Abstract
Background
There have been few genome-wide association studies (GWAS) of prostate cancer among diverse populations. To search for novel prostate cancer risk variants, we conducted GWAS of prostate cancer in Japanese and Latinos. In addition, we tested prostate cancer risk variants and developed genetic risk models of prostate cancer for Japanese and Latinos.
Methods
Our first stage GWAS of prostate cancer included Japanese (cases/controls=1,033/1,042) and Latino (cases/controls=1,043/1,057) from the Multiethnic Cohort. Significant associations from stage 1 (P < 1.0×10−4) were examined in silico in GWAS of prostate cancer (stage 2) in Japanese (cases/controls=1,583/3,386) and Europeans (cases/controls=1,854/1,894).
Results
No novel stage 1 SNPs outside of known risk regions reached genome-wide significance. For Japanese, in stage 1, the most notable putative novel association was seen with 10 SNPs (P<8.0. x10−6) at chromosome 2q33; however, this was not replicated in stage 2. For Latinos, the most significant association was observed with rs17023900 at the known 3p12 risk locus (stage 1: OR=1.45; P=7.01×10−5 and stage 2: OR=1.58; P =3.05×10−7). The majority of the established risk variants for prostate cancer, 79% and 88%, were positively associated with prostate cancer in Japanese and Latinos (stage I), respectively. The cumulative effects of these variants significantly influence prostate cancer risk (OR per allele=1.10; P = 2.71×10−25 and OR=1.07; P = 1.02×10−16 for Japanese and Latinos, respectively).
Conclusion and Impact
Our GWAS of prostate cancer did not identify novel genome-wide significant variants. However, our findings demonstrate that established risk variants for prostate cancer significantly contribute to risk among Japanese and Latinos.
INTRODUCTION
Prostate cancer displays dramatic differences in incidence rates across racial/ethnic populations. In the United States, African-Americans have the highest incidence rate of prostate cancer followed by European Americans, Latinos, and Asians. The contribution of genetic variants to prostate cancer risk likely varies across race/ethnicity and may play a key role in the unequal burden of disease across racial/ethnic groups (1). The first wave of genome-wide association studies (GWAS) of prostate cancer were heavily weighted by studies of European men (2-10), revealing more than 40 prostate cancer risk variants, many of which replicated in subsequent studies of non-European populations (1, 11-17). More recently, GWAS of prostate cancer have been conducted in non-Europeans (18, 19), identifying 5 new risk variants in Japanese (19) and one novel risk variant in men of African ancestry (18). Identifying the full spectrum of prostate cancer risk alleles, in terms of numbers and frequencies, requires conducting GWAS of prostate cancer in all possible racial/ethnic populations. With differences in allele frequencies, linkage disequilibrium (LD) patterns, and population-specific risk of disease across race/ethnicities, evaluating the generalizability of known risk variants is important to increase our understanding of the genetic contributions to prostate cancer. Equally important is defining the genetic risk profiles relevant for each racial/ethnic group.
In this study, we conducted two-stage GWAS to search for novel risk variants for prostate cancer in Japanese and Latinos, respectively. We also tested known risk variants for prostate cancer and utilized these variants to develop genetic risk models of prostate cancer for Japanese and Latinos.
METHODS
Stage 1 of the GWAS included Japanese and Latino prostate cancer cases and controls from the Multiethnic Cohort (MEC). In silico replication of the most significant associations from stage 1 were conducted in GWAS of prostate cancer in Japanese (19) and Europeans (7). Below is a brief description of the first and second stage study populations.
The MEC is a large population-based cohort study of over 215,000 individuals from Hawaii and California (20). Further methodological details of this cohort are provided elsewhere (20). Briefly, incident prostate cancer cases were identified by cohort linkage to Surveillance, Epidemiology and End Results cancer registries covering Hawaii and California. Controls had no diagnosis of prostate cancer, were randomly selected from the random control pool of participants, and provided blood specimens for genetic analysis. Controls were frequency matched to cases by age (5 year categories) and ethnicity. Through January 1, 2008, the Japanese and Latino nested case-control studies of prostate cancer included 1,033 cases and 1,042 controls and 1,043 cases and 1,057 controls, respectively.
In silico replication of findings in Japanese men was conducted in a GWAS of prostate cancer of 1,583 Japanese with prostate cancer and 3,386 controls, who were part of the BioBank Japan at the Institute of Medical Science at the University of Tokyo (19). The 1,583 cases were diagnosed as having prostate cancer based on the pathological evaluation of prostatic biopsy. The controls were 2,480 individuals registered in the BioBank Japan as subjects with 13 diseases other than prostate cancer and 906 healthy volunteers collected at the Osaka-Midosuji Rotary Club. All participants provided written informed consent. Study subjects were genotyped using either the Illumina Infinium Human610-Quad BeadChip or Infinium HumanHap550v3 BeadChip.
