Abstract
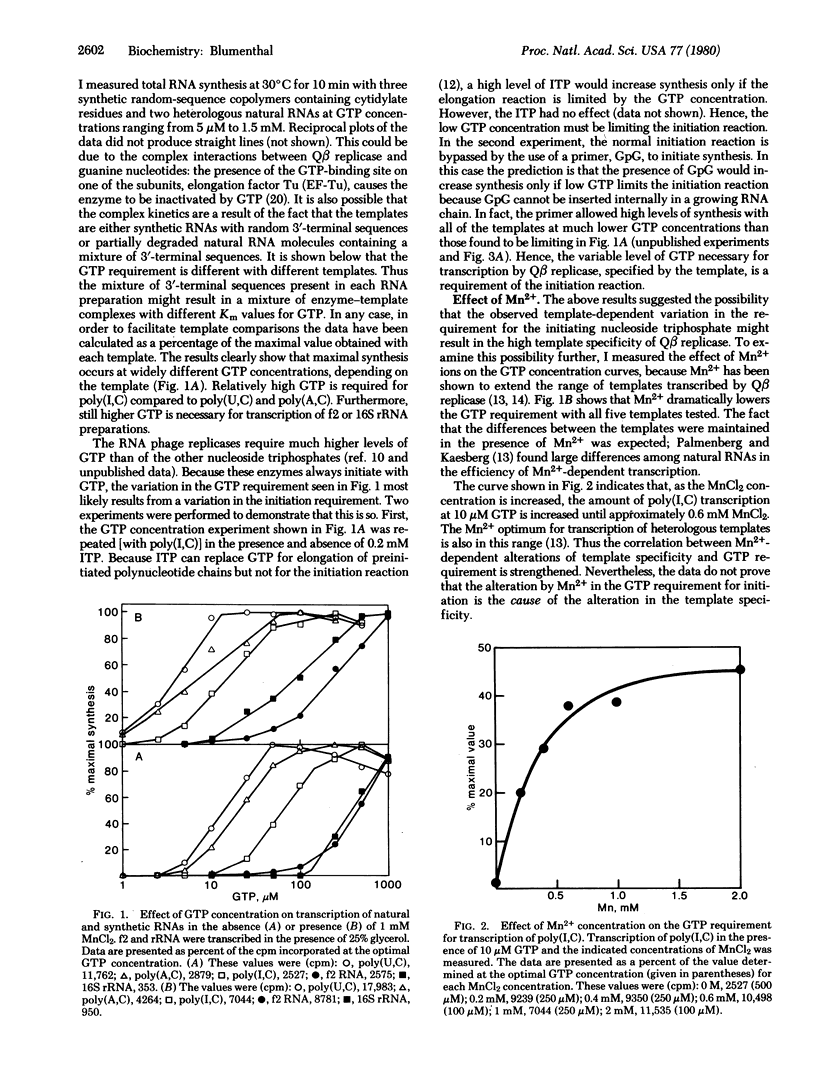
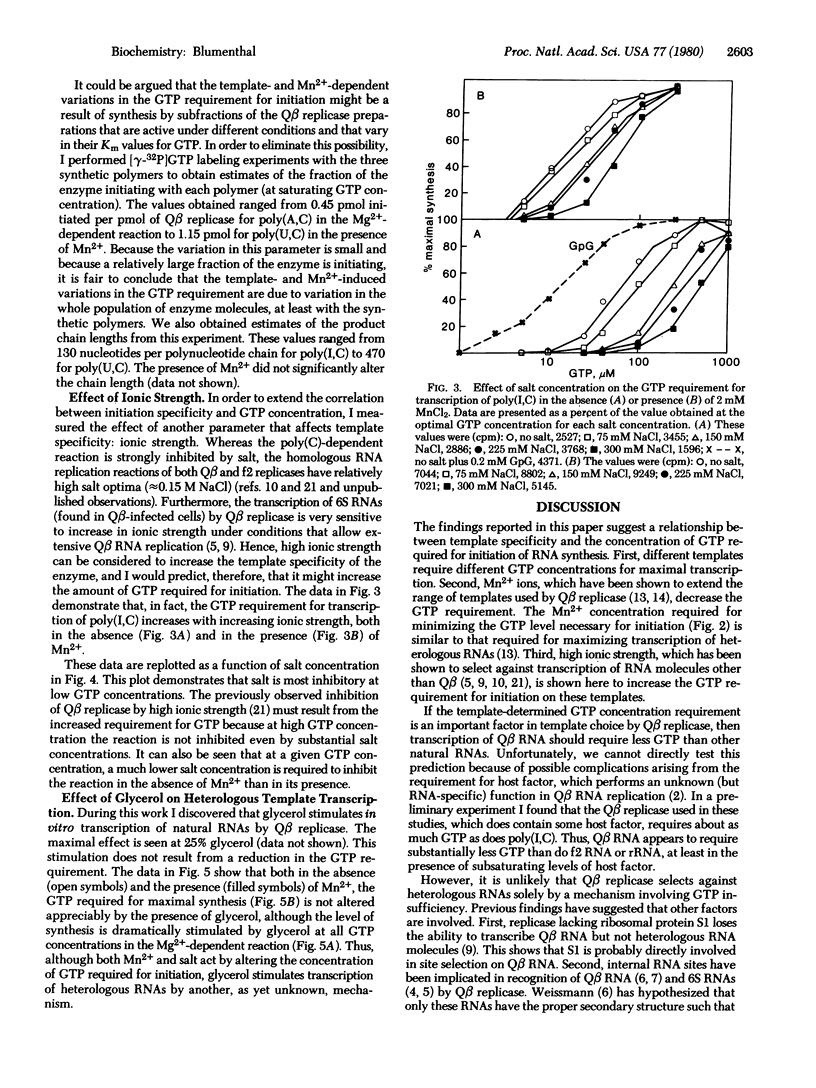
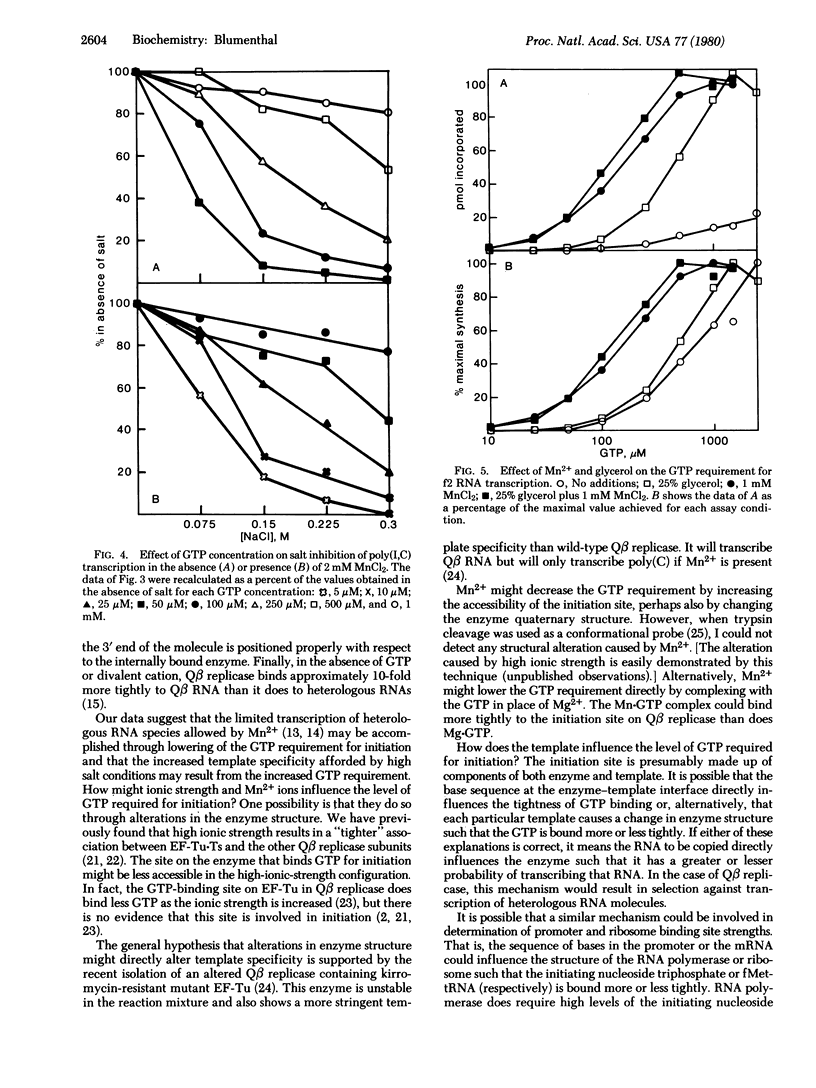
Qbeta replicase is notable for its high degree of template specificity. It has been shown to transcribe Qbeta RNA and synthetic polymers containing cytidylate. However, other natural RNAs are not transcribed unless Mn2+ ions are present. The enzyme initiates all RNA synthesis with GTP. In this report it is shown that Qbeta replicase can transcribe heterologous natural RNA species in the absence of Mn2+ if sufficient GTP is present. Each RNA tested requires a different GTP concentration for initiation. These results indicate that the site for the initiating nucleoside triphosphate on Qbeta replicase is strongly influenced by the template. It is proposed that the high degree of template specificity is a consequence of the fact that different templates induce initiation sites with varying affinities for GTP. Two lines of evidence support this idea. First, Mn2+ ions, which reduce template specificity, reduce the concentration of GTP required for initiation. Second, high ionic strength, which decreases transcription of all templates except Qbeta RNA, increases the GTP requirement. The possibility is considered that variable promoter or ribosome binding site strengths could result from a mechanism similar to that proposed here.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Banerjee A. K., Rensing U., August J. T. Replication of RNA viruses. X. Replication of a natural 6 s RNA by the Q-beta RNA polymerase. J Mol Biol. 1969 Oct 28;45(2):181–193. doi: 10.1016/0022-2836(69)90098-9. [DOI] [PubMed] [Google Scholar]
