Abstract
In autophagic processes a variety of cargos is delivered to the degradative compartment of cells. Recent progress in autophagy research has provided support for the notion that when autophagic processes are operating in selective mode, a receptor protein complex will process the cargo. Here we present a concept of receptor protein complexes as comprising a functional tetrad of components: a ligand, a receptor, a scaffold and an Atg8 family protein. Our current understanding of each of the four components and their interaction in the context of cargo selection are considered in turn.
Keywords: autophagic cargo, ligand, receptor, scaffold protein, Atg8 family protein, phagophore
Autophagy is a constellation of quality and quantity degradation pathways by which cells may nonselectively or selectively capture, deliver and, in most cases, digest their internal components in a homeostatic function and in response to a diverse range of cellular emergencies. The cargos include nonspecific cytoplasmic substrates, as well as vacuolar hydrolase precursors, protein aggregates, unwanted or damaged organelles and invasive microorganisms.1-7 The signature of such a sophisticated and tightly regulated autophagic degradation pathway, is that, in selective mode, almost all (if not all) autophagic cargos destined for degradation will be processed by a receptor protein complex.8-11 Therein lies a conundrum: How is a given autophagic cargo selected from many others, and what parameters are required to guide it to its ultimate degradation and the subsequent reuse of the degradative products by the cell? We suggest that the choice of autophagic cargo for selective macroautophagy (hereafter autophagy) is orchestrated by a functional tetrad of components: a ligand, a receptor, a scaffold and an Atg8 family protein (Fig. 1A).8-11
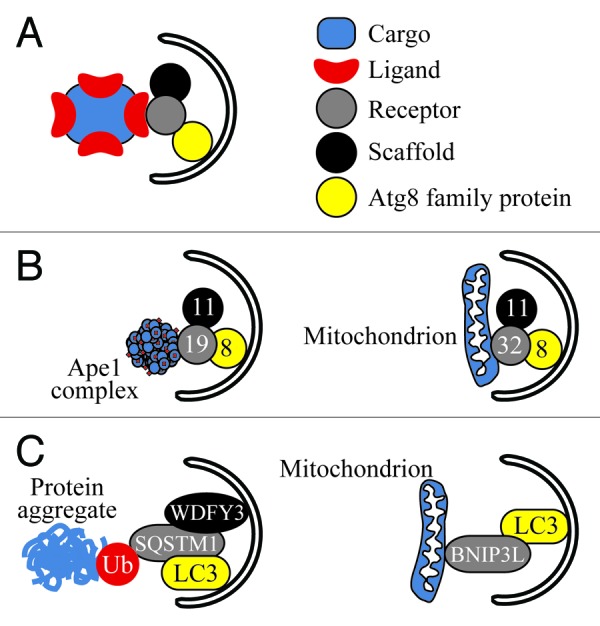
Figure 1. Receptor protein complexes in macroautophagy. (A) A general model of the receptor protein complex. (B) The Cvt pathway (left) and mitophagy (right) in yeast with their respective cargos (prApe1 complex, mitochondrion), ligand (prApe1 propeptide), receptors (Atg19 and Atg32), scaffold (Atg11) and Atg8 family protein (Atg8). (C) Aggrephagy (left) and mitophagy (right) in mammalian cells and their receptor protein complexes. BNIP3L might act as a “mammalian Atg32” being integral to the mitochondrial outer membrane and interacting with LC3. SQSTM1 binds to ubiquitin (Ub) conjugated to aggregated proteins and mediates their interaction with the autophagic scaffold (WDFY3) and Atg8 family protein (LC3); SQSTM1 might play a similar role during pexophagy, mitophagy and xenophagy. Note that SQSTM1 might also directly recognize several ligands for aggrephagy and xenophagy in a Ub-independent manner (Table 1).
In autophagic terms, a ligand can be defined as a molecular entity on the surface of a cargo recognized by a receptor. In order to be ultimately degraded, cargo with ligand must go through a cascade of sequentially regulated steps involving a cohort of molecular players (e.g., receptors and Atg proteins) and intricate membrane dynamic events (e.g., autophagosome formation, and their subsequent fusion with the vacuole/lysosome membrane).1,4,5,7,8,10-12 The specific ligands responsible for targeting various cargos for degradation by autophagy are just beginning to be revealed, but ligands for some autophagy cargos have been identified. Examples include mitochondria [addition of ubiquitin (Ub) to one or more outer membrane proteins, including VDAC1, MFN1/2 and BNIP1, by the E3 Ub ligase PARK2/PARKIN in mammals],13-15 peroxisomes (Pex3 and Pex14 in yeasts)16-18 or protein aggregates (Ub, mutant SOD1 or STAT5A_ΔE18 in mammals)9,10,19-22 (Table 1). However, the specific ligands responsible for selective targeting of certain types of autophagic cargo, such as the endoplasmic reticulum or lipid droplets, have not yet been identified.4
Table 1. Components of the receptor protein complexes involved in autophagy.