In silico replication of findings in Latinos and those from the combined analysis of Japanese and Latinos was conducted in the United Kingdom GWAS of 1,854 prostate cancer cases diagnosed at age 60 years or younger with a family history of disease, and 1,894 controls aged >50 years with a PSA of <0.5ng/ml (7).
Genotyping
Genotyping of the Japanese and Latinos in the MEC was conducted using the Illumina.Human660W_Quad_v1 bead array at the Broad Institute. Samples with DNA concentrations <18.8ng/ul were not scanned (53 Japanese and 52 Latinos). Samples were removed based on the following exclusion criteria: 1) call rates <95% (5 Japanese and 4 Latinos); 2) ancestry outliers (21 Japanese and 25 Latinos, discussed below), and; 3) related samples (88 Japanese and 57 Latinos, discussed below). We also removed SNPs with minor allele frequencies <1% (n=16,793). To assess genotyping reproducibility, we included 9 replicate samples; the average concordance rate was 99.99% (≥99.3% for all pairs). The final analysis included 528,023 SNPs evaluated in 2,075 Japanese and 2,100 Latinos.
Statistical Analysis
Ancestry Estimation
The EIGENSTRAT software (21) was used to calculate eigenvectors that explained genetic differences in ancestry. The analysis included data from HapMap Phase 3 populations and our study, so that comparisons to reference populations of known ethnicity could be made. An individual was subject to filtering from the analysis if this value along eigenvector 1 or 2 was outside of 4 SDs of the mean of each respective eigenvector. Twenty-one self-reported Japanese and 25 self-reported Latinos met this filtering criterion. Together the top 10 eigenvectors (used in the analysis) explained 8% of the global genetic variability among subjects.
Relatedness Inference
We used PLINK (22) to calculate the probabilities of sharing 0, 1, and 2 alleles (Z = Z0,Z1,Z2) across all possible pairs of samples to determine individuals who were likely to be related to others. We identified 1 pair of monozygotic twins (confirmed), 57 half siblings, and 129 first degree relative pairs (parent offspring/full siblings) based on the values of their observed probability vector Z being within 1 SD of the expected values of Z for their respective relationship. For the 187 pairs, one individual was removed from analysis. The criterion for removal was such that individuals that were related with a higher number of pairs were chosen for removal. In all other cases, one of the two members was randomly selected for removal.
SNP Imputation
We carried out genome-wide imputation using the software MACH. Phased haplotype data from the founders of the JPT, CEU, and YRI. HapMap Phase 2 samples were used to infer LD patterns in order to impute untyped markers. The Rsq metric, defined as the observed variance divided by the expected variance, provides a measure of the quality of the imputation at any SNP and was used as a threshold in determining which SNPs to filter from analysis (Rsq <0.3). For all imputed SNPs reported, Rsq was ≥0.3.
Association Testing
In stage 1, we examined the observed versus the expected distribution of the Chi square test statistics from the 1-degree-of-freedom (d.f.) trend test, comparing genotype counts in cases and controls. All tests of statistical significance were two-sided. Odds ratios (OR) and 95% confidence intervals (95% CI) were estimated using unconditional logistic regression adjusting for age and the first 10 ancestry eigenvalues. For each SNP, we tested for a gene dosage effect through a 1 d.f. Wald chi-square trend test. To address the hypothesis that the same variants could be informative across populations as shown for the 8q24 locus (1) and combine risk estimates between Japanese and Latinos, we conducted a meta-analysis of stage 1 results for SNPs genotyped in Japanese and Latinos, using the inverse variance method (METAL) (23). The genomic control value for the meta-analyzed results was 1.007.
For the replication studies, statistical tests for the association with each SNP were performed by a 1 d.f. Cochrane-Armitage trend test. Per-allele ORs were estimated using logistic regression.
Risk Modeling
In each population in stage 1, we examined the association of 56 known risk variants for prostate cancer—45 independent variants and 11 risk variants at 8q24 that had an association with prostate cancer risk in previous European, African, and Japanese studies (r2<0.16 in Europeans and r2<0.27 in Asians with the exception of r2=0.52 between rs1016343 and rs6983561 at 8q24) (1, 3, 10, 24-31). SNP rs10090154 was used in place of rs11986220 as it is located in a predicted enhancer site (24). The risk SNP BD11934905 (1) is not on the Illumina 660W array and was not genotyped in this study. To model the cumulative genetic risk for the 56 variants, we summed the number of risk alleles for each individual and estimated the OR per allele for this aggregate unweighted allele count variable, serving as an approximate risk score appropriate for unlinked variants with independent effects of roughly the same magnitude for each allele. For individuals missing genotypes (2.7%) for a given SNP (range=0-1.27%; mean=0.05%), we assigned the average number of risk alleles (2 x risk allele frequency) to replace the missing value for that SNP. We also tested for differences in the effect of the risk score by race/ethnicity, age group (median of 64 <years vs. 64 ≥ years), family history of prostate cancer, and stage of disease (localized vs. regional/distant disease).
