Abstract
Infiltration of plasma cells is associated with better prognosis in breast, lung and colon cancer. Immunoglobulin κ chain (IGKC) is now available as a single, robust immune marker predicting metastasis-free survival and response to chemotherapy. This will facilitate a deeper understanding of the role of the humoral immune system in cancer development.
Keywords: breast cancer prognosis, colon cancer, humoral immune system, immunoglobulin kappa C, lung cancer, prediction of chemotherapy response, prognosis
Breast Cancer Prognosis and the Relevance Biomarkers
Defining risk categories among cancer patients is of considerable clinical relevance and will avoid over, as well as under-treatment. The most important factor for risk stratification in primary breast cancer is nodal status. Age, tumor size, estrogen receptor (ER) status and histological grade are all useful factors to further improve the outcome prediction. These “traditional” prognostic factors have been included into several outcome classification systems, e.g., the NNBC-3 risk algorithm.1 The advent of genome-wide gene expression analysis has undeniably improved the possibility to identify breast carcinomas that progress to metastasis.2–5 Some of the established gene expression signatures have been included as part of the clinical routine. Most of these predictive classification algorithms predominantly rely on ERα-regulated genes and/or on genes involved in proliferation.6–11
The Discovery of Biological Motifs in Gene Expression Patterns
Gene expression profiling also helped researchers recognize the heterogeneous nature of breast cancer.12–14 As a result, breast cancer is now classified into the five molecular subtypes: luminal A, luminal B, basal-like, normal-like and HER2-like.12, 15 A subsequent milestone was the discovery of biological motifs.16 Unsupervised clustering of gene expression data of breast carcinomas demonstrated the existence of highly correlative sets of genes representing either specific biological processes or specific cell types. The already well known influence of proliferation and ER receptor status was confirmed by the presence of two gene clusters consisting of genes involved in cell cycle progression and genes controlled by ERα.16 Interestingly, the importance of a further, so far unrecognized, gene cluster consisting of immunoglobulins and other B cell/plasma cell-associated genes was discovered. A normalized mean of this 60-gene B -cell/plasma cell cluster was introduced and named the “B-cell metagene”16–17. High expression of the B-cell metagene is associated with better prognosis in breast cancer, particularly in highly proliferating carcinomas.
Immunoglobulin κ Chain Surfaces to the Top
A limitation of the B-cell metagene is that the analysis of 60 genes is excessively laborious for routine clinical practice. Moreover, fresh frozen tissue for RNA isolation is often not available. Therefore, biostatistical analyses were performed to identify the top single biomarkers among the 60 gene of the B-cell metagene, considering prognostic power and the dynamic range (the range between low and high expressing carcinomas). Immunoglobulin κ chain (IGKC) was identified as one the top genes,18 and when validated in further cohorts, was found to be as predictive as the entire B-cell metagene. The availability of IGKC antibodies that could be used to immunostain paraffin slices also supported the use of IGKC use as a biomarker. With these immunostainings, carcinomas with strong IGKC immunostaining (3+, 2+) can easily be differentiated from negative (0) tissues (Fig. 1A). IGKC immunoreactivity, particularly IGKC 3+/2+ vs. 0, predicts prognosis that is consistent with the predictions based on RNA expression (Fig. 1B). Co-staining with the plasma cell/plasma blast marker MUM1 demonstrates that tumor-infiltrating plasma cells or plasma blasts are the source of IGKC expression (Fig. 1C). In contrast, CD20+ B cells, T cells and tumor cells are IGKC-.18 It should be considered that it is the presence of infiltrating plasma cells that is relevant for the association with prognosis, and not the expression of IGKC itself, since other immunoglobulins, e.g., immunoglobulin λ, are also associated with prognosis.16 However, IGKC is particularly convenient as a biomarker for statistical reasons, as reflected by its high dynamic range, and practical reasons, due to the availability of a reliable antibody. We also tested antibodies directed against other plasma cell constituents (e.g., CD138) and obtained significant associations with prognosis. However, the evaluation is more difficult, because the anti-CD138 antibody also stains tumor cells. In contrast, the anti-IGKC antibody exclusively labels infiltrating plasma cells/plasma blasts, leading to unequivocal and easy-to-interpret results (Fig. 1).
