Abstract
L-arginine (L-Arg) deficiency results in decreased T-cell proliferation and impaired T-cell function. Here we have found that L-Arg depletion inhibited expression of different membrane antigens, including CD247 (CD3ζ), and led to an ER stress response, as well as cell cycle arrest at G0/G1 in both human Jurkat and peripheral blood mitogen-activated T cells, without undergoing apoptosis. By genetic and biochemical approaches, we found that L-Arg depletion also induced autophagy. Deprivation of L-Arg induced EIF2S1 (eIF2α), MAPK8 (JNK), BCL2 (Bcl-2) phosphorylation, and displacement of BECN1 (Beclin 1) binding to BCL2, leading to autophagosome formation. Silencing of ERN1 (IRE1α) prevented the induction of autophagy as well as MAPK8 activation, BCL2 phosphorylation and XBP1 splicing, whereas led T lymphocytes to apoptosis under L-Arg starvation, suggesting that the ERN1-MAPK8 pathway plays a major role in the activation of autophagy following L-Arg depletion. Autophagy was required for survival of T lymphocytes in the absence of L-Arg, and resulted in a reversible process. Replenishment of L-Arg made T lymphocytes to regain the normal cell cycle profile and proliferate, whereas autophagy was inhibited. Inhibition of autophagy by ERN1, BECN1 and ATG7 silencing, or by pharmacological inhibitors, promoted cell death of T lymphocytes incubated in the absence of L-Arg. Our data indicate for the first time that depletion of L-Arg in T lymphocytes leads to a reversible response that preserves T lymphocytes through ER stress and autophagy, while remaining arrested at G0/G1. Our data also show that the L-Arg depletion-induced ER stress response could lead to apoptosis when autophagy is blocked.
Keywords: autophagy, endoplasmic reticulum stress, L-arginine, arginine depletion, survival, apoptosis, signaling, T cell, Jurkat cell
Introduction
L-arginine (L-Arg) is a conditionally essential amino acid for adult mammals, and thereby it must be supplied in the diet during certain physiological or pathological conditions, in which the requirements exceed the production capacity. This amino acid is used for the biosynthesis of proteins, creatine and agmatine, and it is mainly catabolized by the enzymes arginase and nitric oxide synthase to produce urea and L-ornithine, and nitric oxide and L-citrulline, respectively.1L-Arg levels are profoundly reduced in cancer patients,2 following liver transplantation,3 or in severe trauma,4 correlating with increased arginase I serum levels. L-Arg deficiency results in decreased T-cell proliferation and impaired T-cell function.5,6 Many of the adverse effects of L-Arg deficiency can be reversed by enteral or parenteral supplementation of L-Arg.7 An increasing number of pathologies as well as physiological conditions are being associated to an arginase-mediated T cell hyporesponsiveness. Arginase level has been reported to be significantly increased in the peripheral blood of pregnant women and placenta, leading to the temporary suppression of the maternal immune response during human pregnancy.8 High levels of arginase activity have been suggested to contribute to T cell dysfunction in human immunodeficiency virus-seropositive patients.9 High arginase activity, a hallmark of nonhealing leishmaniasis, is primarily expressed locally at the site of pathology and causes local depletion of L-Arg, resulting in impaired T cell responses.10
Two major cell types have been reported to be particularly abundant in arginase I activity, namely human neutrophils11 and the so-called myeloid-derived suppressor cells (MDSCs),12 which turned out to be a subpopulation of activated neutrophils.13 Recent studies demonstrated that activated T cells cultured in medium without L-Arg,14 co-cultured with MDSCs isolated from tumors,5 or exposed to neutrophil lysate,15 showed decreased proliferation, low expression of T-cell receptor CD3ζ (CD247) chain and impaired production of cytokines. However, the mechanisms by which L-Arg starvation blocks T-cell proliferation and function have not yet been determined.
Because the endoplasmic reticulum (ER) is the primary site where proteins are synthesized, folded and sorted, and arginine plays an important role in protein synthesis and folding,16 it could be envisaged that a deficiency in this amino acid might affect ER-related functions. Protein folding in the ER is a highly regulated process. Only properly folded proteins can pass the quality control surveillance and traffic to the Golgi complex. When misfolded proteins accumulate in the ER, cells activate a self-protective mechanism, termed the ER stress response, which is signaled through the unfolded protein response (UPR).17 In mammalian cells, EIF2AK3 (PERK), ERN1 and ATF6 are activated after sensing the presence of unfolded proteins in the ER. EIF2AK3 activation leads to phosphorylation of the α-subunit of eukaryotic translation initiation factor 2 (EIF2S1/eIF2α) and inhibits protein synthesis. Activation of ERN1 and ATF6 promotes transcription of UPR target genes. ERN1 induces X-box binding protein 1 (XBP1) mRNA processing to generate mature spliced XBP1 (sXBP1) mRNA, which in turn activates transcription of ER molecular chaperones, such as HSPA5 (GRP78/BiP), or ER-associated degradation (ERAD)-related genes, such as ER degradation-enhancing alpha-mannosidase-like 1 (EDEM1). ATF6 is cleaved by site 1 and site 2 proteases (S1P and S2P) in response to ER stress. ERN1 recruits TNF receptor-associated factor 2 (TRAF2), which in turn recruits apoptosis signal-regulating kinase 1 (MAP3K5/ASK1) that activates c-Jun amino terminal kinase (MAPK8). The cleaved ATF6 N-terminal fragment migrates to the nucleus to activate the transcription of HSPA5 through direct binding to the ER stress response element.
A number of reports have shown that autophagy is induced by ER stress and is critical for cell survival under conditions of ER stress.18,19 Autophagy is the primary cellular pathway by which long-lived proteins, cytoplasmic organelles and intracellular pathogens undergo degradation. Autophagy involves sequestration of these cellular constituents in double- or multimembrane cytoplasmic vesicles called autophagosomes, which are subsequently delivered to the lysosome, where they are degraded and recycled. During this process, autophagosomes enclose cytosol as well as organelles. Induction of autophagy is regulated by the complex of class III PtdIns3K with BECN1 (yeast Vps30/Atg6). Later steps required for the formation of autophagosomes involve the autophagy-related proteins ATG3, ATG5, ATG7, microtubule-associated protein 1 light chain 3 (MAP1LC3A/LC3, yeast Atg8), ATG10 and ATG12 in two ubiquitin-like conjugation pathways.20 Autophagy promotes cell survival by enabling cells to keep macromolecule synthesis and the energy requirements during nutrient deprivation and other forms of cellular stress. Autophagy also plays a role in differentiation and development, aging, innate and adaptive immunity and cancer.21
Our present study shows that depletion of L-Arg downregulates a number of proteins in human T-cells, in addition to CD247, leading to a ER stress response. Subsequent triggering of autophagy keeps T-cells alive under ER stress conditions, thus preventing the onset of apoptosis.
Results
Depletion of L-Arg inhibits expression of CD247 and additional membrane antigens, arrests cells in G0/G1 and blocks cell proliferation in Jurkat cells and peripheral blood mitogen-activated T cells
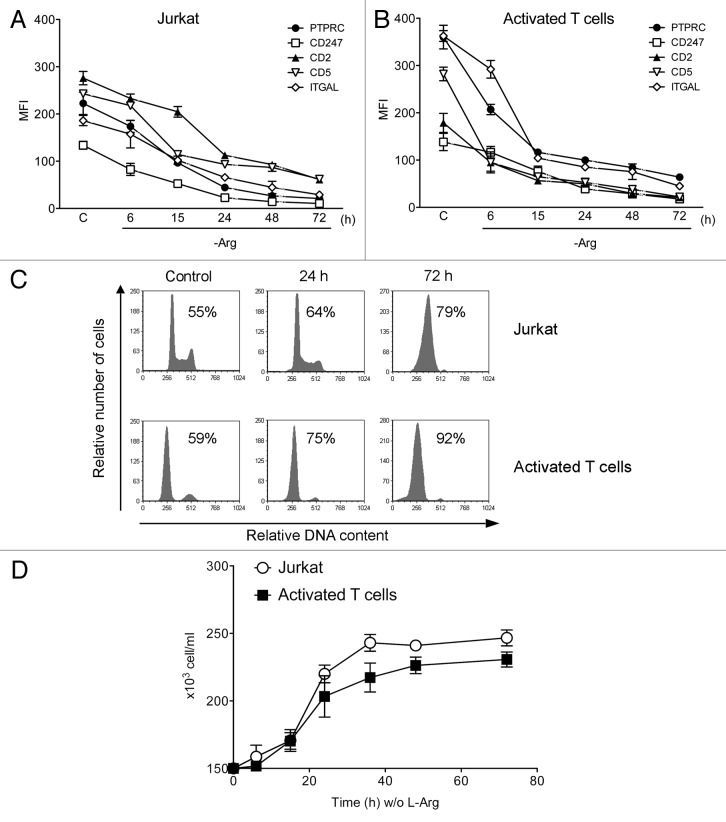
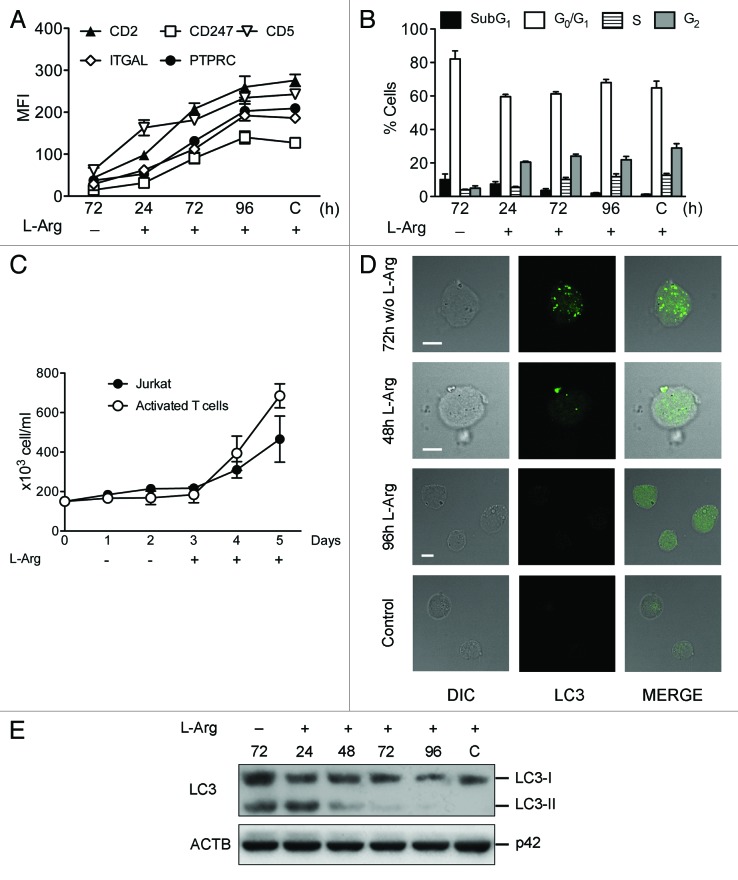
L-Arg deficiency has been reported to downregulate CD247 in activated T cells, thus preventing the normal expression of the T cell receptor (TCR).22 This downregulation of CD247 was reported not being accompanied by any alteration in the expression of other surface molecules, such as CD4 and CD69.23 However, we found that the effect of L-Arg deficiency on cell surface protein expression in activated lymphocytes was more general than previously reported. Incubation of both human Jurkat T-cell line and peripheral blood mitogen-activated T cells in a L-Arg-deficient medium for 72 h led not only to TCR CD247 downregulation, but additional membrane antigens, including CD2, CD5, ITGAL (CD11A) and PTPRC (CD45), were also downregulated (Fig. 1A and B), as assessed by fluorescence flow cytometry. Jurkat and peripheral blood mitogen-activated T cells, cultured in the absence of L-Arg, underwent an arrest in G0/G1 and cell proliferation was blocked (Fig. 1C and D). After 72 h in culture without L-Arg, most lymphocytes (~80–90%) were arrested at G0/G1 (Fig. 1C), and cells stopped proliferation (Fig. 1D). In contrast, Jurkat and activated T cells remained growing (to about 700,000 cells/ml) in the presence of L-Arg after 72-h incubation, with a normal cell cycle profile (data not shown). However, no apoptotic response was observed in the absence of L-Arg, as assessed by the lack of any increase in the sub-G0/G1 population in cell cycle analysis by flow cytometry (Fig. 1C).
Figure 1. L-Arg depletion induces downregulation of CD247 and additional membrane antigens, arrests cells in G0/G1 and inhibits cell proliferation in human Jurkat and peripheral blood mitogen-activated T cells. Jurkat (A) and peripheral blood mitogen-activated T cells (B) were incubated in the absence of L-Arg for the indicated times, and then cells were subjected to flow cytometry to determine the expression of the indicated membrane antigens, measured as mean fluorescence intensity (MFI), and compared with respect to control cells (C) grown for the same periods of time in the presence of L-Arg. (C) Cells incubated in the presence (Control) and in the absence of L-Arg for 24 and 72 h were analyzed for cell cycle profiling. Representative histograms, from at least five different experiments, show cell cycle arrest in G0/G1 following L-Arg deprivation, and the percentages of T cells at G0/G1 phase are shown in each cell cycle profile. (D) Cells incubated without L-Arg were counted at the indicated times as a measurement of cell proliferation. Data in (A, B and D) are means ± SD of three independent experiments.
L-Arg depletion induces ER stress in Jurkat and peripheral blood mitogen-activated T cells
Because the absence of L-Arg in the culture medium had a rather general effect on downregulating a number of different cell surface proteins, and ER is critical for protein synthesis as well as for post-translational protein modification, folding and export, we analyzed whether L-Arg deprivation could lead to an ER stress response.