Results
Genome-wide Association Study of Prostate Cancer
Study characteristics of the 2,075 prostate cancer cases and 2,100 controls in the MEC are presented in Supplemental Table 1 (Japanese cases/controls=1,033/1,042; Latinos cases/controls=1,043/1,057). The mean age for Japanese cases and controls was 64.0 years and 63.9 years, respectively, and the mean age for both Latino cases and controls was 62.6 years. As expected, cases were more likely than controls (~1.7 times) to report a family history of prostate cancer. Among cases, approximately 47.6% and 37.7% of Japanese and Latinos presented with regional/distant disease, respectively.
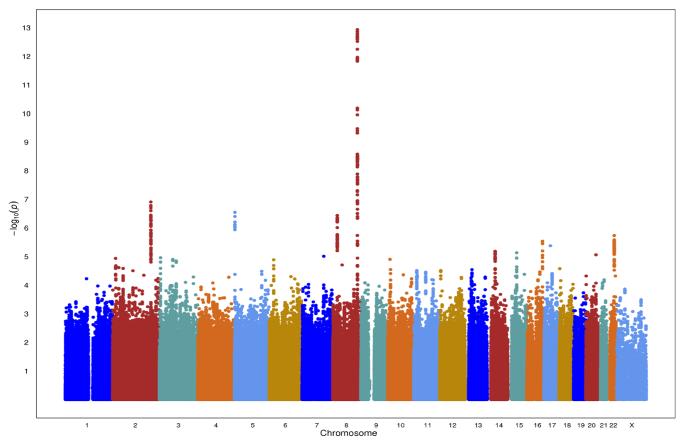
Quantile-quantile plots of the distribution of test statistics for the comparison of genotype frequencies in prostate cancer cases versus controls showed no evidence of over-inflation; the genomic inflation factor lambda (λ) was 0.986 and 1.008 in Japanese and Latinos, respectively (Supplemental Figure 1A, 1B). For Japanese, in stage 1, 69 SNPs (including 10 genotyped SNPs) had P <5×10−8 (Figure 1A). All of these genome-wide significant SNPs were at the 8q24 risk locus between 128.16 and 128.61 Mb and were correlated with the known risk variants in this region. No novel SNPs reached genome-wide significance (P <5×10−8) in these stage 1 samples. The most notable putative novel association was seen with a cluster of 10 SNPs (P < 8.0 ×10−6; Table 1), spanning 802 kb at chromosome 2q33 that includes the genes BOLL, PLCL1, COQ10B, and RFTN2. For stage 2, we selected the 69 genotyped SNPs with P < 1.0 × 10−4 and located outside of known risk regions for in silico replication in 1,583 Japanese prostate cancer cases and 3,386 Japanese controls (Supplemental Table 2). None of the associations with these 69 SNPs replicated with P<0.05 and effect estimates in the same direction as in stage 1. The results for the most significant SNPs in stage 1 (n=13 with P<1×10−5) are shown in Table 1.
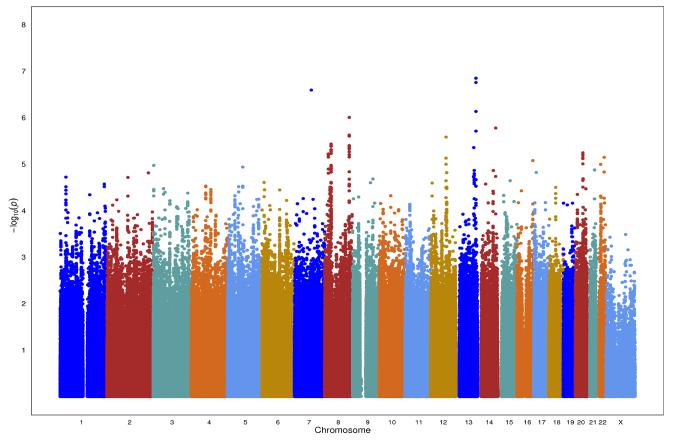
Figure 1.
A. Manhattan Plot for Japanese
B. Manhattan Plot for Latinos
Table 1.
