Abstract
Background
Dengue is the most important mosquito-borne viral disease worldwide. Dengue virus comprises four antigenically related viruses named dengue virus type 1 to 4 (DENV1-4). DENV-3 was re-introduced into the Americas in 1994 causing outbreaks in Nicaragua and Panama. DENV-3 was introduced in Brazil in 2000 and then spread to most of the Brazilian States, reaching the neighboring country, Paraguay in 2002. In this study, we have analyzed the phylogenetic relationship of DENV-3 isolated in Brazil and Paraguay with viruses isolated worldwide. We have also analyzed the evolutionary divergence dynamics of DENV-3 viruses.
Results
The entire open reading frame (ORF) of thirteen DENV-3 isolated in Brazil (n = 9) and Paraguay (n = 4) were sequenced for phylogenetic analysis. DENV-3 grouped into three main genotypes (I, II and III). Several internal clades were found within each genotype that we called lineage and sub-lineage. Viruses included in this study belong to genotype III and grouped together with viruses isolated in the Americas within the lineage III. The Brazilian viruses were further segregated into two different sub-lineage, A and B, and the Paraguayan into the sub-lineage B. All three genotypes showed internal grouping. The nucleotide divergence was in average 6.7% for genotypes, 2.7% for lineages and 1.5% for sub-lineages. Phylogenetic trees constructed with any of the protein gene sequences showed the same segregation of the DENV-3 in three genotypes.
Conclusion
Our results showed that two groups of DENV-3 genotypes III circulated in Brazil during 2002–2009, suggesting different events of introduction of the virus through different regions of the country. In Paraguay, only one group DENV-3 genotype III is circulating that is very closely related to the Brazilian viruses of sub-lineage B. Different degree of grouping can be observed for DENV-3 and each group showed a characteristic evolutionary divergence. Finally, we have observed that any protein gene sequence can be used to identify the virus genotype.
Background
Dengue is the most important mosquito-borne viral disease in tropical and subtropical regions. An estimated 50 million dengue infections occur annually and approximately 2.5 billion people live in dengue endemic countries [1]. Dengue virus (DENV) infection can be assyntomatic or lead to a wide spectrum of clinical manifestation, ranging from an undifferentiated fever, the self-limiting non-severe dengue fever (DF), to the severe dengue haemorrhagic fever (DHF), sometimes with fatal outcomes.
DENV is an enveloped virus and comprise four distinct serotypes (DENV1-4) that belong to the genus Flavivirus, family Flaviviridae. The viral genome is constituted by a single copy of a single-stranded, positive-sense RNA of approximately 11 kb in size, containing a single open reading frame (ORF), flanked by untranslated regions (5′UTR and 3′UTR) [2]. The ORF encodes a single polyprotein, which is co- and pos-translationally cleaved into 3 structural (C, prM and E) and 7 nonstructural proteins (NS1-NS2A-NS2B-NS3-NS4A-NS4B-NS5) [3].
DENV-3 was re-introduced into the Americas in 1994, specifically in Nicaragua and Panama, and then spread to other Central American countries, Mexico, the Caribbean countries, and finally South America [4-10]. In 2001/2002, a large outbreak of DENV-3 occurred in Rio de Janeiro [9,11]. DENV-3 demonstrated its greatest epidemic potential, spreading into most of the Brazilian States and, by March 2002, into the neighboring country, Paraguay [9,12]. Our previous phylogenetic study based on the E protein gene and the 3′UTR has suggested that DENV-3 was introduced into Brazil through Rio de Janeiro as well as by the Northern Region, at least in three different occasions and subsequently has spread to Paraguay [12]. Several phylogenetic studies using partial genomic sequences of DENV were carried out to analyze its molecular epidemiology [13-16]. However, it is believed that a better picture of the dynamics of viral populations could be analyzed by sequencing the entire viral genome. Recently, complete genome analyses have been performed to study DENV phylodinamics in Singapore and India [17,18]. In the present study, we have analyzed the phylogenetic relationship of 13 DENV-3 isolated in Brazil and Paraguay with viruses isolated worldwide, using their entire RNA genome sequences.
Results
Genome sequencing and phylogenetic analysis of viruses isolated in Brazil and Paraguay
Six fragments with overlapping regions amplified by RT-PCR were subjected to direct nucleotide sequencing to obtain the full-length genome sequence of the 13 viruses included in this study (Table 1). The assembled sequences showed that most of the viruses have a genome of 10,707 nucleotides, with the exception of D3BR/MR9/2003 isolate, which showed a deletion of 8 nucleotides between positions 10,276 and 10,284 at the 3′UTR of the viral genome. The same deletion was also observed in sequences retrieved from the GenBank for 7 viruses isolated in Brazil, 1 in Puerto Rico, and 25 in Vietnam (Additional file 1).
Table 1.
DENV-3 from Brazil and Paraguay used in this study
| Virus | Passage (No.) | City/State/Region | Country/year of isolation |
|---|---|---|---|
| D3BR/SL3/02 |
2 |
São Luis/Maranhão/Northeast |
Brazil/2002 |
| D3BR/BV4/02 |
2 |
Boa Vista/Roraima/North |
Brazil/2002 |
| D3BR/CU6/02 |
2 |
Cuiabá/Mato Grosso/Midwest |
Brazil/2002 |
| D3BR/RP1/03 |
2 |
Ribeirão Preto/São Paulo/Southeast |
Brazil/2003 |
| D3BR/PV1/03 |
2 |
Porto Velho/Rondônia/North |
Brazil/2003 |
| D3BR/MR9/03 |
2 |
Marituba/Pará/North |
Brazil/2003 |
| D3BR/BR8/04 |
2 |
Bragança/Pará/North |
Brazil/2004 |
| D3BR/ACN/2007 |
2 |
Ribeirão Preto/São Paulo/Southeast |
Brazil/2007 |
| D3BR/AL95/2009 |
2 |
Ribeirão Preto/São Paulo/Southeast |
Brazil/2009 |
| D3PY/AS12/02 |
2 |
Asunción/Central/Eastern |
Paraguay/2002 |
| D3PY/PJ4/03 |
2 |
Pedro Juan Caballero/Amambay/Eastern |
Paraguay/2003 |
| D3PY/AS10/03 |
2 |
Asunción/Central/Eastern |
Paraguay/2003 |
| D3PY/SUS/2003 | 2 | Asunción/Central/Eastern | Paraguay/2003 |
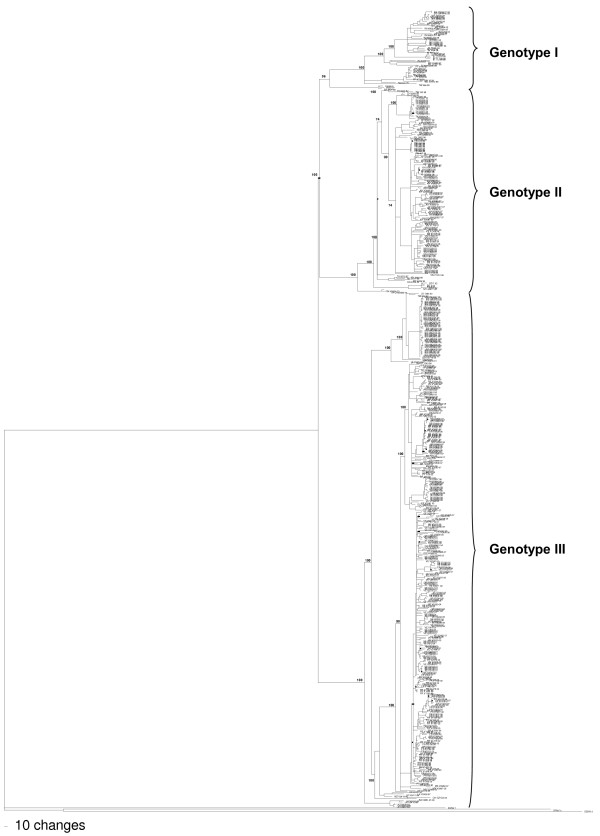
For phylogenetic analysis, we retrieved from the GenBank the so-called complete DENV-3 sequences. However, several sequences lack 5′ and 3′ ends or had the UTRs not validated. Therefore, in the alignment we included only the open reading frame sequences (10,168 nucleotides long). Thus, the alignment included our 13 sequences and 527 sequences of DENV-3 deposited in the GenBank. Based on this alignment, a phylogenetic tree was constructed (Figure 1), showing that DENV-3 comprises three genetic groups or genotypes (I, II and III). The 13 isolates described in this study were grouped within genotype III, together with viruses isolated in the Americas.
Figure 1.
Neighbor-joining phylogenetic trees based on the entire ORFs derived from 542 global samples of the DENV-3 inferred with PAUP program. The bootstrap are indicated at important nodes. The best-fit model of nucleotide substitution for phylogenetic reconstruction used was TrN + I + G model with gamma-distributed rate variation (G = 0.9908) and a proportion of invariable sites (I = 0.3678). Branch lengths are proportional to percentage of divergence.