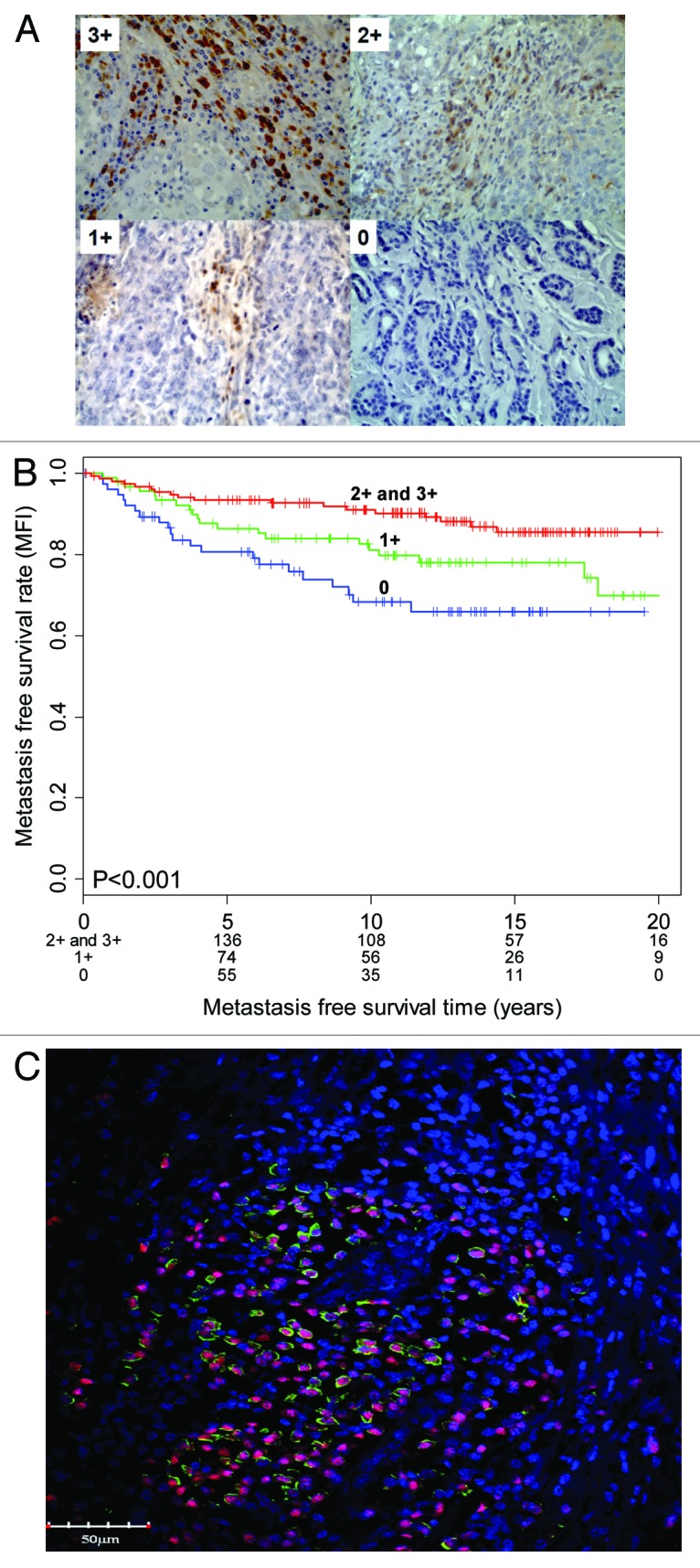
Figure 1. (A and B) Immunoglobulin κ chain (IGKC) expression (A) is associated with metastasis-free survival in breast cancer (B). (C) IGKC (green) is expressed in tumor-infiltrating plasma cells. The plasma blast/plasma cell marker MUM1 is visualized by red nuclear staining (adapted from ref.18).
After analyzing data from over 1800 breast cancers, 1000 non-small cell lung cancers and 500 colorectal carcinomas, it is now quite evident that the prognostic relevance of IGKC is not limited to breast cancer. It represents indeed a universal, single, robust immune marker for clinical-scale testing.19
IGKC Predicts Response to Neoadjuvant Chemotherapy in Breast Cancer
Biomarkers that help to decide whether patients benefit from chemotherapy are urgently needed in oncology.19 Today, only a few such markers are available, and none fits into the immune marker category.19 Importantly, IGKC does not only predict metastasis, but is also associated with response to chemotherapy, as demonstrated by the analysis of 845 breast cancer patients receiving anthracycline-based chemotherapy.18 Again, a comparison of IGKC as a single marker compared with the entire B-cell metagene yielded similar results. A possible explanation for this observation is that tumor cell killing by chemotherapy releases antigens that trigger immune responses. In conclusion, the introduction of IGKC into the clinical practice may fulfill the urgent need to identify patients who will profit from chemotherapy.