SNPs outside of known risk regions at p<10−5 in Japanese and Latinos GWAS of prostate cancer
| MEC Japanese (cases/controls=1033/1042) |
Japanese (cases/controls=1583/3386) |
||||||||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| chromosome | locus | SNP | position | Coded Allele/Ref Allele | CAF | OR | p | OR | p |
| 2 | intergenic | rs10931777 | 197851836 | T/C | 0.84 | 0.66 | 1.23×10−7 | 1.04 | 0.48 |
| 2 | intergenic | rs16824376 | 198392587 | T/C | 0.17 | 1.49 | 5.00×10−7 | 0.97 | 0.61 |
| 2 | BOLL | rs1851779 | 198355353 | A/G | 0.17 | 1.49 | 5.41×10−7 | 0.97 | 0.61 |
| 2 | intergenic | rs6707521 | 198421476 | A/G | 0.17 | 1.49 | 5.47×10−7 | 0.97 | 0.56 |
| 2 | intergenic | rs996427 | 198393854 | T/C | 0.17 | 1.49 | 5.51×10−7 | 0.97 | 0.61 |
| 2 | PLCL1 | rs1595825 | 198583709 | A/G | 0.16 | 1.47 | 1.47×10−6 | 0.97 | 0.54 |
| 2 | COQ10B | rs3754822 | 198035559 | T/C | 0.17 | 1.45 | 1.84×10−6 | 0.96 | 0.43 |
| 2 | PLCL1 | rs1016883 | 198589913 | T/C | 0.16 | 1.46 | 2.56×10−6 | 0.98 | 0.64 |
| 2 | RFTN2 | rs2045244 | 198214711 | A/G | 0.18 | 1.45 | 2.68×10−6 | 0.97 | 0.62 |
| 16 | intergenic | rs7202575 | 81005610 | T/C | 0.08 | 1.62 | 2.92×10−6 | 0.90 | 0.13 |
| 16 | intergenic | rs7202694 | 81005355 | G/A | 0.08 | 1.59 | 6.33×10−6 | -- | -- |
| 22 | GTPBP1 | rs4821815 | 37435653 | A/G | 0.53 | 0.75 | 6.47×10−6 | 0.95 | 0.24 |
| 2 | PLCL1 | rs6434955 | 198654796 | A/G | 0.16 | 1.43 | 8.00×10−6 | 0.96 | 0.49 |
|
| |||||||||
| MEC Latinos (cases/controls=1043/1057) |
UK (cases/controls=1854/1894) |
||||||||
|
| |||||||||
| chromosome | locus | SNP | position | Coded Allele/Ref Allele | CAF | OR | P | OR | p |
|
| |||||||||
| 12 | intergenic | rs4240731 | 80786862 | G/A | 0.28 | 1.36 | 2.60×10−6 | 1.00 | 0.94 |
| 20 | ZHX3 | rs6102322 | 39306182 | T/C | 0.36 | 1.33 | 5.70×10−6 | 0.99 | 0.81 |
| 20 | TOP1 | rs6129760 | 39179817 | G/A | 0.39 | 1.32 | 9.83×10−6 | 0.98 | 0.74 |
For Latinos, in stage 1, we observed no genome-wide significant associations (P<5×10−8) (Figure 1B). We selected the 56 genotyped SNPs with P < 1.0 × 10−4 in stage 1 (Supplemental Table 3) to evaluate in stage 2 samples of 1,854 prostate cancer cases and 1,894 controls of European ancestry. The most significant association in stage 1 was with the imputed SNP, rs12873332 at 3q33 (P=1.41×10−7); a genotyped proxy for this SNP (rs12874523; r2=0.7 in HapMap MEX) did not replicate at P < .05 in stage 2. SNP, rs17023900, at the known risk locus at 3p12 (6, 32), was associated with prostate cancer (stage 1; OR=1.45; P=7.01×10−5) and replicated in the European population (stage 2; OR=1.58; P =3.05×10−7). SNP rs17023900 was not correlated with the known risk variant, rs17181170 at 3p12, identified in Europeans (6) (r2=0.07 in CEU and r2=0.07 in JPT) Yet, rs17023900 was somewhat correlated with the other 3p12 risk variant, rs9284813, identified in Japanese (32) (r2=0.14 in CEU and r2= 0.65 in JPT). The three top-ranked genotyped SNPs [rs4240731 (12q21), rs6102322 (ZHX3), rs6129760 (TOP1); P < 1.0 × 10−5] for Latinos in stage 1, outside of known risk regions, were not significantly associated with prostate cancer in stage 2 (Table 1). Of the remaining 53 stage 1 SNPs, only two SNPs (chromosome 13-rs9514490 and chromosome 8-rs11306015) were associated with prostate cancer in stage 2, however, the effect estimates were in the opposite direction (Supplemental Table 3).
From the meta-analysis of stage 1 results for Japanese and Latinos, all genome-wide significant SNPs (n=10; P <5×10−8) were located at chromosome 8q24 (Supplemental Figure 2). For SNPs with P <10−6 in the combined analysis (Supplemental Table 4), only one SNP, rs4999155 at 9q21 (OR = 1.32; P -7 meta meta= 6.17×10 ), was located outside of known risk regions. This SNP, rs4999155, was significantly associated with risk in both Japanese (OR=1.31; P=9.08×10−4) and Latinos (OR=1.33; P=1.73×10−4), but it did not replicate at P < .05 in the European GWAS of prostate cancer.
