Abstract
Background
Horizontal gene transfer (HGT) is traditionally considered to be rare in multicellular eukaryotes such as animals. Recently, many genes of miscellaneous algal origins were discovered in choanoflagellates. Considering that choanoflagellates are the existing closest relatives of animals, we speculated that ancient HGT might have occurred in the unicellular ancestor of animals and affected the long-term evolution of animals.
Results
Through genome screening, phylogenetic and domain analyses, we identified 14 gene families, including 92 genes, in the tunicate Ciona intestinalis that are likely derived from miscellaneous photosynthetic eukaryotes. Almost all of these gene families are distributed in diverse animals, suggesting that they were mostly acquired by the common ancestor of animals. Their miscellaneous origins also suggest that these genes are not derived from a particular algal endosymbiont. In addition, most genes identified in our analyses are functionally related to molecule transport, cellular regulation and methylation signaling, suggesting that the acquisition of these genes might have facilitated the intercellular communication in the ancestral animal.
Conclusions
Our findings provide additional evidence that algal genes in aplastidic eukaryotes are not exclusively derived from historical plastids and thus important for interpreting the evolution of eukaryotic photosynthesis. Most importantly, our data represent the first evidence that more anciently acquired genes might exist in animals and that ancient HGT events have played an important role in animal evolution.
Keywords: Gene transfer, Endosymbiosis, Plastids, Animal evolution
Background
Gene transfer, including those occurred horizontally between distinct species (horizontal gene transfer, HGT) or those intracellularly from organelles (mitochondria or plastids) to the nucleus (endosymbiotic gene transfer, EGT), has now been widely recognized as an important force in organismal and genome evolution [1-3]. In particular, HGT may rapidly spread evolutionary success across lineages and allow recipient organisms to access new niches or other resources. Although numerous cases of HGT have been documented in prokaryotes and unicellular eukaryotes, a common belief is that HGT is rare in multicellular eukaryotes such as animals and plants [4,5]. Such a belief, however, largely ignores the dynamic nature of HGT and the more ancient HGT events to the unicellular ancestors of multicellular eukaryotes [6].
Gene transfer, although usually disruptive in phylogenetic reconstruction, may also serve as molecular fossils or footprints for historical evolutionary events [7,8]. An intriguing case in this respect is the distribution of algal/cyanobacterial genes in eukaryotes. Because eukaryotic photosynthesis is derived from a primary endosymbiosis with a cyanobacterium as well as secondary and tertiary endosymbioses with miscellaneous algae [9-12], these genes are frequently interpreted as relicts of earlier cyanobacterial/algal endosymbionts and, therefore, evidence of plastid losses in some aplastidic eukaryotes [13-15]. As eukaryotic photosynthesis spans multiple major lineages whose relationships critically rely on the plastid existence in them [16-18], an accurate understanding of the distribution and origin of algal genes in eukaryotes is essential.
Several recent studies indicated that algal genes also exist in organisms that are generally considered to be plastid-lacking throughout their evolution [15,19-21]. In particular, over 100 genes of algal origins were found in the choanoflagellate Monosiga brevicollis[19,21]. Because of the widespread distribution of algae and the phagotrophic nature of choanoflagellates, it was suggested that algal genes in Monosiga might have been acquired from food sources [21]. However, because choanoflagellates are the closest unicellular relatives of animals [22], it is unclear whether such acquisition of algal genes represents species-specific events or might have occurred also in the unicellular ancestor of animals and been inherited by diverse extant animals.
To address the above question, we performed phylogenomic analyses of the tunicate Ciona intestinalis to search for algal genes, in particular those anciently acquired by the ancestral animal. We choose C. intestinalis as a query to detect algal genes in animals because of its relatively small genome size and frequent physical contact with phytoplanktons. We here report 14 gene families of likely algal origin in C. intestinalis and other animals. We also further discuss the potential mechanisms of algal gene acquisition in animals and the importance of HGT in animal evolution.
Results and discussion
Algae-related genes identified in C. intestinalis and other animals
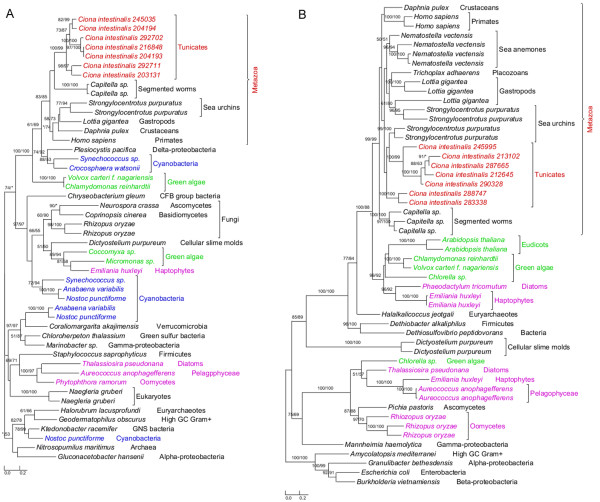
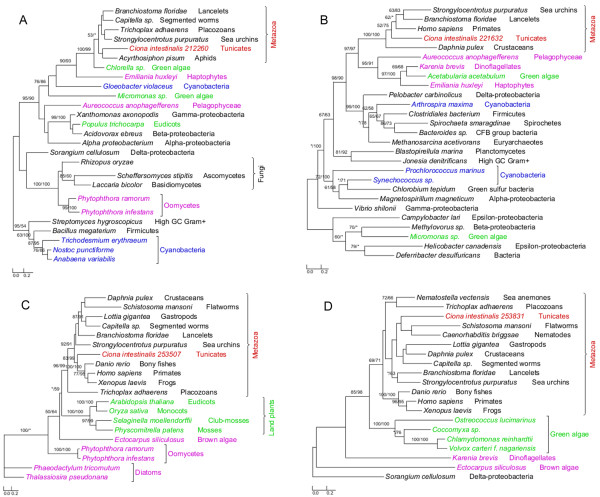
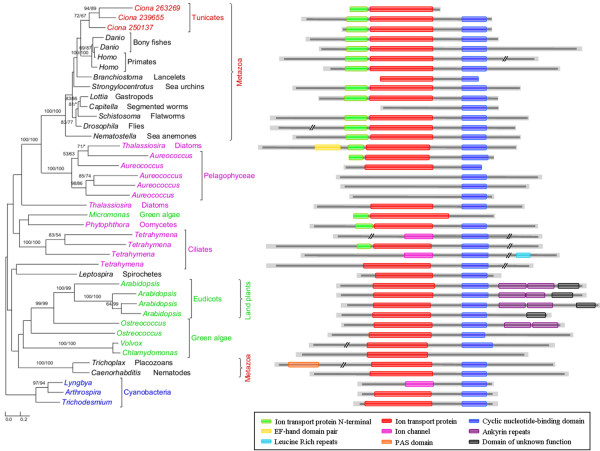