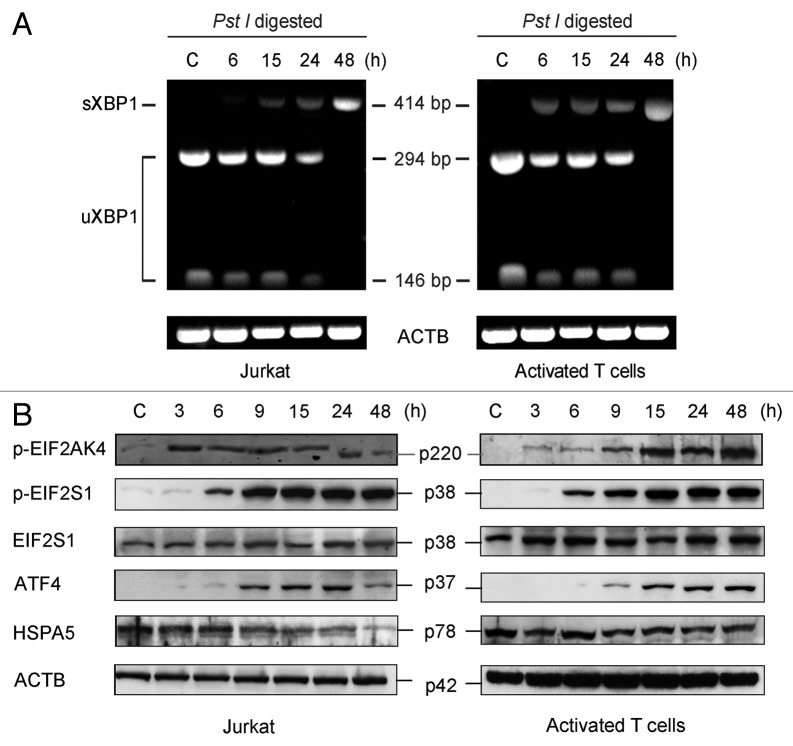
ER stress signaling cascades lead to oligomerization of ERN1, which results in trans-autophosphorylation and activation of its RNase domain, thus acting on the mRNA of XBP1 to excise a 26-nucleotide internal sequence from uXBP1 (unspliced XBP1) and producing mature sXBP1 mRNA (spliced XBP1). The presence of a PstI restriction site in this 26-nucleotide fragment allows to distinguish between both forms by restriction analysis of PCR-amplified cDNA, and thus to assay for ER stress response activation.24 After culturing both Jurkat and peripheral blood mitogen-activated T cells with and without L-Arg, we observed an equilibrium between the two splicing variants of XBP1 between 6 and 24 h, whereas at 48 h only sXBP1 was present (Fig. 2A). These data suggest that L-Arg depletion triggers an ERN1-mediated ER stress response.
Figure 2. L-Arg depletion induces ER stress in Jurkat cells and peripheral blood mitogen-activated T cells. (A) Cells were incubated with (C) or without L-Arg for the indicated times, and then total RNA was isolated and subjected to RT-PCR using specific primers for the XBP1 gene. PCR amplicons were incubated with PstI, and then run in agarose gel electrophoresis and stained with ethidium bromide. Only cDNA derived from sXBP1 mRNA was not cut with PstI, because of the loss of a 26-bp intron in response to ER stress. The positions of the amplification products uXBP1 and sXBP1 are indicated. (B) Jurkat and peripheral blood mitogen-activated T cells were incubated with (C) or without L-Arg for the times shown, and then cells were analyzed by immunoblotting with antibodies directed against the indicated proteins. Total ACTB was used as a loading control for western blots. Data shown are representative of three experiments performed.
sXBP1 encodes an active leucine zipper transcription factor that regulates the transcription of several genes involved in ER quality control mechanisms, ER/Golgi biogenesis, as well as ERAD components.25 We next analyzed the effect of L-Arg deprivation on the expression of XBP1 downstream genes in both Jurkat cells and peripheral blood mitogen-activated T cells by semiquantitative RT-PCR. We found upregulation of EDEM1 (Fig. S1), which has been reported to increase degradative rates of certain misfolded proteins.26 We also detected a slight downregulation of HSPA5 and upregulation of DDIT3 (DNA damage-inducible transcript 3, also named CHOP, C/EBP homologous protein, or GADD153, growth arrest- and DNA damage-inducible gene 153), both genes being involved in ER stress signaling (Fig. S1).
Another consequence of ER stress is translational arrest, signaled through phosphorylation of EIF2S1 at Ser51, which leads to decreased formation of the ternary complex required for the binding of Met-tRNA to the 40S ribosomal subunit.27 The protein kinase EIF2AK4 (GCN2) is activated when the level of any amino acid diminishes sufficiently to cause the accumulation of uncharged tRNAs, which are direct activators of the EIF2AK4 kinase. EIF2AK4 phosphorylates EIF2S1 at Ser51, resulting in the sequestration of EIF2B, which greatly reduces the rate of general translational initiation, while upregulating the translation of a subset of mRNAs. As shown in Figure 2B, depletion of L-Arg induced phosphorylation of EIF2AK4 and EIF2S1 in both Jurkat and peripheral blood mitogen-activated T cells. Phosphorylation of EIF2S1 results in preferential translation of selected mRNAs, such as activating transcription factor 4 (ATF4),28 which promotes expression of an array of genes including those coding for amino acid biosynthetic proteins and autophagy genes like ATG12.29 In this regard, we found that the absence of L-Arg induced the synthesis of ATF4 in both Jurkat and peripheral blood mitogen-activated T cells (Fig. 2B), as well as upregulation of ATG12 expression in microarray analysis (data not shown). In addition, the absence of L-Arg promoted the downregulation of the HSPA5 protein in Jurkat cells, but not in peripheral blood mitogen-activated T cells (Fig. 2B).
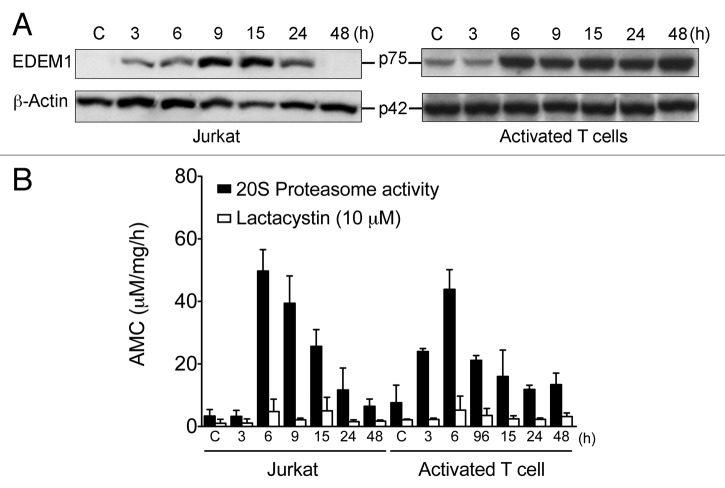
EDEM1 is an ER stress-inducible regulator of glycoprotein disposal from the ER.30 Here, we found that L-Arg depletion induced upregulation of EDEM1 in Jurkat and peripheral blood mitogen-activated T cells at both mRNA and protein levels (Fig. S1; Fig. 3A).
Figure 3. L-Arg depletion induces EDEM1 expression and 20S proteasome activity. (A) Jurkat and activated T cells were incubated with (C) and without L-Arg for selected times and the expression of EDEM 1 was determined by western blot. ACTB was used as a loading control. Data shown are representative of three experiments performed. (B) 20S proteasome activity was measured in 20 µg of protein extract of Jurkat and activated T cells incubated with (C) and without L-Arg for the indicated times, in the absence or presence of the proteasome inhibitor lactacystin (10 μM), and performed in triplicate. 20S proteasome activity was measured as the hydrolysis of the fluorogenic peptidyl substrate LLVY-AMC, and reported as μmol/l AMC per mg protein per hour. Data are means ± SD of three independent experiments performed in triplicate.
In addition, 20S proteasome activity was assayed in lysates of Jurkat and peripheral blood mitogen-activated T cells incubated in the presence and absence of L-Arg. 20S proteasome activity was increased after 3-6 h incubation without L-Arg, followed by a gradual decrease in both Jurkat and peripheral blood mitogen-activated T cells (Fig. 3B). This proteasome activity was inhibited by lactacystin in both Jurkat and activated T cells (Fig. 3B).
L-Arg depletion induces autophagy in Jurkat and peripheral blood mitogen-activated T cells
We found that the rate of apoptosis in Jurkat cells and peripheral blood mitogen-activated T cells, maintained in L-Arg-deficient RPMI-1640 culture medium for 7 days, was lower than 7% (data not shown). On these grounds, we hypothesized that T lymphocytes adapted to this new situation of L-Arg deprivation through the ER stress response we have found here. The initial intent of the UPR is to adapt to the changing environment, and restore normal ER function. Cell adaptation to these changes is provided by transcriptional programs that upregulate genes, including the prosurvival UPR marker HSPA5, that enhance the protein folding capacity of the ER, and promote ER-associated protein degradation to remove misfolded proteins.17 However, we found that L-Arg deprivation did not induce HSPA5 expression (Fig. S1; Fig. 2B). In addition, recent studies show that ER stress induces autophagy,18 as a major mechanism for survival.19 A primary role of autophagy in a variety of organisms is to adapt to nutrient starvation by releasing amino acids through the catabolism of existing proteins, and in this regard, autophagy is a cellular response to adverse environment and stress. On these grounds, we wondered whether cells undergoing ER stress upon L-Arg deprivation might survive through the triggering of an autophagic program.
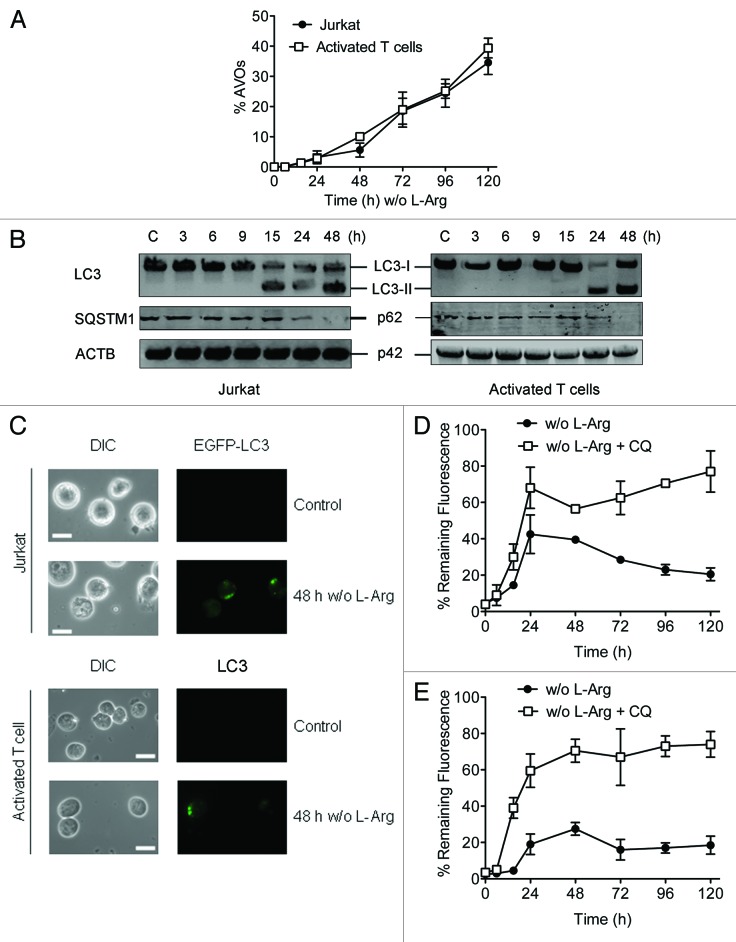
Autophagy is featured by the formation of autophagosomes and autolysosomes. Acridine orange (AO) is a fluorescent weak base that accumulates in acidic spaces, such as autolysosomes and lysosomes, which are called acid vesicular organelles (AVOs) and fluoresce bright red, whereas the cytoplasm and nucleolus fluoresce bright green and dim red. After incubating the cells with or without L-Arg, AO was added, and the increment of red fluorescence was quantified by flow cytometry. As shown in Figure 4A, AVO fluorescence was increased in both Jurkat and activated T cells following L-Arg depletion, suggesting the induction of autophagy. A major marker of autophagy activation is the conversion of the microtubule-associated protein 1 light chain 3 (LC3) from the unconjugated form (LC3-I), which is in the cytosol, to the phosphatidylethanolamine-conjugated form (LC3-II), that targets to the autophagosomal membrane.31 We found that incubation of Jurkat T cells and peripheral blood mitogen-activated T cells in L-Arg-deficient culture medium elicited a strong formation of the lipidated LC3-II along the incubation time (Fig. 4B). Sequestosome 1 (SQSTM1/p62) is a polyubiquitin-binding protein that binds autophagosomal membrane protein LC3, bringing SQSTM1-containing protein aggregates to the autophagosome and thus facilitating degradation of ubiquinated protein aggregates by autophagy.32 Lysosomal degradation of autophagosomes leads to a decrease in SQSTM1 protein level, and so this protein can be considered as a substrate of autophagy. In this regard, we found a decrease in the SQSTM1 protein level in Jurkat T and peripheral blood mitogen-activated T cells incubated in the absence of L-Arg (Fig. 4B), supporting the involvement of autophagy. In order to further assess the autophagic response, Jurkat cells were transfected with EGFP-LC3, and peripheral blood mitogen-activated T cells were immunostained with specific anti-LC3, and then cells were analyzed by fluorescence microscopy. Jurkat-EGFP-LC3 cells and peripheral blood mitogen-activated T cells were incubated with and without L-Arg for 48 h, before fixation for microscopy analysis. Consistently with the above data, we found that LC3 displayed a transition from the diffusive cytoplasm pattern to the punctated membrane pattern following L-Arg depletion, as assessed by confocal microscopy (Fig. 4C), suggesting the generation of autophagosomes. Next, we analyzed the autophagic flux by using a previously reported method33 involving saponin extraction, which is specific for nonautophagosome-associated LC3, coupled to flow cytometry. Jurkat-EGFP-LC3 cells and peripheral blood mitogen-activated T cells were incubated without L-Arg in the presence and absence of 10 µM chloroquine (CQ), and then cells were treated with 0.05% saponin, that results in extraction of the soluble LC3-I form, and subsequently they were subjected to flow cytometry analysis. Because saponin treatment selectively extracted nonautophagosome-associated LC3, the remaining fluoresecence allowed determination of autophagosome formation by flow cytometry (Fig. 4D and E). We found an increase in fluorescence until 24 and 48 h, and then a decrease over time, indicating degradation of autophagosome-associated LC3-II protein, measured by using the EGFP-LC3-II reporter in Jurkat cells (Fig. 4D), and immunostaining with an anti-LC3 antibody in peripheral blood mitogen-activated T cells (Fig. 4E). In contrast, this loss of fluorescence was not observed in CQ-treated cells, indicating an accumulation of autophagosomes (Fig. 4D and E).