Future Perspectives: Understanding the Janus-Faced Immune System
Considering the complexity of mechanisms controlling metastasis and chemosensitivity, the hunt for useful biomarkers has just begun. Biomarkers of other possibly relevant mechanisms, such as redox control,20 mechanoactivity21 and the tumor metabolome and lipidome17, 22, 23 still need to be evaluated. However, a perspective for the near future is that we are close to fully understanding the mechanisms responsible for the Janus-faced nature of the immune system. With respect to prognosis of tumor patients, a favorable but also detrimental influence of the humoral immune system has been described (reviewed in ref. 10) The favorable influence has recently been harnessed by therapy with tumor antigen-specific antibodies, such as trastuzumab, rituximab or cetuximab.19 Similarly, antibodies produced by tumor-infiltrating plasma cells may bind to tumor antigens and mediate antibody-dependent cellular cytotoxicity.19 On the other hand, experimental studies and clinical data indicate that humoral immune responses may also mediate pro-tumor effects.24, 25 This is likely due to cytokines released from immune cells that stimulate the proliferation of tumor cells. As described in this article, both the entire B-cell metagene (defined as a normalized mean of 60 genes) and its single representative IGKC are associated with improved prognosis. Interestingly, a minority of genes within the B-cell metagene are clearly associated with worse (!) survival. Do they indicate a subtype of B cells that have turned “evil” and now initiate pro-tumor effects? Differentiating good and evil components of the humoral immune system in the future will further help in the selection of treatments that are most beneficial to cancer patients.
Footnotes
Previously published online: www.landesbioscience.com/journals/oncoimmunology/article/21653
References
- 1.Schmidt M, Victor A, Bratzel D, Boehm D, Cotarelo C, Lebrecht A, et al. Long-term outcome prediction by clinicopathological risk classification algorithms in node-negative breast cancer--comparison between Adjuvant! St Gallen, and a novel risk algorithm used in the prospective randomized Node-Negative-Breast Cancer-3 (NNBC-3) trial. Ann Oncol. 2009;20:258–64. doi: 10.1093/annonc/mdn590. [DOI] [PubMed] [Google Scholar]
- 2.van ’t Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–6. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 3.Wang Y, Klijn JG, Zhang Y, Sieuwerts AM, Look MP, Yang F, et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005;365:671–9. doi: 10.1016/S0140-6736(05)17947-1. [DOI] [PubMed] [Google Scholar]
- 4.Kammers K, Lang M, Hengstler JG, Schmidt M, Rahnenführer J. Survival models with preclustered gene groups as covariates. BMC Bioinformatics. 2011;12:478. doi: 10.1186/1471-2105-12-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hellwig B, Hengstler JG, Schmidt M, Gehrmann MC, Schormann W, Rahnenführer J. Comparison of scores for bimodality of gene expression distributions and genome-wide evaluation of the prognostic relevance of high-scoring genes. BMC Bioinformatics. 2010;11:276. doi: 10.1186/1471-2105-11-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351:2817–26. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 7.Sotiriou C, Wirapati P, Loi S, Harris A, Fox S, Smeds J, et al. Gene expression profiling in breast cancer: understanding the molecular basis of histologic grade to improve prognosis. J Natl Cancer Inst. 2006;98:262–72. doi: 10.1093/jnci/djj052. [DOI] [PubMed] [Google Scholar]
- 8.Fan C, Oh DS, Wessels L, Weigelt B, Nuyten DS, Nobel AB, et al. Concordance among gene-expression-based predictors for breast cancer. N Engl J Med. 2006;355:560–9. doi: 10.1056/NEJMoa052933. [DOI] [PubMed] [Google Scholar]