Testing of Known Risk Variants
We tested, in stage 1 samples, 56 known prostate cancer risk variants located in 37 regions and in chromosome 8q24 (1-8, 18, 19, 26, 33-37); 51 were genotyped and 5 were imputed with high accuracy. The risk allele frequency ranged from 0.01 to 0.90 in Japanese and 0.12 to 0.92 in Latinos (Supplemental Figure 3). Positive associations were observed with the majority of variants in each population (44 in Japanese and 49 in Latinos; Tables 2 and 3). Of the 56 risk variants, 18 SNPs were positively and significantly associated with prostate cancer risk in either Japanese or Latinos (Tables 2 and 3), with rs1512268 at 8p21, rs10993994 at 10q11 and five SNPs at 8q24 (rs10086908, rs13254738, rs6983561, rs10090154) reaching statistical significance (P<0.05) in both populations. For both Japanese and Latinos, the strongest associations were noted at 8q24, albeit with different SNPs: rs6983561 in Japanese, OR=1.87; 95% CI: 1.58-2.22; P =3.8×10−13; and, rs10090154 in Latinos, OR=1.68; 95% CI: 1.35-2.09; P=3.4×10−6. The strongest association outside of chromosome 8q24 was with rs12653946 at 5p15 in Japanese (OR=1.39; 95% CI: 1.22-1.57; P =3.4×10−7) and rs5759167 at 22q13 in Latinos (OR=1.22; 95% CI: 1.08-1.39; P=2.0×10−3). At 8q24, all 11 of the risk variants, except for rs12543663, were positively associated with risk in Japanese and Latinos. In Japanese, 9 of 11 variants were significantly associated with risk (Table 3) with four remaining demonstrating statistically significant independent genetic associations (rs10086908-region 1, rs13254738-region 2, rs6983561-region 2, and rs6983267-region 4). In Latinos, 5 of the 11 8q24 risk variants were significantly associated with prostate cancer and 3 demonstrated independent significant associations (rs10086908-region 1, rs13254738-region 2, and rs6983561-region 2). Notably, the per allele OR for rs6983561 was ~1.5 in both populations, which is considerably larger compared to effect estimates observed for other known risk alleles for prostate cancer (OR ~1.1 – 1.2).
Table 2.
Associations with established risk variants for prostate cancer in Japanese (1033 cases, 1042 controls) and Latinos (1043 cases, 1057 controls).
| Japanese | Latinos | |||||||
|---|---|---|---|---|---|---|---|---|
| Chr., Marker | Position, Allelesd | RAFa Europeans | RAF | Per allele OR (95% CI)b | P-valuec | RAF | Per allele OR (95% CI)b | P-valuec |
| 2p24, rs13385191 | 20751746, G/A | 0.2 | 0.57 | 1.03(0.91-1.17) | 0.66 | 0.27 | 1.16(1.01-1.32) | 0.037 |
| 2p21, rs1465618 | 43407453, T/C | 0.23 | 0.66 | 1.19(1.04-1.35) | 1.0×10−2 | 0.41 | 1.03(0.91-1.17) | 0.63 |
| 2p15, rs721048e,g | 62985235, A/G | 0.19 | 0.04 | 0.93(0.68-1.27) | 0.65 | 0.16 | 1.26(1.07-1.48) | 5.2×10−3 |
| 2p15, rs2710647 | 63067474, C/T | 0.55 | 0.73 | 1.05(0.91-1.2) | 0.50 | 0.58 | 1.07(0.95-1.21) | 0.28 |
| 2p11, rs10187424 | 85647807, T/C | 0.55 | 0.64 | 0.98(0.86-1.11) | 0.73 | 0.64 | 1.16(1.02-1.33) | 0.026 |
| 2q21, rs12621278 | 173019799, A/G | 0.94 | 0.77 | 1.13(0.97-1.31) | 0.11 | 0.92 | 1.34(1.05-1.71) | 0.018 |
| 2q37, rs2292884 | 238107965, G/A | 0.26 | 0.25 | 1.11(0.96-1.27) | 0.16 | 0.28 | 1.10(0.96-1.27) | 0.16 |