The annotated genome of C. intestinalis consists of 14,002 protein-coding genes and genome screening using AlienG[23] identified 169 of them as candidates of algal origin. We performed detailed analyses of taxonomic distribution, gene structure and molecular phylogeny for each of the identified candidate genes. One of these genes (GenBank accession number: XP_002127943) has identifiable homologs only in animals and plastid-containing eukaryotes. Another identified gene (GenBank accession number: XP_002122576) is restricted to animals, plastid-containing eukaryotes and ciliates, which also contain algal genes because of their possible historical plastids or phagotrophic lifestyle [14]. For additional 90 genes, although their molecular phylogenies often lack sufficient resolution in some branches, algal and animal sequences form monophyletic groups with over 70% bootstrap support (Figures 1,2 and 3). Therefore, 92 of the 169 genes are considered to be algae-related. Additional evidence from Pfam domain analyses [24] also indicates animal/algae affinity. For instance, three genes encoding potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 contain three identical tandem domains only in animals and algae (Figure 3). These 92 genes can be further classified into 14 gene families or subfamilies, including those encoding solute carriers, short-chain dehydrogenase/reductase, and UDP galactopyranose mutase etc (Table 1), based on their sequence relatedness (30-80% protein sequence identities and similar tree topologies) and GO annotation information (Additional file 1). Except the genes encoding UDP galactopyranose mutase, all other algae-related genes are present in multiple groups of animals (Figures 1,2 and 3; Additional file 2).
Figure 1.
Phylogeny of algal genes identified in Ciona . Numbers above branches show bootstrap values for maximum likelihood and distance analyses respectively. Asterisks indicate values lower than 50%. Other bootstrap values below 50% in both methods are not shown. Red: tunicates; green: Plantae; blue: cyanobacteria; pink: other plastid-containing eukaryotes. (A) Short-chain dehydrogenase/reductase family and (B) solute carrier family 23.
Figure 2.
Phylogeny of algal genes identified in Ciona. Numbers above branches show bootstrap values for maximum likelihood and distance analyses respectively. Asterisks indicate values lower than 50%. Other bootstrap values below 50% in both methods are not shown. Red: tunicates; green: Plantae; blue: cyanobacteria; pink: other plastid-containing eukaryotes. (A) Taurine dioxygenase; (B) cytidine monophospho-N-acetylneuraminic acid synthetase; (C) arginine N-methyltransferase 7; and (D) peptidylglycine alpha-hydroxylating monooxygenase.
Figure 3.
Phylogenetic and Pfam domain analyses of potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2. Numbers above branches show bootstrap values for maximum likelihood and distance analyses respectively. Asterisks indicate values lower than 50%. Other bootstrap values below 50% in both methods are not shown. Red: tunicates; green: Plantae; blue: cyanobacteria; pink: other plastid-containing eukaryotes. Three identical tandem domains (Ion transport protein N-terminal/Ion transport protein/Cyclic nucleotide-binding domain) are restricted to animals and algae. Several animal and algal sequences contain only one or two domains, suggestive of potential loss over time.
Table 1.
Putative algal genes identified in Ciona intestinalis
| Gene familya | Putative donor | Putative function |
|---|---|---|
| Short-chain dehydrogenase/reductase family (7) |
Green algae/ cyanobacteria |
Cellular differentiation and signaling |
| Solute carrier family 23 (7) |
Algae/land plants |
Na+-dependent vitamin C transport |
| Taurine dioxygenase (1) |
Green algae |
Cell redox homeostasis |
| Cytidine monophospho-N-acetylneuraminic acid synthetase (1) |
Algae |
Signaling regulation |
| Arginine N-methyltransferase 7 (1) |
Land plants/ stramenopiles |
Protein methylation |
| Peptidylglycine alpha-hydroxylating monooxygenase (1) |
Green algae |
Hormone biosynthesis |
| Aminoglycoside phosphotransferase (1) |
Algae |
Protein amino acid phosphorylation |
| Solute carrier family 22 (27) |
Algae/land plants |
Ion transport |
| Solute carrier family 34 (6) |
Diatoms |
Phosphate ion transport |
| Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (3) |
Stramenopiles |
Potassium ion transport |
| UDP galactopyranose mutase (2)* |
Algae |
Uridine diphosphogalactofuranose biosynthesis |
| Biphenyl/valacyclovir hydrolase (3) |
Dinoflagellates/haptophytes |
Aromatic compound metabolism |
| Solute carrier family 6 (31) |
Algae |
Neurotransmitter transport |
| Alkylation repair homolog 5 (1) | Algae/land plants | Alkylation repair |
aNumbers in the brackets show paralogous gene copies within each family. *The gene transfer event happened only between algae and C. intestinalis. All other gene transfers happened between plastid-containing organisms and the ancestral animal.