- 9.Petry IB, Fieber E, Schmidt M, Gehrmann M, Gebhard S, Hermes M, et al. ERBB2 induces an antiapoptotic expression pattern of Bcl-2 family members in node-negative breast cancer. Clin Cancer Res. 2010;16:451–60. doi: 10.1158/1078-0432.CCR-09-1617. [DOI] [PubMed] [Google Scholar]
- 10.Schmidt M, Hengstler JG, von Törne C, Koelbl H, Gehrmann MC. Coordinates in the universe of node-negative breast cancer revisited. Cancer Res. 2009;69:2695–8. doi: 10.1158/0008-5472.CAN-08-4013. [DOI] [PubMed] [Google Scholar]
- 11.Schmidt M, Petry IB, Böhm D, Lebrecht A, von Törne C, Gebhard S, et al. Ep-CAM RNA expression predicts metastasis-free survival in three cohorts of untreated node-negative breast cancer. Breast Cancer Res Treat. 2011;125:637–46. doi: 10.1007/s10549-010-0856-5. [DOI] [PubMed] [Google Scholar]
- 12.Perou CM, Sørlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–52. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 13.Sørlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98:10869–74. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rouzier R, Perou CM, Symmans WF, Ibrahim N, Cristofanilli M, Anderson K, et al. Breast cancer molecular subtypes respond differently to preoperative chemotherapy. Clin Cancer Res. 2005;11:5678–85. doi: 10.1158/1078-0432.CCR-04-2421. [DOI] [PubMed] [Google Scholar]
- 15.Cadenas C. Prognostic signatures of breast cancer: Perou's molecular subtypes and Schmidt's metagenes. EXCLI J. 2012;11:204–7. [PMC free article] [PubMed] [Google Scholar]
- 16.Schmidt M, Böhm D, von Törne C, Steiner E, Puhl A, Pilch H, et al. The humoral immune system has a key prognostic impact in node-negative breast cancer. Cancer Res. 2008;68:5405–13. doi: 10.1158/0008-5472.CAN-07-5206. [DOI] [PubMed] [Google Scholar]
- 17.Marchan R, Stewart JD, Lesjak M. EDI3, a key enzyme of choline metabolism controls tumour cell migration. EXCLI J. 2012;11:260–2. [PMC free article] [PubMed] [Google Scholar]
- 18.Schmidt M, Hellwig B, Hammad S, Othman A, Lohr M, Chen Z, et al. A comprehensive analysis of human gene expression profiles identifies stromal immunoglobulin κ C as a compatible prognostic marker in human solid tumors. Clin Cancer Res. 2012;18:2695–703. doi: 10.1158/1078-0432.CCR-11-2210. [DOI] [PubMed] [Google Scholar]
- 19.Whiteside TL, Ferrone S. For breast cancer prognosis, immunoglobulin kappa chain surfaces to the top. Clin Cancer Res. 2012;18:2417–9. doi: 10.1158/1078-0432.CCR-12-0566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cadenas C, Franckenstein D, Schmidt M, Gehrmann M, Hermes M, Geppert B, et al. Role of thioredoxin reductase 1 and thioredoxin interacting protein in prognosis of breast cancer. Breast Cancer Res. 2010;12:R44. doi: 10.1186/bcr2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martin M, Müller K, Cadenas C, Hermes M, Zink M, Hengstler JG, et al. ERBB2 overexpression triggers transient high mechanoactivity of breast tumor cells. Cytoskeleton (Hoboken) 2012;69:267–77. doi: 10.1002/cm.21023. [DOI] [PubMed] [Google Scholar]
- 22.Stewart JD, Marchan R, Lesjak MS, Lambert J, Hergenroeder R, Ellis JK, et al. Choline-releasing glycerophosphodiesterase EDI3 drives tumor cell migration and metastasis. Proc Natl Acad Sci USA. 2012;109:8155–60. doi: 10.1073/pnas.1117654109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cadenas C, Vosbeck S, Hein EM, Hellwig B, Langer A, Hayen H, et al. Glycerophospholipid profile in oncogene-induced senescence. Biochim Biophys Acta. 2012;1821:1256–68. doi: 10.1016/j.bbalip.2011.11.008. [DOI] [PubMed] [Google Scholar]
- 24.Tan TT, Coussens LM. Humoral immunity, inflammation and cancer. Curr Opin Immunol. 2007;19:209–16. doi: 10.1016/j.coi.2007.01.001. [DOI] [PubMed] [Google Scholar]
- 25.Inoue S, Leitner WW, Golding B, Scott D. Inhibitory effects of B cells on antitumor immunity. Cancer Res. 2006;66:7741–7. doi: 10.1158/0008-5472.CAN-05-3766. [DOI] [PubMed] [Google Scholar]