Phylogenetic relationship of genotype III viruses
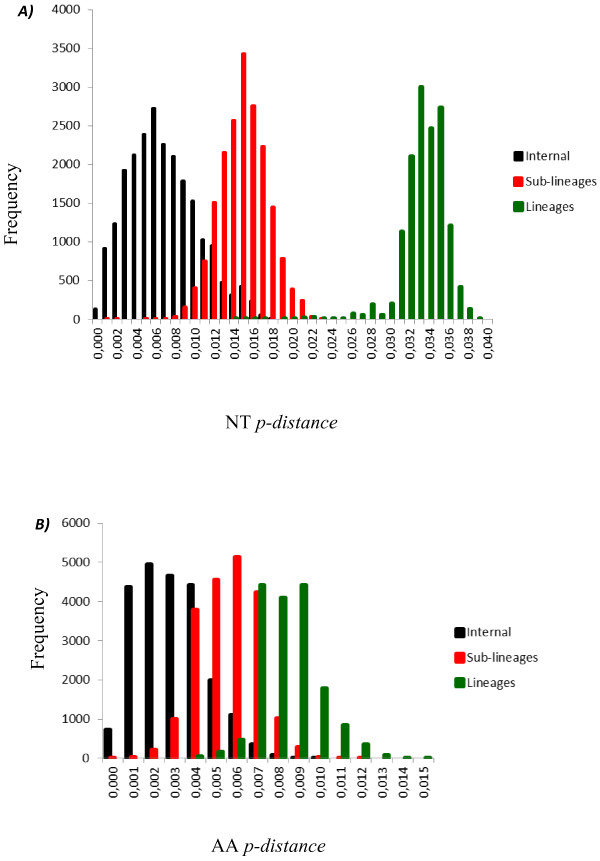
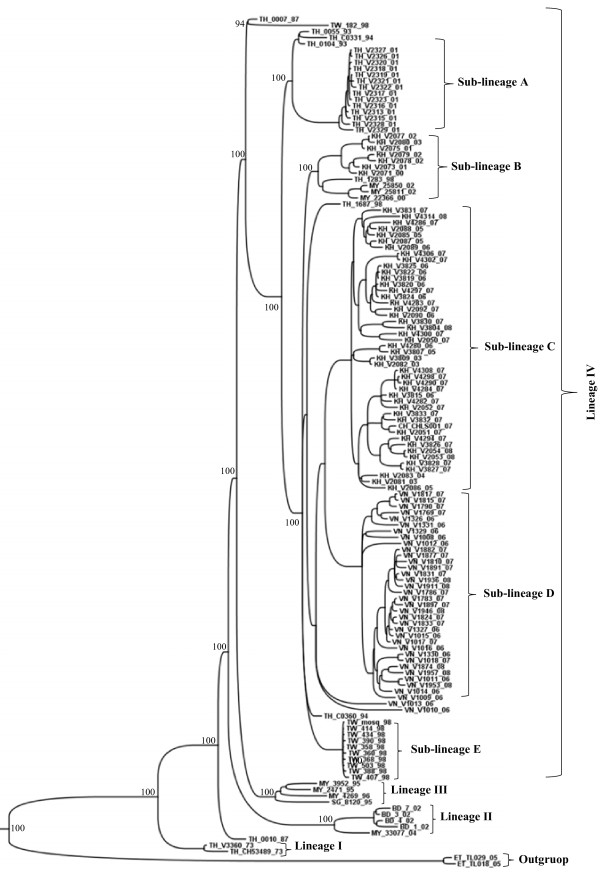
To perform a more accurate analysis of the phylogenetic relationships of viruses isolated in Brazil and Paraguay, ORF sequences of genotype III viruses (n = 347) were used to construct other phylogenetic trees using distance and Bayesian methods; these phylogenetic trees showed similar topology (Figure 2). In addition, the evolutionary divergence among sequences and the presence of amino acid motif were analyzed. Based on the topology of the tree, the frequency distribution profile of the divergence among sequences (Figure 3) and the presence of characteristic amino acid motifs (Additional file 2), genotype III viruses were clustered into three lineages (I, II and III) (Figure 2). Viruses of lineage III were further clustered into four monophyletic groups (sub-lineages A, B, C, and D) (Figure 2). The mean divergence among lineages ranged from 2.9 to 3.4% for nucleotide sequences (Table 2), and from 0.6 to 1.0% for amino acid sequences (Table 2). While the mean divergence among sub-lineages ranged from 1.0 to 1.6% for nucleotide sequences, and from 0.5 to 0.6% for amino acid sequences (Table 2). All the mean values coincided with the highest peaks in the frequency distribution profile (Figure 3). The viruses described in this study grouped together with viruses isolated in the Americas within the lineage III, distributed into two different groups, sub-lineages A and B. The Brazilian viruses isolated in the Northern Region, BR_BV4_02 from Boa Vista, Roraima, and BR_BR8_04 from Belen, Para, grouped with other Brazilian viruses isolated in the same region, with viruses isolated in the Caribbean islands (Martinique, Trinidad and Tobago, St. Lucia, Anguilla and Puerto Rico) and in the north of South America (French Guyana and Venezuela) (Figure 2, sub-lineage A). The other Brazilian isolates (BR_PV1_03, BR_CU6_02, BR_MR9_03, BR_SL3_02, BR_ACN_07, BR_AL95_09, and BR_RP1_2003) and the Paraguayan isolates (PY_AS10_03, PY_AS12_02, PY_SUS_03, PY_PJ4_03) grouped with viruses isolated in other regions of Brazil within the sub-lineage B (Figure 2, sub-lineage B). Sub-lineage C includes viruses isolated in Nicaragua, Puerto Rico, Venezuela, Peru and Ecuador. Most of the viruses isolated in Venezuela and Puerto Rico belong to the sub-lineage D, which also include viruses isolated in Colombia. Interestingly, BR_V2386_03 is the only Brazilian virus that grouped with viruses of sub-lineage D. Finally, the lineage I is composed only by old isolates of SriLanka (1983–1989) and lineage II by viruses isolated in Asia (1993–2005). In addition, viruses from Asian (two viruses from SriLanka isolated in 1989 and 1997; one virus from China isolated in 2009) and East Africa (one virus from Mozambique isolated in 1985 and one virus from India isolated in 2003) are located basally in the branch that containing the American isolates (Lineage III) and do not form a monophyletic group.
Figure 2.
Bayesian phylogenetic trees based on the entire ORFs derived from 347 global samples of the genotype III DENV-3 inferred with MrBayes program. The posterior probabilities are expressed in percent and indicated at important nodes. The best fit-model of nucleotide substitution for Bayesian phylogenetic reconstruction used was under a General Time Reversible model with gamma-distributed rate variation (G = 1.2923) and a proportion of invariable sites (I = 0.4777) (GTR + G + I).
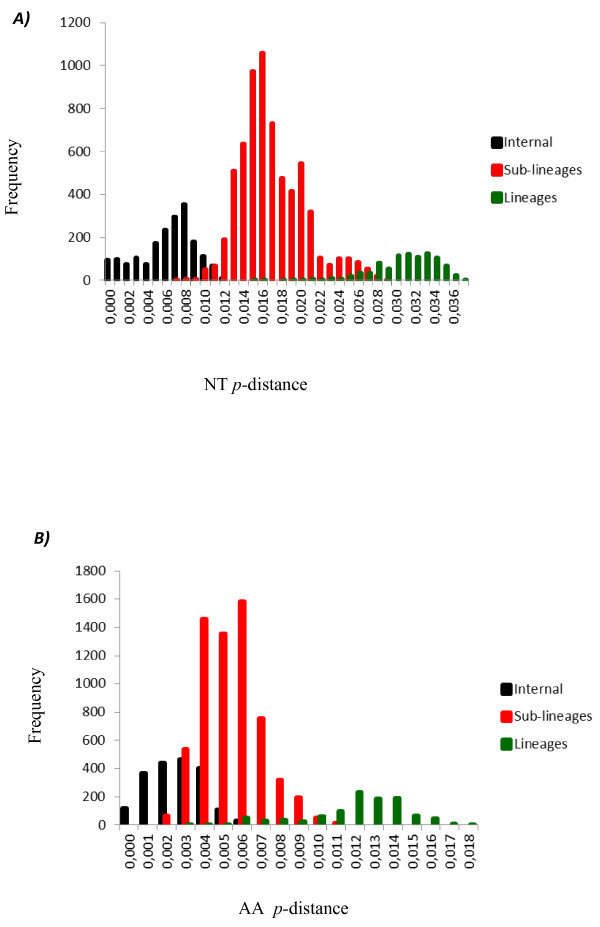
Figure 3.
The frequency distribution profile of pairwise distances among sequences of genotype III, based on the entire ORFs derived from 347 global samples of the genotype III DENV-3. A ). The frequency distribution profile distance of nucleotide (NT). B) The frequency distribution profile distance of amino acids (AA). The NT and AA p-distance are represented on the X axis and the frequency are represented on the Y axis.
Table 2.