Each of the 14 gene families identified in our analyses shows a complex evolutionary history that differs from perceived organismal relationships in three domains of life. In many cases, multiple eukaryotic sequences are sporadically clustered within bacterial homologs. Such a complex evolutionary history for individual gene (or gene family) is somewhat expected, considering frequent HGT events both within domains (e.g., bacteria and archaea) [25,26] and between domains (e.g., bacteria or archaea to unicellular eukaryotes) [27-29], EGT following primary and higher level endosymbioses [3,30], and differential genes losses or replacements [31,32] etc. The dynamic occurrence of these factors over evolutionary time may further compound their effects. Therefore, aside from potential phylogenetic artifacts (see next paragraph), the monophyletic relationship between algal and animal sequences observed in this study can largely be explained by two alternative scenarios: (i) gene losses in most protists and fungi and (ii) gene acquisition by animals. For example, genes encoding UDP galactopyranose mutase are restricted to C. intestinalis, algae, and bacteria (including cyanobacteria). Phylogenetic analyses show that Ciona UDP galactopyranose mutase sequences group with homologs from multiple algae (Additional file 2). This may be attributed to either extensive gene losses from other eukaryotes or unique gene acquisition by Ciona. The scenario of extensive gene losses from other eukaryotes, though theoretically possible, is not parsimonious [6]. Gene acquisition followed by duplication appears to be a more plausible explanation for the presence of this gene family in Ciona. Other phylogenetic trees provide similar topologies, where Ciona sequences form monophyletic groups with homologs from algae and other animals (Figures 1,2 and 3; Additional file 2). For some identified gene families, other lines of evidence also argue strongly against the scenario of differential gene losses in other eukaryotes; such evidence includes 1) the presence of identifiable homologs only in animals and plastid-containing eukaryotes; 2) the monophyly of animal sequences within algal homologs; 3) uniquely shared domain structures between animal and algal sequences e.g. (Figure 3); 4) the availability of a large number of fungal genomes and the absence of some identified genes in fungi. Furthermore, two of the 14 gene families show cyanobacterial/plastidic affiliation (Figures 1A and 2)A, suggesting likely algal origin. Several other gene families also encode proteins that are targeted to chloroplasts in green plants (e.g., Arabidopsis thalianaChlamydomonas reinhardtii and Micromonas pusilla) (Additional file 1).
Accurate identification of HGT may be affected by multiple factors. Improper methodology, poor data quality, and insufficient or biased taxonomic sampling are among the factors leading to incorrect gene tree topologies [33]. Especially, long-branch attraction, which may occur when two sequences evolved much faster or slower than others, demands particular attention [34]. In order to reduce the potential complications resulting from these issues, we searched the NCBI non-redundant (nr) protein sequence database, ESTs and other sequence data to ensure a broad sequence sampling. We further examined the data quality and performed careful phylogenetic analyses by selecting the optimal substitution matrix for each dataset and using different computational algorithms. Nevertheless, with improved phylogenetic algorithms and more sequence data from other eukaryotes becoming available, we expect that the tree topologies for the identified genes may vary. It should also be noted that, because the phylogenetic approach tends to underestimate the number of acquired genes and because the Ciona genome was the sole query in our analyses, it is likely that other algae-related genes may exist in animals. Therefore, the identified genes in our analyses should not be considered as a definitive list of algae-related genes in animals.
Algal genes and the historical distribution of plastids
Given their presence and relatedness in multiple animal groups, the 14 genes families identified in this study, if indeed of algal origin (algal genes hereafter), most likely were acquired by the ancestral animal. This ancestral animal was probably unicellular, like its extant closest relatives choanflagellates [22]. The identified gene families might have been acquired through either symbioses or feeding activities. In the first scenario, the ancestral animal acquired algal genes as a result of endosymbioses with algal cells, followed by loss of plastids. The second scenario suggests that algal genes were routed to the nucleus of the ancestral animal after algal cells were phagocytosed. In most symbioses between algae and animals, algal symbionts either form loose extracellular association with animals or are sequestered in vacuoles of host cells [35]. Thus far, neither scenario is known to lead to gene transfers. Corals and salamander Ambystoma maculatum, for example, enjoy an endosymbiotic relationship with the dinoflagellate Symbiodinium and the green alga Oophila amblystomatis, respectively [36,37]. But neither EGT between corals and dinoflagellates nor between salamanders and green algal endosymbionts has been reported. By far, only the endosymbioses in sea slug Elysia chlorotica and cnidarian Hydra viridis are reportedly accompanied by transfer of algal genes [35,38-40]. Because such endosymbioses-mediated gene transfer is unknown in other animals, it might be extremely rare. Given the number of algal gene families identified in our study and their miscellaneous donor groups (Table 1; Figures 1,2 and 3; Additional file 2), it is highly improbable that these gene families are derived from such rare events. On the other hand, because most of the algal gene families identified in animals are derived from microscopic phytoplanktons that are widely distributed in aquatic environments, it is plausible that the ancestral animal obtained these algal genes via food capture.
The algal gene families identified in animals and their possible acquisition from food sources bear important implications for interpreting the historical distribution of plastids and eukaryotic photosynthesis. The origin of plastids is one of the most important evolutionary events in eukaryotic evolution. However, the historical distribution of plastids and photosynthesis in eukaryotes is still heatedly debated [14,16-18,41,42]. Because of the potential loss of plastids, algal genes in aplastidic eukaryotes have often been interpreted as derived from historical plastids. For example, algal genes in apicomplexan Cryptosporidium, heterokont Phytophthora, and ciliates have been cited as evidence for plastid losses in these taxa [14,33,43]. Similar arguments were also made for kinetoplastids based on the presence of “plant-like” genes [44], though whether these genes are indeed of plant origin is questionable [45,46]. Historical existence of plastids was also suggested for some protozoan species such as heterolobosean Naegleria[15,47]. Although endosymbiosis indeed appears to be a reasonable explanation for the observed data in some cases, recent findings of widespread HGT events in eukaryotes raise serious questions about whether all algal genes are derived from historical algal endosymbionts or plastids.
Many studies show that HGT from bacteria is frequent in unicellular eukaryotes [28,29,48]. There is also convincing evidence for HGT between eukaryotes [4,49-51]. In particular, recent studies identified over 100 algal genes in the choanoflagellate Monosiga[19,21]. The number of algal genes in Monosiga is considerably greater than those in many other aplastidic eukaryotes (e.g., ciliates, CryptosporidiumNaegleria etc) that have been suggested to contain historical plastids. Although the possibility that algal genes in Monosiga are derived from an obsolete algal endosymbiont or plastid cannot be confidently excluded, the phagotrophic lifestyle of Monosiga points to the possibility that these algal genes are derived from food sources. Considering that genes from other prokaryotes can be horizontally acquired by eukaryotes, it should not be surprising to see cyanobacterial (or plastidic) genes in aplastidic eukaryotes. Likewise, it should not be expected that algal genes are immune to HGT, considering that HGT events do occur between other eukaryotes. This may be particularly true for phagotrophic organisms feeding on miscellaneous microbes including unicellular algae. In such cases, algal genes in aplastidic eukaryotes may not necessarily represent relics of historical plastids. These genes should be treated equally as other HGT-derived genes when they are used to infer historical endosymbionts [41], unless other independent evidence (e.g. plastid existence in closely related taxa and compatible phylogenetic signals from multiple genes) is available. Our finding of algal genes in animals provides additional evidence about the distribution of algal genes in aplastidic eukaryotes. Additionally it is consistent with the earlier suggestion that algal genes are expected in phagotrophic eukaryotes or descendants of phagotrophic ancestors [21].