| Process | Cargo | Ligand | Receptor | Scaffold | Atg8 family protein | Refs. |
|---|---|---|---|---|---|---|
| Cvt pathway (yeasts) |
prApe1 complex |
prApe1 propeptide |
Atg19 |
Atg11 |
Atg8 |
23, 24, 26, 27 |
| Ams1 complex |
Ams1 |
Atg19, Atg34 |
Atg11 |
Atg8 |
38, 39 |
|
| Glycophagy (mammals) |
Glycogen particles |
Glycogen |
STBD1 |
- |
GABARAP |
58, 59 |
| Aggrephagy (worms) |
PGL granules |
PGL-3 |
SEPA-1 |
EPG-2 |
LGG-1 |
60, 61 |
| Aggrephagy (mammals) |
Mutant SOD1 aggregates |
Mutant SOD1 |
SQSTM1 |
- |
LC3 |
20, 21 |
| STAT5A_ΔE18 aggregates |
STAT5A_ΔE18 |
SQSTM1 |
- |
- |
22 |
|
| Ubiquitinated protein aggregates |
Ub |
SQSTM1, NBR1 |
WDFY3 |
LC3 |
43–52 |
|
| Midbophagy (mammals) |
Midbodies |
Ub |
SQSTM1 |
- |
LC3 |
62 |
| CEP55 |
NBR1 |
- |
- |
63 |
||
| Pexophagy (yeasts) |
Peroxisomes |
Pex3, Pex14 |
Atg30 |
Atg11, Atg17 |
- |
17, 18 |
| Pexophagy (mammals) |
Peroxisomes |
Pex14 |
- |
- |
LC3 |
64 |
| Ub |
SQSTM1 |
- |
LC3 |
65 |
||
| Mitophagy (yeasts) |
Mitochondria |
Outer membrane |
Atg32 |
Atg11 |
Atg8 |
28, 29 |
| Mitophagy (mammals) |
Mitochondria |
Outer membrane |
BNIP3, BNIP3L, FUNDC1 |
- |
LC3, GABARAP |
30–33 |
| Ub |
SQSTM1 |
- |
LC3 |
66–72 |
||
| Xenophagy (mammals) | Viruses |
Viral capsid proteins |
SQSTM1 |
- |
LC3 |
73 |
| Ubiquitinated bacteria |
Ub |
SQSTM1, CALCOCO2, OPTN |
- |
LC3 |
34–36, 74–76 |
|
| Bactericidal factors |
FAU, Ub |
SQSTM1 |
- |
LC3 |
77 |
|
| Membrane remnants, damaged vesicles | Ub, LGALS8 | SQSTM1, CALCOCO2 | - | LC3 | 78, 79 |
In general, components of the autophagic cargo may be either recognized directly by a receptor, or first modified with Ub by an E3 Ub ligase and then recognized by a Ub-binding receptor. Ubiquitination of cargo proteins often triggers selective autophagy in mammalian cells. However, this regulatory mechanism is not found in yeast (Table 1). In yeast, the best-characterized ligand is the propeptide of precursor aminopeptidase I (prApe1). This ligand is recognized by a soluble receptor, Atg19, as the first step of import of the prApe1 oligomer to the vacuole through the cytoplasm-to-vacuole targeting (Cvt) pathway (Fig. 1B).23-27 Similarly, pexophagy in Pichia pastoris requires the Atg30 receptor that interacts with the peroxisomal membrane proteins Pex3 and Pex14.16,17 Atg19 has some structural similarities to SQSTM1/p62 and NBR1 in mammalian cells.9 Although SQSTM1 and NBR1 function as Ub-binding receptors for the autophagic elimination of ubiquitinated protein aggregates, organelles and bacteria, recently it was found that SQSTM1 can directly recognize some protein aggregates, bactericidal factors and viruses, like a yeast receptor, strengthening the possibility of its common origin with Atg19 (Table 1).5,7,10 In contrast to the receptors of the Cvt pathway and pexophagy, the mitophagy receptors, Atg32 (yeasts), BNIP3, BNIP3L/NIX and FUNDC1 (mammals), are integral components of the mitochondrial outer membrane. However, ligand proteins recognized by Atg32, BNIP3, BNIP3L and FUNDC1 in the mitochondrial outer membrane, if they exist, have not been identified (Fig. 1B and C).3,8,11-14,28-33
It is interesting to briefly consider the evolution of the receptor mechanism with regard to bacteria. SQSTM1, CALCOCO2/NDP52 and OPTN are all required for efficient xenophagy, and they appear to bind the same bacteria at various microdomains. Specifically, CALCOCO2 and OPTN bind to the same microdomain, which is distinct from the microdomain bound by SQSTM1.34-36 At this time we can only speculate as to the reason for involving multiple receptors, but it is possible that each one contributes a unique component or function to the autophagic process. The tissue-specificity and the distribution of receptor proteins have not been fully evaluated, but could be an important contributor to the different observations that have been reported.
