Abstract
Macroautophagy is an important process for removing misfolded and aggregated protein in cells, the dysfunction of which has been directly linked to an increasing number of neurodegenerative disorders. However, the details of macroautophagy in prion diseases remain obscure. Here we demonstrated that in the terminal stages of scrapie strain 263K-infected hamsters and human genetic prion diseases, the microtubule-associated protein 1 light chain 3 (LC3) was converted from the cytosolic form to the autophagosome-bound membrane form. Macroautophagy substrate sequestosome 1 (SQSTM1) and polyubiquitinated proteins were downregulated in the brains of sick individuals, indicating enhanced macroautophagic protein degradation. The levels of mechanistic target of rapamycin (MTOR) and phosphorylated MTOR (p-MTOR) were significantly decreased, which implies that this enhancement of the macroautophagic response is likely through the MTOR pathway which is a negative regulator for the initiation of macroautophagy. Dynamic assays of the autophagic system in the brains of scrapie experimental hamsters after inoculation showed that alterations of the autophagic system appeared along with the deposits of PrPSc in the infected brains. Immunofluorescent assays revealed specific staining of autophagosomes in neurons that were not colocalized with deposits of PrPSc in the brains of scrapie infected hamsters, however, autophagosome did colocalize with PrPSc in a prion-infected cell line after treatment with bafilomycin A1. These results suggest that activation of macroautophagy in brains is a disease-correlative phenomenon in prion diseases.
Keywords: transmissible spongiform encephalopathies, autophagy, neurodegenerative diseases
Introduction
Many neurodegenerative diseases, such as Alzheimer (AD), Huntington (HD), Parkinson (PD) and prion diseases, are characterized by misfolded protein intra- and extracellular deposition and aggregation.1 In order to avert and fight disease proteins, cells have developed powerful quality control systems that assure the maintenance of cellular homeostasis and counteract damage caused by disease proteins. The ubiquitin-proteasome and autophagy-lysosome pathways are two main quality control systems in eukaryotic cells.2 Proteasomes predominantly degrade short-lived nuclear and cytosolic proteins, while autophagy tends to digest long-lived protein and organelle.3,4 The process of autophagy is initiated with the formation of double-membrane-bounded structures named autophagosomes, which are generated from phagophores. The outer membrane of the autophagosome fuses with a lysosome in the cytoplasm to form an autolysosome, where the contents are degraded via acidic lysosomal hydrolases. Although the precise mechanism of autophagy initiation still needs to be elucidated, several signal pathways show abilities to regulate autophagy. Among them the best characterized ones are the mechanistic target of rapamycin (MTOR) and BECN1-PIK3C3/Vps34 (phosphatidylinositol 3-kinase, class 3)-PIK3R4/Vps15 (phosphatidylinositol 3-kinase, regulatory subunit 4) class III phosphatidylinositol 3-kinase core complex.5,6 Autophagy can function as a cellular protective activity, while it can also lead to cell death in some circumstances, namely autophagic cell death (ACD) that is referred to as type II programmed cell death, in contrast to type I programmed cell death (apoptosis).7 This double-edged sword mechanism probably affects various diseases as well as the different stages of these diseases, which subsequently results in different outcomes. An increased number of autophagosomes has been detected in a number of neurological diseases. However, in many cases, it has not been verified if this phenomenon is due to the induction of autophagosome formation (increasing autophagic activity) or the inhibition of autophagosome-lysosome fusion and lysosomal hydrolases (decreasing autophagic activity). Increasing direct and indirect evidence shows that autophagic activity tends to decrease in some neurodegenerative diseases, such as reduction of BECN1 expression in Alzheimer disease and inefficient cargo recognition in Huntington disease.8,9 Autophagy inhibition (Atg5 and Atg7 deletion in neurons) also results in progressive neurodegeneration associated with ubiquitinated protein aggregates.10,11
Prion diseases, also known as transmissible spongiform encephalopathies (TSEs) are a group of fatal neurodegenerative diseases affecting humans and animals. Human prion diseases consist of Creutzfeldt-Jakob disease (CJD), Gerstmann-Sträussler-Scheinker disease (GSS), fatal familial insomnia (FFI) and kuru. CJD is the most common form of human TSEs, exhibiting characteristic lesions in the central nervous system (CNS) with spongiform degeneration, neuronal loss, reactive gliosis and amyloid plaque formation. Like Alzheimer, Huntington and Parkinson diseases, prion diseases are related to a misfolded protein; specifically, the normal prion protein (PrPC) is converted into a protease-resistant isoform prion protein (PrPSc). With the help of electron microscopy, autophagic vacuoles have been observed in the neurons of experimental hamsters with prion diseases, as well as in synapses of human CJD and FFI patients.12,13 These data highlight the fact that, like other neurodegenerative diseases, alteration of the autophagic system in neurons also happens in prion diseases. However, the details and possible functions of autophagy in prion diseases are poorly understood.
In this report, we investigated the alterations of the autophagic system in the brains of scrapie-infected hamsters and patients with human genetic prion diseases. The remarkable phenomenon of autophagosome formation in neurons in the cortex and Purkinje cells in the cerebellum was detected in the brains infected with 263K scrapie agent. The autophagy negative regulator (MTOR signaling pathway) and autophagy substrates (SQSTM1 and polyubiquitinated proteins) were downregulated gradually during disease progression. In addition, colocalization of autophagosomes and PrPSc in SMB-S15 cells was observed after treatment with bafilomycin A1. Our data here demonstrate an enhanced autophagic activity in prion diseases.
Results
LC3-II, a marker of cellular autophagosomes, emerged in the brains of 263K-infected hamsters and human patients with genetic prion diseases
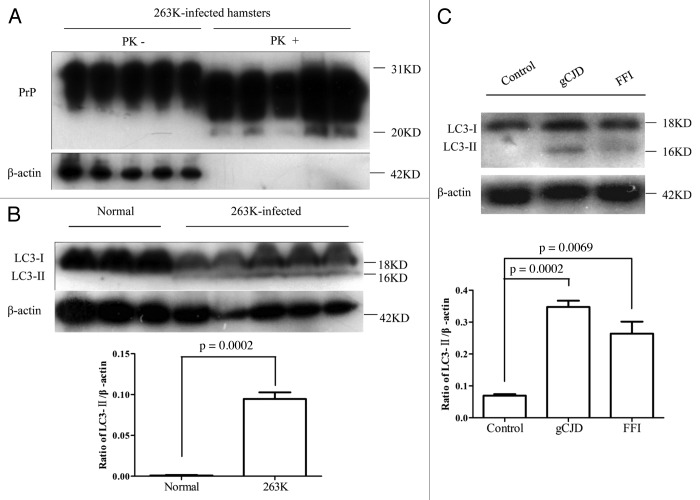
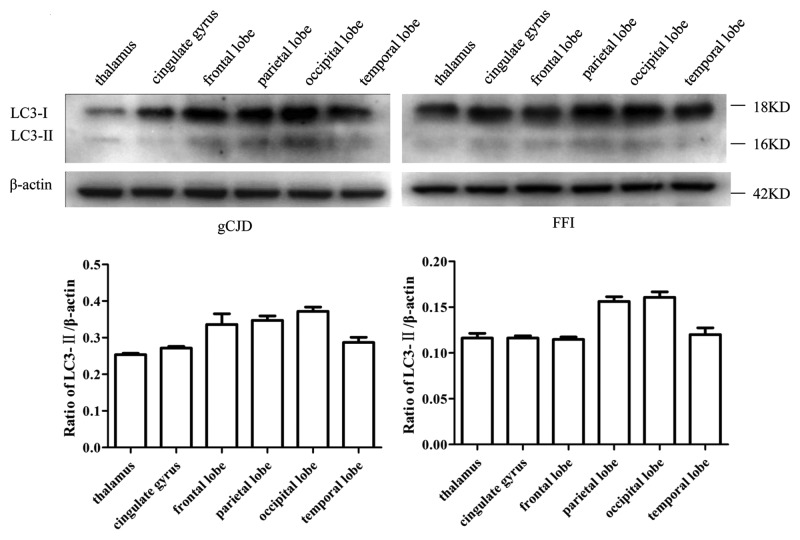
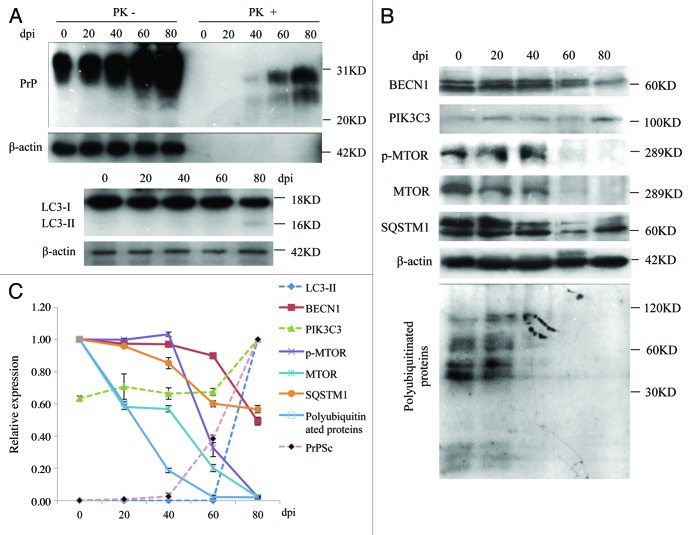
The presence of PrPSc in brain tissues of five enrolled scrapie-infected hamsters were screened by PK digested western blots with PrP-specific monoclonal antibody (mAb) 3F4. PK-resistant PrP bands were identified in all infected brains, with molecular mass ranging from 21 to 27 kDa (Fig. 1A). PrPSc deposits in various brain regions of scrapie agent 263K-infected hamsters were described in previous studies.14,15 To assess the presence of autophagy in the brains of scrapie-infected hamsters, the amounts of lipidated LC3 protein in brains was evaluated by western blots with a LC3B-specific polyclonal antibody (pAb). As shown in Figure 1B, a roughly 18-kDa band representing LC3-I was observed in all preparations. The autophagosome-associated lipidation form LC3-II with apparent molecular mass 16 kDa was detectable in the brain homogenates of five 263K-infected hamsters, but not in that of three normal hamsters. To observe the presence of LC3-II in human TSEs, the brain homogenates from a healthy adult donor, a G114V gCJD case and a FFI case were analyzed using LC3-specific western blots. As expected, LC3-II-specific signals were observed in the brain homogenates of the FFI and gCJD cases, but not in that of the normal control (Fig. 1C). Furthermore, six different regions of the autopsy brains from these two cases were analyzed. LC3-II-specific signals were detected in all tested brain regions of the patients. Among them, the signals in the occipital lobe and parietal lobe were relatively stronger (Fig. 2). This suggests that the levels of LC3-II were obviously upregulated in the prion-infected brain tissue. Interestingly, as our previous studies have shown,16 large amounts of PrPSc signals were detected in the temporal lobe, occipital lobe, parietal lobe, frontal lobe, thalamus and midbrain of G114V gCJD, but only markedly weak PrPSc signals were observed in the frontal lobe and thalamus of FFI. It seems that appearance of LC3-II most likely does not closely associate with PrPSc accumulation in the brain samples of human genetic prion diseases. We speculate that neurons of gCJD and FFI patients may chronically suffer from stress caused by mutant prion protein, and this possibly induces autophagy, although there is no obvious PrPSc accumulation.