Mean of the evolutionary divergence (NT and AA) from all sequence pairs between genotypes III groups
| Genotype III groups | NTp-distance | % | AAp-distance | % |
|---|---|---|---|---|
| Lineage I vs Lineage II |
0.029 |
2.9 |
0.006 |
0.6 |
| Lineage I vs Lineage III |
0.034 |
3.4 |
0.010 |
1.0 |
| Lineage II vs Lineage III |
0.030 |
3.0 |
0.008 |
0.8 |
| Lineage III [Sub-lineage A vs Sub-lineage B] |
0.010 |
1.0 |
0.005 |
0.5 |
| Lineage III [Sub-lineage A vs Sub-lineage C] |
0.013 |
1.3 |
0.005 |
0.5 |
| Lineage III [Sub-lineage A vs Sub-lineage D] |
0.014 |
1.4 |
0.005 |
0.5 |
| Lineage III [Sub-lineage B vs Sub-lineage C] |
0.014 |
1.4 |
0.005 |
0.5 |
| Lineage III [Sub-lineage B vs Sub-lineage D] |
0.015 |
1.5 |
0.006 |
0.6 |
| Lineage III [Sub-lineage C vs Sub-lineage D] | 0.016 | 1.6 | 0.005 | 0.5 |
Phylogenetic relationship among viruses from genotypes I and II
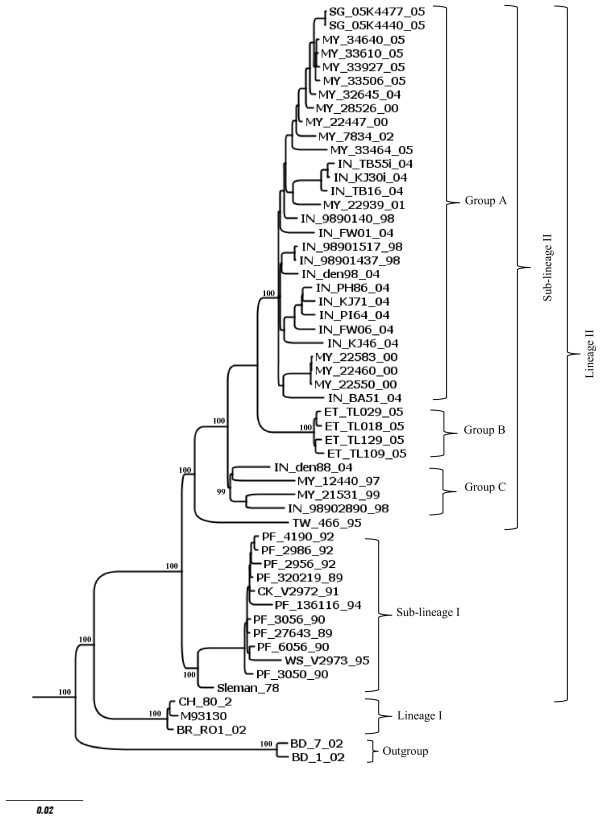
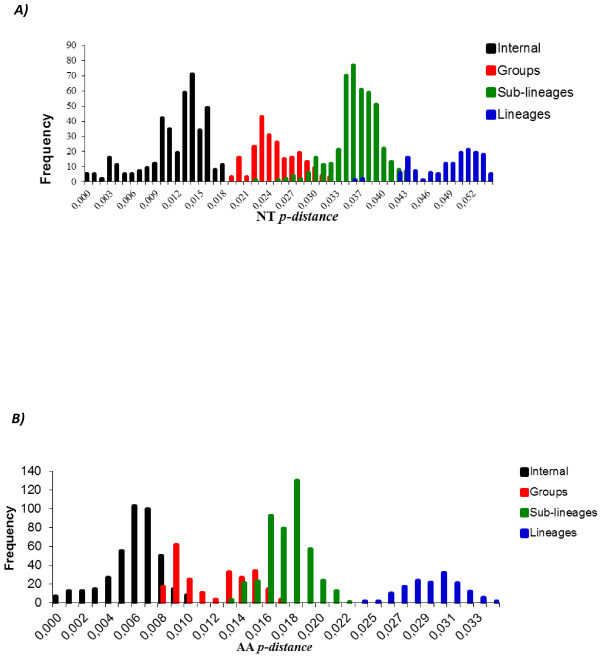
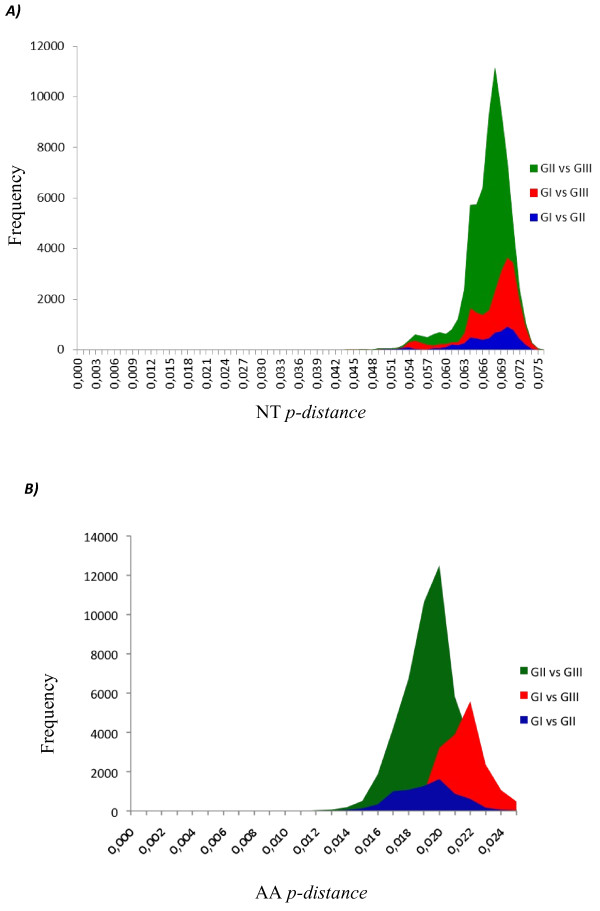
Considering that genotypes III viruses clustered into separate monophyletic groups, which we called lineages and sub-lineage, we analyzed whether a similar clustering could be observed for genotype I and II viruses. The phylogenetic relationship analysis was carried out as mentioned above for genotype III viruses. Genotype I viruses were clustered into two lineages (I and II); viruses of lineage II were segregated into two sub-lineages (I and II) and sub-lineage II includes three internal monophyletic groups (A, B and C) (Figure 4, Additional file 3). The mean nucleotide divergence between lineages I and II was 4.9% (Table 3), while the mean amino acid divergence was 1.8% (Table 3). The mean divergence among sub-lineages I and II was 3.6% for nucleotide sequences and 1.2% for amino acid sequences (Table 3). Finally, the divergence among the internal groups (A, B and C) of sub-lineage II ranged from 2.3 to 2.8% for nucleotides sequences and from 0.6 to 0.9% for amino acid sequences (Table 3). Once again, all the mean values coincided with the highest peaks in the frequency distribution profile (Figure 5).
Figure 4.
Bayesian phylogenetic trees based on the entire ORFs derived from 53 global samples of the genotype I DENV-3 inferred with MrBayes program. The posterior probabilities are expressed in percent and indicated at important nodes. The best fit-model of nucleotide substitution for Bayesian phylogenetic reconstruction used was under a General Time Reversible model with gamma-distributed rate variation (G = 1.1436) and a proportion of invariable sites (I = 0.4716) (GTR + G + I).
Table 3.
Mean of the evolutionary divergence (NT and AA) from all sequence pairs between genotypes I groups
| Genotype I groups | NTp-distance | % | AA p-distance | % |
|---|---|---|---|---|
| Lineage I vs Lineage II |
0,049 |
4,9 |
0,018 |
1,8 |
| Lineage II [Sub-lineage I vs Sub-lineage II] |
0,036 |
3,6 |
0,012 |
1,2 |
| Sub-Lineage II [Group A vs Group B] |
0,023 |
2,3 |
0,006 |
0,6 |
| Sub-Lineage II [Group A vs Group C] |
0,027 |
2,7 |
0,009 |
0,9 |
| Sub-Lineage II [Group B vs Group C] | 0,028 | 2,8 | 0,007 | 0,7 |
Figure 5.
Distribution of NT (A) and AA (B) p-distances between 53 complete ORF sequences of genotype I. NT: nucleotides; AA: amino acids.
Genotype II viruses segregated into four lineages (Figures 6 and 7, Additional file 4). The lineage IV includes four sub-lineages (A, B, C and D). The mean nucleotide divergence among lineages ranged from 2.4 to 3.2% (Table 4), while the mean amino acid divergence ranged from 0.8 to 1.5% (Table 4). The mean divergence among sub-lineages ranged from 1.2 to 1.9% for nucleotide sequences and from 0.4 to 0.7% for amino acid sequences (Table 4).
Figure 6.
Bayesian phylogenetic trees based on the entire ORFs derived from 137 global samples of the genotype II DENV-3 inferred with MrBayes program. The posterior probabilities are expressed in percent and indicated at important nodes. The best fit-model of nucleotide substitution for Bayesian phylogenetic reconstruction used was under a General Time Reversible model with gamma-distributed rate variation (G = 1.4745) and a proportion of invariable sites (I = 0.4956) (GTR + G + I). Branch lengths are proportional to percentage of divergence.