The next step in selective autophagy typically involves binding of the receptor to a scaffold. In autophagic terms, a scaffold can be defined as an autophagic protein that connects a receptor with the rest of the autophagic machinery. Usually, a scaffold is a protein that organizes the autophagic machinery at the phagophore assembly site (PAS). For example, Atg11 acts as a scaffold within the Cvt pathway, pexophagy and mitophagy in yeast (Fig. 1B), binding Atg19/Atg34, Atg30 and Atg32, respectively.11,12,16-18,26,27,37-39 This interaction is necessary to recruit cargo into close proximity with the autophagy machinery, and in particular an Atg8 family protein (see below). Atg11 might also act as a nonconventional tether for Atg9-containing membranes through its interaction with the GTP-bound form of the Ypt1 GTPase.40 Bridging between the prApe1-Atg19 complex and the Atg9-containing membranes might constitute the earliest step in Cvt-specific PAS formation that is accomplished by Atg11. Atg19 contains binding sites for both Atg11 and Atg8.12 prApe1 does not colocalize with Atg8 in the absence of either Atg19 or Atg11. Thus, Atg11 first contributes to the organization of the Cvt-specific PAS and brings prApe1-Atg19 into proximity of Atg8 at the PAS for subsequent interaction with the phagophore and selective sequestration of the prApe1-Atg19 complex. Interaction of Atg19 with Atg8 is considered to be instrumental in achieving the selectivity of this sequestration.37,41 Similarly, Atg32 does not localize to the PAS in the absence of Atg11, whereas in wild-type cells under conditions where mitophagy is induced, Atg32 binds Atg11 and subsequently interacts with Atg8 (Fig. 1B).11,12,28,29,42 In mammalian cells, WDFY3/ALFY functions as a scaffold (Fig. 1C), recruiting aggregated proteins tagged by Ub and recognized by SQSTM1 to PtdIns3P-containing membranes, and facilitating the interaction of SQSTM1 with mammalian Atg8 (LC3).7-10,43-52 At present, there are no known scaffolds that interact with other mammalian receptors (Table 1).
The last player in the process, then, is an Atg8 family protein (often LC3 in mammals) recognized by the ligand-bound receptor on the phagophore membrane through a specific Atg8 family interacting motif (AIM) or LC3-interacting region (LIR). It should be noted that there are multiple Atg8 homologs in mammals, and these are grouped into two major subfamilies, LC3 and GABARAP. The in vivo binding specificities of the different family members still remains elusive but the LC3 proteins have been suggested to participate in an earlier stage of autophagy than GABARAP proteins.53 It should also be pointed out that not all LIR-containing proteins are autophagy receptors.