Figure 4. L-Arg depletion induces autophagy in Jurkat and peripheral blood mitogen-activated T cells. (A) The formation of acidic vesicular organelles (AVOs) was detected in acridine orange-stained cells by flow cytometry. The increment in red fluorescence represents formation of AVOs. (B) Cells were incubated with (C) or without L-Arg for the indicated times, and then conversion of LC3-I to LC3-II and degradation of SQSTM1 was determined by western blot using antibodies specific to LC3 and SQSTM1. ACTB was used as a loading control. (C) Immunofluorescence images of Jurkat-EGFP-LC3 cells and activated T cells incubated in L-Arg-containing (Control) and L-Arg-free medium for 48 h, and then activated T cells were immunostained for intracellular LC3. Scale bar: 10 µm. (D and E) Measurement of autophagic flux. Jurkat-EGFP-LC3 (D) and activated T cells (E) were incubated without L-Arg in the presence or absence of chloroquine (CQ) for the indicated times, washed briefly with 0.05% saponin in PBS, and analyzed for autophagosome-bound LC3 fluorescence. Activated T cells were immunostained with anti-LC3 antibody and a Cy2-conjugated secondary antibody, and then analyzed by flow cytometry for total fluorescence. Data are presented as a percentage of the total fluorescence intensity before saponin treatment (% remaining fluorescence). Data shown are means ± SD or representative of three experiments performed.
Effect of L-Arg deprivation on signaling pathways regulating autophagy
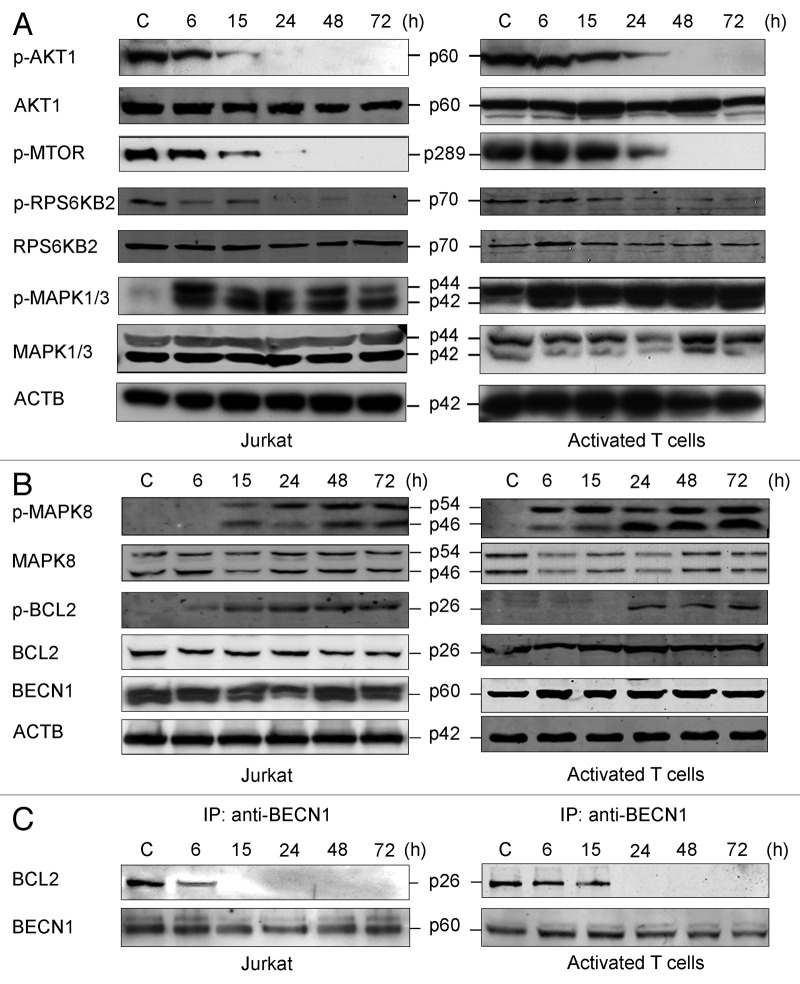
AKT1 and mTOR (MTOR) signaling molecules are major regulators of autophagy.34 As shown in Figure 5A, the basal levels of AKT1 and MTOR phosphorylation were decreased along the incubation of human Jurkat T cells and peripheral blood mitogen-activated T cells in L-Arg-deficient culture medium. Because both AKT1 and MTOR act as major autophagy inhibitors, their down-modulation leads to an increase in autophagy (Fig. 4). Unlike AKT1 and MTOR, extracellular signal-regulated kinase 1/2 (ERK1/2, MAPK1/3) activation has been previously shown to contribute to autophagy-induced prosurvival function.35 Interestingly, we detected an increase in MAPK1/3 phosphorylation when Jurkat cells were incubated in L-Arg-depleted medium (Fig. 5A). In addition, a slight increase in MAPK1/3 phosphorylation was also detected in peripheral blood mitogen-activated T cells, although to a much lesser extent than in Jurkat cells, following L-Arg depletion (Fig. 5A).
Figure 5. Downregulation of AKT1-MTOR signaling and dissociation of the BECN1-BCL2 complex in response to L-Arg depletion. (A) Jurkat and peripheral blood mitogen-activated T cells were incubated with (C) or without L-Arg for the indicated times, and then analyzed by immunoblotting with antibodies directed against the indicated proteins. (B) Lysates from Jurkat and activated T cells treated as above were analyzed by western blot for the total content of MAPK8, p-MAPK8, p-BCL2, BCL2 and BECN1 using specific antibodies. ACTB was used as a loading control in (A and B). (C) Western blot analysis of anti-BECN1 immunoprecipitates (IP), derived from total lysates of Jurkat and activated T cells (500 μg) incubated with (C) or without L-Arg for the indicated times, and then probed with anti-BCL2 and anti-BECN1 antibodies. Data shown are representative of three experiments performed.
Despite the involvement of autophagy in cell death and survival is controversial, there is increasing evidence that suggests a protective role for autophagy against nutrient stress, aggregates of misfolded proteins, organelle damage and microbes.36-39 Because autophagy activity generates energy, autophagy is often induced under nutrient-limiting conditions, providing a mechanism to maintain cell viability, which may be exploited by cancer cells for survival under metabolic stress.40
Protracted activation of ERN1 results in the phosphorylation of MAPK8.41 Depending on the cellular context, activation of MAPK8 can either allow the cells to adapt to ER stress by initiating autophagy, or to promote apoptosis.19 As shown in Figure 5B, L-Arg depletion induced a sustained MAPK8 activation in both Jurkat and peripheral blood mitogen-activated T cells. MAPK8 positively regulates autophagy through direct phosphorylation of BCL2 that leads to the release of the inhibitory lock on BECN1.42 Interestingly, only the ER-localized pool of BCL2 is subjected to regulation by this mechanism.42 BECN1 is essential for the initiation of the early stages of autophagy,43 and its function is inhibited by the interaction of the BECN1 BH3 domain with the BH3-binding groove in BCL2 or BCL2L1/Bcl-XL.44 Although endogenous BECN1 is found at mitochondria, ER and the trans-Golgi network, the interaction between BCL2 and BECN1 takes place exclusively on the surface of the ER. This suggests that the regulatory hold held by BCL2 over BECN1 is particularly responsive to alterations in ER homeostasis.45 Here, we found that L-Arg depletion induced BCL2 phosphorylation (Fig. 5B). Because we found that L-Arg starvation prompted MAPK8 activation and BCL2 phosphorylation (Fig. 5B), whereas BECN1 protein level was preserved, we wondered whether the ER stress response induced by depletion of L-Arg affected the interaction between BCL2 and BECN1. By immunoprecipitation assays, we found that the interaction between BCL2 and BECN1 was lost after incubation of Jurkat and activated T cells in medium deficient in L-Arg (Fig. 5C).
Inhibition of MAPK8 and MAPK1/3 signaling blocks L-Arg depletion-induced autophagy
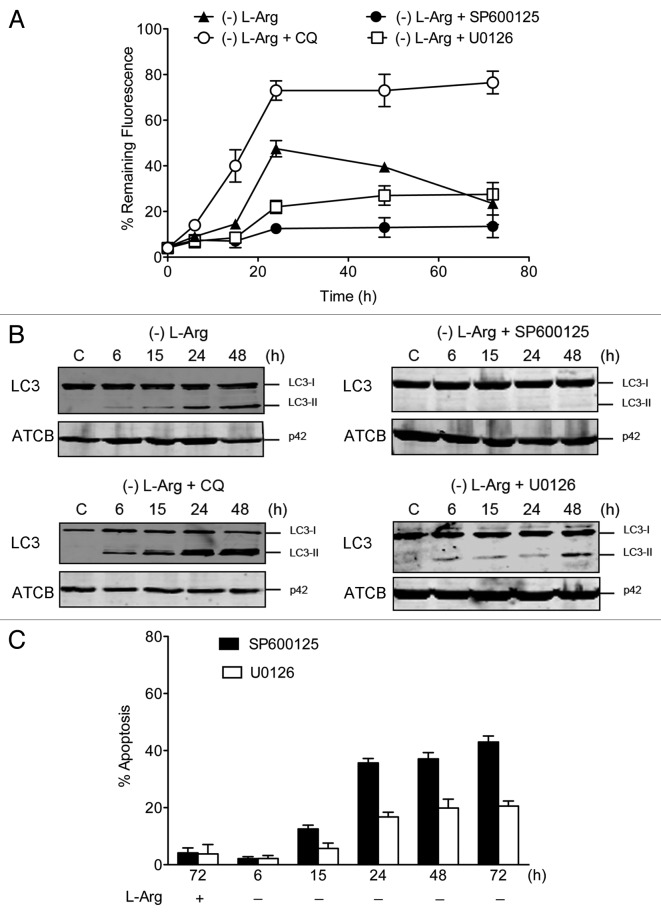
To further investigate the role of MAPK8 and MAPK1/3 in L-Arg depletion-mediated autophagy, Jurkat cells were pretreated with 10 μM of SP600125 (MAPK8 specific inhibitor) and 10 μM U0126 (inhibitor of both MEK1 and MEK2, upstream activators of MAPK1/3) for 1 h, and then incubated in L-Arg-deficient culture medium. Immunoblotting analysis showed that phosphorylation of MAPK8 and MAPK1/3 was inhibited following pretreatment with SP600125 and U0126 (data not shown). We found that both inhibitors prevented the autophagic flux in Jurkat (Fig. 6A) and peripheral blood mitogen-activated T cells (data not shown). Treatment of cells with chloroquine led to the accumulation of LC3-II following L-Arg deprivation, as this compound inhibits the formation of autolysosomes, and therefore LC3-II is not degraded. In addition, we found that preincubation with SP600125 blocked LC3-II formation (Fig. 6B). U0126 was a weaker inhibitor than SP600125 in the initial processes of autophagy, as a low generation of LC3-II was still detected (Fig. 6B). Furthermore, the inhibition of the formation of autolysosomes by SP600125 and U0126 resulted in enhanced apoptotic cell death under L-Arg depletion, as assessed by an increase in the percentage of annexin V-positive cells by flow cytometry (Fig. 6C).
Figure 6. Inhibition of MAPK8 and MAPK1/3 in Jurkat cells leads to inhibition of autophagic flux and induction of apoptosis following L-Arg depletion. (A) Jurkat-EGFP-LC3 cells were incubated without L-Arg in the presence or absence of chloroquine (CQ, 10 µM), and inhibitors of MAPK8 (SP600125, 10 µM) and MAPK1/3 (U0126, 10 µM) signaling pathways for the indicated times, washed briefly with 0.05% saponin in PBS, and then analyzed by flow cytometry for total fluorescence. Data are presented as a percentage of the fluorescence intensity before saponin treatment (% remaining fluorescence), and are means ± SD of three independent experiments. (B) Jurkat cells were incubated with (C) or without L-Arg in the presence or absence of chloroquine (CQ, 10 µM), and inhibitors of MAPK8 (SP600125, 10 µM) and MAPK1/3 (U0126, 10 µM) signaling pathways for the indicated times, and then conversion of LC3-I to LC3-II was determined by western blot. ACTB was used as a loading control. Results shown are representative of three experiments performed. (C) Cells were incubated in RPMI-1640 medium with and without L-Arg in the presence of the indicated inhibitors of MAPK8 (SP600125, 10 µM) and MAPK1/3 (U0126, 10 µM) signaling routes for 6, 15, 24, 48 and 72 h. Control cells were cultured with L-Arg and inhibitors for 72 h. At the indicated times cells were stained with FITC-Annexin V and 7-ADD. Annexin V-positive populations were analyzed by flow cytometry, and represented the percentage of apoptosis. Data are means ± SD (C) of three independent assays.