Evolutionary significance of algal genes in animals
Several cases of HGT between bacterial endosymbionts and their animal hosts have been documented [52-55]. Moreover, HGT from other free-living organisms has also been reported in cnidarians [39,56,57], bdelloid rotifers [58], insects [59], and nematodes [60-62] and other animals [63]. The acquired genes are often linked to novel phenotypes and abilities in recipient organisms, such as improved preying capability in cnidarians [56] and reduced susceptibility to predation in aphids [59]. These HGT cases predominantly occurred more recently in taxa of lower taxonomic ranks and, therefore, do not affect the evolution of entire animal lineage.
Despite the increasing number of HGT-derived genes reported in animals, a common belief is that HGT in animals is rare and its role is limited. This belief largely stems from the fact that the acquired genes need to overcome the germline barrier in order to be transmitted to next generations [4,5]. However, such a germline barrier should not exist in the unicellular ancestor of animals and it is conceivable that the ancestral animal, like its closest relatives choanoflagellates or many other unicellular eukaryotes [29,64], was subject to more frequent HGT. In our study, 13 of the 14 gene families identified in Ciona are most likely derived from ancient HGT events. These anciently acquired genes have been vertically inherited, duplicated and retained in diverse groups of extant animals, and thus have contributed to the long-term evolution of animals. Such frequent duplication following gene acquisition has also been observed in other studies [62,65-68] and, in many cases, associated with positive selection in recipient organisms [66,67].
Compared to Monosiga, the algal gene families identified in animals are considerably fewer and differ distinctly in their functions. Given the limited number of choanoflagellate genomes available, it is unclear what percentage of the identified algal gene families in Monosiga were anciently acquired. However, the lower number of anciently acquired genes in animals is consistent with the suggestion that most acquired genes will eventually be deleted from the recipient genome over time [69]. While the algal genes in Monosiga are predominantly involved in carbohydrate and amino acid metabolism [21], five of the 14 gene families, which account for 80% (74/92) of algal genes identified in Ciona, are functionally related to molecule transport. The vast majority of these transporter genes (72/74) belong to four solute carrier (SLC) families, including SLC6, SLC22, SLC23 and SLC34 (Table 1), which include transporters of neurotransmitter, organic cation, zwitterion/cation, organic anion, Na+−dependent vitamin C transport and phosphate ion, participating in uptake or excretion of numerous important compounds [70-73]. Besides these SLC families, another identified algal gene family is involved in potassium ion transport [74] (Table 1). As these genes encode multiple membrane-bound transporters, substrates such as amino acids, oligopeptides, sugars, inorganic cations and anions, essential metals, biogenic amines, vitamins, nucleosides and ammonia can be transferred across membranes, which in turn accelerates intercellular communication and leads to more efficient cellular metabolism. Several other acquired algal gene families in animals encode proteins that are functionally related to signaling and hormone biosynthesis. For example, the large short-chain dehydrogenase/reductase family (SDR) (Table 1) encodes enzymes involved in cellular differentiation and signaling [75]. Additionally, the biphenyl/valacyclovir hydrolase family is related to aromatic compound metabolism, which may allow the ancestral animal to access diverse digestible food sources.
As all functional information for the algal gene families identified in our analyses is based on investigations of homologs in humans and model organisms, whether these genes have exactly identical functions in other animals remains to be further studied. However, considering the high percent identifies (30%-80%) and close affinities between homologs from humans and other animals (Figures 1,2, and 3, Additional file 1), it is likely that these genes have similar primary functions. Even if the identified genes might have different functions in other animals, they could provide raw stocks for gene and functional differentiation. Extensive intercellular communication is a major feature distinguishing animals from unicellular eukaryotes, and such communication requires proteins related to molecule transport and signal transduction in multicellular animals [76,77]. The acquisition of genes related to molecular transport and signaling and their subsequent duplication might have facilitated the gene renovation and multicellular development in animals. Therefore, our findings, although based solely on analyses of algal genes in a relative small genome of Ciona, point to an important contribution of HGT to animal evolution.
Conclusions
Phylogenomic analyses of the tunicate C. intestinalis provide evidences for the existence of algal genes in animals. However, the existence of algal genes in animals does not necessarily constitute evidence for historical occurrence of plastids. Almost all algal gene families identified in our analyses were likely transferred to the ancestral animal and duplicated afterward. Most identified algal genes are related to molecule transport and signaling, suggesting their important role in intercellular communication and possibly the origin of multicellularity in animals.
Methods
Data sources and genome screening
The annotated genome of the tunicate C. intestinalis was downloaded from the Joint Genome Institute (http://genome.jgi-psf.org/Cioin2/Cioin2.home.html). Expressed sequence tags (ESTs) of 50 diverse eukaryotes (Additional file 3) were downloaded from the NCBI dbEST database and the Taxonomically Broad EST Database (TBestDB) [78]. Assembly of these ESTs was carried out using CAP3 [79]. The resulting consensus sequences were translated in all six frames using transeq of the EMBOSS package [80]. All other sequences used in the analyses were retrieved from the NCBI nr protein sequence database.
A comprehensive database was created by including the nr database and other available eukaryotic genomes (Additional file 3). Genome screening for candidates of HGT-derived genes was performed using AlienG[23]. AlienG presumes that sequence similarity is correlated with sequence relatedness and it identifies candidates of acquired genes by comparing query sequence similarity to homologs from potential donors and those from closely related taxa. Genes with identifiable homologs only in potential donors are also identified by AlienG. In this study, candidate genes of algal origin in C. intestinalis were obtained if they showed significantly higher sequence similarity to algal or cyanobacterial homologs than to those from other eukaryotes; the significant sequence similarity to algal homologs was empirically set to a bit score ratio of over 1.5.
Phylogenetic analyses
For each gene candidate predicted by AlienG, we performed further detailed analyses of taxonomic distribution, gene structure, and molecular phylogeny. Protein sequences were sampled from representative groups within each domain of life (bacteria, archaea, and eukaryotes) based on the blastp results against nr database. To ensure that sequences were sampled as broadly as possible, we also retrieved sequences from other available eukaryotic genomes and EST databases (see above section).