- Blumenthal T., Carmichael G. G. RNA replication: function and structure of Qbeta-replicase. Annu Rev Biochem. 1979;48:525–548. doi: 10.1146/annurev.bi.48.070179.002521. [DOI] [PubMed] [Google Scholar]
- Blumenthal T. Interaction of Qluta RNA replicase with guanine nucleotides. Different modes of inhibition and inactivation. Biochim Biophys Acta. 1977 Sep 20;478(2):201–208. doi: 10.1016/0005-2787(77)90183-6. [DOI] [PubMed] [Google Scholar]
- Blumenthal T. Qbeta RNA replicase and protein synthesis elongation factors EF-Tu and EF-Ts. Methods Enzymol. 1979;60:628–638. doi: 10.1016/s0076-6879(79)60059-9. [DOI] [PubMed] [Google Scholar]
- Blumenthal T., Young R. A., Brown S. Function and structure in phage Qbeta RNA replicase. Association of EF-Tu-Ts with the other enzyme subunits. J Biol Chem. 1976 May 10;251(9):2740–2743. [PubMed] [Google Scholar]
- Carmichael G. G., Landers T. A., Weber K. Immunochemical analysis of the functions of the subunits of phage Qbeta ribonucleic acid replicase. J Biol Chem. 1976 May 10;251(9):2744–2748. [PubMed] [Google Scholar]
- Douglass J., Blumenthal T. Conformational transition of protein synthesis elongation factor Tu induced by guanine nucleotides. Modulation by kirromycin and elongation factor Ts. J Biol Chem. 1979 Jun 25;254(12):5383–5387. [PubMed] [Google Scholar]
- Fedoroff N. V., Zinder N. D. Properties of the phage f2 replicase. II. Comparative studies on the ribonucleic acid-dependent and poly (C)-dependent activities of the replicase. J Biol Chem. 1972 Jul 25;247(14):4586–4592. [PubMed] [Google Scholar]
- Feix G., Hake H. Primer directed initiation of RNA synthesis catalysed by Qbeta replicase. Biochem Biophys Res Commun. 1975 Jul 22;65(2):503–509. doi: 10.1016/s0006-291x(75)80176-8. [DOI] [PubMed] [Google Scholar]