Autophagy protects lymphocytes from apoptosis in the absence of L-Arg
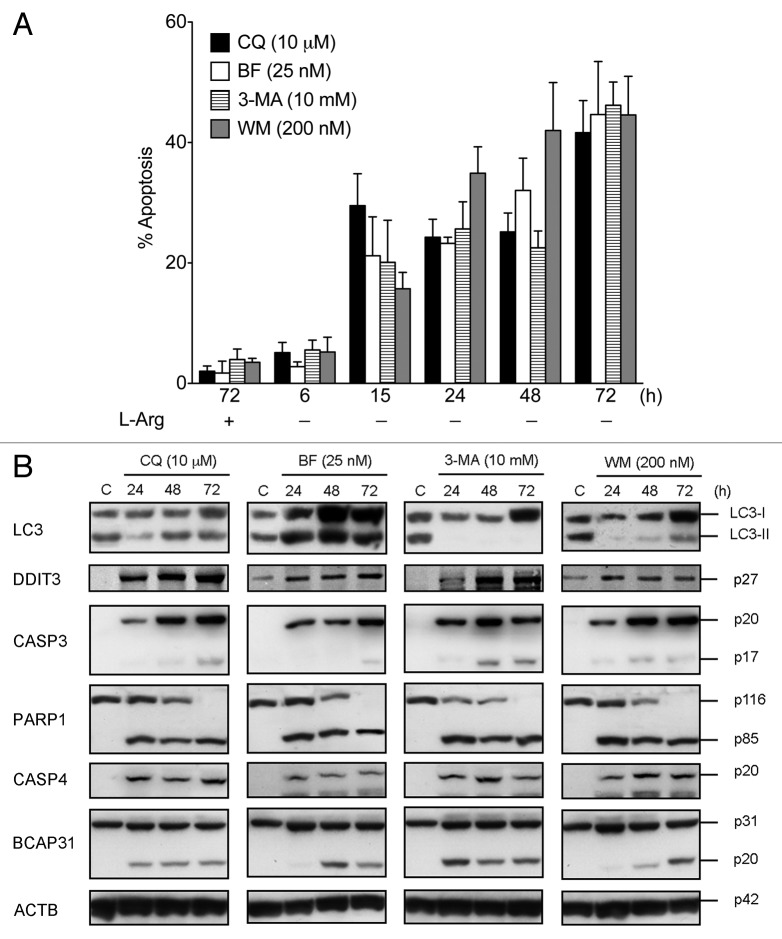
To examine whether the autophagic response induced by ER stress plays a role in cell survival or cell death, Jurkat cells incubated in culture medium without L-Arg were treated with chloroquine, bafilomycin A1 (BF), 3-methyladenine (3-MA) or wortmannin (WM) to block autophagy. The inhibition of the formation of autolysosomes by CQ and BF resulted in enhanced apoptotic cell death under L-Arg depletion, as assessed by an increase in the percentage of annexin V-positive (Fig. 7A) and hypodiploid sub-G0/G1 (data not shown) cells by flow cytometry. Inhibition of the fusion of lysosomes with autophagosomes by CQ or BF resulted in the accumulation of LC3-II, which was detectable by western blot (Fig. 7B). Induction of apoptosis in lymphocytes following L-Arg depletion in the presence of autophagy inhibitors was further assessed by caspase 3 (CASP3) activation and breakdown of its substrate poly(ADP-ribose) polymerase 1 (PARP1) (Fig. 7B). Similarly, inhibition of the early stages of autophagy, using 3-MA or WM, which acted before autophagosome formation and thereby without LC3-I to LC3-II conversion, sensitized cells to undergo ER stress-induced apoptosis following L-Arg depletion, as assessed by an increase of annexin V-positive (Fig. 7A) and hypodiploid sub-G0/G1 (data not shown) cells, as well as by activation of CASP3 and PARP1 cleavage (Fig. 7B). Likewise, three additional markers of ER stress-mediated apoptosis, such as DDIT3 upregulation, caspase 4 (CASP4) activation and BCAP31 (Bap31) cleavage,46 were also observed following L-Arg depletion in the presence of autophagy inhibitors (Fig. 7B). Thus our data indicate that, irrespective of the stage at which autophagy was inhibited, disabled autophagy prompted L-Arg starvation-induced death via a common final pathway involving biochemical features of apoptosis (Fig. 7). These data also indicate that autophagy plays a pivotal role in protecting against cell death induced by ER stress caused by L-Arg depletion.
Figure 7. Inhibition of the autophagic-lysosomal pathway in Jurkat cells leads to apoptosis following L-Arg depletion. (A) Cells were incubated in RPMI-1640 medium without L-Arg in the presence of the indicated inhibitors of the autophagic-lysosomal route for 6, 15, 24, 48 and 72 h. Control cells were grown with L-Arg and inhibitors for 72 h. At the indicated times cells were stained with FITC-annexin V and 7-ADD. Annexin V-positive populations were determined by flow cytometry, and represented the percentage of apoptosis. Data are means ± SD of 3 independent experiments. (B) Cells treated as above were analyzed by western blot for the expression of DDIT3, LC3, activation of CASP3 and CASP4, as well as for the cleavage of PARP1 and BCAP31. Control Jurkat cells (C), incubated for 72 h in medium without L-Arg in the absence of the above inhibitors, were run in parallel. ACTB was used as a loading control. Data shown are representative of three experiments performed.
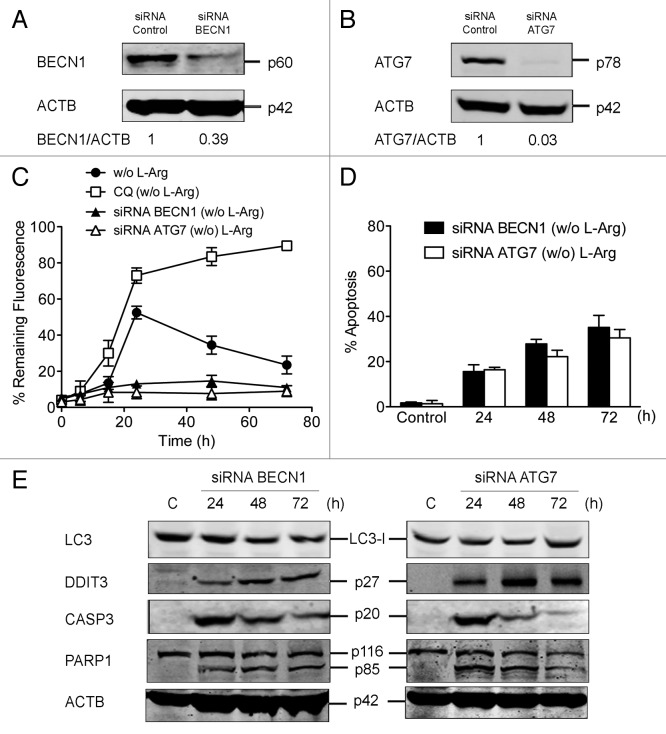
To further investigate if the above apoptotic response was the result of inhibition of the autophagic process, we targeted by small interfering RNA (siRNA) two established autophagic proteins, namely BECN1 and ATG7, in Jurkat cells (Fig. 8A and B). The autophagic flux was largely abrogated following BECN1 and ATG7 silencing in Jurkat cells incubated in L-Arg-deficient culture medium, as assessed by measuring the remaining fluorescence of EGFP-LC3 through a saponin/flow cytometry-based assay (Fig. 8C). Comparison of these data with those obtained when Jurkat cells were cultured in the presence of CQ, indicated the blockage of the initial stages of autophagy, with no formation of autophagosomes, following BECN1 and ATG7 silencing (Fig. 8C). Inhibition of autophagy by BECN1 and ATG7 siRNA induced an apoptotic response when Jurkat cells were incubated in L-Arg-depleted medium (Fig. 8D), which was accompanied by DDIT3 upregulation and CASP3 activation, that in turn led to the cleavage of the CASP3 substrate PARP1 (Fig. 8E). These results suggest that the L-Arg depletion-mediated autophagic response in Jurkat cells is a cell-protective mechanism whose inhibition enables the induction of apoptosis.
Figure 8. Downregulation of BECN1 and ATG7 in Jurkat cells leads to apoptosis and inhibition of autophagic flux upon L-Arg depletion. Jurkat cells were transiently transfected with 50 nM of either control siRNA, siRNA specific to BECN1 (A), or siRNA specific to ATG7 (B), and then analyzed by immunoblotting for BECN1 and ATG7 expression. ACTB was used as a loading control. (C) To evaluate the autophagic flux, Jurkat-EGFP-LC3 cells were transiently transfected with 50 nM either control siRNA or siRNA specific to BECN1 and ATG7. At 24 h after transfection, cells were incubated in medium without L-Arg for the indicated times, washed briefly with 0.05% saponin in PBS, and analyzed by flow cytometry for total fluorescence. Data are shown as the percentage of the fluorescence intensity before saponin treatment (% remaining fluorescence). (D) At 24 h after transfection, cells were incubated in medium without L-Arg for the indicated times cells, and then stained with FITC-Annexin V and 7-ADD. Annexin V-positive populations were analyzed by flow cytometry and represented the percentage of apoptosis. (E) Jurkat cells, transiently transfected with 50 nM either control siRNA or siRNA specific to BECN1 and ATG7, were subjected to either immunoblot analysis for LC3, DDIT3, CASP3 (using an antidody recognizing cleaved CASP3) and PARP1 (using an antidody recognizing both intact and cleaved forms). ACTB was used as a loading control. Data shown are means ± SD or representative experiments of three performed.
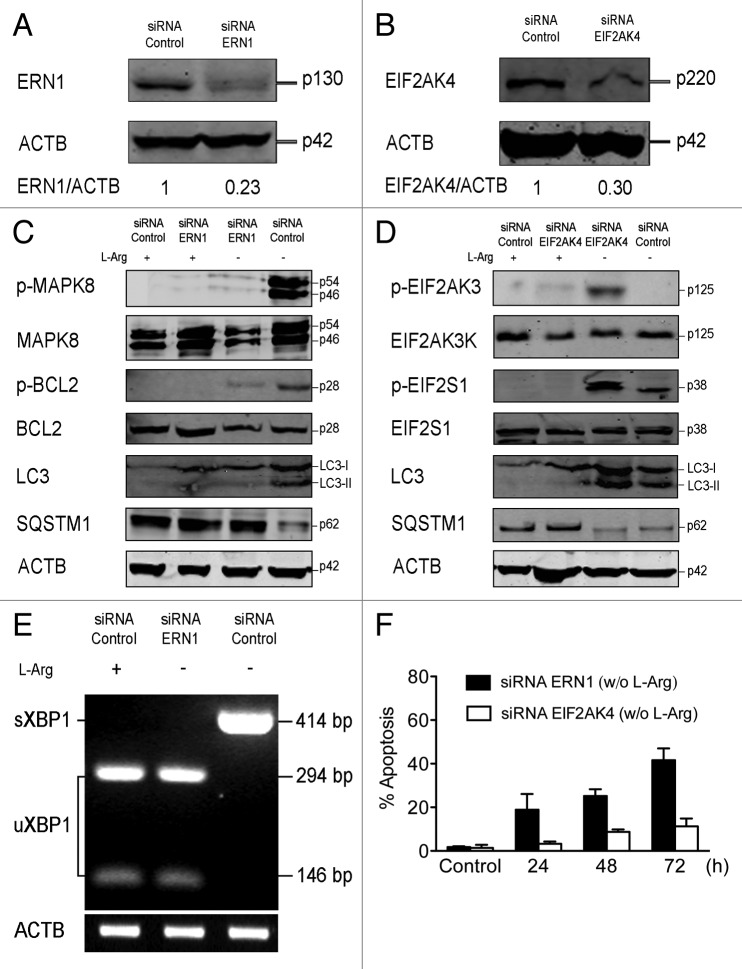
ERN1 signaling pathway, but not EIF2AK4 signaling, is required for activation of L-Arg depletion-induced autophagy
In mammalian cells, ERN1, EIF2AK3 and ATF6 sense the presence of unfolded proteins in the ER lumen and transduce signals to the cytoplasm and nucleus.47 EIF2AK3 activation leads to the phosphorylation of EIF2S1, and EIF2AK4 is another EIF2S1 kinase that is activated by amino acid starvation. A certain controversy has been raised regarding the involvement of either EIF2S1 or ERN1 signaling pathways as the crucial mediator of ER stress-induced autophagy.48 On these grounds, we next analyzed the role of ERN1 and EIF2AK4 in the autophagy response triggered by L-Arg depletion through RNA silencing (Fig. 9A and B). Jurkat cells transfected with ERN1 siRNA largely downregulated ERN1 (Fig. 9A), which led to inhibition of MAPK8 and BCL2 phosphorylation as well as LC3-II formation, whereas SQSTM1 was not degraded following L-Arg starvation (Fig. 9C). In contrast, silencing of EIF2AK4 (Fig. 9B) did not affect the generation of the autophagosome-bound LC3-II form and the degradation of SQSTM1 upon L-Arg depletion (Fig. 9D). EIF2AK3 phosphorylation was hardly detected, if any, when Jurkat cells were incubated in a L-Arg-depleted medium, but phosphorylation of EIF2AK3 was readily observed in EIF2AK4-silenced cells (Fig. 9D). These data suggest that EIF2AK3 could act as an alternative route to EIF2AK4 to phosphorylate EIF2S1 when Jurkat cells were incubated in the absence of L-Arg. ERN1 silencing also blocked XBP1 splicing, thus preventing the generation of sXBP1 (Fig. 9E). Furthermore, we found that downregulation of ERN1, but not of EIF2AK4, induced an increase in apoptosis in Jurkat cells incubated in the absence of L-Arg (Fig. 9F). These results suggest that ERN1 is required for autophagy activation after L-Arg-induced ER stress.