Figure 7.
Distribution of NT (A) and AA (B) p-distances between 137 complete ORF sequences of genotype II. NT: nucleotides; AA: amino acids.
Table 4.
Mean of the evolutionary divergence (NT and AA) from all sequence pairs between genotypes II groups
| Genotype II groups | NTp-distance | % | AAp-distance | % |
|---|---|---|---|---|
| Lineage I vs Lineage II |
0.031 |
3.1 |
0.015 |
1.5 |
| Lineage I vs Lineage III |
0.024 |
2.4 |
0.011 |
1.1 |
| Lineage I vs Lineage IV |
0.029 |
2.9 |
0.013 |
1.3 |
| Lineage II vs Lineage III |
0.027 |
2.7 |
0.012 |
1.2 |
| Lineage II vs Lineage IV |
0.032 |
3.2 |
0.012 |
1.2 |
| Lineage III vs Lineage IV |
0.024 |
2.4 |
0.008 |
0.8 |
| Sub-lineage A vs Sub-lineage B |
0.016 |
1.6 |
0.006 |
0.6 |
| Sub-lineage A vs Sub-lineage C |
0.019 |
1.9 |
0.007 |
0.7 |
| Sub-lineage A vs Sub-lineage D |
0.019 |
1.9 |
0.006 |
0.6 |
| Sub-lineage A vs Sub-lineage E |
0.015 |
1.5 |
0.006 |
0.6 |
| Sub-lineage B vs Sub-lineage C |
0.016 |
1.6 |
0.006 |
0.6 |
| Sub-lineage B vs Sub-lineage D |
0.017 |
1.7 |
0.005 |
0.5 |
| Sub-lineage B vs Sub-lineage E |
0.012 |
1.2 |
0.005 |
0.5 |
| Sub-lineage C vs Sub-lineage D |
0.015 |
1.5 |
0.004 |
0.4 |
| Sub-lineage C vs Sub-lineage E |
0.014 |
1.4 |
0.005 |
0.5 |
| Sub-lineage D vs Sub-lineage E | 0.015 | 1.5 | 0.004 | 0.4 |
Evolutionary divergence among genotypes
We have shown above the evolutionary divergence among the different monophyletic groups within each genotype. In this section, we analyzed the evolutionary divergence among genotypes using the entire ORF sequence and, individually, each protein gene sequence (Table 5 and Figure 8). The mean divergence among genotypes ranged from 6.6 to 6.8% for nucleotide and from 3.1 to 3.4% for amino acid when the entire ORF was analyzed (Figure 8). Analyzing each viral protein, the mean divergence among genotypes varied from 4.1 to 8.8% for nucleotide and from 0.5 to 5.3% for amino acid (Table 5). The lowest divergence was observed for C protein (4.1 to 4.9%) and the highest for NS2a (8.4 to 8.8%), NS4a (7.7 to 8.5) and E (7.0 to 7.6) proteins when the nucleotide sequences were analyzed (Table 4). While NS2b (0.5 to 1.0%), NS4b (0.8 to 1.1%) and NS3 (1.2 to 1.5%) proteins showed the lowest divergence and NS2a (3.0 to 3.4%) NS4a (2.8 to 3.6%) and E (2.3 to 3.0%) protein the highest divergences when the amino acid sequences were analyzed (Table 5). The means p-distance of the each of the genomic regions are coinciding with the highest peaks of the frequency distribution profile (Additional files 5 to 14).
Table 5.
Mean of the evolutionary divergence from all sequence pairs between genotypes for each genomic region
|
A) | |||||||||||
|
Genotypes |
NTp-distance (%) |
||||||||||
|
ORF |
C |
prM |
E |
NS1 |
NS2A |
NS2B |
NS3 |
NS4A |
NS4B |
NS5 |
|
| I vs II |
6.6 |
4.1 |
6.4 |
7.0 |
6.7 |
8.8 |
6.2 |
6.9 |
8.5 |
5.7 |
6.0 |
| I vs III |
6.8 |
4.9 |
5.3 |
7.3 |
6.7 |
8.4 |
6.0 |
6.8 |
8.0 |
6.5 |
6.7 |
| II vs III |
6.7 |
4.5 |
5.6 |
7.6 |
6.4 |
8.5 |
5.9 |
6.7 |
7.7 |
6.6 |
6.3 |
|
B) | |||||||||||
|
Genotypes |
AAp-distance (%) |
||||||||||
|
ORF |
C |
prM |
E |
NS1 |
NS2A |
NS2B |
NS3 |
NS4A |
NS4B |
NS5 |
|
| I vs II |
1.9 |
2.8 |
1.8 |
2.5 |
2.4 |
3.4 |
0.5 |
1.5 |
2.8 |
1.0 |
1.6 |
| I vs III |
2.2 |
5.3 |
1.8 |
3.0 |
1.6 |
3.3 |
1.0 |
1.2 |
3.6 |
1.1 |
2.3 |
| II vs III | 1.9 | 2.9 | 1.8 | 2.3 | 1.9 | 3.0 | 0.7 | 1.3 | 3.6 | 0.8 | 2.1 |
Figure 8.
Distribution of NT (A) and AA (B) p-distances between different genotypes based on the entire ORF of 537 DENV-3. NT: nucleotides; AA: amino acids.
Genotypes identification
To identify which genomic region is responsible for the segregation of DENV-3 into different genotypes, several phylogenetic trees were constructed using, individually, the sequences coding for each viral protein (Additional files 15 to 24). All trees showed the same segregation of the viruses in three genotypes viruses segregated to the same genetic groups as observed when the entire ORF was used (Figure 1), except for the prM sequences, which the genotype I not formed a monophyletic group. The constructed tree based on the NS4A protein coding sequence, however, showed that CH_80_2, DENV-3 isolated from a patient in China in 1980, and, therefore, others viruses with identical sequences (M93130 and BR/RO1/02, not included in this tree), grouped with viruses from genotype II and not with those from genotype I, as observed in all other phylogenetic trees.
Discussion
DENV-3 was introduced into the Americas via Panama and Nicaragua in 1994 [4], spreading rapidly to neighboring countries and reaching Brazil through Rio de Janeiro in December 2000 [9]. Interestingly, DENV-1 and DENV-2 were also introduced into Brazil through Rio de Janeiro [19]. Thus, it seems that the main route of entrance for DENV in Brazil was always Rio de Janeiro. Analyzing the E gene and the 3′UTR sequences, we have previously found that isolates BR/BV4/02 isolated in Boa Vista, Roraima, and BR8/04 isolated Belem, Para, in the Northern Region of the country, were phylogenetically more closely related to viruses isolated in the Caribbean islands rather than to those isolated in Rio de Janeiro, suggesting that DENV-3 was also introduced into Brazil by the northern region [12]. In the present study, now analyzing the entire ORF, the isolates BR/BV4/02 and BR/BR8/04, as well as new viruses isolated between 2006 and 2007 by other groups, were phylogenetically more closely related to viruses isolated in the Caribbean islands than to viruses circulating in Brazil, supporting the hypothesis that DENV-3 was introduced by the Norther Region of the country in addition to the well documented introduction by Rio de Janeiro. According to our phylogenetic analysis, at least two groups of DENV-3 genotype III are circulating in Brazil (Figure 2, sub-lineages A and B). A single virus (BR_V2387_03, FJ850079) isolated in the Northern Region of Brazil was more closely related to viruses isolated in Venezuela, suggesting that was an imported case. In addition to the DENV-3 genotype III, recent studies have shown that genotype I viruses are also circulating in Brazil [20-22]. The Paraguayan isolates were closely related to viruses isolated in Brazil within sub-lineage B of lineage III, suggesting that these viruses were introduced into Paraguay from Brazil as previously described [12].
A previous phylogenetic analysis of DENV-3 genotype III isolated in Sri Lanka, based on a 966 nt fragment spanning part of capsid, preM/M and part of E genes, identified the emergence, after 1989, of a new sub-type, which was correlated with severe disease epidemics that spread to Africa and then to the Americas [15]. Our phylogenetic analysis showed a similar distribution of DENV-3 genotype III isolates (Figure 2). Thus, our lineage I corresponds to DENV-3 genotype III circulating in SriLanka before 1989, which was called as sub-lineage A by Messer and colleagues (2003). On the other hand, LK_V2411_89 and LK_V2409_97 viruses, located basally in the branch of lineage III, could correspond to the new sub-type described by Messer and colleagues (2003) that they called Group B. The topology of our phylogenetic tree suggests that this last sub-type migrated to East Africa and Indian subcontinent and later to the Americas in agreement with observations made by Messer and colleagues (2003). Interestingly, phylogentic analysis of genotype III showed also that NI/V2420/1994 virus isolated in Nicaragua is located at the base of the branch containing the American isolates (Lineage III), suggesting that the introduction of genotype III in the Americas occurred through Nicaragua in 1994, in agreement with the epidemiological data [4,23].