Multiple protein sequence alignments were performed using ClustalX [81,82] under the default settings, followed by manual refinement. Misaligned sequences, gaps and ambiguous sites were removed manually. The alignment data are available upon request. Phylogenetic analyses were performed with a maximum likelihood method using PhyML 3.0 [83] and a distance method using PHYLIP 3.69 [84]. ModelGenerator [85] was used to select the available model of protein substitution and rate heterogeneity that best fit each dataset. Bootstrap analyses with 100 pseudoreplicates were performed in both methods. Programs from the PHYLIP 3.69 package were used to create pseudoreplicate datasets (SEQBOOT), compute distance matrix (PROTDIST), calculate distance trees (NEIGHBOR), and generate the bootstrap consensus tree (CONSENSE). Gene trees were depicted by combining phylogenies from PhyML 3.0 and PHYLIP 3.69 using TreeGraph 2 [86,87].
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
JFW and JH conceived and designed the study and wrote manuscript. TN performed the analyses and wrote the manuscript. JY participated in the data interpretation and wrote the manuscript. GLS and YZ contributed to the data generation. All authors read and approved the final manuscript.
Supplementary Material
Table S1 and Table S2. Gene identifiers, chromosomal locations and functions for algal gene families identified in this study.
Figure S1-S8. Molecular phylogenies of algal genes identified in Ciona intestinalis.
Document. List of 50 eukaryotes whose ESTs were used and other 14 complete genome sequences were used in this study, in addition to the NCBI nr database.
Contributor Information
Ting Ni, Email: niting@mail.kiz.ac.cn.
Jipei Yue, Email: yuej@ecu.edu.
Guiling Sun, Email: sung@ecu.edu.
Yong Zou, Email: zouyong@mail.kiz.ac.cn.
Jianfan Wen, Email: wenjf@mail.kiz.ac.cn.
Jinling Huang, Email: huangj@ecu.edu.
Acknowledgements
We thank two anonymous reviewers for their helpful comments and suggestions. This work is supported in part by 973 Program (2007CB815705), the Knowledge Innovation Program (KSCX2-EW-J-23), grant (30830018) from the NSFC, and grant (2008GA002) from Yunnan province to JW and a NSF Assembling the Tree of Life (ATOL) grant (DEB 0830024) to JH.
References
- Keeling PJ, Palmer JD. Horizontal gene transfer in eukaryotic evolution. Nat Rev Genet. 2008;9(8):605–618. doi: 10.1038/nrg2386. [DOI] [PubMed] [Google Scholar]
- Gogarten JP, Doolittle WF, Lawrence JG. Prokaryotic evolution in light of gene transfer. Mol Biol Evol. 2002;19(12):2226–2238. doi: 10.1093/oxfordjournals.molbev.a004046. [DOI] [PubMed] [Google Scholar]
- Martin W, Rujan T, Richly E, Hansen A, Cornelsen S, Lins T, Leister D, Stoebe B, Hasegawa M, Penny D. Evolutionary analysis of Arabidopsis, cyanobacterial, and chloroplast genomes reveals plastid phylogeny and thousands of cyanobacterial genes in the nucleus. Proc Natl Acad Sci U S A. 2002;99(19):12246–12251. doi: 10.1073/pnas.182432999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson JO. Lateral gene transfer in eukaryotes. Cell Mol Life Sci. 2005;62(11):1182–1197. doi: 10.1007/s00018-005-4539-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurland CG, Canback B, Berg OG. Horizontal gene transfer: A critical view. Proc Natl Acad Sci U S A. 2003;100(17):9658–9662. doi: 10.1073/pnas.1632870100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang JL, Gogarten JP. Ancient horizontal gene transfer can benefit phylogenetic reconstruction. Trends in Genet. 2006;22(7):361–366. doi: 10.1016/j.tig.2006.05.004. [DOI] [PubMed] [Google Scholar]
- Huang JL, Gogarten JP. Did an ancient chlamydial endosymbiosis facilitate the establishment of primary plastids? Genome Biol. 2007;8(6):R99. doi: 10.1186/gb-2007-8-6-r99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moustafa A, Beszteri B, Maier UG, Bowler C, Valentin K, Bhattacharya D. Genomic Footprints of a Cryptic Plastid Endosymbiosis in Diatoms. Science. 2009;324(5935):1724–1726. doi: 10.1126/science.1172983. [DOI] [PubMed] [Google Scholar]
- Delwiche CF. Tracing the thread of plastid diversity through the tapestry of life. Am Nat. 1999;154:S164–S177. doi: 10.1086/303291. [DOI] [PubMed] [Google Scholar]
- Keeling PJ. Diversity and evolutionary history of plastids and their hosts. Am J Bot. 2004;91(10):1481–1493. doi: 10.3732/ajb.91.10.1481. [DOI] [PubMed] [Google Scholar]
- Bhattacharya D, Yoon HS, Hackett JD. Photosynthetic eukaryotes unite: endosymbiosis connects the dots. Bioessays. 2004;26(1):50–60. doi: 10.1002/bies.10376. [DOI] [PubMed] [Google Scholar]
- Lane CE, Archibald JM. The eukaryotic tree of life: endosymbiosis takes its TOL. Trends in Ecol & Evol. 2008;23(5):268–275. doi: 10.1016/j.tree.2008.02.004. [DOI] [PubMed] [Google Scholar]
- Maruyama S, Misawa K, Iseki M, Watanabe M, Nozaki H. Origins of a cyanobacterial 6-phosphogluconate dehydrogenase in plastid-lacking eukaryotes. BMC Evol Biol. 2008;8:151. doi: 10.1186/1471-2148-8-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Prieto A, Moustafa A, Bhattacharya D. Multiple genes of apparent algal origin suggest ciliates may once have been photosynthetic. Curr Biol. 2008;18(13):956–962. doi: 10.1016/j.cub.2008.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama S, Matsuzaki M, Misawa K, Nozaki H. Cyanobacterial contribution to the genomes of the plastid-lacking protists. BMC Evol Biol. 2009;9:197. doi: 10.1186/1471-2148-9-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier-Smith T. Principles of protein and lipid targeting in secondary symbiogenesis: Euglenoid, dinoflagellate, and sporozoan plastid origins and the eukaryote family tree. J Eukaryot Microbiol. 1999;46(4):347–366. doi: 10.1111/j.1550-7408.1999.tb04614.x. [DOI] [PubMed] [Google Scholar]