Figure 1. Upregulation of LC3-II in the brains in prion diseases at terminal stage. (A) Western blot analysis of PrPSc in the brain tissues of scrapie agents 263K-infected hamsters collected at moribund stage. All tested brain homogenates were digested with a final concentration of 30 μg/ml PK at 37°C for 30 min. (B) Western blot for LC3 in the brain tissues of normal and scrapie agent 263K-infected hamsters. The same amounts of individual brain homogenate were loaded and analyzed by 15% SDS-PAGE. The scrapie agents as well as normal control are shown on the top of the graphs. (C) Western blots for LC3 in human brains with gCJD and FFI. A homogenate from whole brains of normal human adult donors (Sigma) and homogenate of cortex tissues from individuals with FFI and gCJD were loaded and analyzed by 15% SDS-PAGE. Molecular mass markers are indicated at the right. The densities of signals are determined by densitometry and shown relative to ACTB/β-actin. Graphical data denote mean ± SD.
Figure 2. Analyses of the levels of LC3 in the brain tissues in human prion diseases. Western blots of six brain regions of a G114V gCJD patient (left panel) and a FFI patient (right panel). Various brain regions are indicated at the top. Molecular mass markers are indicated at the right. The densities of signals are determined by densitometry and shown relative to ACTB. p = 0.0006 (gCJD), p < 0.0001 (FFI). Graphical data denote mean ± SD.
Autophagosomes localized in the cytoplasm of neurons
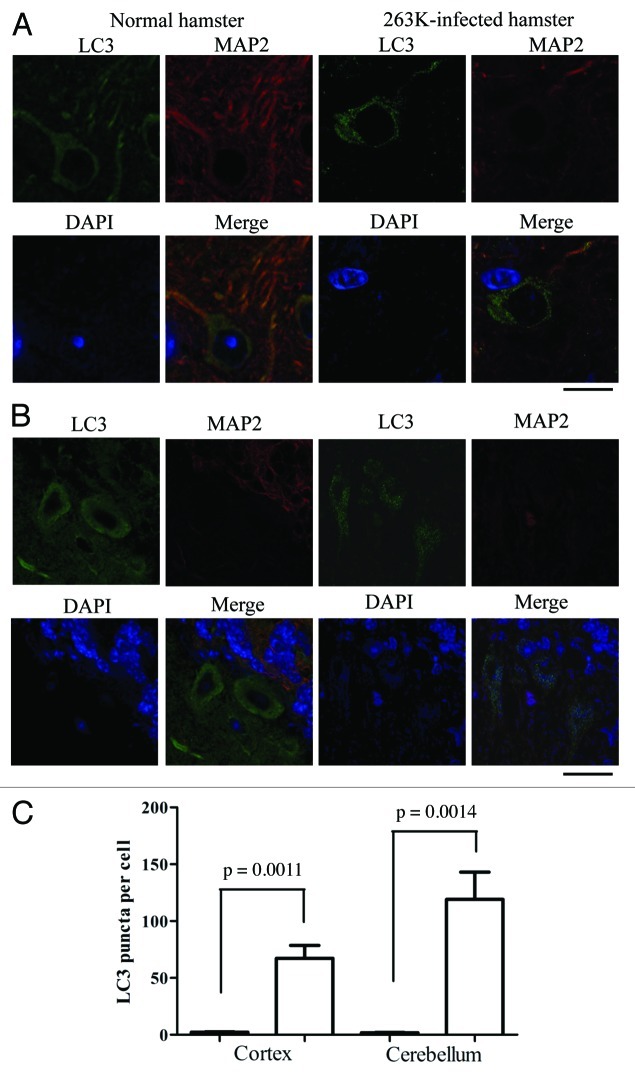
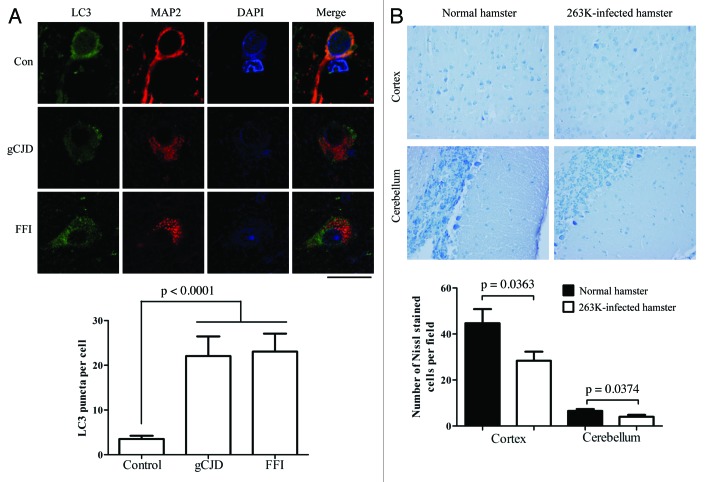
The formation of autophagic vacuoles in neurons of experimental hamster scrapie has previously been proposed based on electron microscopy.12 To see the distribution of LC3 in the brain tissues, the brain sections of normal and 263K-infected hamsters were immunofluorescently double-stained with specific antibodies to LC3 and microtubule-associated protein 2 (MAP2), which is a well-documented neuron marker exclusively expressed in soma and dendrites. Confocal microscopy assays revealed the different patterns of the LC3 signals (green) in those two groups. Large amounts of small bright green granular substances were observed in the cytoplasm of neurons in the cortex and Purkinje cells in the cerebellum of 263K-infected hamsters, whereas evenly distributed green LC3 signals with a few bright green puncta were seen in the normal control (Fig. 3A and B), revealing significant differences after counting the LC3 puncta per cell (p < 0.01, Fig. 3C). Such LC3 puncta signals were also observed in other brain regions, showing statistical differences in the hippocampus, brainstem and thalamus (p < 0.05) compared with the normal control (Fig. S2). The relevant negative controls were shown in Figure S1. To observe the autophagosomes in human TSE cases, the sections of the parietal lobes from the patients of gCJD and FFI, as well as the control, were LC3- and MAP2-specific antibody double-stained. Similarly, significant bright green LC3 punctate signals were observed in the cytoplasm of the brain tissues of those cases, showing a significant difference in LC3 puncta per cell compared with the normal control (Fig. 4A). These results illustrate a TSEs-related profile of LC3 bodies in neurons, which indicates that autophagosome formation is induced in neurons in prion diseases.
Figure 3. Autophagosomes localized in cytoplasm of neurons in the 263K-infected brains at terminal stage. (A) Double immunofluorescence staining for LC3 and MAP2 in the cortex region of brain sections of scrapie agents 263K-infected and normal hamsters. Scale bar: 20 μm. (B) Double immunofluorescence staining for LC3 and MAP2 in the cerebellum region of brain sections of scrapie agents 263K-infected and normal hamsters. Scale bar: 20 μm. (C) The number of LC3 puncta per neuron in the cortex and the cerebellum regions. Graphical data denote mean ± SD.
Figure 4. Autophagosomes localized in cytoplasm of neurons in brains of FFI and gCJD cases. (A) Top: Double immunofluorescence staining for LC3 and MAP2 in the parietal lobe region of brain sections of control and genetic prion diseases patients. The images of LC3 (green), MAP2 (red), DAPI (blue) and merge are indicated above. Scale bar: 20 μm. Bottom: The numbers of LC3 puncta per neuron in the parietal lobe regions of brain sections of control and genetic prion diseases patients. The images of LC3 (green), MAP2 (red), DAPI (blue) and merge are indicated above. (B) Top: Nissl staining for neurodegeneration in the cortex and the cerebellum regions of 263K-infected and normal hamsters (×40). Bottom: Quantity of neurons in cortex and Purkinje cells. Graphical data denote mean ± SD.