Previous studies analyzing the C, PrM, E and NS3 genes have shown that DENV-3 segregated into four genotypes [13,14,24,25]. In this study, we have found that DENV-3 segregated in the same genotypes I, II and III as shown in the previous studies mentioned above, analyzing either the entire ORF or each protein gene sequence. Thus, DENV-3 genotype can be determined by sequencing any part of the ORF. The genotype IV was not observed in our analysis because no complete genome sequence of any of these viruses is available in the GenBank. Klungthong and colleagues (2008) have also suggested that any genomic region can be sequenced to determine the genotype [26]; however, these authors used the entire genomic sequences of only 12 isolates, while our results were supported by the analysis of more than 500 isolates.
Similar to other RNA viruses, DENV exhibit a high degree of genetic variation due to the non-proofreading activity of its RNA polymerase, the high rates of mutation, the immense population size, and the immunological pressure, leading to the emergence of new subtypes of DENV [27]. Recently, we have described the existence of various taxa or viral sub-types within DENV-3 genotypes by analyzing of the E protein gene [28]. In this study, we have found similar, and even new, internal groups within each genotype. The segregation of the viruses into genotypes, lineage, sub-lineage and groups as suggested in this study were supported by high posterior probability, by nucleotide divergence, and by the presence of characteristic amino acid motifs.
The first studies that identify different genotypes within each DENV serotypes were based on the topology of the trees, supported by bootstrap values [13,24,29-32]. The nucleotide divergence among the genotypes of DENV-1 and DENV-2 was in average 6% when the genomic sequence corresponding to the junction E/NS1 (240 bp) was analyzed [13], and 7% in mean when the E protein gene (1,485 bp) was analyzed for DENV-1 and DENV-2 [31,32]. In this study, we carried out a more detailed analysis of the nucleotide divergence among the different genetic groups of DENV-3. We have observed that nucleotide divergence varied in average 6.7% for genotypes, 2.7% for lineages and 1.5% for sub-lineages when the complete ORF of genotypes II and III was analyzed. For genotype I, a higher nucleotide divergence rate was observed among lineages and sub-lineages.
In addition, our analysis showed that the nucleotide divergence among genotypes varied depending on the genomic region, ranging from 4.1% for C protein gene to 8.8% for NS2a protein gene. These comparisons showed also that C protein gene sequence is the most conserved, and NS2a, NS4a, E protein genes the more variables. However, when the amino acid divergence was analyzed, a different picture was observed; NS2b, NS4b and NS3 were the more conserved proteins, while NS2a, NS4a, E proteins were the more variables. Knowledge of the rates of divergence among the different taxonomic levels, are an important tool for the detection of new viral groups, as well as, information about the variability of each of the genes among different viral groups, could be used to select targets for: the design of probes for diagnosis, antiviral therapies and the construction of candidate vaccines.
Recently, Wittke and colleagues have suggested the existence of an additional genotype (genotype V) within DENV-3 [33]. Our previous phylogenetic analysis based on the E protein, however, has suggested that the genotype V corresponds to a lineage within the genotype I [28]. In this study, the nucleotide divergence among lineage I (called genotype V by Wittke and colleagues) and lineage II of genotype I was 4.9% in average, lower than the 6% observed among genotypes. Therefore, we suggest the maintenance of the classification of DENV-3 into four genotypes as previously proposed [14,24].
In this work, we performed phylogenetic analysis and evolutionary divergence dynamics of DENV-3 and provided data related to the processes that control the viral evolution. These data will be useful to better characterize the DENV-3 epidemics in future and might even be used for selection of vaccine candidates.
Methods
Virus
Thirteen DENV-3 isolated from different regions of Brazil (n = 9) and Paraguay (n = 4) were included in this study. All viruses were isolated and passed twice in C6/36 [34]. Ten of them (D3BR/RP1/03, D3BR/PV1/03, D3BR/SL3/02, D3BR/BV4/02, D3BR/CU6/02, D3BR/BR8/04, D3BR/MR9/03, D3PY/PJ4/03, D3PY/AS10/03, D3PY/AS12/02) belong to different clusters in the phylogenetic trees constructed based on the of E gene and 3′UTR region sequences as previously described [12]. Two more recently isolates (D3BR/ACN/07 and D3BR/AL95/09) describe in other study were also included [12,35]. The Paraguayan isolate D3PY/SUS/2003 was also included because it was never characterized by sequencing.
RNA extraction
Viral RNA was extracted from 140 μl of supernatant of infected C6/36 cells using the QIAamp Viral RNA kit (Qiagen, Germany), following the manufacturer’s recommendations. The RNA was eluted with 80 μl of DNase/RNase free water.
RT-PCR
cDNA synthesis
The reaction of cDNA synthesis contained 24 μl of RNA, 200 ng of random primers (Invitrogen, USA), 0.25 mM dNTPs mix (Invitrogen, USA), 80 U of inhibitor RNAse (RNAseOUT, Invitrogen, USA), 400 U of M-MLV Reverse Transcriptase (USB, USA) and 8 μl 5X buffer (250 mM Tris–HCl [pH 8.3], 375 mM KCl, 15 mM MgCl2) in a final volume of 40 μl. The mixture was incubated at 25°C for 10 min, followed by incubation at 37°C for 4 h, and a final incubation of 5 min at 85°C. Subsequently, the cDNA was treated with 1 U RNase H (GIBCO, USA) at 37°C for 30 min, and stored at −20°C until use.
Polymerase Chain Reaction (PCR)
To amplify the entire viral genomes, the primers used in PCR reactions were designed based on the D3_BR74886_02 isolated (AY679147.1) (Additional file 25A). Six overlapping fragments representing the full viral genome were amplified. The master mix contained: 1 μl of cDNA, 0.2 mM dNTP, 0.3 mM of each primer, 1.5 U of Platinum® Taq DNA Polymerase High Fidelity (Invitrogen, USA), 5 μl of 10X buffer (600 mM Tris-SO4 [pH 8.9], 180 mM Ammonium Sulfate), and 2 mM of MgSO4 in a final volume of 50 μl. The amplification was performed using the MyCycler™ Thermal Cycler (BIO-RAD, USA). The reaction mixture was heated at 94°C for 2 min followed by 45 amplification cycles: 94°C for 10 s, 56°C for 1 min, 68°C for 5 min, and a final extension at 68°C for 7 min. The PCR products were subjected to electrophoresis in a 1% agarose gel and visualized under UV light after staining with ethidium bromide. Bands of DNA were purified from agarose gels using a QIAquick Gel Extraction Kit (Qiagen®, Germany) following the manufacturer’s specifications.
Nucleotide sequencing
The gel-purified DNA fragments were sequenced using an Applied Biosystems BigDye ddNTP capillary sequencer ABI 3130 (Applied Biosystems, USA) following the manufacturer’s specifications. Both strain of each DNA fragments were sequenced at least three times using walking primers (Additional file 25B). Viral genome sequences generated in this study were deposited in the GenBank, under the accession numbers: BR_BV4_02 (JF808118), BR_BR8_04 (JF808119), BR_AL95_09 (JF808120), BR_ACN_07 (JF808121), PY_SUS_02 (JF808122), PY_AS12_02 (JF808123), BR_MR9_03 (JF808124), BR_SL3_03 (JF808125), BR_PV1_03 (JF808126), BR_CU6_02 (JF808127), PY_PJ4_03 (JF808128), PY_AS10_02 (JF808129) and D3BR_RP1_2003 (EF643017).
The electropherograms were analyzed using the MEGA 5.0 program and the consensus assemble were carried out using the BioEdit v.7.0.9 program [36,37]. The 30 nucleotides at 5′ and 3′ ends of each fragment were deleted to avoid the influence of primers used for amplification. However, the sequences at 5′ and 3′ ends of the viral genome represent the sequences of the primers used to amplify these regions.
Phylogenetic and evolutionary analysis
Database of DENV-3 sequences
A database containing complete genome sequences of DENV-3 retrieved from GenBank was prepared for phylogenetic analysis. Representative sequences of DENV-1 (AB074760), DENV-2 (M20558) and DENV-4 (AF326573) were also included. The database contained the following information: GenBank access number, isolated name, country and year of isolation. A total of 563 sequences of DENV-3 were retrieved from GenBank until March 27, 2010 (Additional file 26), which were indicated as complete genome. However, several of these sequences did not contain the first 39 and last 135 nucleotides of the viral genome, or did not have the UTRs sequences validated. Therefore, in order to use the largest number of sequences possible, we have included only the open reading frame (ORF). The sequences were analyzed using the program DAMBE 5.2.6 in order to identify identical sequences, which were excluded from the analysis (Additional file 27) [38]. In addition, mutants and clones were also excluded, resulting in a total of 527 sequences of DENV-3 available for phylogenetic analysis (Additional file 26).