The phosphorylation of autophagy receptors (e.g., Atg30, Atg32, and OPTN) might be a general mechanism for the regulation of selective autophagy.17,35,54 The Atg8/LC3 proteins themselves are also phosphorylated, and recent studies have identified specific phosphorylation sites for protein kinase A (PKA) and protein kinase C (PKC) in the N terminus of LC3. Interestingly, this part of LC3 is involved in its binding to autophagic receptors.55,56 It is therefore tempting to speculate that phosphorylation at the PKA and PKC sites might facilitate or prevent the interaction of LC3 with autophagic receptors such as SQSTM1. Along these lines, phosphorylation of the PKA site, which is conserved in all mammalian LC3 isoforms, but not in GABARAP, inhibits recruitment of LC3 into autophagosomes.55
Before concluding, we present some speculative musings that suggest the potential for biological complexity. One advantage of a receptor recruiting a scaffold is that a scaffold can specifically organize the autophagic machinery on the receptor-tagged cargos. The possibility exists that under different conditions a scaffold can be mono- or multi-specific, meaning that it could be recognized by either a single autophagy receptor (e.g., WDFY3 recognized by SQSTM1) or multiple receptors (e.g., Atg11 recognized by Atg19, Atg30, Atg32 and Atg34). Accordingly, it is feasible that the same scaffold could be recruited by multiple receptors at the same time (e.g., under starvation conditions). Moreover, the receptors may recruit multiple scaffolds (e.g., Atg30 recognizes both Atg11 and Atg17). The receptors can be soluble or membrane-bound, yet must be able to interact with their cognate scaffolds and/or Atg8 family members as well as retain the potential to form an aggregated structure under appropriate conditions (e.g., SQSTM1 involvement in the selective autophagy of bacteria, xenophagy7,10).
Rapid progress in autophagy research has strengthened the notion of the receptor protein complex and its role in the mechanism of selective autophagy. We anticipate the results of further studies designed to identify and characterize more autophagic cargos, ligands, receptors, scaffolds and phagophore proteins, and their respective roles under both physiological and pathological settings. Along these lines, a recent genome-wide siRNA screen aimed at identifying mammalian genes required for selective autophagy found 141 candidate genes that are required for viral autophagy, and 96 of those were also required for PARK2-mediated mitophagy.57
Acknowledgments
This work was supported by grant GM069373 (T.Y.N.), NSC 98-2311-B-002-004-MY3 (W.-P.H.), GM53396 (D.J.K.) and DP0986937 (R.J.D).
Footnotes
Previously published online: www.landesbioscience.com/journals/autophagy/article/21332
References
- 1.van der Vaart A, Mari M, Reggiori F. A picky eater: exploring the mechanisms of selective autophagy in human pathologies. Traffic. 2008;9:281–9. doi: 10.1111/j.1600-0854.2007.00674.x. [DOI] [PubMed] [Google Scholar]
- 2.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–75. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Komatsu M, Ichimura Y. Selective autophagy regulates various cellular functions. Genes Cells. 2010;15:923–33. doi: 10.1111/j.1365-2443.2010.01433.x. [DOI] [PubMed] [Google Scholar]
- 4.Kiel JAKW. Autophagy in unicellular eukaryotes. Philos Trans R Soc Lond B Biol Sci. 2010;365:819–30. doi: 10.1098/rstb.2009.0237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol. 2011;27:107–32. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- 6.Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature. 2011;469:323–35. doi: 10.1038/nature09782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weidberg H, Shvets E, Elazar Z. Biogenesis and cargo selectivity of autophagosomes. Annu Rev Biochem. 2011;80:125–56. doi: 10.1146/annurev-biochem-052709-094552. [DOI] [PubMed] [Google Scholar]
- 8.Yu L, Strandberg L, Lenardo MJ. The selectivity of autophagy and its role in cell death and survival. Autophagy. 2008;4:567–73. doi: 10.4161/auto.5902. [DOI] [PubMed] [Google Scholar]