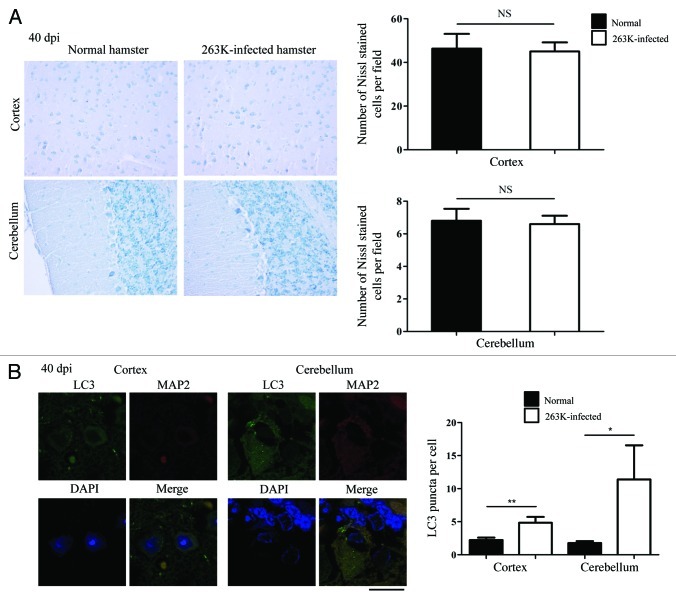
Neuron loss is a salient feature of prion diseases. To examine a possible relationship between neurodegeneration and autophagy activation, various brain sections of normal and 263K-infected hamsters underwent Nissl staining. Compared with the normal control, more severe neuron loss occurred in the regions of cortex and the cerebellum of the 263K-infected hamsters, showing statistical differences in the numbers of Nissl-stained cells per field (Fig. 4B). Neuron loss was also observed in the hippocampus, brainstem and thalamus of 263K-infected cells, but did not show statistical differences (Fig. S3). These data might highlight a speculation that there is a linkage between autophagy activation and neuron loss in 263K-infected hamsters.
To see the potential influence of blocking autophagy in the survival of prion infection, the scrapie-infected cell line SMB-S15 [Scrapie (Chandler) Mouse Brain cells, cloned] and its healthy control cell line SMB-PS (SMB-S15 cured with pentosan polysulphate) were treated with 3-methyladenine (3-MA), a small molecule inhibitor of macroautophagy. The cell survival was evaluated with CCK-8 assays. When compared with the mock cells without treatment, the relative cell survival rates of SMB-S15 were slightly higher after exposure to 3-MA, which revealed the improvement of cell viability in the preparations of 2- to 6 d incubation, while that of SMB-PS exhibited the opposite (Fig. S4). These data may indicate that the blockage of autophagy will improve the viability of prion-infected cells, which partially supports the speculation of the relationship between autophagy activation and neuron loss in prion diseases.
Autophagic activity was obviously enhanced in the brains of 263K-infected hamsters and human patients with genetic prion diseases
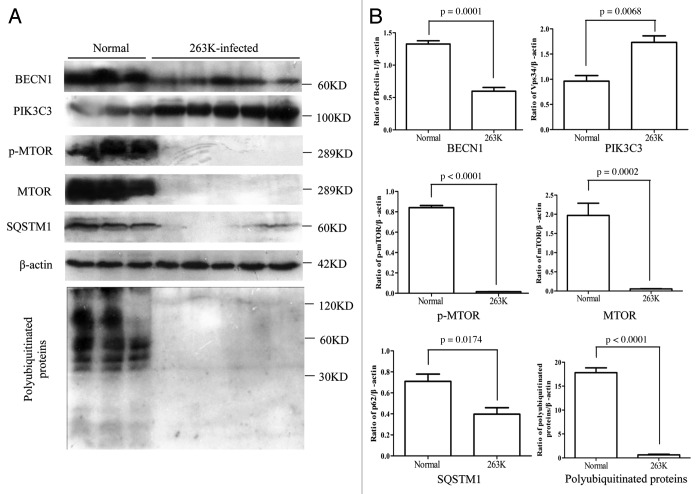
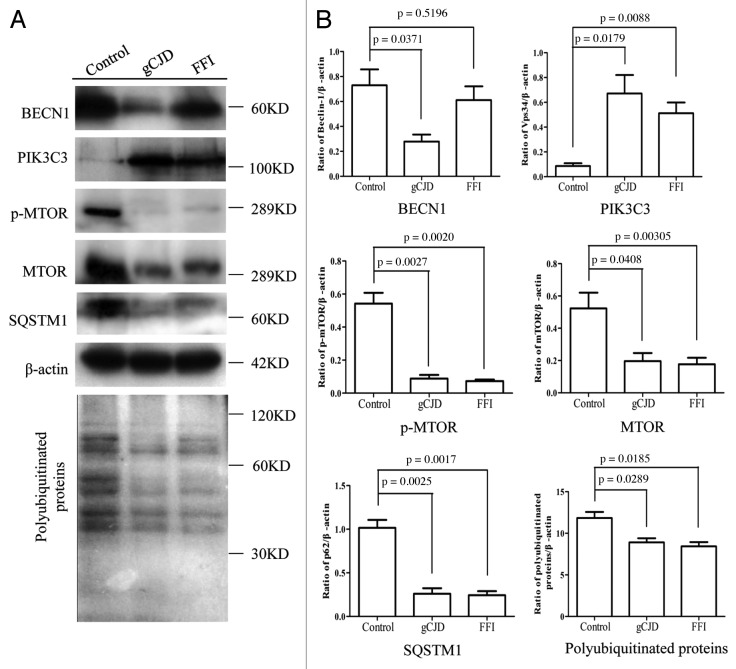
To determine the activity of the autophagic system during prion pathogenesis, the alterations of the SQSTM1 protein, which serves as a link between LC3 and ubiquitinated substrates and is able to be incorporated into, and then degraded via autophagosomes,17 were evaluated in brains of 263K-infected hamsters and human TSEs. Western blots showed obvious weak signals of SQSTM1 in brain homogenates of five 263K-infected hamsters when compared with that of normal hamsters (Fig. 5A and B). Similarly, the signals of SQSTM1 in the brains of G114V gCJD and FFI were markedly weaker than that of the normal control (Fig. 6A and B). Subsequently, the levels of total polyubiquitinated proteins, which served as another marker of autophagic activity, were comparatively analyzed through western blots. This showed that the levels of polyubiquitinated proteins were substantially lower in the brain samples from either scrapie-infected hamsters (Fig. 5A and B) or human prion diseases (Fig. 6A and B). Significantly downregulated levels of SQSTM1 and polyubiquitinated proteins in the brains with prion diseases may reflect an upregulated autophagic activity.
Figure 5. Comparative analyses of levels of BECN1, PIK3C3, p-MTOR, MTOR, SQSTM1 and polyubiquitinated proteins in the brains of 263K-infected hamsters at terminal stage. (A) Western blots of autophagic markers in brain homogenates of normal and scrapie agents 263K-inefcted hamsters. (B) Quantitative analyses of (A). Molecular mass markers are indicated at the right. The densities of signals are determined by densitometry and shown relative to β-actin. Graphical data denote mean ± SD.
Figure 6. Comparative analyses of levels of BECN1, PIK3C3, p-MTOR, MTOR, SQSTM1 and polyubiquitinated proteins in a FFI and a gCJD cases. (A) Brain homogenates of normal human adult donors and the parietal cortexes of gCJD and FFI. (B) Quantitative analyses of (A). Molecular mass markers are indicated at the right. The densities of signals are determined by densitometry and shown relative to β-actin. Graphical data denote mean ± SD.
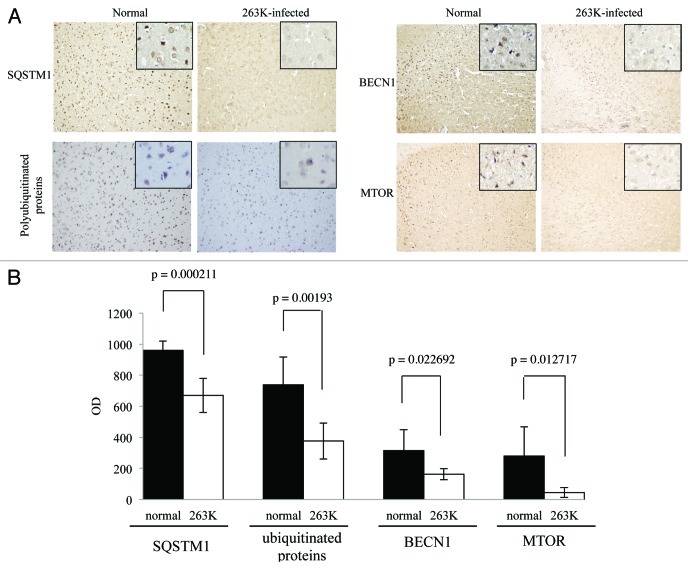
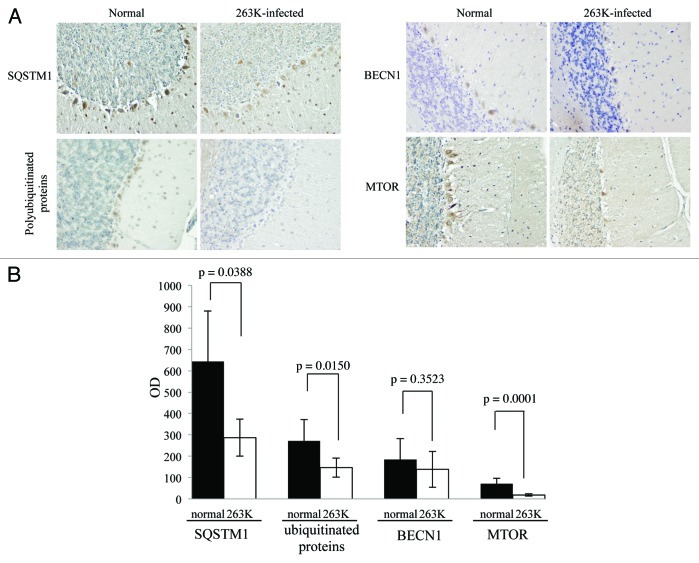
In order to verify the decrease of SQSTM1 and polyubiquitinated proteins in TSE, the brain slides of 263K-infected hamsters were subjected to immunohistochemical analysis with individual antibodies. Compared with normal hamsters, many fewer SQSTM1 and polyubiquitinated protein-positive signals were detected in the cortex and the cerebellum of scrapie-infected animals, showing statistical differences in MOD (Figs. 7 and 8). The relevant negative controls are shown in Figure S5. Moreover, the patterns of polyubiquitinated protein distribution seemed different between the brains of scrapie-infected animals and normal controls, where the inclusion body-like structures of polyubiquitinated proteins were seen in the brains of 263K-infected hamsters, while relatively uniform staining of polyubiquitinated proteins were observed in the brains of controls (Fig. S6). These data highly suggest an enhanced autophagic flux and more active autophagic protein degradation in the CNS tissues during prion pathogenesis.