Figure 9. Downregulation of ERN1, but not EIF2AK4, leads apoptosis and inhibition of autophagic flux in Jurkat cells upon L-Arg depletion. (A) Jurkat cells were transiently transfected with 50 nM either control siRNA or siRNA specific to ERN1, and then analyzed by immunoblotting for ERN1 expression. (B) Jurkat cells were transiently transfected with 50 nM either control siRNA or siRNA specific to EIF2AK4, and then analyzed by immunoblotting for EIF2AK4 expression. (C and D) Cells transiently transfected with control siRNA, siRNAs specific to ERN1 (C) or EIF2AK4 (D) were incubated with or without L-Arg for 24 h, and then analyzed by immunoblotting with antibodies directed against the indicated proteins. ACTB was used as a loading control. (E) Cells transiently transfected with control siRNA or siRNA specific to ERN1 were incubated with or without L-Arg for 24 h, and then total RNA was isolated and subjected to semiquantitative RT-PCR using specific primers for the XBP1 gene. PCR amplicons were incubated with PstI, and then run in agarose gel electrophoresis and stained with ethidium bromide. Only cDNA derived from sXBP1 mRNA was not cut with PstI, because of the loss of a 26-bp intron in response to ER stress. The positions of the amplification products uXBP1 and sXBP1 are indicated. Expression of ACTB was used as a loading control. (F) At 24 h after transfection, cells transiently transfected with siRNA to ERN1 and siRNA to EIF2AK4 were incubated in medium without L-Arg for 24, 48 and 72 h. At the indicated times cells were stained with FITC-annexin V and 7-ADD. Annexin V-positive populations were determined by flow cytometry, and represented the percentage of apoptosis. Data shown are means ± SD or representative experiments of three performed.
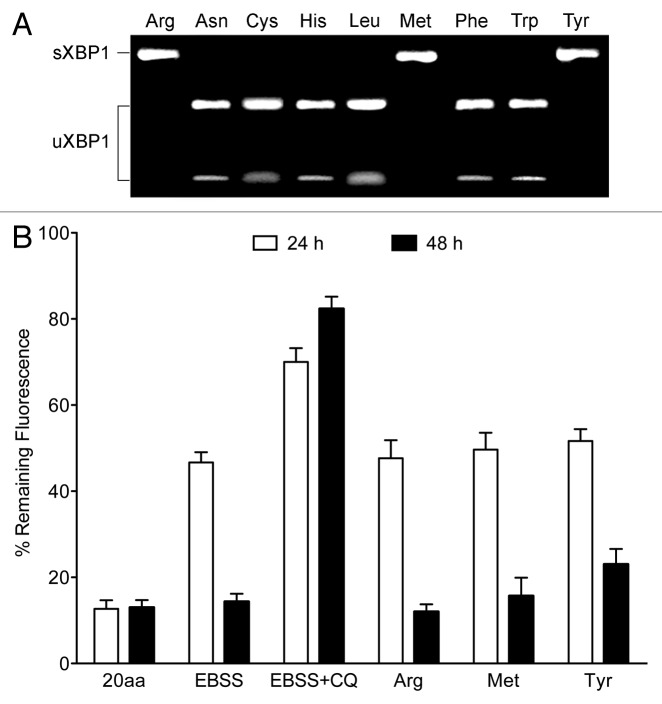
Depletion of L-Arg, L-Met and L-Tyr leads to XBP1 splicing and autophagy
We next analyzed whether the above induction of ER stress on T lymphocytes was specific to L-Arg depletion. To this aim, we examined the capacity of nine different amino acids to elicit an ER stress response in Jurkat cells. We found that depletion of L-Arg, L-Met and L-Tyr, out of nine amino acids tested, induced splicing of XBP1 (Fig. 10A). This suggests that the ability to prompt ER stress was not a general response to deprivation of any single amino acid, and that cells show a differential ability to mount an ER stress response depending on the depleted amino acid. Furthermore, we found that depletion of the above three amino acids L-Arg, L-Met and L-Tyr led to autophagic flux as assessed with Jurkat-EGFP-LC3 cells (Fig. 10B). However, we also detected that deprivation of some additional amino acids induced autophagy without triggering an ER stress response (data not shown).
Figure 10. Induction of ER stress response and induction of autophagy by removal of specific single amino acids from the medium. (A) Jurkat cells were incubated in the absence of the indicated amino acids, and analyzed for XBP1 splicing in order to determine induction of ER stress. Data shown are representative of three experiments. (B) Jurkat EGFP-LC3 cells were incubated for 24 and 48 h in amino acid-rich medium (20aa), in EBSS medium in the absence of all amino acids (EBSS), in EBSS + chloroquine (CQ), or in the absence of the indicated amino acids. Autophagic flux was then measured by flow cytometry of EGFP-LC3 as indicated in the Materials and Methods section. Data shown are means ± SD of three experiments performed.
Replenishment of L-Arg restores cell proliferation and antigen expression, and inhibits autophagy induced by L-Arg deficiency
In order to assess whether the above effects of depletion of L-Arg on autophagy induction were reversible, we replaced the L-Arg-deficient culture medium for L-Arg-containing RPMI-1640 culture medium, and then we performed a time-course assay to analyze the expression of membrane antigens in Jurkat cells, cell cycle, cell proliferation and autophagy. We found that after 96 h of restoring L-Arg to the medium, cells recovered the expression of membrane antigens (Fig. 11A), the normal cell cycle distribution profile (Fig. 11B), and their proliferation capacity (Fig. 11C). In addition, LC3-II formation was downregulated following L-Arg replenishment, as assessed by both confocal microscopy (Fig. 11D) and western blot (Fig. 11E). Thus, these results indicate that the autophagic stress response caused by the absence of L-Arg is a reversible process in T lymphocytes.
Figure 11. Replenishment of L-Arg restores expression of membrane antigens, cell cycle profile, cell proliferation, and blocks autophagosome formation. (A) Jurkat cells incubated in medium without L-Arg for 72 h were washed with PBS, and then resuspended in culture medium with L-Arg for the indicated times. Membrane antigen expression was determined by flow cytometry at different incubation times, and shown as mean fluorescence intensity (MFI), and compared with respect to control cells (C) grown for the same periods of time in the presence of L-Arg. (B) For cell cycle analysis, cells treated as above were fixed, stained with propidium iodide and the percentage of cells in each cell cycle phase was assessed by flow cytometry. Control cells (C) always grown in the presence of L-Arg were run in parallel. (C) Jurkat cells were counted to determine cell proliferation when incubated for the indicated times in a medium without L-Arg and then after L-Arg replenishment. (D) Confocal images of Jurkat cells immunostained for intracellular LC3 to detect autophagy. Scale bar: 10 µm. (E) Induction of autophagy was also determined in Jurkat cells, incubated in the absence of L-Arg and after L-Arg replenishment, through conversion of LC3-I to LC3-II by western blot, using an antibody specific to LC3. Data in (A–C) are means ± SD of three independent experiments. Data shown in (D and E) are representative of three experiments performed.
Discussion
Here we report for the first time that depletion of L-Arg leads to a reversible response that preserves the life of the T lymphocytes through ER stress and autophagy, while cells remain arrested at G0/G1. Replenishment of L-Arg drives T lymphocytes to proliferate again and they recover the normal cell cycle profile, whereas autophagy is inhibited and no further required. Our results suggest that depletion of L-Arg induces ER stress through ERN1 signaling, which results in activation of its RNase domain and removal of a 26-nt sequence from uXBP1, producing mature sXBP1 mRNA, which in turn activates genes responsible for ER-associated degradation of misfolded glycoproteins, like EDEM1.
Amino acid starvation has been shown to promote autophagy through the EIF2S1 kinase signaling pathway,49 and ER stress induced by protein aggregates can lead to autophagy through EIF2AK3 and EIF2S1 phosphorylation.50 Our data reported here show phosphorylation of EIF2S1 following depletion of L-Arg, which could lead to the upregulation of ER stress markers and the induction of autophagy promoting proteins, such as ATF4 and ATG12. There are four distinct EIF2S1-kinases in mammals, namely EIF2AK4, PLK3/PKR, EIF2AK3 and EIF2AK1/HRI, which are activated by amino acid starvation, viral infection, ER stress and heme depletion, respectively.51 Here, we observed EIF2AK4 phosphorylation, whereas EIF2AK3 phosphorylation was only detected upon EIF2AK4 downregulation following L-Arg deprivation. However, we found that EIF2AK4 silencing did not affect the generation of the autophagosome-bound LC3-II form and SQSTM1 degradation upon L-Arg depletion. Taken together, these results suggest that EIF2AK4 activation is not critical for the induction of autophagy following L-Arg depletion, and when EIF2AK4 is absent, its putative role in EIF2S1 phosphorylation could be carried out by alternative routes, such as the EIF2AK3-mediated pathway, which otherwise remains inactive. In addition, we also found that depletion of L-Arg increased the proteasome activity, and this might be a consequence of the herein reported ER stress response.
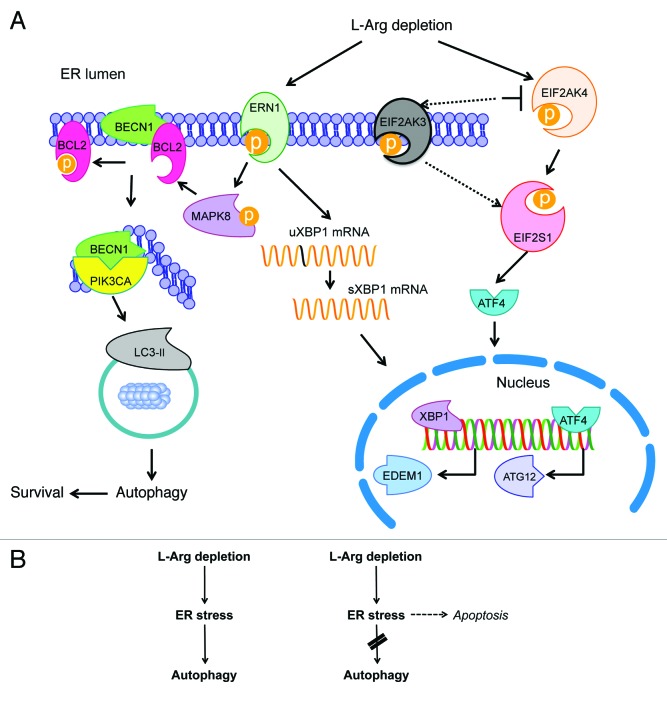
There is a growing body of evidence supporting the notion that ER stress is a potent inducer of autophagy, however the precise molecular pathways linking ER stress to autophagy remain to be fully elucidated. Furthermore, a persistent bone of contention is whether autophagy acts as a cytoprotective mechanism or a precursor to cell death.52 Sustained autophagy may be detrimental to cell survival because of excessive organelle and macromolecule catabolism, but conversely, autophagy may play an important role in cytoprotection by degrading protein aggregates during ER stress. BECN1, the mammalian ortholog of yeast Atg6 and essential for autophagy, was initially identified as a BCL2-interacting protein in a yeast two-hybrid screen.53 A point of convergence in the regulation of apoptotic and autophagic pathways was provided for the first time through the finding that BCL2 suppressed starvation-induced, BECN1-dependent autophagy in both yeast and mammalian cells.54 The interaction between BCL2 and BECN1 takes place exclusively on the surface of the ER, and thereby the ability of BCL2 to reduce BECN1’s capacity to induce autophagy is particularly responsive to alterations in ER homeostasis.55 We found here that the interaction between BECN1 and BCL2 is lost following L-Arg depletion, leading to the formation of the autophagosome. Activation of MAPK8 by L-Arg deprivation might be related to the displacement of BCL2 from the complex BECN1-BCL2. Figure 12A depicts a model for the involvement of ER stress in the induction of autophagy by L-Arg depletion in T lymphocytes based on the data reported here. In addition to the ERN1 and EIF2AK4-EIF2S1 signaling pathways shown in Figure 12A, we found here that L-Arg depletion might also promote autophagy through inhibition of AKT1 and MTOR.
Figure 12. Schematic model of ER involvement in L-Arg depletion-induced autophagy, survival and apoptosis. (A) This is a schematic outline to portray a plausible mechanism of how L-Arg deprivation induces autophagy via ER stress. Activated ERN1 splices a 26-nucleotide sequence from XBP-1, resulting in a translational frame-shift to generate sXBP1, a potent transcription factor involved in the expression of ER-associated degradation (ERAD) genes, such as ER degradation-enhancing mannosidase-like protein 1 (EDEM1). At the same time, MAPK8 is phosphorylated, allowing the release of BECN1 from the BECN1-BCL2 complex and the generation of the autophagosome. In addition, phosphorylation of EIF2S1 by EIF2AK4, or by EIF2AK3 in the absence of EIF2AK4, induces the expression of genes as ATF4 and other genes involved in the activation of the autophagy pathway. (B) Apoptosis (right) and survival (left) responses triggered by L-Arg depletion are dependent on ER stress and autophagy. Depletion of L-Arg induces an ER stress response that triggers autophagy as a way for survival (left). When autophagy is inhibited, ER stress signals lead to apoptosis (right). See the text for further details.