- 9.Kraft C, Peter M, Hofmann K. Selective autophagy: ubiquitin-mediated recognition and beyond. Nat Cell Biol. 2010;12:836–41. doi: 10.1038/ncb0910-836. [DOI] [PubMed] [Google Scholar]
- 10.Johansen T, Lamark T. Selective autophagy mediated by autophagic adapter proteins. Autophagy. 2011;7:279–96. doi: 10.4161/auto.7.3.14487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang K, Klionsky DJ. Mitochondria removal by autophagy. Autophagy. 2011;7:297–300. doi: 10.4161/auto.7.3.14502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kanki T, Klionsky DJ, Okamoto K. Mitochondria autophagy in yeast. Antioxid Redox Signal. 2011;14:1989–2001. doi: 10.1089/ars.2010.3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tolkovsky AM. Mitophagy. Biochim Biophys Acta. 2009;1793:1508–15. doi: 10.1016/j.bbamcr.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 14.Tolkovsky AM. New gene on the block: Atg32--a specific receptor for selective mitophagy in S. cerevisiae. Autophagy. 2009;5:1077–8. doi: 10.4161/auto.5.8.10124. [DOI] [PubMed] [Google Scholar]
- 15.Youle RJ, Narendra DP. Mechanisms of mitophagy. Nat Rev Mol Cell Biol. 2011;12:9–14. doi: 10.1038/nrm3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Manjithaya R, Nazarko TY, Farré JC, Subramani S. Molecular mechanism and physiological role of pexophagy. FEBS Lett. 2010;584:1367–73. doi: 10.1016/j.febslet.2010.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farré JC, Manjithaya R, Mathewson RD, Subramani S. PpAtg30 tags peroxisomes for turnover by selective autophagy. Dev Cell. 2008;14:365–76. doi: 10.1016/j.devcel.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nazarko TY, Farré JC, Subramani S. Peroxisome size provides insights into the function of autophagy-related proteins. Mol Biol Cell. 2009;20:3828–39. doi: 10.1091/mbc.E09-03-0221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kirkin V, McEwan DG, Novak I, Dikic I. A role for ubiquitin in selective autophagy. Mol Cell. 2009;34:259–69. doi: 10.1016/j.molcel.2009.04.026. [DOI] [PubMed] [Google Scholar]
- 20.Gal J, Ström AL, Kilty R, Zhang F, Zhu H. p62 accumulates and enhances aggregate formation in model systems of familial amyotrophic lateral sclerosis. J Biol Chem. 2007;282:11068–77. doi: 10.1074/jbc.M608787200. [DOI] [PubMed] [Google Scholar]
- 21.Gal J, Ström AL, Kwinter DM, Kilty R, Zhang J, Shi P, et al. Sequestosome 1/p62 links familial ALS mutant SOD1 to LC3 via an ubiquitin-independent mechanism. J Neurochem. 2009;111:1062–73. doi: 10.1111/j.1471-4159.2009.06388.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Watanabe Y, Tanaka M. p62/SQSTM1 in autophagic clearance of a non-ubiquitylated substrate. J Cell Sci. 2011;124:2692–701. doi: 10.1242/jcs.081232. [DOI] [PubMed] [Google Scholar]
- 23.Scott SV, Guan J, Hutchins MU, Kim J, Klionsky DJ. Cvt19 is a receptor for the cytoplasm-to-vacuole targeting pathway. Mol Cell. 2001;7:1131–41. doi: 10.1016/S1097-2765(01)00263-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shintani T, Huang W-P, Stromhaug PE, Klionsky DJ. Mechanism of cargo selection in the cytoplasm to vacuole targeting pathway. Dev Cell. 2002;3:825–37. doi: 10.1016/S1534-5807(02)00373-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shintani T, Klionsky DJ. Cargo proteins facilitate the formation of transport vesicles in the cytoplasm to vacuole targeting pathway. J Biol Chem. 2004;279:29889–94. doi: 10.1074/jbc.M404399200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leber R, Silles E, Sandoval IV, Mazón MJ. Yol082p, a novel CVT protein involved in the selective targeting of aminopeptidase I to the yeast vacuole. J Biol Chem. 2001;276:29210–7. doi: 10.1074/jbc.M101438200. [DOI] [PubMed] [Google Scholar]
- 27.Kim J, Huang W-P, Stromhaug PE, Klionsky DJ. Convergence of multiple autophagy and cytoplasm to vacuole targeting components to a perivacuolar membrane compartment prior to de novo vesicle formation. J Biol Chem. 2002;277:763–73. doi: 10.1074/jbc.M109134200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Okamoto K, Kondo-Okamoto N, Ohsumi Y. Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy. Dev Cell. 2009;17:87–97. doi: 10.1016/j.devcel.2009.06.013. [DOI] [PubMed] [Google Scholar]