| 3p12, rs2660753 | 87193364, T/C | 0.11 | 0.24 | 1.14(0.98-1.31) | 0.08 | 0.19 | 1.23(1.06-1.44) | 7.2×10−3 |
| 3q21, rs10934853 | 129521063, A/C | 0.28 | 0.52 | 0.93(0.82-1.05) | 0.24 | 0.40 | 0.97(0.86-1.10) | 0.65 |
| 3q23, rs6763931 | 142585522, A/G | 0.43 | 0.35 | 1.16(1.02-1.32) | 0.019 | 0.40 | 1.03(0.91-1.16) | 0.68 |
| 3q26, rs10936632g | 171612795, A/C | 0.52 | 0.42 | 1.16(0.99-1.36) | 0.06 | 0.39 | 1.04(0.90-1.21) | 0.60 |
| 4q22, rs12500426 | 95733632, A/C | 0.46 | 0.44 | 1.05(0.93-1.19) | 0.43 | 0.54 | 1.13(1.00-1.28) | 0.046 |
| 4q22, rs17021918 | 95781900, C/T | 0.66 | 0.62 | 0.99(0.88-1.13) | 0.93 | 0.72 | 1.08(0.94-1.24) | 0.27 |
| 4q24, rs7679673f,g | 106280983, C/A | 0.55 | 0.27 | 1.02(0.87-1.21) | 0.79 | 0.47 | 1.01(0.88-1.15) | 0.94 |
| 5p15, rs401681 | 1375087, C/T | 0.55 | 0.67 | 0.97(0.85-1.1) | 0.59 | 0.61 | 0.93(0.82-1.05) | 0.25 |
| 5p15, rs12653946 | 1948829, T/C | 0.42 | 0.43 | 1.39(1.22-1.57) | 3.4×10−7 | 0.49 | 1.02(0.90-1.15) | 0.78 |
| 5p12, rs2121875 | 44401301, C/A | 0.35 | 0.47 | 1.02(0.9-1.16) | 0.71 | 0.55 | 0.98(0.87-1.12) | 0.80 |
| 6p21, rs130067 | 31226489, G/T | 0.23 | 0.36 | 0.99(0.87-1.12) | 0.82 | 0.29 | 0.92(0.80-1.06) | 0.23 |
| 6p21, rs1983891 | 41644405, T/C | 0.27 | 0.42 | 1.04(0.92-1.18) | 0.52 | 0.38 | 1.16(1.02-1.31) | 0.025 |
| 6q22, rs339331 | 117316745, T/C | 0.64 | 0.63 | 1.14(1-1.29) | 0.046 | 0.73 | 1.03(0.89-1.18) | 0.72 |
| 6q25, rs9364554 | 160753654, T/C | 0.29 | 0.34 | 1.09(0.95-1.24) | 0.22 | 0.21 | 1.06(0.91-1.23) | 0.44 |
| 7p15, rs10486567 | 27943088, G/A | 0.77 | 0.09 | 1.22(0.99-1.5) | 0.06 | 0.53 | 1.13(1.00-1.28) | 0.052 |
| 7q21, rs6465657 | 97654263, C/T | 0.46 | 0.90 | 1.06(0.86-1.3) | 0.61 | 0.70 | 0.96(0.84-1.10) | 0.53 |
| 8p21, rs2928679 | 23494920, A/G | 0.42 | 0.09 | 0.90(0.73-1.12) | 0.34 | 0.32 | 1.03(0.91-1.18) | 0.61 |
| 8p21, rs1512268 | 23582408, T/C | 0.45 | 0.36 | 1.35(1.19-1.53) | 4.5×10−6 | 0.44 | 1.21(1.07-1.37) | 2.8×10−3 |
| 10q11, rs10993994 | 51219502, T/C | 0.4 | 0.46 | 1.19(1.05-1.34) | 6.4×10−3 | 0.35 | 1.19(1.05-1.35) | 6.4×10−3 |
| 10q26, rs4962416 | 126686862, C/T | 0.27 | 0.01 | 1.11(0.61-2.08) | 0.73 | 0.24 | 1.05(0.91-1.22) | 0.47 |
| 11p15, rs7127900 | 2190150, A/G | 0.2 | 0.08 | 1.18(0.95-1.47) | 0.14 | 0.31 | 1.17(1.02-1.33) | 0.025 |
| 11q13, rs12418451g | 68691995, A/G | 0.28 | 0.09 | 0.90(0.67-1.21) | 0.49 | 0.22 | 1.08(0.92-1.27) | 0.34 |
| 11q13, rs11228565g | 68735156, A/G | 0.2 | 0.04 | 1.03(0.75-1.41) | 0.88 | 0.12 | 1.26(1.05-1.52) | 0.015 |
| 11q13, rs7931342 | 68751073, G/T | 0.51 | 0.22 | 1.00(0.86-1.16) | 0.99 | 0.38 | 1.11(0.98-1.26) | 0.10 |
| 11q13, rs10896449 | 68751243, G/A | 0.52 | 0.04 | 1.17(0.88-1.56) | 0.27 | 0.35 | 1.10(0.97-1.25) | 0.14 |
| 12q13, rs10875943 | 47962276, C/T | 0.28 | 0.81 | 1.13(0.96-1.32) | 0.14 | 0.32 | 1.06(0.93-1.21) | 0.42 |
| 13q22, rs9600079 | 72626140, T/G | 0.47 | 0.36 | 1.2(1.06-1.36) | 4.1×10−3 | 0.39 | 1.02(0.90-1.16) | 0.71 |
| 17p12, rs4054823 | 13565749, T/C | 0.56 | 0.57 | 1.08(0.96-1.22) | 0.20 | 0.49 | 1.09(0.97-1.23) | 0.17 |
| 17q12, rs11649743 | 33149092, G/A | 0.8 | 0.71 | 1.08(0.94-1.23) | 0.28 | 0.82 | 1.27(1.08-1.50) | 3.8×10−3 |
| 17q12, rs4430796 | 33172153, A/G | 0.53 | 0.64 | 1.13(0.99-1.29) | 0.06 | 0.59 | 1.09(0.96-1.24) | 0.16 |