Figure 7. Comparative analyses of levels of BECN1, MTOR, SQSTM1 and polyubiquitinated proteins in the cortex of 263K-infected hamsters at terminal stage with IHC. (A) IHC assays of cortex of normal and 263K-infected hamsters (×20). The magnification views were shown in the upper right of each picture. (B) The MOD values of different proteins in cortex. Graphical data denote mean ± SD.
Figure 8. Comparative analyses of levels of BECN1, MTOR, SQSTM1 and polyubiquitinated proteins in the cerebellum of 263K-infected hamsters at terminal stage with IHC. (A) IHC assays of the cerebellum of normal and 263K-infected hamsters (×40). (B) The MOD values of different proteins in the cerebellum. Graphical data denote mean ± SD.
Alterations of the MTOR pathway and class III phosphatidylinositol 3-kinase pathway
The MTOR pathway and BECN1-PIK3C3-PIK3R4 class III phosphatidylinositol 3-kinase core complex pathway are the most important negative and positive regulators for autophagy in mammalian cells, respectively. To explore the possible changes of those pathways, the levels of MTOR, p-MTOR (Ser2448) and BECN1 in brain tissues were individually tested with western blots and IHC. Both MTOR and p-MTOR were reduced significantly to an almost undetectable level in the brains of scrapie experimental hamsters (Fig. 5A) and human TSE cases (Fig. 6A) based on western blots, showing a statistical difference compared with that of normal controls (Fig. 5B and Fig. 6B). IHC assays of the cortex (Fig. 7) and the cerebellum (Fig. 8) of 263K-infected hamsters identified remarkably less MTOR-positive signals. Furthermore, MTOR-specific real time-PCR assays showed that the levels of Mtor mRNAs in the brain samples of 263K-infected hamsters were significantly lower (p = 0.0025) than that of normal ones (Fig. S7). Meanwhile, lower levels of BECN1 were also identified in the brain samples of 263K-infected hamsters and human gCJDs in western blots (Fig. 5 and Fig. 6). IHC assays of the brain sections of 263K-infected hamsters revealed less BECN1-positive stained signal in the cortex (p = 0.022692, Fig. 7, but not in the cerebellum (p = 0.3523, Fig. 8). The relevant negative controls are shown in Figure S5. To get more detailed information about the changes of the class III phosphatidylinositol 3-kinase complex, the levels of PIK3C3 were evaluated by western blots. As shown in Figures 5 and 6, the levels of PIK3C3 were significantly upregulated in brains of 263K-infected hamsters (p = 0.0068) and two gCJD patients (p < 0.05). It seems that both regulatory pathways for the autophagic system, BECN1 and MTOR, are influenced in the CNS of TSEs at the terminal stages, in which the repressive regulatory pathway MTOR is more deeply affected. The positive regulatory pathway involving BECN1 might still maintain its function, since the increased PIK3C3 possibly compensates for the potential change of class III PtdIns3K activity caused by the decreased BECN1.
Functional alterations of the autophagic system appeared when PrPSc was deposited
To observe the possible dynamic alterations of the autophagic system in the brains of scrapie experimental hamsters after inoculation, various biomarkers involved in the autophagic system were comparatively analyzed in the brain samples collected on day 0 and on the 20th, 40th, 60th and 80th day post inoculation (dpi). Coincident with our previous study with the same samples,18 weak PrPSc signals began in the 40th dpi sample and became gradually stronger by the 60th and 80th dpi (Fig. 9A and C). The signals of lipidated LC3 protein were maintained at an undetectable level in the early stages, but were finally detected in the 80th dpi sample (Fig. 9A and C). The levels of SQSTM1 decreased gradually with the extension of the incubation period and reached roughly 40% of that prior to inoculation at the terminal stage. In parallel, the levels of polyubiquitinated protein decreased from the 20th dpi and almost disappeared at the 40th dpi (Fig. 9B and C). The levels of MTOR started to decrease from the 20th dpi, while that of p-MTOR showed clear downregulation from the 60th dpi. p-MTOR and MTOR declined to undetectable levels at the terminal stages (Fig. 9B and C). Meanwhile, the levels of BECN1 remained fairly stable in the samples collected at the early stage and decreased rapidly at the end of the disease, albeit still maintaining a level 60% of that prior to inoculation (Fig. 9B and C). Coincidental with the decrease of BECN1, the levels of PIK3C3 remained almost unchanged until the 60th dpi, but increased clearly in the 80th dpi sample (Fig. 9B and C). These dynamic results of various biomarkers for autophagy and its regulatory systems indicate that activation of the autophagic system occurs at the early stage of scrapie infection, which coincides well with the deposits of PrPSc and neurodegeneration change in the 263K-infected hamsters’ brains.15
Figure 9. Dynamic assays of autophagic system and PrPSc in the brain tissues of 263K-infected hamsters during incubation period. (A) Western blots for PrPSc and LC3. (B) Western blots for BECN1, PIK3C3, p-MTOR, MTOR, SQSTM1 and polyubiquitinated proteins. The same amounts of individual brain homogenate were loaded and analyzed by 6% or 15% SDS-PAGE. The data of dpi are showed at the top. β-actin was used as an internal control. Molecular mass markers are indicated at the right. (C) Quantitative analysis of each gray numerical value of PrPSc, LC3-II, BECN1, PIK3C3, p-MTOR, MTOR, SQSTM1 and polyubiquitinated proteins vs that of individual β-actin. The relative expressing data of each marker are normalized to that of 0th dpi (Beclin-1, p-MTOR, MTOR, SQSTM1 and polyubiquitinated proteins) or that of 80th dpi (PrPSc, LC3-II and PIK3C3). Graphical data denote mean ± SD.
To address the possibility that the delayed appearance of LC3-II in the brains of the infected hamsters was due to neuron loss during the incubation period, the brain sections of the 40th dpi underwent Nissl staining and LC3- and MAP2-specific double staining. As shown in Figure 10A, there was no remarkable neuron loss in the cortex and the cerebellum at the 40th dpi compared with the normal control. Small quantities of, but clearly identified, LC3 puncta were detected in the cortical neurons and Purkinje of the brain sections at the 40th dpi, revealing statistical differences in LC3 puncta per cell compared with that of the normal control (Fig. 10B). These data suggest that the appearance of autophagosomes seems to occur prior to marked neuron loss. The undetectable LC3-II in the western blot at the 40th dpi is probably due to the detection limit of the western blot assay, rather than a large scale of neuron loss leading to autophagosome invisibility.
Figure 10. Autophagosome formation and neurodegeneration at 40th dpi. (A) Nissl staining for neurons in the cortex and the cerebellum regions of the scrapie agents 263K-infected hamsters at 40th dpi (×40). Left: Representative images for Nissl staining (×40). Right: Quantity of neurons in cortex and Purkinje cells. (B) Left: Double immunofluorescent staining for LC3 and MAP2 in the cortex region of brain sections of scrapie agents 263K-infected hamsters at 40th dpi. Scale bar: 20 μm. Right: The number of LC3 puncta per neuron in the cortex and the cerebellum regions at 40th dpi. Graphical data denote mean ± SD.
Autophagosomes seemed to not colocalize with PrPSc in the brain tissues of infected hamsters, but did colocalize with PrPSc in SMB cells after treatment with bafilomycin A1.
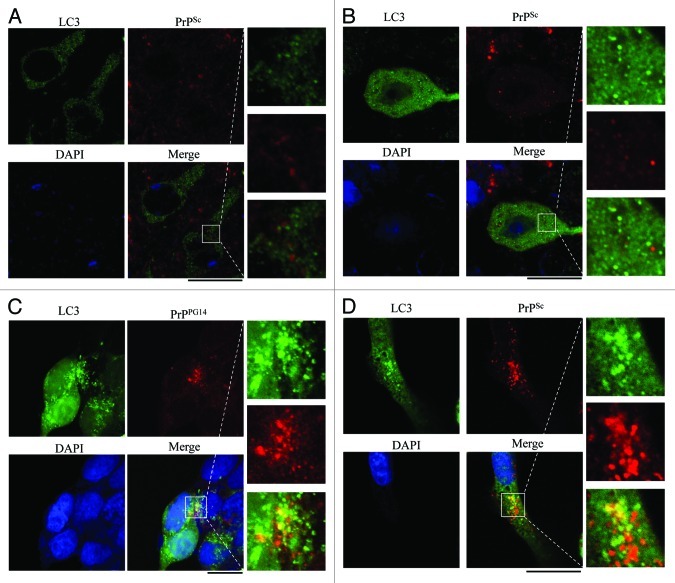
In order to determine whether autophagosomes were involved in PrPSc trafficking or processes of metabolism, brain sections of 263K-infected hamsters were immunofluorescently stained with LC3- and PrP-specific antibodies, respectively, in which the sections were treated with 6 M GdnHCl in order to remove the normal PrPC prior to PrP staining. Confocal microscopy assays revealed PrPSc signals (bright red particles), which distributed either outside or inside of neurons and abnormal LC3 granular substances (bright green) which were concentrated in the cytoplasm of neurons in the cortex (Fig. 11A) and the cerebellum (Fig. 11B) of the scrapie-infected hamsters. However, the intracellular PrPSc particles did not seem to colocalize with the LC3 granules after merging the two images, although they appeared to be close to each other (Fig. 11A and B, magnified figures on the right).