Phylogenetic analysis
DENV-3 sequences were aligned using the program CLUSTAL X 5.2 [39]. The distance based Neighbor-joining (NJ) and/or the Bayesian inference (BI) methods were used to construct the phylogenetic trees. For NJ method, the sequences were first analyzed using the Modeltest 3.7.MacX program to identify the best nucleotide substitution model [40]. The best nucleotide substitution model was selected under the criterion hierarchical likelihood ratio tests (hLRT). Phylogenetic trees was constructed using the NJ method as implemented in the PAUP * 4.0 b10 program and statistically supported by bootstrap method using 1,000 replicates [41]. For BI method, the aligned sequences were first analyzed using the MrModeltest 2.3 for Mac OS X 10.4.11 program to identify the best model of nucleotide substitution [42]. The best nucleotide substitution model was selected under the criterion hierarchical likelihood ratio tests (hLRT). Five runs of 4 chains each (one cold and tree heated, temperature = 0.20), generating a total of 1.5 × 106 generations (10% removed as burn-in) were done to ensure statistical convergence. The phylogenetic trees were inferred with MrBayes program and statistically supported by calculating the posterior probability, which was expressed in percentage [43].
Analysis of evolutionary divergence between genetic groups
The nucleotide and amino acid divergences were calculated using p-distance model with 1,000 replicates using the MEGA 5 program [36]. The frequency distribution profiles of divergences between populations were calculated as described previously [44]. The mean (%) p-distances between sequences were also calculated.
Identification of amino acid motif for each genetic group
The polyprotein sequences of each genetic group were aligned for the identification of the amino acid motifs, which were defined as amino acid substitutions present in at least 90% of the sequences within each genetic group.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
Conceived and designed the experiments: HLA, AAA and VHA. Performed the experiments: HLA, AAA, PFG, MTB. Analyzed the data: HLA, AAA and VHA. Contributed reagents/materials/analysis tools: FTDA, TRS, EVDS, MTN, PFCV, DSV, WCB, MLB, CV, MM, LTF. Wrote the paper: HLA, AAA and VHA. All authors read and approved the final manuscript.
Supplementary Material
Comparison of the 3′UTR variable region between the position 10,264 to 1,287 of several isolates belongs to genotypes I, II and III. Sequence of isolates from genotypes I, II and III, which showed differences in 3′UTR region.
Motifs of amino acids for the genotype III. The file provides details on amino acid substitutions present within each genetic group of genotype III.
Motifs of amino acids for the genotype I. The file provides details on amino acid substitutions present within each genetic group of genotype I.
Motifs of amino acids for the genotype II. The file provides details on amino acid substitutions present within each genetic group of genotype II.
Distribution of NT (A) and AA (B) p-distances between different genotypes based on the C protein gene of 537 DENV-3. NT: nucleotides; AA: amino acids.
Distribution of NT (A) and AA (B) p-distances between different genotypes based on the prM protein gene of 537 DENV-3. NT: nucleotides; AA: amino acids.
Distribution of NT (A) and AA (B) p-distances between different genotypes based on the E protein gene of 424 DENV-3. NT: nucleotides; AA: amino acids.
Distribution of NT (A) and AA (B) p-distances between different genotypes based on the NS1 protein gene of 372 DENV-3. NT: nucleotides; AA: amino acids.
Distribution of NT (A) and AA (B) p-distances between different genotypes based on the NS2A protein gene of 328 DENV-3. NT: nucleotides; AA: amino acids.
Distribution of NT (A) and AA (B) p-distances between different genotypes based on the NS2B protein gene of 233 DENV-3. NT: nucleotides; AA: amino acids.
Distribution of NT (A) and AA (B) p-distances between different genotypes based on the NS3 protein gene of 431 DENV-3. NT: nucleotides; AA: amino acids.
Distribution of NT (A) and AA (B) p-distances between different genotypes based on the NS4A protein gene of 268 DENV-3. NT: nucleotides; AA: amino acids.
Distribution of NT (A) and AA (B) p-distances between different genotypes based on the NS4B protein gene of 323 DENV-3. NT: nucleotides; AA: amino acids.
Distribution of NT (A) and AA (B) p-distances between different genotypes based on the NS5 protein gene of 537 DENV-3. NT: nucleotides; AA: amino acids.
Neighbor-joining phylogenetic trees based on C gene derived from 537 global samples of the DENV-3 inferred with MEGA 5 program. The bootstrap are indicated at important nodes. The best-fit model of nucleotide substitution for phylogenetic reconstruction used was TrN + G model with gamma-distributed rate variation (G = 1.2). Branch lengths are proportional to percentage of divergence.
Neighbor-joining phylogenetic trees based on prM gene derived from 283 global samples of the DENV-3 inferred with MEGA 5 program. The bootstrap are indicated at important nodes. The best-fit model of nucleotide substitution for phylogenetic reconstruction used was TrN + G model with gamma-distributed rate variation (G = 1.2). Branch lengths are proportional to percentage of divergence.
Neighbor-joining phylogenetic trees based on E gene derived from 424 global samples of the DENV-3 inferred with MEGA 5 program. The bootstrap are indicated at important nodes. The best-fit model of nucleotide substitution for phylogenetic reconstruction used was TrN + G model with gamma-distributed rate variation (G = 1.2). Branch lengths are proportional to percentage of divergence.
Neighbor-joining phylogenetic trees based on NS1 gene derived from 372 global samples of the DENV-3 inferred with MEGA 5 program. The bootstrap are indicated at important nodes. The best-fit model of nucleotide substitution for phylogenetic reconstruction used was TrN + G model with gamma-distributed rate variation (G = 1.2). Branch lengths are proportional to percentage of divergence.
Neighbor-joining phylogenetic trees based on NS2A gene derived from 328 global samples of the DENV-3 inferred with MEGA 5 program. The bootstrap are indicated at important nodes. The best-fit model of nucleotide substitution for phylogenetic reconstruction used was TrN + G model with gamma-distributed rate variation (G = 1.2). Branch lengths are proportional to percentage of divergence.
Neighbor-joining phylogenetic trees based on NS2B gene derived from 233 global samples of the DENV-3 inferred with MEGA 5 program. The bootstrap are indicated at important nodes. The best-fit model of nucleotide substitution for phylogenetic reconstruction used was TrN + G model with gamma-distributed rate variation (G = 1.2). Branch lengths are proportional to percentage of divergence.
Neighbor-joining phylogenetic trees based on NS3 gene derived from 431 global samples of the DENV-3 inferred with MEGA 5 program. The bootstrap are indicated at important nodes. The best-fit model of nucleotide substitution for phylogenetic reconstruction used was TrN + G model with gamma-distributed rate variation (G = 1.2). Branch lengths are proportional to percentage of divergence.
Neighbor-joining phylogenetic trees based on NS4A gene derived from 268 global samples of the DENV-3 inferred with MEGA 5 program. The bootstrap are indicated at important nodes. The best-fit model of nucleotide substitution for phylogenetic reconstruction used was TrN + G model with gamma-distributed rate variation (G = 1.2). Branch lengths are proportional to percentage of divergence.
Neighbor-joining phylogenetic trees based on NS4B gene derived from 323 global samples of the DENV-3 inferred with MEGA 5 program. The bootstrap are indicated at important nodes. The best-fit model of nucleotide substitution for phylogenetic reconstruction used was TrN + G model with gamma-distributed rate variation (G = 1.2). Branch lengths are proportional to percentage of divergence.
Neighbor-joining phylogenetic trees based on NS5 gene derived from 464 global samples of the DENV-3 inferred with MEGA 5 program. The bootstrap are indicated at important nodes. The best-fit model of nucleotide substitution for phylogenetic reconstruction used was TrN + G model with gamma-distributed rate variation (G = 1.2). Branch lengths are proportional to percentage of divergence.
Primers used for amplification and sequencing. A) Primers used in the PCR of overlapping regions representing the entire genome of DENV3. B) Primers used for nucleotide sequencing of the genome. The file provides details on all the sequences of primers used for amplification and sequencing including in this study.
The database of the all viruses analyzed. The file provides details on all the sequences including in this study.
Identical virus. The virus that were excluded for each data set because they are identical.
Contributor Information
Helda Liz Alfonso, Email: alfonso_helda@yahoo.com.ar.
Alberto Anastacio Amarilla, Email: alberilla@usp.br.
Paula Fernanda Gonçalves, Email: paulaferbio@yahoo.com.br.
Matheus Takatuba Barros, Email: matheusja@hotmail.com.
Flavia Tremeschin de Almeida, Email: flavia_tche@yahoo.com.br.
Telma R Silva, Email: telmarrsilva@yahoo.com.br.
Eliana V da Silva, Email: elianapinto@iec.pa.gov.br.
Marcio T Nunes, Email: marcionunes@iec.pa.gov.br.
Pedro F C Vasconcelos, Email: pedrovasconcelos@iec.pa.gov.br.
Deusilene S Vieira, Email: deusylenebio@hotmail.com.
Weber Cheli Batista, Email: weber_cepem@hotmail.com.
Maria Liz Bobadilla, Email: bobadillaml@gmail.com.
Cynthia Vazquez, Email: cynthiavlm@yahoo.com.
Mirian Moran, Email: miryammoran5@gmail.com.
Luiz Tadeu Moraes Figueiredo, Email: ltmfigue@fmrp.usp.br.
Victor Hugo Aquino, Email: vhugo@fcfrp.usp.br.