- Nozaki H, Matsuzaki M, Takahara M, Misumi O, Kuroiwa H, Hasegawa M, Shin-i T, Kohara Y, Ogasawara N, Kuroiwa T. The phylogenetic position of red algae revealed by multiple nuclear genes from mitochondria-containing eukaryotes and an alternative hypothesis on the origin of plastids. J Mol Evol. 2003;56(4):485–497. doi: 10.1007/s00239-002-2419-9. [DOI] [PubMed] [Google Scholar]
- Nozaki H. A new scenario of plastid evolution: plastid primary endosymbiosis before the divergence of the "Plantae," emended. J Plant Res. 2005;118(4):247–255. doi: 10.1007/s10265-005-0219-1. [DOI] [PubMed] [Google Scholar]
- Nedelcu AM, Miles IH, Fagir AM, Karol K. Adaptive eukaryote-to-eukaryote lateral gene transfer: stress-related genes of algal origin in the closest unicellular relatives of animals. J Evol Biol. 2008;21(6):1852–1860. doi: 10.1111/j.1420-9101.2008.01605.x. [DOI] [PubMed] [Google Scholar]
- Nedelcu AM, Blakney AJC, Logue KD. Functional replacement of a primary metabolic pathway via multiple independent eukaryote-to-eukaryote gene transfers and selective retention. J Evol Biol. 2009;22(9):1882–1894. doi: 10.1111/j.1420-9101.2009.01797.x. [DOI] [PubMed] [Google Scholar]
- Sun GL, Yang ZF, Ishwar A, Huang JL. Algal Genes in the Closest Relatives of Animals. Mol Biol Evol. 2010;27(12):2879–2889. doi: 10.1093/molbev/msq175. [DOI] [PubMed] [Google Scholar]
- Lang BF, O'Kelly C, Nerad T, Gray MW, Burger G. The closest unicellular relatives of animals. Curr Biol. 2002;12(20):1773–1778. doi: 10.1016/s0960-9822(02)01187-9. [DOI] [PubMed] [Google Scholar]
- Tian J, Sun GL, Ding Q, Huang JL, Oruganti S, Xie B. AlienG: an effective computational tool for phylogenetic identification of horizontally transferred genes. New Orleans, Lousiana: The third International Conference on Bioinformatics and Computational Biology (BICoB); 2011. [Google Scholar]
- Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K. et al. The Pfam protein families database. Nucleic Acids Res. 2010;38:D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman H, Lawrence JG, Groisman EA. Lateral gene transfer and the nature of bacterial innovation. Nature. 2000;405(6784):299–304. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- Jain R, Rivera MC, Moore JE, Lake JA. Horizontal gene transfer in microbial genome evolution. Theor Popul Biol. 2002;61(4):489–495. doi: 10.1006/tpbi.2002.1596. [DOI] [PubMed] [Google Scholar]
- Andersson JO, Sarchfield SW, Roger AJ. Gene transfers from Nanoarchaeota to an ancestor of diplomonads and parabasalids. Mol Biol Evol. 2005;22(1):85–90. doi: 10.1093/molbev/msh254. [DOI] [PubMed] [Google Scholar]
- Huang JL, Mullapudi N, Sicheritz-Ponten T, Kissinger JC. A first glimpse into the pattern and scale of gene transfer in the Apicomplexa. Int J Parasitol. 2004;34(3):265–274. doi: 10.1016/j.ijpara.2003.11.025. [DOI] [PubMed] [Google Scholar]
- Ricard G, McEwan NR, Dutilh BE, Jouany JP, Macheboeuf D, Mitsumori M, McIntosh FM, Michalowski T, Nagamine T, Nelson N. et al. Horizontal gene transfer from Bacteria to rumen Ciliates indicates adaptation to their anaerobic, carbohydrates-rich environment. Bmc Genomics. 2006;7:22. doi: 10.1186/1471-2164-7-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett JD, Yoon HS, Soares MB, Bonaldo MF, Casavant TL, Scheetz TE, Nosenko T, Bhattacharya D. Migration of the plastid genome to the nucleus in a peridinin dinoflagellate. Curr Biol. 2004;14(3):213–218. doi: 10.1016/j.cub.2004.01.032. [DOI] [PubMed] [Google Scholar]
- Dagan T, Martin W. Ancestral genome sizes specify the minimum rate of lateral gene transfer during prokaryote evolution. Proc Natl Acad Sci U S A. 2007;104(3):870–875. doi: 10.1073/pnas.0606318104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snel B, Bork P, Huynen MA. Genomes in flux: The evolution of archaeal and proteobacterial gene content. Genome Res. 2002;12(1):17–25. doi: 10.1101/gr.176501. [DOI] [PubMed] [Google Scholar]
- Huang JL, Mullapudi N, Lancto CA, Scott M, Abrahamsen MS, Kissinger JC. Phylogenomic evidence supports past endosymbiosis, intracellular and horizontal gene transfer in Cryptosporidium parvum. Genome Biol. 2004;5(11):R88. doi: 10.1186/gb-2004-5-11-r88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe H, Zhou Y, Brinkmann H, Rodrigue N, Delsuc F. Heterotachy and long-branch attraction in phylogenetics. BMC Evol Biol. 2005;5:50. doi: 10.1186/1471-2148-5-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanten JJ, Pierce SK. Synthesis of several light-harvesting complex I polypeptides is blocked by cycloheximide in symbiotic chloroplasts in the sea slug, Elysia chlorotica (Gould): A case for horizontal gene transfer between alga and animal? Biol Bull. 2001;201(1):34–44. doi: 10.2307/1543523. [DOI] [PubMed] [Google Scholar]
- Wooldridge SA. A new conceptual model for the warm-water breakdown of the coral-algae endosymbiosis. Mar Freshw Res. 2009;60(6):483–496. [Google Scholar]
- Petherick A. Salamander's egg surprise. Nature. 2010;466(7307):675–675. doi: 10.1038/466675a. [DOI] [PubMed] [Google Scholar]
- Rumpho ME, Worful JM, Lee J, Kannan K, Tyler MS, Bhattacharya D, Moustafa A, Manhart JR. Horizontal gene transfer of the algal nuclear gene psbO to the photosynthetic sea slug Elysia chlorotica. Proc Natl Acad Sci U S A. 2008;105(46):17867–17871. doi: 10.1073/pnas.0804968105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habetha M, Bosch TCG. Symbiotic Hydra express a plant-like peroxidase gene during oogenesis. J Exp Biol. 2005;208(11):2157–2164. doi: 10.1242/jeb.01571. [DOI] [PubMed] [Google Scholar]