Recently there has been an increasing number of studies showing that the activation of the MAPK1/3 pathway induces autophagy.56 Our data showed that Jurkat cells incubated without L-Arg induced the phosphorylation of MAPK1/3, and inhibition of this signaling pathway inhibited autophagy, suggesting a role for MAPK1/3 route in the process. In addition, we have found here that MAPK8 activation, mediated by ERN1 in the early phase of ER stress, is required for autophagosome formation after ER stress and is sufficient for the induction of autophagy. Furthermore, in ERN1-deficient cells, we were unable to detect LC3-II formation, indicating that a signaling pathway ERN1-MAPK8 plays a major role in the activation of autophagy following L-Arg depletion.
Our present study demonstrates that L-Arg triggers an ER stress-dependent autophagic response as a means for survival in T lymphocytes. This prosurvival autophagic response can be maintained for rather protracted times, and it is reversed once L-Arg levels are back to normal. Interestingly, we also found that inhibition of autophagy, at early or late stages, promoted cell death of T lymphocytes cultured in the absence of L-Arg. Thus, the ER stress response triggered by deprivation of L-Arg leads preferentially to autophagy-dependent survival, but the final outcome can be diverted to apoptosis when autophagy is blocked (Fig. 12B). In this regard, it is worthy to note that L-Arg depletion in T cells hardly affects or slightly downregulates the expression of the survival UPR marker HSPA5, whereas it upregulates the death-promoting transcription factor DDIT3. Thus, the ER stress induced by L-Arg depletion is ready to promote cell death when autophagy is prevented. Autophagy is a cellular defense mechanism associated with ER stress, but prolonged ER stress may induce autophagic and apoptotic cell death.57,58 Here, as stated above, we have found that the ER stress response in T lymphocytes induced by L-Arg depletion leads to apoptosis when autophagy is blocked (Fig. 12B), highlighting a major role for autophagy in T cell survival under L-Arg starvation. These results square with a major role of autophagy in T cell survival during T lymphocyte development, function and homeostasis.59-62
Our results provide an explanation for the arrest and survival of activated T lymphocytes, as well as for the inhibition of the lymphocyte-dependent immune response in the absence of L-Arg. Our data also provide new insights on how T cell survival or death is regulated, through modulation of ER stress and autophagy following L-Arg depletion, which could be of relevance for autoimmune and immune-dependent diseases.
Materials and Methods
Cell culture and reagents
Human T-cell leukemia Jurkat cells (DSMZ, ACC 282) and human peripheral blood T lymphocytes were cultured in RPMI-1640 (GIBCO-BRL, 21875-034) supplemented with 10% (v/v) heat-inactivated fetal bovine serum (FBS, GIBCO-BRL, 10091-148), 2 mM L-glutamine (GIBCO-BRL, 25030024), 100 U/ml of penicillin and 100 μg/ml streptomycin (GIBCO-BRL, 15070-063) at 37ºC in a humidified atmosphere of air containing 5% CO2. Jurkat-EGFP-LC3 cells were generated by transfection of Jurkat cells with pEGFP-LC3 (Addgene, plasmid 21073) using Lipofectamine 2000 (Invitrogen, 11668019), according to the manufacturer’s instructions. Transfected cells were selected with 1 mg/ml G418 (Sigma-Aldrich, A1720), and the EGFP-positive population cells were maintained under G418 selection. RPMI-1640 medium without L-Arg (GIBCO-BRL, P04-16598) was supplemented with MnCl2 (Sigma-Aldrich, 203734) to a physiologic concentration (4 µM) and 5% dialyzed (10 kDa cutoff) serum (Sigma-Aldrich, 12105). Medium deficient in one single amino acid, apart from L-Arg, was prepared by addition of all amino acids, except the one of interest, into Earle’s balanced salt solution (EBSS, GIBCO-BRL, 14155048) at concentrations as in αMEM medium. The following inhibitors were used: 3-methyladenine (Sigma-Aldrich, M9281), bafilomycin A1 (Sigma-Aldrich, B1793), chloroquine (Sigma-Aldrich, C6628), wortmannin (Sigma-Aldrich, W1628), MEK1/MEK2 inhibitor U0126 (Sigma-Aldrich, U120) and JNK Inhibitor II SP600125 (Calbiochem, 420119).
Cell transfection
107 Jurkat cells were transfected using Lipofectamine 2000 according to the manufacturer’s instructions (Invitrogen). BECN1 (L-010552-00-0005) and ATG7 (L-020112-00-0005) siRNA SMARTpool oligonucleotide mixtures, and scrambled siRNA (D-001810-01-05), used as control, were from Dharmacon. EIF2AK4 siRNA (SC-45644) and nontargeting siRNA (SC-37007), used as control, were from Santa Cruz Biotechnology. ERN1 siRNA (RI11837) was from AbGent. Transfection with siRNAs (50 nM) was performed using Lipofectamine 2000, according to the manufacturer’s instructions (Invitrogen).
Isolation of peripheral blood lymphocytes and mitogen activation of T cells
Mononuclear cells were isolated from fresh human peripheral blood by dextran sedimentation and centrifugation on Ficoll-Paque PLUS (Amersham Biosciences, 17-1440-02) density gradients as previously described.63 Mononuclear cells were saved, washed twice with phosphate-buffered saline (PBS), and resuspended in RPMI-1640 culture medium containing 10% heat-inactivated FBS, 2 mM L-glutamine, 100 μg/ml streptomycin and 100 U/ml penicillin, and incubated overnight at 37°C in a humidified atmosphere of 5% CO2 and 95% air. Monocytes were depleted by culture dish adherence. After overnight incubation at 37°C, the nonadherent cells (lymphocytes) were washed with PBS and collected. Lymphocyte preparations were typically 65–71% CD3+, 27–29% CD19+ and < 0.4% CD14+. To further purify T-cells, the nonadherent cells were washed with PBS and passed twice through a nylon wool column to deplete residual B cells and monocytes as previously described.63 T-cell purity was checked by flow cytometry analysis. These purified T-cell preparations were typically > 95% CD3+, < 0.3% CD14+ and < 4% CD25+. Proliferation of T lymphocytes was induced by incubation of peripheral blood lymphocytes for 4 days with 0.5 μg/ml phytohemagglutinin (PHA, Sigma-Aldrich, L1668), which primarily stimulates T-cell proliferation, followed by one day treatment with both 0.5 μg/ml PHA and 50 U/ml interleukin-2 (IL2, Sigma-Aldrich, SRP3085) in RPMI-1640 culture medium containing 10% heat-inactivated FBS, 2 mM L-glutamine, 100 μg/ml streptomycin and 100 U/ml penicillin. Activated T cells were more than 80% CD25+.
Antigen detection by flow cytometry
Cells were then incubated with monoclonal antibodies against PTPRC (BD Pharmingen, 555486), CD2 (BD Pharmingen, 555324), CD5 (BD Pharmingen, 555350), CD247 (BD Pharmingen, 558402) and ITGAL (BD Pharmingen, 555378) at 1:100 dilution in PBS for 15 min at room temperature, washed with PBS, stained for 15 min with Cy2-conjugated anti-mouse IgG antibody (Jackson ImmunoResearch Laboratories, 200-222-037) at 1:50 dilution, and analyzed in a Becton Dickinson fluorescence-activated cell sorting (FACS)Calibur flow cytometer. For intracellular proteins, cells were fixed and permeabilized by means of the Fix & Perm cell permeabilization kit (CALTAG Laboratories, GAS004) following the manufacturer’s instructions. Mean fluorescence intensity was measured for each antigen, which was corrected for unspecific staining by subtracting the fluorescence of cells stained with negative controls. P3X63 myeloma culture supernatant, kindly provided by F. Sánchez-Madrid (Hospital de La Princesa, Madrid, Spain), and an isotype-matched nonrelevant IgG monoclonal antibody were used as negative controls, leading to virtually identical background values.
20S proteasome activity
Proteasome activity in Jurkat and activated T cells were measured in triplicate using a 20S Proteasome Activity Assay Kit (Chemicon International, Millipore, APT280). 20S proteasome chymotrypsin activity was measured by incubating 20 μg of each cell lysate with fluorophore 7-amino-4-methylcoumarin (AMC)-labeled peptide substrate LLVY-AMC at 37°C for 60 min. The free AMC released by proteasome activity was quantified using a 380/460-nm filter set in a fluorometer (ULTRA Evolution; XFLUOR4 Version: V 4.50). The AMC standard curve was generated with a series dilution of AMC standard solution. Proteasome activity was confirmed using purified 20S proteasome as the positive control, and was reported as μmol/l AMC per mg protein per h. Each sample/substrate combination was measured both in the presence and in the absence of lactacystin (10 μM) to account for any nonproteasomal degradation of the substrate.
Autophagic flux by flow cytometry
Autophagic flux was measured as previously described.33 Briefly, Jurkat-EGFP-LC3 cells were harvested by centrifugation, washed with PBS, and then washed with either PBS containing 0.05% saponin (Sigma-Aldrich, 47036) or with PBS alone, and analyzed by flow cytometry. For intracellular staining of endogenous LC3 in peripheral blood activated T cells, these latter were harvested by centrifugation, washed with culture medium and PBS, and washed with PBS containing 0.05% saponin. Cells were then incubated with mouse anti-LC3 monoclonal antibody (Cell Signaling, 2775) for 30 minutes, rinsed with PBS, incubated with Cy2-conjugated anti-mouse IgG antibody at 1:50 dilution (Jackson ImmunoResearch Laboratories, 200-222-037), and washed twice with PBS. More than 30,000 events were captured for each analysis. FACS data were collected using a FACSCalibur flow cytometer (Becton Dickinson) with CellQuest Pro software. Data analysis was carried out with FlowJo. For detection of acidic vesicular organelles (AVOs), cells were stained with 2.5 mg/ml acridine orange (Molecular Probes, Invitrogen Corporation, A3568) for 10 min at 37ºC, and analyzed for both green and red fluorescence by flow cytometry.
Cell cycle and apoptosis
Quantification of cells in each phase of cell cycle and of apoptotic cells was determined by flow cytometry as previously described.64,65 Apoptotic cells were quantitated as the percentage of cells in the sub-G0/G1 region (hypodiploidy) in cell cycle analysis as previously described.64,65 Briefly, cells (5 × 105) were centrifuged and fixed overnight in 70% ethanol (MERK, 1085430250) at 4°C. Then, cells were washed three times with PBS, incubated for 1 h with 1 mg/ml RNase A (Sigma-Aldrich, R4875) and 20 μg/ml propidium iodide (Sigma-Aldrich, P4170) at room temperature, and analyzed for the distinct cell cycle phases with a Becton Dickinson FACSCalibur flow cytometer. Apoptosis was also assessed using the Annexin V/7-ADD kit (BD Biosciencies, 559763), and the whole cell population was labeled with fluorescein isothiocyanate (FITC)-conjugated Annexin V/7-ADD without prior fixation, according to the manufacturer’s instructions. Cells were analyzed using a FACSCalibur flow cytometer (Becton Dickinson) with CellQuest Pro software. At least 10,000 events were analyzed for each sample. Data analysis was carried out with FlowJo.
Reverse transcriptase-polymerase chain reaction (RT-PCR)
Total RNA from 107 cells was extracted using the TRIZOL (Invitrogen, 15596018) reagent following the manufacturer’s instructions. RNA preparations were carefully checked by gel electrophoresis and found to be free of DNA contamination. Total RNA (5 µg), primed with oligo-dT, was reverse-transcribed into cDNA with SuperScriptTM III First-Strand Synthesis System for RT-PCR (Invitrogen, 18080051). The PCR mixture (25 µl) contained the template cDNA (1–2 µl), 10 pmol of the corresponding primers, 0.2 mM dNTP (ECOGEN, BIO39028), 2.5 mM MgCl2 (ECOGEN, BIO21040) and 5 units of EcoTaq DNA polymerase (ECOGEN, BIO21040) derived from Thermus aquaticus. PCR reactions were performed in GeneAmp PCR System model 9600 (PerkinElmer). The primers used are listed below, where the nucleotide numbers indicate the primer location in the corresponding human sequences obtained from the GenBank/EMBL database:
XBP1 (accession number: NM_001079539)
(forward; nt 423–432) 5'-CCTTGTAGTTGAGAACCAGG -3'
(reverse; nt 819–838) 5'-GGGGCTTGGTATATATGTGG -3'
EDEM1 (accession number: NM_014674)
(forward; nt 776–796) 5'- TTGACAAAGATTCCACCGTCC -3'
(reverse; nt 1075–1094) 5'- TCCCAAATTCCACCAGGAGG -3'
HSPA5 (GRP78) (accession number: NM_005347)
(forward; nt 1824–1846) 5'- AGATCACAATCACCAATGACC -3'
(reverse; nt 2129–2151) 5'- CTTCCAGTTCCTTCTTCTTAGC -3'
DDIT3 (accession number: NM_004083)
(forward; nt 194–215) 5'- CATTGCCTTTCTCCTTCGGAC -3'
(reverse; nt 555–575) 5'- GCAACTAAGTCATAGTCCGC -3'
ACTB (accession number: X00351)
(forward; nt 936–955) 5'- CTGTCTGGCGGCACCACCAT -3'
(reverse; nt 1170–1189) 5'- GCAACTAAGTCATAGTCCGC -3'
Primers were designed by using the DS Gene 1.5 program for DNA analysis from Accelrys Scientific. The conditions for semiquantitative PCR amplification using a thermal cycler were as follows: 1 cycle at 95°C for 5 minutes as an initial denaturation step, then denaturation at 95°C for 30 seconds, annealing at 58°C for 30 seconds, and extension at 72°C for 90 seconds (18 cycles for ACTB, 25 cycles for DDIT3 and HSPA5, 35 cycles for XBP1, 20 cycles for EDEM1), followed by further incubation for 15 minutes at 72°C (1 cycle). An aliquot of the PCR reaction was analyzed on a 2% agarose (Sigma-Aldrich, A4718) gel in 1 × TAE (40 mM Tris-acetate, 1 mM EDTA, pH 8.0, Sigma-Aldrich, T8280) and checked for the expected PCR products.