Acknowledgments
The authors thank Emilio Espínola by the suggestions made to the manuscript. This work was supported by the Fundacão de Amparo à Pesquisa do Estado de São Paulo (FAPESP PRONEX REDE DENGUE, Processo 2010/50432-6; and by the Instituto Nacional de Ciência e Tecnologia (INCT) em Dengue (http://memoria.cnpq.br/programas/inct/_apresentacao/inct_dengue.html).
References
- WHO. Dengue: guidelines for diagnosis, treatment, prevention and control. Geneva: World Health Organization; 2009. [PubMed] [Google Scholar]
- Henchal E, Putnak J. The dengue viruses. Clin Microbiol Rev. 1990;3:376–396. doi: 10.1128/cmr.3.4.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie J, Gubler D, Petersen L. Emerging flaviviruses: the spread and resurgence of Japanese encephalitis, West Nile and dengue viruses. Nat Med. 2004;10:S98–S109. doi: 10.1038/nm1144. [DOI] [PubMed] [Google Scholar]
- CDC. From the Centers for Disease Control and Prevention. Dengue type 3 infection--Nicaragua and Panama, October-November 1994. JAMA. 1995;273:840–841. [PubMed] [Google Scholar]
- Guzmán MG, Vázquez S, Martínez E, Alvarez M, Rodríguez R, Kourí G, de los Reyes J, Acevedo F. [Dengue in Nicaragua, 1994: reintroduction of serotype 3 in the Americas] Bol Oficina Sanit Panam. 1996;121:102–110. [PubMed] [Google Scholar]
- Usuku S, Castillo L, Sugimoto C, Noguchi Y, Yogo Y, Kobayashi N. Phylogenetic analysis of dengue-3 viruses prevalent in Guatemala during 1996–1998. Arch Virol. 2001;146:1381–1390. doi: 10.1007/s007050170098. [DOI] [PubMed] [Google Scholar]
- Balmaseda A, Sandoval E, Pérez L, Gutiérrez CM, Harris E. Application of molecular typing techniques in the 1998 dengue epidemic in Nicaragua. Am J Trop Med Hyg. 1999;61:893–897. doi: 10.4269/ajtmh.1999.61.893. [DOI] [PubMed] [Google Scholar]
- Peyrefitte CN, Couissinier-Paris P, Mercier-Perennec V, Bessaud M, Martial J, Kenane N, Durand JP, Tolou HJ. Genetic characterization of newly reintroduced dengue virus type 3 in Martinique (French West Indies) J Clin Microbiol. 2003;41:5195–5198. doi: 10.1128/JCM.41.11.5195-5198.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogueira R, Miagostovich M, de Filippis A, Pereira M, Schatzmayr H. Dengue virus type 3 in Rio de Janeiro, Brazil. Mem Inst Oswaldo Cruz. 2001;96:925–926. doi: 10.1590/S0074-02762001000700007. [DOI] [PubMed] [Google Scholar]
- Uzcategui NY, Comach G, Camacho D, Salcedo M, Cabello de Quintana M, Jimenez M, Sierra G, Cuello de Uzcategui R, James WS, Turner S. et al. Molecular epidemiology of dengue virus type 3 in Venezuela. J Gen Virol. 2003;84:1569–1575. doi: 10.1099/vir.0.18807-0. [DOI] [PubMed] [Google Scholar]
- Nogueira R, Schatzmayr H, de Filippis A, dos Santos F, da Cunha R, Coelho J, de Souza L, Guimarães F, de Araújo E, De Simone T. et al. Dengue virus type 3, Brazil, 2002. Emerg Infect Dis. 2005;11:1376–1381. doi: 10.3201/eid1109.041043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aquino VH, Anatriello E, Goncalves PF, da Silva EV, Vasconcelos PFC, Vieira DS, Batista WC, Bobadilla ML, Vazquez C, Moran M, Figueiredo LTM. Molecular epidemiology of dengue type 3 virus in Brazil and Paraguay, 2002–2004. Am J Trop Med Hyg. 2006;75:710–715. [PubMed] [Google Scholar]
- Rico-Hesse R. Molecular evolution and distribution of dengue viruses type 1 and 2 in nature. Virology. 1990;174:479–493. doi: 10.1016/0042-6822(90)90102-W. [DOI] [PubMed] [Google Scholar]
- Chungue E, Deubel V, Cassar O, Laille M, Martin P. Molecular epidemiology of dengue 3 viruses and genetic relatedness among dengue 3 strains isolated from patients with mild or severe form of dengue fever in French Polynesia. J Gen Virol. 1993;74(Pt 12):2765–2770. doi: 10.1099/0022-1317-74-12-2765. [DOI] [PubMed] [Google Scholar]
- Messer W, Gubler D, Harris E, Sivananthan K, de Silva A. Emergence and global spread of a dengue serotype 3, subtype III virus. Emerg Infect Dis. 2003;9:800–809. doi: 10.3201/eid0907.030038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochel T, Aguilar P, Felices V, Comach G, Cruz C, Alava A, Vargas J, Olson J, Blair P. Molecular epidemiology of dengue virus type 3 in Northern South America: 2000–2005. Infect Genet Evol. 2008;8:682–688. doi: 10.1016/j.meegid.2008.06.008. [DOI] [PubMed] [Google Scholar]
- Schreiber M, Holmes E, Ong S, Soh H, Liu W, Tanner L, Aw P, Tan H, Ng L, Leo Y. et al. Genomic epidemiology of a dengue virus epidemic in urban Singapore. J Virol. 2009;83(9):4163–73. doi: 10.1128/JVI.02445-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Dash PK, Agarwal S, Shukla J, Parida MM, Rao PV. Comparative complete genome analysis of dengue virus type 3 circulating in India between 2003–2008. J Gen Virol. 2011;92:1595–1600. doi: 10.1099/vir.0.030437-0. [DOI] [PubMed] [Google Scholar]
- Figueiredo L. The Brazilian flaviviruses. Microbes Infect. 2000;2:1643–1649. doi: 10.1016/S1286-4579(00)01320-4. [DOI] [PubMed] [Google Scholar]
- Vilela A, Figueiredo L, dos Santos J, Eiras A, Bonjardim C, Ferreira P, Kroon E. Dengue virus 3 genotype I in Aedes aegypti mosquitoes and eggs, Brazil, 2005–2006. Emerg Infect Dis. 2010;16:989–992. doi: 10.3201/eid1606.091000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barcelos Figueiredo L, Batista Cecílio A, Portela Ferreira G, Paiva Drumond B, Germano de Oliveira J, Bonjardim C, Peregrino Ferreira P, Kroon E. Dengue virus 3 genotype 1 associated with dengue fever and dengue hemorrhagic fever, Brazil. Emerg Infect Dis. 2008;14:314–316. doi: 10.3201/eid1402.070278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aquino V, Amarilla A, Alfonso H, Batista W, Figueiredo L. New genotype of dengue type 3 virus circulating in Brazil and Colombia showed a close relationship to old Asian viruses. PLoS One. 2009;4:e7299. doi: 10.1371/journal.pone.0007299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araújo J, Nogueira R, Schatzmayr H, Zanotto P, Bello G. Phylogeography and evolutionary history of dengue virus type 3. Infect Genet Evol. 2009;9:716–725. doi: 10.1016/j.meegid.2008.10.005. [DOI] [PubMed] [Google Scholar]
- Lanciotti R, Lewis J, Gubler D, Trent D. Molecular evolution and epidemiology of dengue-3 viruses. J Gen Virol. 1994;75(Pt 1):65–75. doi: 10.1099/0022-1317-75-1-65. [DOI] [PubMed] [Google Scholar]
- Chow V, Seah C, Chan Y. Comparative analysis of NS3 sequences of temporally separated dengue 3 virus strains isolated from southeast Asia. Intervirology. 1994;37:252–258. doi: 10.1159/000150386. [DOI] [PubMed] [Google Scholar]
- Klungthong C, Putnak R, Mammen MP, Li T, Zhang C. Molecular genotyping of dengue viruses by phylogenetic analysis of the sequences of individual genes. J Virol Methods. 2008;154:175–181. doi: 10.1016/j.jviromet.2008.07.021. [DOI] [PubMed] [Google Scholar]
- Twiddy S, Holmes E, Rambaut A. Inferring the rate and time-scale of dengue virus evolution. Mol Biol Evol. 2003;20:122–129. doi: 10.1093/molbev/msg010. [DOI] [PubMed] [Google Scholar]
- Amarilla A, de Almeida F, Jorge D, Alfonso H, de Castro-Jorge L, Nogueira N, Figueiredo L, Aquino V. Genetic diversity of the E protein of dengue type 3 virus. Virol J. 2009;6:113. doi: 10.1186/1743-422X-6-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis J, Chang G, Lanciotti R, Kinney R, Mayer L, Trent D. Phylogenetic relationships of dengue-2 viruses. Virology. 1993;197:216–224. doi: 10.1006/viro.1993.1582. [DOI] [PubMed] [Google Scholar]
- Lanciotti R, Gubler D, Trent D. Molecular evolution and phylogeny of dengue-4 viruses. J Gen Virol. 1997;78(Pt 9):2279–2284. doi: 10.1099/0022-1317-78-9-2279. [DOI] [PubMed] [Google Scholar]
- Goncalvez A, Escalante A, Pujol F, Ludert J, Tovar D, Salas R, Liprandi F. Diversity and evolution of the envelope gene of dengue virus type 1. Virology. 2002;303:110–119. doi: 10.1006/viro.2002.1686. [DOI] [PubMed] [Google Scholar]
- Twiddy S, Farrar J, Vinh Chau N, Wills B, Gould E, Gritsun T, Lloyd G, Holmes E. Phylogenetic relationships and differential selection pressures among genotypes of dengue-2 virus. Virology. 2002;298:63–72. doi: 10.1006/viro.2002.1447. [DOI] [PubMed] [Google Scholar]
- Wittke V, Robb T, Thu H, Nisalak A, Nimmannitya S, Kalayanrooj S, Vaughn D, Endy T, Holmes E, Aaskov J. Extinction and rapid emergence of strains of dengue 3 virus during an interepidemic period. Virology. 2002;301:148–156. doi: 10.1006/viro.2002.1549. [DOI] [PubMed] [Google Scholar]