- Pierce SK, Fang X, Schwartz JA, Jiang X, Zhao W, Curtis NE, Kocot KM, Yang B, Wang J. Transcriptomic Evidence for the Expression of Horizontally Transferred Algal Nuclear Genes in the Photosynthetic Sea Slug. Molecular biology and evolution: Elysia chlorotica; 2012. [DOI] [PubMed] [Google Scholar]
- Stiller JW, Huang JL, Ding Q, Tian J, Goodwillie C. Are algal genes in nonphotosynthetic protists evidence of historical plastid endosymbioses? Bmc Genomics. 2009;10:484. doi: 10.1186/1471-2164-10-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodyl A. Do plastid-related characters support the chromalveolate hypothesis? J Phycol. 2005;41(3):712–719. [Google Scholar]
- Tyler BM, Tripathy S, Zhang XM, Dehal P, Jiang RHY, Aerts A, Arredondo FD, Baxter L, Bensasson D, Beynon JL. et al. Phytophthora genome sequences uncover evolutionary origins and mechanisms of pathogenesis. Science. 2006;313(5791):1261–1266. doi: 10.1126/science.1128796. [DOI] [PubMed] [Google Scholar]
- Hannaert V, Saavedra E, Duffieux F, Szikora JP, Rigden DJ, Michels PAM, Opperdoes FR. Plant-like traits associated with metabolism of Trypanosoma parasites. Proc Natl Acad Sci U S A. 2003;100(3):1067–1071. doi: 10.1073/pnas.0335769100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Sayed NM, Myler PJ, Blandin G, Berriman M, Crabtree J, Aggarwal G, Caler E, Renauld H, Worthey EA, Hertz-Fowler C. et al. Comparative genomics of trypanosomatid parasitic protozoa. Science. 2005;309(5733):404–409. doi: 10.1126/science.1112181. [DOI] [PubMed] [Google Scholar]
- Rogers M, Keeling PJ. Lateral transfer and recompartmentalization of Calvin cycle enzymes of plants and algae. J Mol Evol. 2004;58(4):367–375. doi: 10.1007/s00239-003-2558-7. [DOI] [PubMed] [Google Scholar]
- Andersson JO, Roger AJ. A cyanobacterial gene in nonphotosynthetic protists - An early chloroplast acquisition in eukaryotes? Curr Biol. 2002;12(2):115–119. doi: 10.1016/s0960-9822(01)00649-2. [DOI] [PubMed] [Google Scholar]
- Whitaker JW, McConkey GA, Westhead DR. The transferome of metabolic genes explored: analysis of the horizontal transfer of enzyme encoding genes in unicellular eukaryotes. Genome Biol. 2009;10(4):R36. doi: 10.1186/gb-2009-10-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obornik M, Green BR. Mosaic origin of the heme biosynthesis pathway in photosynthetic eukaryotes. Mol Biol Evol. 2005;22(12):2343–2353. doi: 10.1093/molbev/msi230. [DOI] [PubMed] [Google Scholar]
- Richards TA, Dacks JB, Jenkinson JM, Thornton CR, Talbot NJ. Evolution of filamentous plant pathogens: Gene exchange across eukaryotic kingdoms. Curr Biol. 2006;16(18):1857–1864. doi: 10.1016/j.cub.2006.07.052. [DOI] [PubMed] [Google Scholar]
- Richards TA, Soanes DM, Foster PG, Leonard G, Thomton CR, Talbot NJ. Phylogenomic analysis demonstrates a pattern of rare and ancient horizontal gene transfer between plants and fungi. Plant Cell. 2009;21(7):1897–1911. doi: 10.1105/tpc.109.065805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotopp JCD, Clark ME, Oliveira DCSG, Foster JM, Fischer P, Torres MC, Giebel JD, Kumar N, Ishmael N, Wang SL. et al. Widespread lateral gene transfer from intracellular bacteria to multicellular eukaryotes. Science. 2007;317(5845):1753–1756. doi: 10.1126/science.1142490. [DOI] [PubMed] [Google Scholar]
- Klasson L, Kambris Z, Cook PE, Walker T, Sinkins SP. Horizontal gene transfer between Wolbachia and the mosquito Aedes aegypti. Bmc Genomics. 2009;10:33. doi: 10.1186/1471-2164-10-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikoh N, Tanaka K, Shibata F, Kondo N, Hizume M, Shimada M, Fukatsu T. Wolbachia genome integrated in an insect chromosome: Evolution and fate of laterally transferred endosymbiont genes. Genome Res. 2008;18(2):272–280. doi: 10.1101/gr.7144908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotopp JCD. Horizontal gene transfer between bacteria and animals. Trends in Genetics. 2011;27(4):157–163. doi: 10.1016/j.tig.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denker E, Bapteste E, Le Guyader H, Manuel M, Rabet N. Horizontal gene transfer and the evolution of cnidarian stinging cells. Curr Biol. 2008;18(18):R858–R859. doi: 10.1016/j.cub.2008.07.031. [DOI] [PubMed] [Google Scholar]
- Chapman JA, Kirkness EF, Simakov O, Hampson SE, Mitros T, Weinmaier T, Rattei T, Balasubramanian PG, Borman J, Busam D. et al. The dynamic genome of hydra. Nature. 2010;464(7288):592–596. doi: 10.1038/nature08830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladyshev EA, Meselson M, Arkhipova IR. Massive horizontal gene transfer in bdelloid rotifers. Science. 2008;320(5880):1210–1213. doi: 10.1126/science.1156407. [DOI] [PubMed] [Google Scholar]
- Moran NA, Jarvik T. Lateral transfer of genes from fungi underlies carotenoid production in aphids. Science. 2010;328(5978):624–627. doi: 10.1126/science.1187113. [DOI] [PubMed] [Google Scholar]
- Craig JP, Bekal S, Hudson M, Domier L, Niblack T, Lambert KN. Analysis of a horizontally transferred pathway involved in vitamin B(6) biosynthesis from the soybean cyst nematode Heterodera glycines. Mol Biol Evol. 2008;25(10):2085–2098. doi: 10.1093/molbev/msn141. [DOI] [PubMed] [Google Scholar]