| 17q12, rs7501939 | 33175269, C/T | 0.58 | 0.68 | 1.21(1.06-1.38) | 4.9×10−3 | 0.67 | 1.04(0.91-1.18) | 0.56 |
| 17q24, rs1859962 | 66620348, G/T | 0.46 | 0.25 | 1.06(0.92-1.21) | 0.43 | 0.60 | 1.14(1.00-1.29) | 0.045 |
| 19q13, rs8102476 | 43427453, C/T | 0.54 | 0.37 | 0.87(0.77-0.99) | 0.030 | 0.49 | 1.07(0.95-1.21) | 0.29 |
| 19q13, rs266849 | 56040902, A/G | 0.8 | 0.64 | 1.15(1.01-1.31) | 0.034 | 0.76 | 1.13(0.98-1.31) | 0.10 |
| 19q13, rs2735839 | 56056435, G/A | 0.85 | 0.59 | 1.20(1.05-1.36) | 5.6×10−3 | 0.76 | 1.15(0.99-1.33) | 0.07 |
| 22q13, rs5759167 | 41830156, G/T | 0.53 | 0.66 | 1.11(0.97-1.27) | 0.13 | 0.58 | 1.22(1.08-1.39) | 2.0×10−3 |
| Xp11, rs5945572 | 51246423, A/G | 0.35 | 0.08 | 1.10(0.94-1.27) | 0.23 | 0.16 | 1.09(0.97-1.22) | 0.15 |
RAF, risk allele frequency in populations of European ancestry from previous reports or HapMap CEU population.
Adjusted for age and the 1st 10 eigenvalues.
Test of trend (1-d.f.).
Risk allele/reference allele.
rs721048 not typed in Japanese and Latinos. Results for rs17432497 are shown for these groups (r2=0.98 with rs721048 in HapMap CEU)
rs7679673 not on Illumina 1M/660.
Imputed SNP.
Table 3.
Associations with known risk variants at 8q24 in Japanese and Latinos.
| Japanese (1033 cases, 1042 controls) | |||||||
|---|---|---|---|---|---|---|---|
| Blocka, Position | Marker, Allelesb | RAFc | RAF | OR (95% CI)d | P-valuee | OR (95% CI) Adjustedf |
P-value |
| 1, 127,993,841 | rs12543663, C/A | 0.31 | 0.08 | 0.98(0.78-1.23) | 8.30×10−1 | 1.02(0.79- 1.31) |
8.77×10−1 |
| 1, 128,081,119 | rs10086908, T/C | 0.70 | 0.80 | 1.26(1.08-1.48) | 3.92×10−3 | 1.27(1.07- 1.51) |
7.11×10-3 |
| 2, 128,162,479 | rs1016343, T/C | 0.20 | 0.26 | 1.48(1.30-1.69) | 5.23×10−9 | 1.10(0.90- 1.34) |
3.61×10−1 |
| 2, 128,164,338 | rs13252298, A/G | 0.70 | 0.63 | 1.41(1.24-1.61) | 3.25×10−7 | 0.97(0.79- 1.19) |
7.92×10−1 |
| 2, 128,173,525 | rs13254738, C/A | 0.35 | 0.54 | 1.59(1.38-1.84) | 4.23×10−10 | 1.34(1.06- 1.69) |
1.35×10−2 |
| 2, 128,176,062 | rs6983561, C/A | 0.04 | 0.18 | 1.87(1.58-2.22) | 3.84×10−13 | 1.52(1.21- 1.92) |
3.88×10−4 |
| 3, 128,404,855 | rs620861h, G/A | 0.61 | 0.53 | 1.05(0.93-1.19) | 4.29×10−1 | 1.08(0.92- 1.26) |
3.42×10−1 |
| 3, 128,410,090 | rs16902104, T/C | 0.14 | 0.24 | 1.13(0.99-1.30) | 8.02×10−2 | 1.06(0.89- 1.26) |
5.13×10−1 |
| 4, 128,482,487 | rs6983267, G/T | 0.51 | 0.31 | 1.25(1.10-1.42) | 5.69×10−4 | 1.21(1.04- 1.40) |
1.13×10−2 |
| 4, 128,510,352 | rs7000448, T/C | 0.36 | 0.22 | 1.20(1.01-1.41) | 3.33×10−2 | 1.09(0.90- 1.31) |
3.79×10−1 |
| 5, 128,601,319 | rs10090154, T/C | 0.09 | 0.16 | 1.62(1.38-1.92) | 5.56×10−9 | 1.06(0.63- 1.78) |
6.89×10−2 |
| Latinos (1043 cases, 1057 controls) | |||||||
| Blocka, Position | Marker, Allelesb | RAFc | RAF | OR (95% CI)d | P-valuee |
OR (95% CI)
Adjustedf |
P-value |
| 1, 127,993,841 | rs12543663, C/A | 0.31 | 0.37 | 0.92(0.81-1.04) | 1.93×10−1 | 0.97(0.84- 1.11) |
6.43×10−1 |
| 1, 128,081,119 | rs10086908, T/C | 0.70 | 0.64 | 1.24(1.09-1.41) | 1.10×10−3 | 1.22(1.07- 1.40) |
3.91×10−3 |
| 2, 128,162,479 | rs1016343, T/C | 0.20 | 0.13 | 1.19(1.00-1.42) | 5.66×10−2 | 1.05(0.86- 1.29) |
6.26×10−1 |
| 2, 128,164,338 | rs13252298, A/G | 0.70 | 0.66 | 1.03(0.91-1.18) | 6.14×10−1 | 0.96(0.82- 1.11) |