Figure 11. Assays of the colocalization between autophagosome and PrPSc in the brain sections of 263K-infected hamsters at terminal stage and in the prion cell models. The paraffin sections of scrapie 263K-infected hamster’s brain [(A) cortex, (B) the cerebellum] were double-stained immunofluorescently for LC3 and PrPSc. (C) HEK293 cells transiently cotransfected with pcDNA-PrP-PG14 and pEGFP-LC3 were stained immunofluorescently for PrP after treated with 400 nM bafilomycin A1. (D) SMB-S15 cells transfected with pEGFP-LC3 were stained for PrPSc after treated with 400 nM bafilomycin A1. The magnification views were shown in the right of each picture. The images of LC3 (green), PrPSc or PrPPG14 (red), DAPI (blue) and merge are indicated above. Scale bar: 20 μm.
To address whether blocking fusion between autophagosomes and lysosomes will influence the potential colocalization of abnormal PrP with autophagosomes, HEK293 cells that had been previously proven to not contain detectable endogenous PrP19 were transiently cotransfected with plasmid pcDNA3.1-PrP-PG14, a familial CJD-associated PrP construct with 9-extra octarepeat insertion, together with a plasmid encoding EGFP-LC3. Meanwhile, SMB-S15 cells were also transiently transfected with the EGFP-LC3-encoding plasmid. Forty-eight hours post-transfection, the cells were exposed to 400 nM bafilomycin A1 for 6 h. Before staining with PrP-specific antibody, SMB-S15 cells were treated with 6 M GdnHCl. Confocal microscopy showed many red PrP-specific particles and bright green LC3-specific particles in the cytoplasm of either the PrP-PG14-expressing cells or SMB-S15 cells, which turned to yellow after the images were merged (right panels, Fig. 11C and D). To ensure that the yellow signals did not come from superposition in the Z-axis, serial optical sections per 0.5 μm were automatically performed by confocal microscopy. As shown in Figure S8, the yellow signals were only present in some scanning layers of the graphs, which definitely excludes the possibility of artificial superposition in the Z-axis. This implies that blocking the fusion of autophagosomes with lysosomes results in retention of the abnormal PrP in autophagosomes.
Discussion
Autophagosomal dysfunction has been observed in an increasing number of diseases, especially in neurodegenerative diseases.20 One of the general pathological features of most neurodegenerative diseases is the formation of intracytoplasmic aggregates within neurons, such as Aβ1–42 and tau accumulations in AD, Lewy bodies in PD, polyglutamine accumulation in HD and PrPSc aggregates in TSEs. Theoretically, intracytoplasmic aggregate-prone proteins are suitable autophagy substrates, whereas nonaggregate-prone proteins, such as wild-type HTT/huntingtin and SNCA/α-synuclein, show a much lower dependency on autophagy for their degradation.21 For the following reasons, the host autophagy system rather than the ubiquitin-proteasome system might be the primary clearance route for PrPSc and its aggregates. First, PrPSc can form oligomeric aggregates that may become inaccessible to the narrow barrel of the proteasome; second, PrPSc can inhibit the proteolytic activity of the 26S proteasome, which results in ubiquitin-proteasome system dysfunction; third, several autophagy pharmaceutical agonists, such as rapamycin, lithium, trehalose and imatinib mesylate, have been proven to be able to enhance PrPSc clearance in vitro and in vivo.22-26 Additionally, the combination of PRND/Doppel (a paralog of prion) overexpression and the prion protein deficit in neurons also inhibits autophagic flux and induces neuron death.27,28 Hence, these connections between prion proteins and autophagy prompted us to determine whether prion infection can induce the host autophagy response. In this report, through the use of western blots, immunohistochemistry and immunofluorescence assays for specific markers of the autophagic system, autophagosome formation was detected in brains during the pathogenesis of scrapie experimental hamsters and human genetic prion diseases.
Several possible steps of autophagy are altered by aggregated proteins, leading to an increasing number of autophagosomes in neurodegenerative diseases, such as the decrease of autophagosome-lysosome fusion in AD and HD,29,30 ineffective lysosome enzyme activity in lysosomal storage disorders, and even inefficient cargo recognition in PD and HD.9,31 In these situations, the increase of autophagic vacuoles does not appear to be associated with the enhanced autophagic flux, but with autophagic flux inhibition. SQSTM1 can form shells around aggregates and serves as a link between LC3 and ubiquitinated substrates.17 SQSTM1 can be incorporated into the completed autophagosome and is degraded in the autolysosome, hence polyubiquitinated proteins and SQSTM1 are usually used as markers for autophagic activity.32 In neurodegenerative diseases, SQSTM1 and ubiquitinated proteins are usually increased and associated with neuronal inclusion.33-35 In this study, the polyubiquitinated inclusion body deposits in the cytoplasm are repeatedly observable in the scrapie-infected hamsters’ brains, which are coincident with a previous report,23 but the levels of autophagic substrates SQSTM1 and polyubiquitinated proteins in the brains of TSEs are obviously downregulated as incubation times increase. The reduction of polyubiquinated proteins in TSE brains contradicts a previous report36 in which the levels of ubiquitinated proteins increase in the brains of scrapie-infected mice when measured by an indirect enzyme-linked immunosorbent assay (ELISA). However, in their results, the OD values of ubiquitinated proteins ranged from 0.05 to 0.15 in brain homogenates of controls and sick mice, which seems too low to establish the linear relationship between quantity of ubiquitinated proteins and OD values. In addition, some nuclear ubiquitinated proteins, such as histone, which is not correlated with intracytoplasmic inclusion body, may influence the results. However, in our study, the reduction of the protein level of ubiquitinated proteins is observed by IHC assays and western blots, which may avoid those problems. Taken together, the evidence concerning formation of autophagosomes and decrease of SQSTM1 and polyubiquitinated proteins suggests an upregulation of autophagic activity during disease progression.
MTOR is a key negative regulator controlling autophagic activity in mammalian cells.37 The MTOR pathway has recently become a validated drug target for some neurodegenerative disorders, since altered MTOR activity has been identified in PD and AD.5,38 Rapamycin, an inhibitor of the MTOR pathway, which has been proven to be able to enhance clearance of a series of aggregated proteins, such as HTT, SNCA, MAPT/tau, ATXN/ataxin and PRNP/prion, is the most promising candidate.5,24,39-43 Unlike other neurodegenerative disorders, decreases of both MTOR and p-MTOR in the brain tissues after inoculation of scrapie agent was observed in this study, which illustrates a close relationship between downregulation of the MTOR pathway and upregulation of autophagic flux in prion diseases. It highlights a distinct functioning mechanism of autophagy in prion disease, which raises the therapeutic potential of MTOR inhibitor in prion diseases. BECN1 works as positive regulator for autophagic activity.44 In this study, the level of BECN1 was maintained at a relatively high level at the early and middle stages during the incubation period of scrapie agent. Only during the very late stage of scrapie infection did the level of BECN1 become downregulated, but even then, it still maintained a value roughly 50% of that observed in normal hamsters. This phenomenon possibly represents an activated autophagic activity in brain tissues at the early stage of prion infection. The reduction of BECN1 protein in our study and BECN1 mRNA in other studies45 in the late stages of prion diseases may reflect a comprehensive depression of neuronal function due to severe damage of brain tissues. Additionally, PIK3C3, another positive regulator of autophagy related to BECN1, is upregulated at the terminal stage in the brains of 263K-infected hamsters and genetic prion diseases. BECN1 functions through interaction with a series of other proteins, including BCL2 and PIK3C3. BCL2 shows an inhibitory effect on BECN1-dependent autophagy.44 During the nucleation process of autophagosome membrane, BECN1 has to dissociate from the BECN1-BCL2 complex and interacts with PIK3C3 to form the class III PtdIns3K complex, subsequently inducing autophagy.44 In the brains of scrapie-infected hamsters46 and a series of cell models expressing PrP mutants,47,48 the levels of BCL2 are usually decreased. Therefore, we may speculate that the upregulation of PIK3C3 and the downregulation of BCL2 may compensate for the potential repression of class III PtdIns3K activity caused by the decrease of BECN1 in the terminal stage of prion infection.
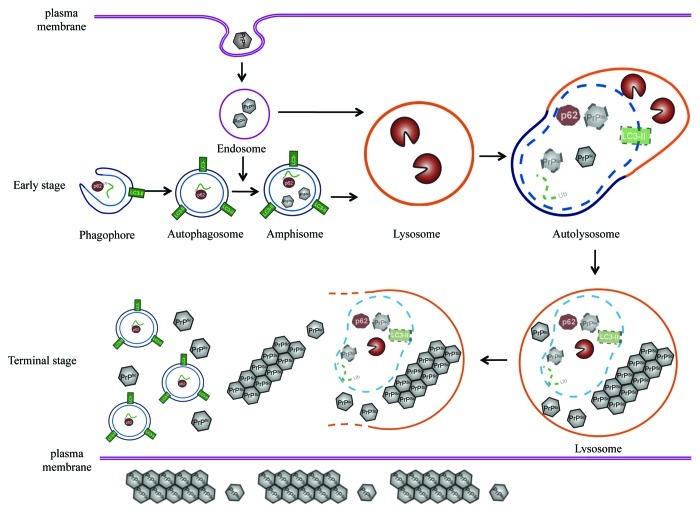
Based on the observations in this study, we can hypothesize about the possible role of autophagy and relevant pathways in neurons during prion infection (Fig. 12). After entering the host cells, PrPSc is delivered via clathrin- or caveolin-coated vesicles to the endosomes, which could then fuse directly with lysosomes or autophagosomes to form amphisomes and finally fuse with lysosomes.50 At that stage the colocalization of PrPSc and autophagosomes is hard to detect, since the level of PrPSc is too low to be observed, or due to the rapid fusion of amphisomes and lysosomes induced by strong autophagic flux. Partially supporting this speculation, colocalization of Aβ1–42 and LC3 will be more easily observed when exposing cells to bafilomycin A1 that can block the fusion of autophagosomes and lysosomes and induce accumulation of autophagosomes.51 PrPSc forms deposits in abnormal lysosomes, in which lysosome-related structures are presumed to act as the bioreactor for processing of PrPC to PrPSc.52 The roles of lysosomes in prion diseases are still a controversial topic. Treatment with a series of lysosomal inhibitors inhibits PrPSc accumulation,24 while autophagy-lysosome system agonists enhance PrPSc clearance.24,26,53 Under the treatment with bafilomycin A1, we have observed the colocalization of the expressed PrP-PG14 with LC3 in cultured cells and colocalization of PrPSc with LC3 in a prion-infected cell line in this study. Therefore, we speculate that the host neurons expect to clear out the aggregations of misfolded prion and inhibit the replication of PrPSc via autophagy-lysosome degradation, while PrPSc may sustainably utilize the autophagy-lysosome system to achieve the conversion of PrPC into PrPSc. When more PrPSc accumulates and deposits in lysosomes, these lysosomes will disrupt and deliver the aggregated PrPSc into the cytoplasm, which may induce cell stress, and at same time, induce an overactive autophagic response. That may explain the activated autophagic flux in the brain tissues of prion disease as well as the lack of colocalization of PrPSc with LC3 within neurons at the terminal stage of the disease.