- 29.Kanki T, Wang K, Cao Y, Baba M, Klionsky DJ. Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev Cell. 2009;17:98–109. doi: 10.1016/j.devcel.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Novak I, Kirkin V, McEwan DG, Zhang J, Wild P, Rozenknop A, et al. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep. 2010;11:45–51. doi: 10.1038/embor.2009.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Novak I, Dikic I. Autophagy receptors in developmental clearance of mitochondria. Autophagy. 2011;7:301–3. doi: 10.4161/auto.7.3.14509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rikka S, Quinsay MN, Thomas RL, Kubli DA, Zhang X, Murphy AN, et al. Bnip3 impairs mitochondrial bioenergetics and stimulates mitochondrial turnover. Cell Death Differ. 2011;18:721–31. doi: 10.1038/cdd.2010.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu L, Feng D, Chen G, Chen M, Zheng Q, Song P, et al. Mitochondrial outer-membrane protein FUNDC1 mediates hypoxia-induced mitophagy in mammalian cells. Nat Cell Biol. 2012;14:177–85. doi: 10.1038/ncb2422. [DOI] [PubMed] [Google Scholar]
- 34.Cemma M, Kim PK, Brumell JH. The ubiquitin-binding adaptor proteins p62/SQSTM1 and NDP52 are recruited independently to bacteria-associated microdomains to target Salmonella to the autophagy pathway. Autophagy. 2011;7:341–5. doi: 10.4161/auto.7.3.14046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wild P, Farhan H, McEwan DG, Wagner S, Rogov VV, Brady NR, et al. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science. 2011;333:228–33. doi: 10.1126/science.1205405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mostowy S, Sancho-Shimizu V, Hamon MA, Simeone R, Brosch R, Johansen T, et al. p62 and NDP52 proteins target intracytosolic Shigella and Listeria to different autophagy pathways. J Biol Chem. 2011;286:26987–95. doi: 10.1074/jbc.M111.223610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yorimitsu T, Klionsky DJ. Atg11 links cargo to the vesicle-forming machinery in the cytoplasm to vacuole targeting pathway. Mol Biol Cell. 2005;16:1593–605. doi: 10.1091/mbc.E04-11-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suzuki K, Kondo C, Morimoto M, Ohsumi Y. Selective transport of α-mannosidase by autophagic pathways: identification of a novel receptor, Atg34p. J Biol Chem. 2010;285:30019–25. doi: 10.1074/jbc.M110.143511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Watanabe Y, Noda NN, Kumeta H, Suzuki K, Ohsumi Y, Inagaki F. Selective transport of α-mannosidase by autophagic pathways: structural basis for cargo recognition by Atg19 and Atg34. J Biol Chem. 2010;285:30026–33. doi: 10.1074/jbc.M110.143545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lipatova Z, Belogortseva N, Zhang XQ, Kim J, Taussig D, Segev N. Regulation of selective autophagy onset by a Ypt/Rab GTPase module. Proc Natl Acad Sci U S A. 2012;109:6981–6. doi: 10.1073/pnas.1121299109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chang CY, Huang W-P. Atg19 mediates a dual interaction cargo sorting mechanism in selective autophagy. Mol Biol Cell. 2007;18:919–29. doi: 10.1091/mbc.E06-08-0683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Okamoto K, Kondo-Okamoto N, Ohsumi Y. A landmark protein essential for mitophagy: Atg32 recruits the autophagic machinery to mitochondria. Autophagy. 2009;5:1203–5. doi: 10.4161/auto.5.8.9830. [DOI] [PubMed] [Google Scholar]
- 43.Bjørkøy G, Lamark T, Brech A, Outzen H, Perander M, Øvervatn A, et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171:603–14. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–45. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 45.Komatsu M, Waguri S, Koike M, Sou YS, Ueno T, Hara T, et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131:1149–63. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- 46.Ichimura Y, Kumanomidou T, Sou YS, Mizushima T, Ezaki J, Ueno T, et al. Structural basis for sorting mechanism of p62 in selective autophagy. J Biol Chem. 2008;283:22847–57. doi: 10.1074/jbc.M802182200. [DOI] [PubMed] [Google Scholar]
- 47.Kirkin V, Lamark T, Sou YS, Bjørkøy G, Nunn JL, Bruun JA, et al. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol Cell. 2009;33:505–16. doi: 10.1016/j.molcel.2009.01.020. [DOI] [PubMed] [Google Scholar]
