducible barley gene. J Biol Chem. 1993;268:23652–23660. [PubMed] [Google Scholar]
- Shen Q, Zhang P, Ho T-HD. Modular nature of abscisic acid (ABA) response complexes: composite promoter units that are necessary and sufficient for ABA induction of gene expression in barley. The Plant Cell. 1996;8:1107–1119. doi: 10.1105/tpc.8.7.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieburth LE, Meyerowitz EM. Molecular dissection of the AGAMOUS control region show that cis elements for spatial regulation are located intragenically. Plant Cell. 1997;9:355–365. doi: 10.1105/tpc.9.3.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh NK, LaRosa PC, Handa AD, Hasegawa PM, Bressan RA. Hormonal regulation of protein synthesis associated with salt tolerance in plant cell. Proc Natl Acad Sci USA. 1987;84:739–743. doi: 10.1073/pnas.84.3.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarczynski MC, Jensen RG, Bohnert HJ. Stress-protection of transgenic tobacco by production of the osmolyte mannitol. Science. 1993;259:508–510. doi: 10.1126/science.259.5094.508. [DOI] [PubMed] [Google Scholar]
- Wang B, Wu R. A vector for inserting foreign genes and selection of transformed rice plants. Rice Biotech Quart. 1995;22:8. [Google Scholar]
- Xu D, Duan X, Wang B, Hong B, Ho T-HD, Wu R. Expression of a late embryogenesis abundant protein gene, HVA1, from barley confers tolerance to water deficit and salt stress in transgenic rice. Plant Physiol. 1996;110:249–257. doi: 10.1104/pp.110.1.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeevaart JAD, Creelmann RA. Metabolism and physiology of abscisic acid. Annu Rev Plant Physiol Plant Mol Biol. 1988;39:439–473. [Google Scholar]
- Zhang W, Wu R. Efficient regeneration of transgenic plants from rice protoplasts and correctly regulated expression of the foreign gene in the plants. Theor Appl Genet. 1988;76:835–840. doi: 10.1007/BF00273668. [DOI] [PubMed] [Google Scholar]
- Zhao X, Wu T, Xie Y, Wu R. Genome-specific repetitive sequences in the genus Oryza. Theor Appl Genet. 1989;78:201–209. doi: 10.1007/BF00288800. [DOI] [PubMed] [Google Scholar]