- Haruna I., Spiegelman S. Recognition of size and sequence by an RNA replicase. Proc Natl Acad Sci U S A. 1965 Oct;54(4):1189–1193. doi: 10.1073/pnas.54.4.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haruna I., Spiegelman S. Specific template requirments of RNA replicases. Proc Natl Acad Sci U S A. 1965 Aug;54(2):579–587. doi: 10.1073/pnas.54.2.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori K., Eoyang L., Banerjee A. K., August J. T. Replication of RNA viruses. V. Template activity of synthetic ribopolymers in the Q-beta RNA polymerase reaction. Proc Natl Acad Sci U S A. 1967 Jun;57(6):1790–1797. doi: 10.1073/pnas.57.6.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamen R., Kondo M., Römer W., Weissmann C. Reconstitution of Q replicase lacking subunit with protein-synthesis-interference factor i. Eur J Biochem. 1972 Nov 21;31(1):44–51. doi: 10.1111/j.1432-1033.1972.tb02498.x. [DOI] [PubMed] [Google Scholar]
- Landers T. A., Blumenthal T., Weber K. Function and structure in ribonucleic acid phage Q beta ribonucleic acid replicase. The roles of the different subunits in transcription of synthetic templates. J Biol Chem. 1974 Sep 25;249(18):5801–5808. [PubMed] [Google Scholar]
- McClure W. R., Cech C. L., Johnston D. E. A steady state assay for the RNA polymerase initiation reaction. J Biol Chem. 1978 Dec 25;253(24):8941–8948. [PubMed] [Google Scholar]
- Mitsunari Y., Hori K. Qbeta replicase-associated, polycytidylic acid-dependent polyguanylic acid polymerase. I. Characterization of the reaction. J Biochem. 1973 Aug;74(2):263–271. [PubMed] [Google Scholar]