- 48.Waters S, Marchbank K, Solomon E, Whitehouse C, Gautel M. Interactions with LC3 and polyubiquitin chains link nbr1 to autophagic protein turnover. FEBS Lett. 2009;583:1846–52. doi: 10.1016/j.febslet.2009.04.049. [DOI] [PubMed] [Google Scholar]
- 49.Clausen TH, Lamark T, Isakson P, Finley K, Larsen KB, Brech A, et al. p62/SQSTM1 and ALFY interact to facilitate the formation of p62 bodies/ALIS and their degradation by autophagy. Autophagy. 2010;6:330–44. doi: 10.4161/auto.6.3.11226. [DOI] [PubMed] [Google Scholar]
- 50.Filimonenko M, Isakson P, Finley KD, Anderson M, Jeong H, Melia TJ, et al. The selective macroautophagic degradation of aggregated proteins requires the PI3P-binding protein Alfy. Mol Cell. 2010;38:265–79. doi: 10.1016/j.molcel.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gao C, Cao W, Bao L, Zuo W, Xie G, Cai T, et al. Autophagy negatively regulates Wnt signalling by promoting Dishevelled degradation. Nat Cell Biol. 2010;12:781–90. doi: 10.1038/ncb2082. [DOI] [PubMed] [Google Scholar]
- 52.Hocking LJ, Mellis DJ, McCabe PS, Helfrich MH, Rogers MJ. Functional interaction between sequestosome-1/p62 and autophagy-linked FYVE-containing protein WDFY3 in human osteoclasts. Biochem Biophys Res Commun. 2010;402:543–8. doi: 10.1016/j.bbrc.2010.10.076. [DOI] [PubMed] [Google Scholar]
- 53.Weidberg H, Shvets E, Shpilka T, Shimron F, Shinder V, Elazar Z. LC3 and GATE-16/GABARAP subfamilies are both essential yet act differently in autophagosome biogenesis. EMBO J. 2010;29:1792–802. doi: 10.1038/emboj.2010.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Aoki Y, Kanki T, Hirota Y, Kurihara Y, Saigusa T, Uchiumi T, et al. Phosphorylation of Serine 114 on Atg32 mediates mitophagy. Mol Biol Cell. 2011;22:3206–17. doi: 10.1091/mbc.E11-02-0145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cherra SJ, III, Kulich SM, Uechi G, Balasubramani M, Mountzouris J, Day BW, et al. Regulation of the autophagy protein LC3 by phosphorylation. J Cell Biol. 2010;190:533–9. doi: 10.1083/jcb.201002108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jiang H, Cheng D, Liu W, Peng J, Feng J. Protein kinase C inhibits autophagy and phosphorylates LC3. Biochem Biophys Res Commun. 2010;395:471–6. doi: 10.1016/j.bbrc.2010.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Orvedahl A, Sumpter R, Jr., Xiao G, Ng A, Zou Z, Tang Y, et al. Image-based genome-wide siRNA screen identifies selective autophagy factors. Nature. 2011;480:113–7. doi: 10.1038/nature10546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jiang S, Heller B, Tagliabracci VS, Zhai L, Irimia JM, DePaoli-Roach AA, et al. Starch binding domain-containing protein 1/genethonin 1 is a novel participant in glycogen metabolism. J Biol Chem. 2010;285:34960–71. doi: 10.1074/jbc.M110.150839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jiang S, Wells CD, Roach PJ. Starch-binding domain-containing protein 1 (Stbd1) and glycogen metabolism: Identification of the Atg8 family interacting motif (AIM) in Stbd1 required for interaction with GABARAPL1. Biochem Biophys Res Commun. 2011;413:420–5. doi: 10.1016/j.bbrc.2011.08.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang Y, Yan L, Zhou Z, Yang P, Tian E, Zhang K, et al. SEPA-1 mediates the specific recognition and degradation of P granule components by autophagy in C. elegans. Cell. 2009;136:308–21. doi: 10.1016/j.cell.2008.12.022. [DOI] [PubMed] [Google Scholar]
- 61.Tian Y, Li Z, Hu W, Ren H, Tian E, Zhao Y, et al. C. elegans screen identifies autophagy genes specific to multicellular organisms. Cell. 2010;141:1042–55. doi: 10.1016/j.cell.2010.04.034. [DOI] [PubMed] [Google Scholar]
- 62.Pohl C, Jentsch S. Midbody ring disposal by autophagy is a post-abscission event of cytokinesis. Nat Cell Biol. 2009;11:65–70. doi: 10.1038/ncb1813. [DOI] [PubMed] [Google Scholar]
- 63.Kuo TC, Chen CT, Baron D, Onder TT, Loewer S, Almeida S, et al. Midbody accumulation through evasion of autophagy contributes to cellular reprogramming and tumorigenicity. Nat Cell Biol. 2011;13:1214–23. doi: 10.1038/ncb2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hara-Kuge S, Fujiki Y. The peroxin Pex14p is involved in LC3-dependent degradation of mammalian peroxisomes. Exp Cell Res. 2008;314:3531–41. doi: 10.1016/j.yexcr.2008.09.015. [DOI] [PubMed] [Google Scholar]