Figure 12. A hypothetical processing schema of autophagy in cytoplasm49 and prion infection. At early stage, PrPSc enter the host cells and is delivered to the endosomes, and then the endosomes can fuse directly with lysosomes or fuse with autophagosomes to form amphisomes and finally fuse with lysosomes. At terminal stage, the accumulation of PrPSc in lysosomes and cytoplasm induced cell stress and overactive autophagic response.
Recently, increasing evidence in fungi and other organisms suggests that prion or prion-like conformational heredity is general and conservative, and its phenotypes are not detrimental, but are beneficial in some cases.54 Hou et al. have reported that in mammalian cells, virus infection could induce a conformational change in MAVS (mitochondrial antiviral signaling) and make MAVS share features of a prion, including resistance to protease digestion, formation of fiber-like polymers, resistance to detergent solubilization and even conformational heredity.55 Interestingly, compared with TSEs prion, the replication and aggregation of physiological endogenous prion-like proteins, such as MAVS, are not harmful to host cells. It is thought that host cells should have certain mechanisms to resolve these aggregates. In the case of MAVS, aggregates are probably degraded by autophagy accompanied by mitochondria clearance, avoiding unrestricted aggregation and further devastating consequences. In our study, the activation of the autophagic system seems to begin at early stages during scrapie infection. Such activated autophagic function seems to reach a saturation state and maintain its activity until the terminal stages of prion diseases. Thus, we presume that once PrPSc replication and deposition reaches a state that is beyond the ability of the host’s clearance system, including autophagy, neuron damage will occur and subsequently the disease will appear which is obviously distinct from other prion-like proteins.
Materials and Methods
Animal brain samples
Chinese golden hamsters inoculated intracerebrally with hamster-adapted scrapie stain 263K described previously14,15 were enrolled in this study. All animals were fed in an Animal Biosafety Level 2 laboratory with human care. Two microliters of 10% brain homogenates of scrapie agent 263K-infected hamsters were injected into the parietal lobes of 70, 14-d old female hamsters at a depth of 4 to 5 mm. Five animals randomly selected on day 0 and the 20th, 40th, 60th and 80th dpi were sacrificed and the brains were removed and frozen at -80°C. The brains of the remaining hamsters infected with agent 263K were collected at the terminal stage (between 76 to 80 dpi at their last gasp) and frozen at -80°C or fixed in 10% buffered formalin solution. Three brains of 80-d-old hamsters intracerebrally injected with 2 μl of physiological saline were used as healthy controls. The incubation time of 263K-infected hamsters was 66.7 ± 1.1 d.15
Human brain samples
A G114V gCJD case and a D178N FFI case, which were diagnosed by experts from the Chinese National Surveillance Network for CJD (CNSNC), were enrolled in this study. The gCJD case was a 47-y-old woman with G114V inherited prion disease and the main clinical and genetic characteristics were reported previously.16 The FFI case was a 55-y-old man with an eight-month-long clinical course. The main clinical manifestations were sleep disturbances, mutism and memory loss. CSF 14-3-3 protein was positive. PRNP gene sequencing revealed a D178N mutation with M129M homozygote. The brain samples of both the gCJD and FFI patients were obtained from the China CJD Surveillance Center, China CDC. Sample collection was done according to the standard protocols enacted by the China CDC. The autopsies of the gCJD and FFI patients were performed by local pathologists within 12 h after death. Half of the brains were fixed in 10% buffered formalin solution and the other half were stored with dry ice and packaged. Subsequently, samples were sent to the China CDC for further pathogen identification and scientific research.
The control brain sections were from a Chinese man who suffered from brain vascular malformation at the age of 49 y old. The patient underwent a neurosurgical operation. The surgically removed part that contained normal brain tissues was used as a control. The control brain was obtained about 8 h after the operation and then was fixed in 10% buffered formalin solution.
Preparations of brain homogenates
Brain samples from normal and scrapie-infected hamsters, as well as human gCJD or FFI patients, were washed in TBS (10 mM TRIS-HCl, 133 mM NaCl, pH 7.4) three times, and then homogenized in lysis buffer (100 mM NaCl, 10 mM EDTA, 0.5% Nonidet P-40, 0.5% Na deoxycholate in 10 mM TRIS-HCl, pH 7.4) containing Cocktail Set III (Merck, 539134). The homogenates were centrifuged at 2,000 g for 10 min, and the supernatant fractions were collected and frozen at -80°C for further experiments. A whole-brain protein sample from healthy adult donors was purchased from Sigma, T6195.
Western blotting
Aliquots of brain homogenates were separated on SDS-PAGE and electroblotted onto a nitrocellulose membrane using a semi-dry blotting system. Membranes were blocked with 5% (w/v) nonfat milk in 1 × TRIS-buffered saline containing 0.1% Tween 20 (TBST) at room temperature for 1 h and probed with various primary antibodies at 4°C overnight, including 1:1000-diluted anti-LC3 pAb (Novus, NB100-2220; Sigma, L8918), 1:1,000-diluted anti-SQSTM1 pAb (Sigma, P0067), 1:500-diluted anti-p-MTOR pAb (Cell Signaling Technology, 2971), 1:500-diluted anti-MTOR pAb (Cell Signaling Technology, 2972), 1:500-diluted anti-BECN1 pAb (Cell Signaling Technology, 3738), 1:500-diluted anti-ubiquitin pAb (Santa Cruz, sc-9133), 1:300-diluted anti-PIK3C3 pAb (Abcam, ab73262), 1:5000-diluted anti-PrP mAb (3F4, Chemicon, MAB1562-K) and 1:1,000-diluted anti-β-actin mAb (Subrray Biotechnology, Sr-25113), respectively. After washing with TBST, blots were incubated with 1:5,000-diluted horseradish peroxidase (HRP)-conjugated goat anti-mouse or rabbit IgG (Jackson ImmunoResearch Labs, 115-035-003 or 111-035-003), at room temperature for 2 h. Blots were developed using the Enhanced ChemoLuminescence system (PerkinElmer, NEL103E001EA) and visualized on autoradiography films. Images were captured by ChemiDocTM XRS+ Imager.
To detect the presence of proteinase K (PK)-resistant PrPSc in brain tissues, the brain homogenates were digested with a final concentration of 30 μg/ml PK at 37°C for 30 min prior to western blots. The PK digestion was terminated by incubating the samples at 100°C for 10 min.
Immunohistochemical (IHC) assays
Brain tissue was fixed in 10% buffered formalin solution and paraffin sections (5-µm in thickness) were prepared routinely. Sections were treated with the antigen retrieval kit (Boster, AR0022) for 1 min at 37°C and quenched for endogenous peroxidases in 3% H2O2 in methanol for 10 min. After blocking in 1% normal goat serum, the sections were incubated with 1:400-diluted pAb for SQSTM1, 1:200-diluted pAb for ubiquitin, 1:200-diluted pAb for BECN1 and MTOR, at 4°C overnight. Rabbit or mouse isotype antibody (Zhongshan Goldenbridge, ZDR-5003 or ZDR-5006) was used as negative controls. Subsequently, the sections were incubated with 1:250-diluted HRP-conjugated goat anti-rabbit or -mouse secondary antibody (Boster, SV0002-12 or SV0001-12) for 60 min, and visualized by incubation with 3,3′-diaminobenzidine tetrahydrochloride (DAB). The slices were counterstained with hematoxylin, dehydrated and mounted in permount.
Photomicrographs were taken with a digital camera DP70, Olympus Optical, attached to the microscope Olympus BX51. Density analysis was performed using Image-Pro Plus 5.0 software. SQSTM1, BECN1, MTOR and ubiquitin staining for each region was separately assessed with the values of mean optical density (MOD). The OD analyses were performed by two investigators in a double-blind manner. Nonspecific background correction in each section was done by subtracting the OD value of the blank area obtained from the same section.
Quantitative real-time PCR (qRT-PCR)
Real-time PCR in this study was performed in an ABI 7900HT Fast sequence detector (Applied Biosystems), with a 96-well microamp optical reaction plate format. Total tissue RNAs were extracted with a commercial RNeasy Lipid Tissue mini kit (Qiagen), and subjected to first-strand cDNA synthesis with Reverse Transcription System (Promega) according to the manufacturer’s protocols. The specific primers for MTOR and ACTB/β-actin were 5′ -TTCACTGACTATGCCTCCC-3′ and 5′-TGTCGCACCAGAACTTTATT-3′, 5′-CTACAATGAGCTGCGTGTGGC-3′ and 5′-CAGGTCCAGACGCAGGATGGC-3′, respectively. PCR amplification was performed in triplicate with the following conditions: 2 min at 50°C and 10 min at 95°C, followed by a total of 40 two-temperature cycles (15 sec at 95°C and 30 sec at 62°C). The comparative Ct (the fractional cycle number at which the amount of amplified target reached a fixed threshold) method was used for the relative quantitative detection of the expressions of the targeting genes. The relative Ct for the target gene was subtracted from that of ACTB gene.