- Igarashi A. Isolation of a Singh’s Aedes albopictus cell clone sensitive to Dengue and Chikungunya viruses. J Gen Virol. 1978;40:531–544. doi: 10.1099/0022-1317-40-3-531. [DOI] [PubMed] [Google Scholar]
- Poloni T, Oliveira A, Alfonso H, Galvao L, Amarilla A, Poloni D, Figueiredo L, Aquino V. Detection of dengue virus in saliva and urine by real time RT-PCR. Virol J. 2010;7:22. doi: 10.1186/1743-422X-7-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;10:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall T. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- Xia X, Xie Z. DAMBE: software package for data analysis in molecular biology and evolution. J Hered. 2001;92:371–373. doi: 10.1093/jhered/92.4.371. [DOI] [PubMed] [Google Scholar]
- Larkin M, Blackshields G, Brown N, Chenna R, McGettigan P, McWilliam H, Valentin F, Wallace I, Wilm A, Lopez R. et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Posada D. ModelTest Server: a web-based tool for the statistical selection of models of nucleotide substitution online. Nucleic Acids Res. 2006;34:W700–W703. doi: 10.1093/nar/gkl042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford D. PAUP*: phylogenetic analysis using parsimony (*and other methods). vol. Version 4.0b10a. Sunderland, Mass: Sinauer Associates; 1998. [Google Scholar]
- Nylander JAA. MrModelTest v2. Uppsala, Sweden: Evolutionary Biology Center, University of Uppsala; 2004. [Google Scholar]
- Ronquist F, Huelsenbeck J. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Grard G, Moureau G, Charrel RN, Lemasson JJ, Gonzalez JP, Gallian P, Gritsun TS, Holmes EC, Gould EA, de Lamballerie X. Genetic characterization of tick-borne flaviviruses: new insights into evolution, pathogenetic determinants and taxonomy. Virology. 2007;361:80–92. doi: 10.1016/j.virol.2006.09.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Comparison of the 3′UTR variable region between the position 10,264 to 1,287 of several isolates belongs to genotypes I, II and III. Sequence of isolates from genotypes I, II and III, which showed differences in 3′UTR region.
Motifs of amino acids for the genotype III. The file provides details on amino acid substitutions present within each genetic group of genotype III.
Motifs of amino acids for the genotype I. The file provides details on amino acid substitutions present within each genetic group of genotype I.
Motifs of amino acids for the genotype II. The file provides details on amino acid substitutions present within each genetic group of genotype II.
Distribution of NT (A) and AA (B) p-distances between different genotypes based on the C protein gene of 537 DENV-3. NT: nucleotides; AA: amino acids.
Distribution of NT (A) and AA (B) p-distances between different genotypes based on the prM protein gene of 537 DENV-3. NT: nucleotides; AA: amino acids.
Distribution of NT (A) and AA (B) p-distances between different genotypes based on the E protein gene of 424 DENV-3. NT: nucleotides; AA: amino acids.
Distribution of NT (A) and AA (B) p-distances between different genotypes based on the NS1 protein gene of 372 DENV-3. NT: nucleotides; AA: amino acids.
Distribution of NT (A) and AA (B) p-distances between different genotypes based on the NS2A protein gene of 328 DENV-3. NT: nucleotides; AA: amino acids.
Distribution of NT (A) and AA (B) p-distances between different genotypes based on the NS2B protein gene of 233 DENV-3. NT: nucleotides; AA: amino acids.
Distribution of NT (A) and AA (B) p-distances between different genotypes based on the NS3 protein gene of 431 DENV-3. NT: nucleotides; AA: amino acids.
Distribution of NT (A) and AA (B) p-distances between different genotypes based on the NS4A protein gene of 268 DENV-3. NT: nucleotides; AA: amino acids.
Distribution of NT (A) and AA (B) p-distances between different genotypes based on the NS4B protein gene of 323 DENV-3. NT: nucleotides; AA: amino acids.
Distribution of NT (A) and AA (B) p-distances between different genotypes based on the NS5 protein gene of 537 DENV-3. NT: nucleotides; AA: amino acids.
Neighbor-joining phylogenetic trees based on C gene derived from 537 global samples of the DENV-3 inferred with MEGA 5 program. The bootstrap are indicated at important nodes. The best-fit model of nucleotide substitution for phylogenetic reconstruction used was TrN + G model with gamma-distributed rate variation (G = 1.2). Branch lengths are proportional to percentage of divergence.
Neighbor-joining phylogenetic trees based on prM gene derived from 283 global samples of the DENV-3 inferred with MEGA 5 program. The bootstrap are indicated at important nodes. The best-fit model of nucleotide substitution for phylogenetic reconstruction used was TrN + G model with gamma-distributed rate variation (G = 1.2). Branch lengths are proportional to percentage of divergence.
Neighbor-joining phylogenetic trees based on E gene derived from 424 global samples of the DENV-3 inferred with MEGA 5 program. The bootstrap are indicated at important nodes. The best-fit model of nucleotide substitution for phylogenetic reconstruction used was TrN + G model with gamma-distributed rate variation (G = 1.2). Branch lengths are proportional to percentage of divergence.
Neighbor-joining phylogenetic trees based on NS1 gene derived from 372 global samples of the DENV-3 inferred with MEGA 5 program. The bootstrap are indicated at important nodes. The best-fit model of nucleotide substitution for phylogenetic reconstruction used was TrN + G model with gamma-distributed rate variation (G = 1.2). Branch lengths are proportional to percentage of divergence.
Neighbor-joining phylogenetic trees based on NS2A gene derived from 328 global samples of the DENV-3 inferred with MEGA 5 program. The bootstrap are indicated at important nodes. The best-fit model of nucleotide substitution for phylogenetic reconstruction used was TrN + G model with gamma-distributed rate variation (G = 1.2). Branch lengths are proportional to percentage of divergence.
Neighbor-joining phylogenetic trees based on NS2B gene derived from 233 global samples of the DENV-3 inferred with MEGA 5 program. The bootstrap are indicated at important nodes. The best-fit model of nucleotide substitution for phylogenetic reconstruction used was TrN + G model with gamma-distributed rate variation (G = 1.2). Branch lengths are proportional to percentage of divergence.
Neighbor-joining phylogenetic trees based on NS3 gene derived from 431 global samples of the DENV-3 inferred with MEGA 5 program. The bootstrap are indicated at important nodes. The best-fit model of nucleotide substitution for phylogenetic reconstruction used was TrN + G model with gamma-distributed rate variation (G = 1.2). Branch lengths are proportional to percentage of divergence.
Neighbor-joining phylogenetic trees based on NS4A gene derived from 268 global samples of the DENV-3 inferred with MEGA 5 program. The bootstrap are indicated at important nodes. The best-fit model of nucleotide substitution for phylogenetic reconstruction used was TrN + G model with gamma-distributed rate variation (G = 1.2). Branch lengths are proportional to percentage of divergence.
Neighbor-joining phylogenetic trees based on NS4B gene derived from 323 global samples of the DENV-3 inferred with MEGA 5 program. The bootstrap are indicated at important nodes. The best-fit model of nucleotide substitution for phylogenetic reconstruction used was TrN + G model with gamma-distributed rate variation (G = 1.2). Branch lengths are proportional to percentage of divergence.
Neighbor-joining phylogenetic trees based on NS5 gene derived from 464 global samples of the DENV-3 inferred with MEGA 5 program. The bootstrap are indicated at important nodes. The best-fit model of nucleotide substitution for phylogenetic reconstruction used was TrN + G model with gamma-distributed rate variation (G = 1.2). Branch lengths are proportional to percentage of divergence.
Primers used for amplification and sequencing. A) Primers used in the PCR of overlapping regions representing the entire genome of DENV3. B) Primers used for nucleotide sequencing of the genome. The file provides details on all the sequences of primers used for amplification and sequencing including in this study.
The database of the all viruses analyzed. The file provides details on all the sequences including in this study.
Identical virus. The virus that were excluded for each data set because they are identical.