Coimmunoprecipitation
107 cells were lysed with 200 μl lysis buffer [20 mM Tris-HCl (Sigma-Aldrich, T5941), 100 mM KCl (Sigma-Aldrich, P9541), 0.9% Triton X-100 (Sigma-Aldrich, T8787), 10% glycerol (Sigma-Aldrich, G5516), 2 mM sodium orthovanadate (Sigma-Aldrich, S6508) and 2 mM PMSF (Sigma-Aldrich, P7626)]. Lysates were precleared with 500 μl protein A-Sepharose (Amersham Biosciences, 17-0780-01) at 4°C for 2 h, and immunoprecipitated by incubation for 2 h at 4°C with anti-BECN1 antibodies, precoupled to protein A-Sepharose. After extensive washing with lysis buffer, the precipitates were subjected to SDS-PAGE and western blot analysis. P3X63 was used as a negative control for immunoprecipitation, and no signal was detected.
Western blot
Cells (5 × 106) were lysed with 60 µl 25 mM Hepes (Sigma-Aldrich, H3375) (pH 7.7), 0.3 M NaCl (Sigma-Aldrich, S3014), 1.5 mM MgCl2 (Sigma-Aldrich, M8266) 0.2 mM EDTA (Sigma-Aldrich, E6758), 0.1% Triton X-100, 20 mM β-glycerophosphate (Sigma-Aldrich, G9422), 0.1 mM sodium orthovanadate supplemented with protease inhibitors [1 mM phenylmethylsulfonyl fluoride (Sigma-Aldrich, P7626), 20 µg/ml aprotinin (Sigma-Aldrich, A1153), 20 µg/ml leupeptin (Sigma-Aldrich, L9783)]. Forty micrograms of proteins were run on SDS-polyacrylamide gels, transferred to nitrocellulose filters, blocked with 5% (w/v) defatted milk in TBST [50 mM Tris-HCl (pH 8.0), 150 mM NaCl and 0.1% Tween 20 (Sigma-Aldrich, P1379)] for 90 min at room temperature, and incubated for 1 h at room temperature or overnight at 4°C with specific antibodies: mouse anti-27 kDa DDIT3 (Cell Signaling, 2895) (1:1000 dilution), anti-44 and 42 kDa phospho-MAPK1/3 (Santa Cruz Biotechnology, SC-81492) (1:1000 dilution), anti-44 and 42 kDa MAPK1/3 (Santa Cruz Biotechnology, SC-135900) (1:1000 dilution), anti-116 and 85 kDa PARP1 (BD Pharmingen, 551024) (1:500 dilution), anti-26 kDa BCL2 (BD Pharmingen, 551098) (1:250 dilution), anti-42 kDa ACTB (Sigma-Aldrich, A5316) (1:5000 dilution) monoclonal antibodies; rabbit anti-38 kDa EIF2S1 (Cell Signaling, 9722) (1:1000 dilution), anti-38 kDa phospho-EIF2S1 (Cell Signaling, 9721) (1:1000 dilution), anti-60 kDa phospho-AKT1 (Cell Signaling, 9275) (1:1000 dilution), anti-54 and 46 kDa MAPK8 (Cell Signaling, 9258) (1:1000), anti-54 and 46 kDa phospho-MAPK8 (Cell Signaling, 9251) (1:1000), anti-289 kDa phospho-MTOR (Cell Signaling, 2971) (1:1000 dilution), anti-70 kDa phospho-RPS6KB2 (Cell Signaling, 9234) (1:1000 dilution), anti-70 kDa RPS6KB2 (Cell Signaling, 2708) (1:1000 dilution), anti-16-14 kDa LC3 (Cell Signaling, 2775) (1:1000 dilution), anti-60 kDa BECN1 (Cell Signaling, 3738) (1:1000 dilution), anti-220 kDa phospho-EIF2AK4 (Cell Signaling, 3301) (1:1000 dilution), anti-62 kDa SQSTM1 (Cell Signaling, 5114) (1:1000), anti-28 kDa phospho-BCL2 (Cell Signaling, 2827) (1:1000 dilution), anti-78 kDa HSPA5 (Santa Cruz Biotechnology, SC-1051) (1:500 dilution), anti-60 kDa AKT1 (Santa Cruz Biotechnology, SC-55523) (1:500 dilution), anti-37 kDa ATF4 (Abcam, ab23760) (1:500 dilution), anti-20 kDa cleaved CASP3 (BD Pharmingen, 559565) (1:500 dilution), anti-65 and 75 kDa EDEM1 (Sigma-Aldrich, E8406) (1:500 dilution), anti-110 kDa ERN1 (Sigma-Aldrich, I6785) dilution, anti-78 kDa ATG7 (Sigma-Aldrich, A2856) (1:500 dilution) polyclonal antibodies; goat anti-31 and 20 kDa BCAP31 (Santa Cruz Biotechnology, SC-18579) (1:500 dilution), anti-20 kDa cleaved CASP4 (Santa Cruz Biotechnology, SC-22174) (1:1000 dilution) anti-125 kDa EIF2AK3 (Santa Cruz Biotechnology, SC-55523) (1:500 dilution) polyclonal antibodies. Anti-mouse (Amersham Biosciences, RPN4201), rabbit (Amersham Biosciences, RPN4301) and goat (Santa Cruz Biotechnology, SC-2022) IgG secondary HRP-antibodies were incubated at 1:5000 dilution in 5% (w/v) nonfat milk in TBST for 1 h at room temperature. Signals were developed using an enhanced chemiluminescence detection kit (Amersham Biosciences, RPN2108).
Immunofluorescence microscopy
T lymphocytes were incubated with or without L-Arg and then settled onto slides coated with poly-L-lysine (Sigma-Aldrich, P4707), fixed in 4% formaldehyde (Sigma-Aldrich, F15587) and permeabilized with 0.1% Triton X-100 for 10 min at room temperature. Activated T cells were then incubated with 1 μg/ml rabbit anti-human LC3 antibody overnight at 4°C. Samples were further processed using FITC-conjugated anti-rabbit antibody (BD Pharmingen, 554020) (diluted 1:200 in PBS) for 1 h at room temperature, and analyzed by immunofluorescence microscopy. Negative controls were prepared by either omitting the primary antibody or by using an irrelevant antibody, showing no fluorescence staining of the samples.
Statistical analysis
Results given are the mean ± SD of the indicated number of experiments.
Supplementary Material
Acknowledgement
We thank M. Modolell (Max-Planck-Institut für Immunbiologie, Freiburg, Germany) for critical reading the manuscript and helpful suggestions. We also thank the blood bank of the University Hospital of Salamanca for blood supply. This work was supported by the Spanish Ministerio de Ciencia e Innovación (SAF2008-02251, SAF2011-30518 and RD06/0020/1037 from Red Temática de Investigación Cooperativa en Cáncer, Instituto de Salud Carlos III, cofunded by the Fondo Europeo de Desarrollo Regional of the European Union), European Community’s Seventh Framework Programme FP7-2007-2013 (grant HEALTH-F2-2011-256986), Junta de Castilla y León (CSI052A11-2, CSI221A12-2, GR15-Experimental Therapeutics and Translational Oncology Program, Biomedicine Project 2009) and Acciones Integradas Spain-Germany (HA2007-0080).
Glossary
Abbreviations:
- AO
acridine orange
- ASK1
apoptosis signal-regulating kinase 1
- ATF4
activating transcription factor 4
- ATF6
activating transcription factor 6
- AVO
acid vesicular organelle
- BCAP31
B-cell receptor-associated protein 31 (Bap31)
- BCL2
B-cell CLL/lymphoma 2, Bcl-2
- BECN1
Beclin 1
- BF
bafilomycin A1
- CASP3
caspase 3
- CASP4
caspase 4
- CD247
CD3ζ
- DDIT3 (DNA damage-inducible transcript 3
also named CHOP, C/EBP homologous protein, or GADD153, growth arrest- and DNA damage-inducible gene 153)
- CQ
chloroquine
- EDEM1
ER degradation-enhancing alpha-mannosidase-like 1
- EIF2AK3
eukaryotic translation initiation factor 2-alpha kinase 3, PERK
- EIF2AK4
eukaryotic translation initiation factor 2 alpha kinase 4, GCN2
- EIF2S1
α-subunit of eukaryotic translation initiation factor 2, eIF2α
- ER
endoplasmic reticulum
- ERAD
ER-associated degradation
- ERK
extracellular signal-regulated kinase
- ERN1
endoplasmic reticulum to nucleus signaling 1, IRE1α
- FITC
fluorescein isothiocyanate
- IRE1α
inositol-requiring enzyme 1α
- HSPA5
heat shock 70 kDa protein 5 (glucose-regulated protein, 78kDa, GRP78)
- ITGAL
CD11a
- JNK
c-Jun amino terminal kinase
- L-Arg
L-arginine
- LC3
microtubule-associated protein 1 light chain 3
- 3-MA
3-methyladenine
- MAPK
mitogen-activated protein kinase
- MAPK8
mitogen-activated protein kinase 8, JNK
- MDSCs
myeloid-derived suppressor cells
- MTOR
mechanistic target of rapamycin (serine/threonine kinase)
- PARP1
poly(ADP-ribose) polymerase 1
- PBS
phosphate-buffered saline
- PERK
protein kinase RNA-like endoplasmic reticulum kinase
- PHA
phytohemagglutinin
- PTPRC
CD45
- RT-PCR
reverse transcriptase-polymerase chain reaction
- siRNA
small interfering RNA
- SQSTM1
sequestosome 1
- sXBP1
spliced XBP1
- TCR
T cell receptor
- UPR
unfolded protein response
- uXBP1
unspliced XBP1
- WM
wortmannin
- XBP1
X-box binding protein 1
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Materials
Supplemental materials may be found here: www.landesbioscience.com/journals/autophagy/article/21315
Footnotes
Previously published online: www.landesbioscience.com/journals/autophagy/article/21315
References
- 1.Bronte V, Zanovello P. Regulation of immune responses by L-arginine metabolism. Nat Rev Immunol. 2005;5:641–54. doi: 10.1038/nri1668. [DOI] [PubMed] [Google Scholar]
- 2.Zea AH, Rodriguez PC, Atkins MB, Hernandez C, Signoretti S, Zabaleta J, et al. Arginase-producing myeloid suppressor cells in renal cell carcinoma patients: a mechanism of tumor evasion. Cancer Res. 2005;65:3044–8. doi: 10.1158/0008-5472.CAN-04-4505. [DOI] [PubMed] [Google Scholar]
- 3.Roth E, Steininger R, Winkler S, Längle F, Grünberger T, Függer R, et al. L-Arginine deficiency after liver transplantation as an effect of arginase efflux from the graft. Influence on nitric oxide metabolism. Transplantation. 1994;57:665–9. doi: 10.1097/00007890-199403150-00006. [DOI] [PubMed] [Google Scholar]
- 4.Angele MK, Smail N, Ayala A, Cioffi WG, Bland KI, Chaudry IH. L-arginine: a unique amino acid for restoring the depressed macrophage functions after trauma-hemorrhage. J Trauma. 1999;46:34–41. doi: 10.1097/00005373-199901000-00006. [DOI] [PubMed] [Google Scholar]
- 5.Rodriguez PC, Quiceno DG, Zabaleta J, Ortiz B, Zea AH, Piazuelo MB, et al. Arginase I production in the tumor microenvironment by mature myeloid cells inhibits T-cell receptor expression and antigen-specific T-cell responses. Cancer Res. 2004;64:5839–49. doi: 10.1158/0008-5472.CAN-04-0465. [DOI] [PubMed] [Google Scholar]
- 6.Rodriguez PC, Quiceno DG, Ochoa AC. L-arginine availability regulates T-lymphocyte cell-cycle progression. Blood. 2007;109:1568–73. doi: 10.1182/blood-2006-06-031856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Popovic PJ, Zeh HJ, 3rd, Ochoa JB. Arginine and immunity. J Nutr. 2007;137(Suppl 2):1681S–6S. doi: 10.1093/jn/137.6.1681S. [DOI] [PubMed] [Google Scholar]
- 8.Kropf P, Baud D, Marshall SE, Munder M, Mosley A, Fuentes JM, et al. Arginase activity mediates reversible T cell hyporesponsiveness in human pregnancy. Eur J Immunol. 2007;37:935–45. doi: 10.1002/eji.200636542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cloke TE, Garvey L, Choi BS, Abebe T, Hailu A, Hancock M, et al. Increased level of arginase activity correlates with disease severity in HIV-seropositive patients. J Infect Dis. 2010;202:374–85. doi: 10.1086/653736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Modolell M, Choi BS, Ryan RO, Hancock M, Titus RG, Abebe T, et al. Local suppression of T cell responses by arginase-induced L-arginine depletion in nonhealing leishmaniasis. PLoS Negl Trop Dis. 2009;3:e480. doi: 10.1371/journal.pntd.0000480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Munder M, Mollinedo F, Calafat J, Canchado J, Gil-Lamaignere C, Fuentes JM, et al. Arginase I is constitutively expressed in human granulocytes and participates in fungicidal activity. Blood. 2005;105:2549–56. doi: 10.1182/blood-2004-07-2521. [DOI] [PubMed] [Google Scholar]
- 12.Rodriguez PC, Hernandez CP, Quiceno D, Dubinett SM, Zabaleta J, Ochoa JB, et al. Arginase I in myeloid suppressor cells is induced by COX-2 in lung carcinoma. J Exp Med. 2005;202:931–9. doi: 10.1084/jem.20050715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rodriguez PC, Ernstoff MS, Hernandez C, Atkins M, Zabaleta J, Sierra R, et al. Arginase I-producing myeloid-derived suppressor cells in renal cell carcinoma are a subpopulation of activated granulocytes. Cancer Res. 2009;69:1553–60. doi: 10.1158/0008-5472.CAN-08-1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rodriguez PC, Zea AH, Culotta KS, Zabaleta J, Ochoa JB, Ochoa AC. Regulation of T cell receptor CD3zeta chain expression by L-arginine. J Biol Chem. 2002;277:21123–9. doi: 10.1074/jbc.M110675200. [DOI] [PubMed] [Google Scholar]