Nissl staining
Brain paraffin sections (5-μm in thickness) were stained with Nissl (1% toluidine blue) for 30 min. After rinsing quickly in distilled water, the sections were differentiated in 95% ethyl alcohol for 0.5 min. After dehydration, the slices were mounted with permount and observed under a microscope (Olympus BX51). The cells containing Nissl bodies were considered as neurons and counted manually.
Confocal microscopy assays
Prior to the staining with PrP mAb, brain sections were treated with 6 M GdnHCl for 10 min. Brain slides were blocked in 1% normal goat serum and then incubated with 1:200-diluted pAb for LC3 and mAb for PrP (hamster brain sections and HEK293 cells transfected with PrP-PG14 were stained with mAb 3F4. SMB-S15 cells were stained with mAb 6D11 (Santa Cruz, sc-58581), or antibody against microtubule-associated protein 2 (MAP2) (Boster, BM1243) at 4°C overnight. Rabbit or mouse isotype antibodies were used as negative controls. The sections were subsequently incubated with 1:500-diluted Alexa Fluor 488-labeled goat anti-rabbit and Alexa Fluor 568-labeled goat anti-mouse secondary antibody (Invitrogen, A-11034 and A-11004) for 60 min. Finally, the sections were incubated with 1 μg/ml DAPI (Invitrogen, D1306) for 10 min. The slices were then mounted in permount and analyzed by confocal microscopy Leica ST2. The numbers of LC3 puncta were counted manually according to previous guidelines.32
Bafilomycin A1 treatment
Human embryonic kidney 293 (HEK293) cells were maintained in DEMD medium supplemented with 10% fetal calf serum. SMB-PS and SMB-S15 cells were obtained from Roslin Institute, UK and maintained in Medium 199 with Earle’s Salts, supplemented with 10% newborn calf serum and 10% fetal calf serum. A mammalian expressing plasmid pcDNA3.1-PrP-PG14 encoding human PrP mutant with 9-extra octareat insertion was constructed previously.19 The EGFP-LC3 plasmid was a gift courtesy of Dr. X.Q. Liu (China CDC). Transfection of cells was conducted with FuGENE® HD reagent (Roche, 04709691001) according to the manufacturer’s instructions. Forty-eight h post-transfection, cells were exposed to 400 nM bafilomycin A1 (Sigma, B1793) and maintained for 6 h. Cells were then subjected to further confocal microscopy assays. SMB-S15 cells were treated with 6 M GdnHCl before blocking with 1% normal goat serum.
Cell viability assay
Cell viability was determined using the CCK-8 cell counting kit (Dojindo, CK04). Briefly, 4,000 cells per well were plated in a 96-well plate and incubated overnight for adherence. The next day, the old medium was replaced with new medium containing a 1 mM concentration of 3-MA. Cell viability assays were performed every day. According to the assigned protocol, 10 μl CCK-8 reagent was added in each well at 37°C for 4 h. Absorbance was measured at 450 nm with a spectrophotometer. Each experiment was performed in triplicate and repeated two times.
Statistical analysis
Statistical analysis was performed using the SPSS 17.0 statistical package. Quantitative analysis of immunoblot images was performed using ImageJ software. The values of each target blot were evaluated. All data were presented as the mean ± SD. The p values among the digitized data from the brain regions of human cases and dynamic alterations of the autophagic system and PrPSc were determined by one-way ANOVA. Other statistical analyses were performed using Student’s t-test. p values less than 0.05 were considered to be statistically significant. p values were described in the figures or expressed as ***p < 0.001; **p < 0.01; *p < 0.05; NS, nonsignificant.
Ethics statement
Usage of human and animal specimens in this study were approved by the Ethical Committee of the National Institute for Viral Disease Prevention and Control, China CDC under protocol 2009ZX10004-101. Animal housing and experimental protocols were in accordance with the Chinese Regulations for the Administration of Affairs Concerning Experimental Animals.
Supplementary Material
Acknowledgment
This work was supported by Chinese National Natural Science Foundation Grant (81100980, 81101302 and 31100117), National Basic Research Program of China (973 Program) (2007CB310505), China Mega-Project for Infectious Disease (2011ZX10004-101) and SKLID Development Grant (2012SKLID102, 2011SKLID211 and 2011SKLID204). We thank Kin-Lung Valentino Wong from the Department of Neurology, New York University School of Medicine for kindly polishing the manuscript.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Materials
Supplemental may be found here: www.landesbioscience.com/journals/autophagy/article/21482
Footnotes
Previously published online: www.landesbioscience.com/journals/autophagy/article/21482
References
- 1.Soto C. Unfolding the role of protein misfolding in neurodegenerative diseases. Nat Rev Neurosci. 2003;4:49–60. doi: 10.1038/nrn1007. [DOI] [PubMed] [Google Scholar]
- 2.Rubinsztein DC. The roles of intracellular protein-degradation pathways in neurodegeneration. Nature. 2006;443:780–6. doi: 10.1038/nature05291. [DOI] [PubMed] [Google Scholar]
- 3.Zhang L, Yu J, Pan H, Hu P, Hao Y, Cai W, et al. Small molecule regulators of autophagy identified by an image-based high-throughput screen. Proc Natl Acad Sci U S A. 2007;104:19023–8. doi: 10.1073/pnas.0709695104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fuertes G, Villarroya A, Knecht E. Role of proteasomes in the degradation of short-lived proteins in human fibroblasts under various growth conditions. Int J Biochem Cell Biol. 2003;35:651–64. doi: 10.1016/S1357-2725(02)00382-5. [DOI] [PubMed] [Google Scholar]
- 5.Ravikumar B, Vacher C, Berger Z, Davies JE, Luo S, Oroz LG, et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat Genet. 2004;36:585–95. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- 6.Tassa A, Roux MP, Attaix D, Bechet DM. Class III phosphoinositide 3-kinase--Beclin1 complex mediates the amino acid-dependent regulation of autophagy in C2C12 myotubes. Biochem J. 2003;376:577–86. doi: 10.1042/BJ20030826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barbieri G, Palumbo S, Gabrusiewicz K, Azzalin A, Marchesi N, Spedito A, et al. Silencing of cellular prion protein (PrPC) expression by DNA-antisense oligonucleotides induces autophagy-dependent cell death in glioma cells. Autophagy. 2011;7:840–53. doi: 10.4161/auto.7.8.15615. [DOI] [PubMed] [Google Scholar]
- 8.Pickford F, Masliah E, Britschgi M, Lucin K, Narasimhan R, Jaeger PA, et al. The autophagy-related protein beclin 1 shows reduced expression in early Alzheimer disease and regulates amyloid beta accumulation in mice. J Clin Invest. 2008;118:2190–9. doi: 10.1172/JCI33585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martinez-Vicente M, Talloczy Z, Wong E, Tang G, Koga H, Kaushik S, et al. Cargo recognition failure is responsible for inefficient autophagy in Huntington’s disease. Nat Neurosci. 2010;13:567–76. doi: 10.1038/nn.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–9. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 11.Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–4. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 12.Boellaard JW, Kao M, Schlote W, Diringer H. Neuronal autophagy in experimental scrapie. Acta Neuropathol. 1991;82:225–8. doi: 10.1007/BF00294449. [DOI] [PubMed] [Google Scholar]
- 13.Sikorska B, Liberski PP, Giraud P, Kopp N, Brown P. Autophagy is a part of ultrastructural synaptic pathology in Creutzfeldt-Jakob disease: a brain biopsy study. Int J Biochem Cell Biol. 2004;36:2563–73. doi: 10.1016/j.biocel.2004.04.014. [DOI] [PubMed] [Google Scholar]
- 14.Zhang J, Chen L, Zhang BY, Han J, Xiao XL, Tian HY, et al. Comparison study on clinical and neuropathological characteristics of hamsters inoculated with scrapie strain 263K in different challenging pathways. Biomed Environ Sci. 2004;17:65–78. [PubMed] [Google Scholar]
- 15.Gao JM, Gao C, Han J, Zhou XB, Xiao XL, Zhang J, et al. Dynamic analyses of PrP and PrP(Sc) in brain tissues of golden hamsters infected with scrapie strain 263K revealed various PrP forms. Biomed Environ Sci. 2004;17:8–20. [PubMed] [Google Scholar]
- 16.Shi Q, Zhang BY, Gao C, Han J, Wang GR, Chen C, et al. The diversities of PrP(Sc) distributions and pathologic changes in various brain regions from a Chinese patient with G114V genetic CJD. Neuropathology. 2012;32:51–9. doi: 10.1111/j.1440-1789.2011.01237.x. [DOI] [PubMed] [Google Scholar]
- 17.Bjørkøy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171:603–14. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo Y, Gong HS, Zhang J, Xie WL, Tian C, Chen C, et al. Remarkable reduction of MAP2 in the brains of scrapie-infected rodents and human prion disease possibly correlated with the increase of calpain. PLoS One. 2012;7:e30163. doi: 10.1371/journal.pone.0030163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jing YY, Li XL, Shi Q, Wang ZY, Guo Y, Pan MM, et al. A novel PrP partner HS-1 associated protein X-1 (HAX-1) protected the cultured cells against the challenge of H₂O₂. J Mol Neurosci. 2011;45:216–28. doi: 10.1007/s12031-011-9498-2. [DOI] [PubMed] [Google Scholar]