- 65.Kim PK, Hailey DW, Mullen RT, Lippincott-Schwartz J. Ubiquitin signals autophagic degradation of cytosolic proteins and peroxisomes. Proc Natl Acad Sci U S A. 2008;105:20567–74. doi: 10.1073/pnas.0810611105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Geisler S, Holmström KM, Skujat D, Fiesel FC, Rothfuss OC, Kahle PJ, et al. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol. 2010;12:119–31. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- 67.Lee JY, Nagano Y, Taylor JP, Lim KL, Yao TP. Disease-causing mutations in parkin impair mitochondrial ubiquitination, aggregation, and HDAC6-dependent mitophagy. J Cell Biol. 2010;189:671–9. doi: 10.1083/jcb.201001039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Okatsu K, Saisho K, Shimanuki M, Nakada K, Shitara H, Sou YS, et al. p62/SQSTM1 cooperates with Parkin for perinuclear clustering of depolarized mitochondria. Genes Cells. 2010;15:887–900. doi: 10.1111/j.1365-2443.2010.01426.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ding W-X, Ni H-M, Li M, Liao Y, Chen X, Stolz DB, et al. Nix is critical to two distinct phases of mitophagy, reactive oxygen species-mediated autophagy induction and Parkin-ubiquitin-p62-mediated mitochondrial priming. J Biol Chem. 2010;285:27879–90. doi: 10.1074/jbc.M110.119537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Narendra D, Kane LA, Hauser DN, Fearnley IM, Youle RJ. p62/SQSTM1 is required for Parkin-induced mitochondrial clustering but not mitophagy; VDAC1 is dispensable for both. Autophagy. 2010;6:1090–106. doi: 10.4161/auto.6.8.13426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gegg ME, Cooper JM, Chau KY, Rojo M, Schapira AH, Taanman JW. Mitofusin 1 and mitofusin 2 are ubiquitinated in a PINK1/parkin-dependent manner upon induction of mitophagy. Hum Mol Genet. 2010;19:4861–70. doi: 10.1093/hmg/ddq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tang F, Wang B, Li N, Wu Y, Jia J, Suo T, et al. RNF185, a novel mitochondrial ubiquitin E3 ligase, regulates autophagy through interaction with BNIP1. PLoS One. 2011;6:e24367. doi: 10.1371/journal.pone.0024367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Orvedahl A, MacPherson S, Sumpter R, Jr., Tallóczy Z, Zou Z, Levine B. Autophagy protects against Sindbis virus infection of the central nervous system. Cell Host Microbe. 2010;7:115–27. doi: 10.1016/j.chom.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yoshikawa Y, Ogawa M, Hain T, Yoshida M, Fukumatsu M, Kim M, et al. Listeria monocytogenes ActA-mediated escape from autophagic recognition. Nat Cell Biol. 2009;11:1233–40. doi: 10.1038/ncb1967. [DOI] [PubMed] [Google Scholar]
- 75.Zheng YT, Shahnazari S, Brech A, Lamark T, Johansen T, Brumell JH. The adaptor protein p62/SQSTM1 targets invading bacteria to the autophagy pathway. J Immunol. 2009;183:5909–16. doi: 10.4049/jimmunol.0900441. [DOI] [PubMed] [Google Scholar]
- 76.Thurston TL, Ryzhakov G, Bloor S, von Muhlinen N, Randow F. The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin-coated bacteria. Nat Immunol. 2009;10:1215–21. doi: 10.1038/ni.1800. [DOI] [PubMed] [Google Scholar]
- 77.Ponpuak M, Davis AS, Roberts EA, Delgado MA, Dinkins C, Zhao Z, et al. Delivery of cytosolic components by autophagic adaptor protein p62 endows autophagosomes with unique antimicrobial properties. Immunity. 2010;32:329–41. doi: 10.1016/j.immuni.2010.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dupont N, Lacas-Gervais S, Bertout J, Paz I, Freche B, Van Nhieu GT, et al. Shigella phagocytic vacuolar membrane remnants participate in the cellular response to pathogen invasion and are regulated by autophagy. Cell Host Microbe. 2009;6:137–49. doi: 10.1016/j.chom.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 79.Thurston TL, Wandel MP, von Muhlinen N, Foeglein A, Randow F. Galectin 8 targets damaged vesicles for autophagy to defend cells against bacterial invasion. Nature. 2012;482:414–8. doi: 10.1038/nature10744. [DOI] [PMC free article] [PubMed] [Google Scholar]