- 15.Munder M, Schneider H, Luckner C, Giese T, Langhans CD, Fuentes JM, et al. Suppression of T-cell functions by human granulocyte arginase. Blood. 2006;108:1627–34. doi: 10.1182/blood-2006-11-010389. [DOI] [PubMed] [Google Scholar]
- 16.Chen J, Liu Y, Wang Y, Ding H, Su Z. Different effects of L-arginine on protein refolding: suppressing aggregates of hydrophobic interaction, not covalent binding. Biotechnol Prog. 2008;24:1365–72. doi: 10.1002/btpr.93. [DOI] [PubMed] [Google Scholar]
- 17.Kim R, Emi M, Tanabe K, Murakami S. Role of the unfolded protein response in cell death. Apoptosis. 2006;11:5–13. doi: 10.1007/s10495-005-3088-0. [DOI] [PubMed] [Google Scholar]
- 18.Yorimitsu T, Nair U, Yang Z, Klionsky DJ. Endoplasmic reticulum stress triggers autophagy. J Biol Chem. 2006;281:30299–304. doi: 10.1074/jbc.M607007200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ogata M, Hino S, Saito A, Morikawa K, Kondo S, Kanemoto S, et al. Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol Cell Biol. 2006;26:9220–31. doi: 10.1128/MCB.01453-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klionsky DJ, Emr SD. Autophagy as a regulated pathway of cellular degradation. Science. 2000;290:1717–21. doi: 10.1126/science.290.5497.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zea AH, Rodriguez PC, Culotta KS, Hernandez CP, DeSalvo J, Ochoa JB, et al. L-Arginine modulates CD3zeta expression and T cell function in activated human T lymphocytes. Cell Immunol. 2004;232:21–31. doi: 10.1016/j.cellimm.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 23.Zabaleta J, McGee DJ, Zea AH, Hernández CP, Rodriguez PC, Sierra RA, et al. Helicobacter pylori arginase inhibits T cell proliferation and reduces the expression of the TCR zeta-chain (CD3zeta) J Immunol. 2004;173:586–93. doi: 10.4049/jimmunol.173.1.586. [DOI] [PubMed] [Google Scholar]
- 24.Hirota M, Kitagaki M, Itagaki H, Aiba S. Quantitative measurement of spliced XBP1 mRNA as an indicator of endoplasmic reticulum stress. J Toxicol Sci. 2006;31:149–56. doi: 10.2131/jts.31.149. [DOI] [PubMed] [Google Scholar]
- 25.Lee AH, Iwakoshi NN, Glimcher LH. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol Cell Biol. 2003;23:7448–59. doi: 10.1128/MCB.23.21.7448-7459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meusser B, Hirsch C, Jarosch E, Sommer T. ERAD: the long road to destruction. Nat Cell Biol. 2005;7:766–72. doi: 10.1038/ncb0805-766. [DOI] [PubMed] [Google Scholar]
- 27.Fritsch RM, Schneider G, Saur D, Scheibel M, Schmid RM. Translational repression of MCL-1 couples stress-induced eIF2 alpha phosphorylation to mitochondrial apoptosis initiation. J Biol Chem. 2007;282:22551–62. doi: 10.1074/jbc.M702673200. [DOI] [PubMed] [Google Scholar]
- 28.Dey S, Baird TD, Zhou D, Palam LR, Spandau DF, Wek RC. Both transcriptional regulation and translational control of ATF4 are central to the integrated stress response. J Biol Chem. 2010;285:33165–74. doi: 10.1074/jbc.M110.167213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hinnebusch AG. Translational regulation of GCN4 and the general amino acid control of yeast. Annu Rev Microbiol. 2005;59:407–50. doi: 10.1146/annurev.micro.59.031805.133833. [DOI] [PubMed] [Google Scholar]
- 30.Calì T, Galli C, Olivari S, Molinari M. Segregation and rapid turnover of EDEM1 by an autophagy-like mechanism modulates standard ERAD and folding activities. Biochem Biophys Res Commun. 2008;371:405–10. doi: 10.1016/j.bbrc.2008.04.098. [DOI] [PubMed] [Google Scholar]
- 31.Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–8. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–45. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 33.Eng KE, Panas MD, Hedestam GB, McInerney GM. A novel quantitative flow cytometry-based assay for autophagy. Autophagy. 2010;6:634–41. doi: 10.4161/auto.6.5.12112. [DOI] [PubMed] [Google Scholar]
- 34.Petiot A, Ogier-Denis E, Blommaart EF, Meijer AJ, Codogno P. Distinct classes of phosphatidylinositol 3′-kinases are involved in signaling pathways that control macroautophagy in HT-29 cells. J Biol Chem. 2000;275:992–8. doi: 10.1074/jbc.275.2.992. [DOI] [PubMed] [Google Scholar]
- 35.Shinojima N, Yokoyama T, Kondo Y, Kondo S. Roles of the Akt/mTOR/p70S6K and ERK1/2 signaling pathways in curcumin-induced autophagy. Autophagy. 2007;3:635–7. doi: 10.4161/auto.4916. [DOI] [PubMed] [Google Scholar]
- 36.Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell. 2004;15:1101–11. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ding WX, Ni HM, Gao W, Yoshimori T, Stolz DB, Ron D, et al. Linking of autophagy to ubiquitin-proteasome system is important for the regulation of endoplasmic reticulum stress and cell viability. Am J Pathol. 2007;171:513–24. doi: 10.2353/ajpath.2007.070188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sasnauskiene A, Kadziauskas J, Vezelyte N, Jonusiene V, Kirveliene V. Apoptosis, autophagy and cell cycle arrest following photodamage to mitochondrial interior. Apoptosis. 2009;14:276–86. doi: 10.1007/s10495-008-0292-8. [DOI] [PubMed] [Google Scholar]
- 39.Harris J, Hope JC, Lavelle EC. Autophagy and the immune response to TB. Transbound Emerg Dis. 2009;56:248–54. doi: 10.1111/j.1865-1682.2009.01069.x. [DOI] [PubMed] [Google Scholar]
- 40.Cheng Y, Li H, Ren X, Niu T, Hait WN, Yang J. Cytoprotective effect of the elongation factor-2 kinase-mediated autophagy in breast cancer cells subjected to growth factor inhibition. PLoS One. 2010;5:e9715. doi: 10.1371/journal.pone.0009715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Urano F, Wang X, Bertolotti A, Zhang Y, Chung P, Harding HP, et al. Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science. 2000;287:664–6. doi: 10.1126/science.287.5453.664. [DOI] [PubMed] [Google Scholar]
- 42.Wei Y, Pattingre S, Sinha S, Bassik M, Levine B. JNK1-mediated phosphorylation of Bcl-2 regulates starvation-induced autophagy. Mol Cell. 2008;30:678–88. doi: 10.1016/j.molcel.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sinha S, Levine B. The autophagy effector Beclin 1: a novel BH3-only protein. Oncogene. 2008;27(Suppl 1):S137–48. doi: 10.1038/onc.2009.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oberstein A, Jeffrey PD, Shi Y. Crystal structure of the Bcl-XL-Beclin 1 peptide complex: Beclin 1 is a novel BH3-only protein. J Biol Chem. 2007;282:13123–32. doi: 10.1074/jbc.M700492200. [DOI] [PubMed] [Google Scholar]
- 45.Szegezdi E, Macdonald DC, Ní Chonghaile T, Gupta S, Samali A. Bcl-2 family on guard at the ER. Am J Physiol Cell Physiol. 2009;296:C941–53. doi: 10.1152/ajpcell.00612.2008. [DOI] [PubMed] [Google Scholar]
- 46.Nieto-Miguel T, Fonteriz RI, Vay L, Gajate C, López-Hernández S, Mollinedo F. Endoplasmic reticulum stress in the proapoptotic action of edelfosine in solid tumor cells. Cancer Res. 2007;67:10368–78. doi: 10.1158/0008-5472.CAN-07-0278. [DOI] [PubMed] [Google Scholar]
- 47.Rutkowski DT, Kaufman RJ. A trip to the ER: coping with stress. Trends Cell Biol. 2004;14:20–8. doi: 10.1016/j.tcb.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 48.Høyer-Hansen M, Jäättelä M. Connecting endoplasmic reticulum stress to autophagy by unfolded protein response and calcium. Cell Death Differ. 2007;14:1576–82. doi: 10.1038/sj.cdd.4402200. [DOI] [PubMed] [Google Scholar]
- 49.Tallóczy Z, Jiang W, Virgin HW, 4th, Leib DA, Scheuner D, Kaufman RJ, et al. Regulation of starvation- and virus-induced autophagy by the eIF2alpha kinase signaling pathway. Proc Natl Acad Sci U S A. 2002;99:190–5. doi: 10.1073/pnas.012485299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kouroku Y, Fujita E, Tanida I, Ueno T, Isoai A, Kumagai H, et al. ER stress (PERK/eIF2alpha phosphorylation) mediates the polyglutamine-induced LC3 conversion, an essential step for autophagy formation. Cell Death Differ. 2007;14:230–9. doi: 10.1038/sj.cdd.4401984. [DOI] [PubMed] [Google Scholar]
- 51.Dever TE. Gene-specific regulation by general translation factors. Cell. 2002;108:545–56. doi: 10.1016/S0092-8674(02)00642-6. [DOI] [PubMed] [Google Scholar]
- 52.Bernales S, McDonald KL, Walter P. Autophagy counterbalances endoplasmic reticulum expansion during the unfolded protein response. PLoS Biol. 2006;4:e423. doi: 10.1371/journal.pbio.0040423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liang XH, Kleeman LK, Jiang HH, Gordon G, Goldman JE, Berry G, et al. Protection against fatal Sindbis virus encephalitis by beclin, a novel Bcl-2-interacting protein. J Virol. 1998;72:8586–96. doi: 10.1128/jvi.72.11.8586-8596.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, et al. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–39. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 55.Ciechomska IA, Goemans GC, Skepper JN, Tolkovsky AM. Bcl-2 complexed with Beclin-1 maintains full anti-apoptotic function. Oncogene. 2009;28:2128–41. doi: 10.1038/onc.2009.60. [DOI] [PubMed] [Google Scholar]
- 56.Ellington AA, Berhow MA, Singletary KW. Inhibition of Akt signaling and enhanced ERK1/2 activity are involved in induction of macroautophagy by triterpenoid B-group soyasaponins in colon cancer cells. Carcinogenesis. 2006;27:298–306. doi: 10.1093/carcin/bgi214. [DOI] [PubMed] [Google Scholar]
- 57.Momoi T. Conformational diseases and ER stress-mediated cell death: apoptotic cell death and autophagic cell death. Curr Mol Med. 2006;6:111–8. doi: 10.2174/156652406775574596. [DOI] [PubMed] [Google Scholar]
- 58.Gajate C, Matos-da-Silva M, Dakir EL-H, Fonteriz RI, Alvarez J, Mollinedo F. Antitumor alkyl-lysophospholipid analog edelfosine induces apoptosis in pancreatic cancer by targeting endoplasmic reticulum. Oncogene. 2012;31:2627–39. doi: 10.1038/onc.2011.446. [DOI] [PubMed] [Google Scholar]
- 59.Pua HH, Dzhagalov I, Chuck M, Mizushima N, He YW. A critical role for the autophagy gene Atg5 in T cell survival and proliferation. J Exp Med. 2007;204:25–31. doi: 10.1084/jem.20061303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pua HH, He YW. Maintaining T lymphocyte homeostasis: another duty of autophagy. Autophagy. 2007;3:266–7. doi: 10.4161/auto.3908. [DOI] [PubMed] [Google Scholar]
- 61.Pua HH, He YW. Autophagy and lymphocyte homeostasis. Curr Top Microbiol Immunol. 2009;335:85–105. doi: 10.1007/978-3-642-00302-8_4. [DOI] [PubMed] [Google Scholar]
- 62.Dunkle A, He YW. Apoptosis and autophagy in the regulation of T lymphocyte function. Immunol Res. 2011;49:70–86. doi: 10.1007/s12026-010-8195-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cabaner C, Gajate C, Macho A, Muñoz E, Modolell M, Mollinedo F. Induction of apoptosis in human mitogen-activated peripheral blood T-lymphocytes by the ether phospholipid ET-18-OCH3: involvement of the Fas receptor/ligand system. Br J Pharmacol. 1999;127:813–25. doi: 10.1038/sj.bjp.0702606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gajate C, Barasoain I, Andreu JM, Mollinedo F. Induction of apoptosis in leukemic cells by the reversible microtubule-disrupting agent 2-methoxy-5-(2′,3′,4′-trimethoxyphenyl)-2,4,6-cycloheptatrien-1 -one: protection by Bcl-2 and Bcl-X(L) and cell cycle arrest. Cancer Res. 2000;60:2651–9. [PubMed] [Google Scholar]
- 65.Gajate C, Santos-Beneit AM, Macho A, Lazaro M, Hernandez-De Rojas A, Modolell M, et al. Involvement of mitochondria and caspase-3 in ET-18-OCH(3)-induced apoptosis of human leukemic cells. Int J Cancer. 2000;86:208–18. doi: 10.1002/(SICI)1097-0215(20000415)86:2<208::AID-IJC10>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.