- Palmenberg A., Kaesberg P. Synthesis of complementary strands of heterologous RNAs with Qbeta replicase. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1371–1375. doi: 10.1073/pnas.71.4.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes G., Chamberlin M. J. Kinetic analysis of ribonucleic acid chain initiation by Escherichia coli Ribonucleic acid polymerase bound to DNA. J Biol Chem. 1975 Dec 10;250(23):9112–9120. [PubMed] [Google Scholar]
- Schaffner W., Rüegg K. J., Weissmann C. Nanovariant RNAs: nucleotide sequence and interaction with bacteriophage Qbeta replicase. J Mol Biol. 1977 Dec 25;117(4):877–907. doi: 10.1016/s0022-2836(77)80004-1. [DOI] [PubMed] [Google Scholar]
- Silverman P. M. Replication of RNA viruses: specific binding of the Q RNA polymerase to Q RNA. Arch Biochem Biophys. 1973 Jul;157(1):222–233. doi: 10.1016/0003-9861(73)90408-6. [DOI] [PubMed] [Google Scholar]
- Stahl S. J., Chamberlin M. J. An expanded transcriptional map of T7 bacteriophage. Reading of minor T7 promoter sites in vitro by Escherichia coli RNA polymerase. J Mol Biol. 1977 Jun 5;112(4):577–601. doi: 10.1016/s0022-2836(77)80165-4. [DOI] [PubMed] [Google Scholar]
- Vollenweider H. J., Koller T. Physical mapping of Qbeta replicase binding sites on Qbeta RNA. J Mol Biol. 1976 Mar 5;101(3):367–377. doi: 10.1016/0022-2836(76)90153-4. [DOI] [PubMed] [Google Scholar]
- Weissmann C. The making of a phage. FEBS Lett. 1974 Mar 23;40(0):suppl–suppl:S18. doi: 10.1016/0014-5793(74)80684-8. [DOI] [PubMed] [Google Scholar]
- Young R. A., Blumenthal T. Phage Q-beta ribonucleic acid replicase. Subunit relationships determined by intramolecular cross-linking. J Biol Chem. 1975 Mar 10;250(5):1829–1832. [PubMed] [Google Scholar]