- 20.Rubinsztein DC, DiFiglia M, Heintz N, Nixon RA, Qin ZH, Ravikumar B, et al. Autophagy and its possible roles in nervous system diseases, damage and repair. Autophagy. 2005;1:11–22. doi: 10.4161/auto.1.1.1513. [DOI] [PubMed] [Google Scholar]
- 21.Ravikumar B, Sarkar S, Davies JE, Futter M, Garcia-Arencibia M, Green-Thompson ZW, et al. Regulation of mammalian autophagy in physiology and pathophysiology. Physiol Rev. 2010;90:1383–435. doi: 10.1152/physrev.00030.2009. [DOI] [PubMed] [Google Scholar]
- 22.Williams A, Jahreiss L, Sarkar S, Saiki S, Menzies FM, Ravikumar B, et al. Aggregate-prone proteins are cleared from the cytosol by autophagy: therapeutic implications. Curr Top Dev Biol. 2006;76:89–101. doi: 10.1016/S0070-2153(06)76003-3. [DOI] [PubMed] [Google Scholar]
- 23.Kristiansen M, Deriziotis P, Dimcheff DE, Jackson GS, Ovaa H, Naumann H, et al. Disease-associated prion protein oligomers inhibit the 26S proteasome. Mol Cell. 2007;26:175–88. doi: 10.1016/j.molcel.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 24.Heiseke A, Aguib Y, Riemer C, Baier M, Schätzl HM. Lithium induces clearance of protease resistant prion protein in prion-infected cells by induction of autophagy. J Neurochem. 2009;109:25–34. doi: 10.1111/j.1471-4159.2009.05906.x. [DOI] [PubMed] [Google Scholar]
- 25.Aguib Y, Heiseke A, Gilch S, Riemer C, Baier M, Schätzl HM, et al. Autophagy induction by trehalose counteracts cellular prion infection. Autophagy. 2009;5:361–9. doi: 10.4161/auto.5.3.7662. [DOI] [PubMed] [Google Scholar]
- 26.Ertmer A, Gilch S, Yun SW, Flechsig E, Klebl B, Stein-Gerlach M, et al. The tyrosine kinase inhibitor STI571 induces cellular clearance of PrPSc in prion-infected cells. J Biol Chem. 2004;279:41918–27. doi: 10.1074/jbc.M405652200. [DOI] [PubMed] [Google Scholar]
- 27.Heitz S, Grant NJ, Leschiera R, Haeberlé AM, Demais V, Bombarde G, et al. Autophagy and cell death of Purkinje cells overexpressing Doppel in Ngsk Prnp-deficient mice. Brain Pathol. 2010;20:119–32. doi: 10.1111/j.1750-3639.2008.00245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heitz S, Grant NJ, Bailly Y. Doppel induces autophagic stress in prion protein-deficient Purkinje cells. Autophagy. 2009;5:422–4. doi: 10.4161/auto.5.3.7882. [DOI] [PubMed] [Google Scholar]
- 29.Lee JH, Yu WH, Kumar A, Lee S, Mohan PS, Peterhoff CM, et al. Lysosomal proteolysis and autophagy require presenilin 1 and are disrupted by Alzheimer-related PS1 mutations. Cell. 2010;141:1146–58. doi: 10.1016/j.cell.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Webb JL, Ravikumar B, Rubinsztein DC. Microtubule disruption inhibits autophagosome-lysosome fusion: implications for studying the roles of aggresomes in polyglutamine diseases. Int J Biochem Cell Biol. 2004;36:2541–50. doi: 10.1016/j.biocel.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 31.Wong E, Cuervo AM. Autophagy gone awry in neurodegenerative diseases. Nat Neurosci. 2010;13:805–11. doi: 10.1038/nn.2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K. at al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8:445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Manetto V, Perry G, Tabaton M, Mulvihill P, Fried VA, Smith HT, et al. Ubiquitin is associated with abnormal cytoplasmic filaments characteristic of neurodegenerative diseases. Proc Natl Acad Sci U S A. 1988;85:4501–5. doi: 10.1073/pnas.85.12.4501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nogalska A, Terracciano C, D’Agostino C, King Engel W, Askanas V. p62/SQSTM1 is overexpressed and prominently accumulated in inclusions of sporadic inclusion-body myositis muscle fibers, and can help differentiating it from polymyositis and dermatomyositis. Acta Neuropathol. 2009;118:407–13. doi: 10.1007/s00401-009-0564-6. [DOI] [PubMed] [Google Scholar]
- 35.Wooten MW, Hu X, Babu JR, Seibenhener ML, Geetha T, Paine MG, et al. Signaling, polyubiquitination, trafficking, and inclusions: sequestosome 1/p62’s role in neurodegenerative disease. J Biomed Biotechnol. 2006;2006:62079. doi: 10.1155/JBB/2006/62079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kang SC, Brown DR, Whiteman M, Li R, Pan T, Perry G, et al. Prion protein is ubiquitinated after developing protease resistance in the brains of scrapie-infected mice. J Pathol. 2004;203:603–8. doi: 10.1002/path.1555. [DOI] [PubMed] [Google Scholar]
- 37.Jung CH, Ro SH, Cao J, Otto NM, Kim DH. mTOR regulation of autophagy. FEBS Lett. 2010;584:1287–95. doi: 10.1016/j.febslet.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pei JJ, Hugon J. mTOR-dependent signalling in Alzheimer’s disease. J Cell Mol Med. 2008;12(6B):2525–32. doi: 10.1111/j.1582-4934.2008.00509.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spilman P, Podlutskaya N, Hart MJ, Debnath J, Gorostiza O, Bredesen D, et al. Inhibition of mTOR by rapamycin abolishes cognitive deficits and reduces amyloid-beta levels in a mouse model of Alzheimer’s disease. PLoS One. 2010;5:e9979. doi: 10.1371/journal.pone.0009979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Malagelada C, Jin ZH, Jackson-Lewis V, Przedborski S, Greene LA. Rapamycin protects against neuron death in in vitro and in vivo models of Parkinson’s disease. J Neurosci. 2010;30:1166–75. doi: 10.1523/JNEUROSCI.3944-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Webb JL, Ravikumar B, Atkins J, Skepper JN, Rubinsztein DC. Alpha-Synuclein is degraded by both autophagy and the proteasome. J Biol Chem. 2003;278:25009–13. doi: 10.1074/jbc.M300227200. [DOI] [PubMed] [Google Scholar]
- 42.Menzies FM, Huebener J, Renna M, Bonin M, Riess O, Rubinsztein DC. Autophagy induction reduces mutant ataxin-3 levels and toxicity in a mouse model of spinocerebellar ataxia type 3. Brain. 2010;133:93–104. doi: 10.1093/brain/awp292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Berger Z, Ravikumar B, Menzies FM, Oroz LG, Underwood BR, Pangalos MN, et al. Rapamycin alleviates toxicity of different aggregate-prone proteins. Hum Mol Genet. 2006;15:433–42. doi: 10.1093/hmg/ddi458. [DOI] [PubMed] [Google Scholar]
- 44.Kang R, Zeh HJ, Lotze MT, Tang D. The Beclin 1 network regulates autophagy and apoptosis. Cell Death Differ. 2011;18:571–80. doi: 10.1038/cdd.2010.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mok SW, Riemer C, Madela K, Hsu DK, Liu FT, Gültner S, et al. Role of galectin-3 in prion infections of the CNS. Biochem Biophys Res Commun. 2007;359:672–8. doi: 10.1016/j.bbrc.2007.05.163. [DOI] [PubMed] [Google Scholar]
- 46.Park SK, Choi SI, Jin JK, Choi EK, Kim JI, Carp RI, et al. Differential expression of Bax and Bcl-2 in the brains of hamsters infected with 263K scrapie agent. Neuroreport. 2000;11:1677–82. doi: 10.1097/00001756-200006050-00017. [DOI] [PubMed] [Google Scholar]
- 47.Wang X, Dong CF, Shi Q, Shi S, Wang GR, Lei YJ, et al. Cytosolic prion protein induces apoptosis in human neuronal cell SH-SY5Y via mitochondrial disruption pathway. BMB Rep. 2009;42:444–9. doi: 10.5483/BMBRep.2009.42.7.444. [DOI] [PubMed] [Google Scholar]
- 48.Xu K, Wang X, Shi Q, Chen C, Tian C, Li XL, et al. Human prion protein mutants with deleted and inserted octarepeats undergo different pathways to trigger cell apoptosis. J Mol Neurosci. 2011;43:225–34. doi: 10.1007/s12031-010-9387-0. [DOI] [PubMed] [Google Scholar]
- 49.Jaeger PA, Wyss-Coray T. All-you-can-eat: autophagy in neurodegeneration and neuroprotection. Mol Neurodegener. 2009;4:16. doi: 10.1186/1750-1326-4-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fader CM, Colombo MI. Autophagy and multivesicular bodies: two closely related partners. Cell Death Differ. 2009;16:70–8. doi: 10.1038/cdd.2008.168. [DOI] [PubMed] [Google Scholar]
- 51.Hung SY, Huang WP, Liou HC, Fu WM. Autophagy protects neuron from Abeta-induced cytotoxicity. Autophagy. 2009;5:502–10. doi: 10.4161/auto.5.4.8096. [DOI] [PubMed] [Google Scholar]
- 52.Laszlo L, Lowe J, Self T, Kenward N, Landon M, McBride T, et al. Lysosomes as key organelles in the pathogenesis of prion encephalopathies. J Pathol. 1992;166:333–41. doi: 10.1002/path.1711660404. [DOI] [PubMed] [Google Scholar]
- 53.Doh-Ura K, Iwaki T, Caughey B. Lysosomotropic agents and cysteine protease inhibitors inhibit scrapie-associated prion protein accumulation. J Virol. 2000;74:4894–7. doi: 10.1128/JVI.74.10.4894-4897.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wickner RB, Edskes HK, Shewmaker F, Nakayashiki T. Prions of fungi: inherited structures and biological roles. Nat Rev Microbiol. 2007;5:611–8. doi: 10.1038/nrmicro1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hou F, Sun L, Zheng H, Skaug B, Jiang QX, Chen ZJ. MAVS forms functional prion-like aggregates to activate and propagate antiviral innate immune response. Cell. 2011;146:448–61. doi: 10.1016/j.cell.2011.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.