- Mayer WE, Schuster LN, Bartelmes G, Dieterich C, Sommer RJ. Horizontal gene transfer of microbial cellulases into nematode genomes is associated with functional assimilation and gene turnover. BMC Evol Biol. 2011;11:13. doi: 10.1186/1471-2148-11-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danchin EGJ, Rosso MN, Vieira P, de Almeida-Engler J, Coutinho PM, Henrissat B, Abad P. Multiple lateral gene transfers and duplications have promoted plant parasitism ability in nematodes. Proc Natl Acad Sci U S A. 2010;107(41):17651–17656. doi: 10.1073/pnas.1008486107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace JK, Gilbert C, Clark MS, Feschotte C. Repeated horizontal transfer of a DNA transposon in mammals and other tetrapods. Proc Natl Acad Sci U S A. 2008;105(44):17023–17028. doi: 10.1073/pnas.0806548105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armbrust EV, Berges JA, Bowler C, Green BR, Martinez D, Putnam NH, Zhou SG, Allen AE, Apt KE, Bechner M. et al. The genome of the diatom Thalassiosira pseudonana: Ecology, evolution, and metabolism. Science. 2004;306(5693):79–86. doi: 10.1126/science.1101156. [DOI] [PubMed] [Google Scholar]
- Hooper SD, Berg OG. Duplication is more common among laterally transferred genes than among indigenous genes. Genome Biol. 2003;4(8):R48. doi: 10.1186/gb-2003-4-8-r48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun G, Yang Z, Kosch T, Summers K, Huang J. Evidence for acquisition of virulence effectors in pathogenic chytrids. BMC Evol Biol. 2011;11:195. doi: 10.1186/1471-2148-11-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novakova E, Moran NA. Diversification of genes for carotenoid biosynthesis in aphids following an ancient transfer from a fungus. Mol Biol Evol. 2012;29(1):313–323. doi: 10.1093/molbev/msr206. [DOI] [PubMed] [Google Scholar]
- Belbahri L, Calmin G, Mauch F, Andersson JO. Evolution of the cutinase gene family: evidence for lateral gene transfer of a candidate Phytophthora virulence factor. Gene. 2008;408(1–2):1–8. doi: 10.1016/j.gene.2007.10.019. [DOI] [PubMed] [Google Scholar]
- Lawrence JG, Ochman H. Molecular archaeology of the Escherichia coli genome. Proc Natl Acad Sci U S A. 1998;95(16):9413–9417. doi: 10.1073/pnas.95.16.9413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koepsell H, Endou H. The SLC22 drug transporter family. Pflugers Archiv-European J Physiology. 2004;447(5):666–676. doi: 10.1007/s00424-003-1089-9. [DOI] [PubMed] [Google Scholar]
- Eraly SA, Monte JC, Nigam SK. Novel slc22 transporter homologs in fly, worm, and human clarify the phylogeny of organic anion and cation transporters. Physiological Genomics. 2004;18(1):12–24. doi: 10.1152/physiolgenomics.00014.2004. [DOI] [PubMed] [Google Scholar]
- Takanaga H, Mackenzie B, Hediger MA. Sodium-dependent ascorbic acid transporter family SLC23. Pflugers Archiv-European J Physiology. 2004;447(5):677–682. doi: 10.1007/s00424-003-1104-1. [DOI] [PubMed] [Google Scholar]
- Murer H, Forster I, Biber J. The sodium phosphate cotransporter family SLC34. Pflugers Archiv-European J Physiology. 2004;447(5):763–767. doi: 10.1007/s00424-003-1072-5. [DOI] [PubMed] [Google Scholar]
- Santoro B, Liu DT, Yao H, Bartsch D, Kandel ER, Siegelbaum SA, Tibbs GR. Identification of a gene encoding a hyperpolarization-activated pacemaker channel of brain. Cell. 1998;93(5):717–729. doi: 10.1016/s0092-8674(00)81434-8. [DOI] [PubMed] [Google Scholar]
- Kallberg Y, Oppermann U, Jornvall H, Persson B. Short-chain dehydrogenase/reductase (SDR) relationships: A large family with eight clusters common to human, animal, and plant genomes. Protein Sci. 2002;11(3):636–641. doi: 10.1110/ps.26902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokas A. The molecular origins of multicellular transitions. Curr Opin Genet Dev. 2008;18(6):472–478. doi: 10.1016/j.gde.2008.09.004. [DOI] [PubMed] [Google Scholar]
- Rokas A. The Origins of multicellularity and the early history of the genetic toolkit for animal development. Annu Rev Genet. 2008;42:235–251. doi: 10.1146/annurev.genet.42.110807.091513. [DOI] [PubMed] [Google Scholar]
- O'Brien EA, Koski LB, Zhang Y, Yang LS, Wang E, Gray MW, Burger G, Lang BF. TBestDB: a taxonomically broad database of expressed sequence tags (ESTs) Nucleic Acids Res. 2007;35:D445–D451. doi: 10.1093/nar/gkl770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang XQ, Madan A. CAP3: A DNA sequence assembly program. Genome Res. 1999;9(9):868–877. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A. EMBOSS: The European molecular biology open software suite. Trends in Genetics. 2000;16(6):276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25(24):4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R. et al. Clustal W and clustal X version 2.0. Bioinformatics. 2007;23(21):2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0. Syst Biol. 2010;59(3):307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP (Phylogeny Inference Package) version 3.65. Seattle (WA): Department of Genome Sciences, University of Washington; 2005. [Google Scholar]
- Keane TM, Creevey CJ, Pentony MM, Naughton TJ, McInerney JO. Assessment of methods for amino acid matrix selection and their use on empirical data shows that ad hoc assumptions for choice of matrix are not justified. BMC Evol Biol. 2006;6:29. doi: 10.1186/1471-2148-6-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller J, Muller K. TREEGRAPH: automated drawing of complex tree figures using an extensible tree description format. Mol Ecol Notes. 2004;4(4):786–788. [Google Scholar]
- Stover BC, Muller KF. TreeGraph 2: Combining and visualizing evidence from different phylogenetic analyses. BMC Bioinforma. 2010;11:7. doi: 10.1186/1471-2105-11-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 and Table S2. Gene identifiers, chromosomal locations and functions for algal gene families identified in this study.
Figure S1-S8. Molecular phylogenies of algal genes identified in Ciona intestinalis.
Document. List of 50 eukaryotes whose ESTs were used and other 14 complete genome sequences were used in this study, in addition to the NCBI nr database.