5.52×10−1 |
| 2, 128,173,525 | rs13254738, C/A | 0.35 | 0.44 | 1.19(1.04-1.37) | 9.93×10−3 | 1.20(1.02- 1.40) |
2.58×10−2 |
| 2, 128,176,062 | rs6983561, C/A | 0.04 | 0.04 | 1.68(1.25-2.26) | 5.78×10−4 | 1.49(1.10- 2.04) |
1.13×10−2 |
| 3, 128,404,855 | rs620861h, G/A | 0.61 | 0.62 | 1.11(0.98-1.26) | 9.95×10−2 | 1.09(0.95- 1.25) |
2.03×10−1 |
| 3, 128,410,090 | rs16902104, T/C | 0.14 | 0.11 | 1.19(0.99-1.44) | 7.09×10−2 | 1.15(0.94- 1.40) |
1.86×10−1 |
| 4, 128,482,487 | rs6983267, G/T | 0.51 | 0.62 | 1.08(0.95-1.23) | 2.23×10−1 | 1.06(0.93- 1.22) |
3.85×10−1 |
| 4, 128,510,352 | rs7000448, T/C | 0.36 | 0.32 | 1.09(0.95-1.25) | 2.39×10−1 | 1.07(0.92- 1.25) |
3.81×10−1 |
| 5, 128,601,319 | rs10090154, T/C | 0.09 | 0.08 | 1.68(1.35-2.09) | 3.40×10−6 | 1.30(0.71- 2.45) |
4.68×10−1 |
As defined in Al Olama et al. (10).
Risk /reference alleles.
RAF, risk allele frequency in populations of European ancestry (EA) as reported previously (2-8, 26, 33-37) and in Japanese (JA) and Latinos (LA).
Adjusted for age and the 1st 10 eigenvalues.
Test of trend (1-d.f.).
From multivariate model. OR adjusted for age and the 1st 10 eigenvalues and all other 8q24 risk variants.
Imputed (R2>0.89). rs445114 was not typed and could not be imputed.
Risk Modeling of Prostate Cancer Variants
Using the 56 prostate cancer risk variants (see Methods), we modeled their cumulative effect in Japanese and Latinos in stage 1 samples (Table 4). For Japanese, a 10% increased risk of prostate cancer was associated with each additional risk allele (P =2.71×10−25). Japanese men at the top quartile of the risk allele distribution had a 3.7-fold increased risk of prostate cancer compared to those at the lowest quartile (P=1.17×10−21). For Latinos, a 7% increased risk of prostate cancer was associated with each additional risk allele (P=1.02×10−16) and those at the highest risk quartile had a 2.8-fold increased risk of disease in comparison to men at the lowest risk quartile (P=1.10×10−14). Heterogeneity in effects of the risk score by race/ethnicity was not statistically significant (Phet=0.06). Stratified analysis of the risk score revealed similar patterns of associations across age groups (Phet ≥ 0.16) and family history of prostate cancer (Phet ≥ 0.77) (data not shown). In addition, similar effects were seen for localized (ORJA=1.10; p=1.11×10−15; ORLA=1.07; P =3.36×10−11) and regional/distant (ORJA=1.08; P =9.47×10−11; ORLA=1.08; P=1.49×10−10) disease for both populations (JA Phet =0.13; LA Phet =0.61).
Table 4.
The association between the total risk score with prostate cancer in Japanese and Latinos.
| Index Markers from GWAS in Japanese (n=56) |
Index Markers from GWAS in Latinos (n=56) |
||
|---|---|---|---|
| Mean No. Risk alleles, (range) | 48(32-67) | 51(35-69) | |
| OR Per Allele (95% CI)a | 1.10(1.08-1.12) | 1.07(1.06-1.09) | |
| P-value | 2.71×10−25 | 1.02×10−16 | |
| Quartiles of Risk Allelesb |
|||
| Q1 | n (cases/controls) |
118/258 | 135/262 |
| OR(95% CI) | 1.0(ref.) | 1.0(ref.) | |
| Q2 | n (cases/controls) |
206/254 | 222/262 |
| OR(95% CI) | 1.77(1.33-2.36) | 1.64(1.24-2.16) | |
| P-value | 8.94×10−5 | 4.61×10−4 | |
| Q3 | n (cases/controls) |
262/264 | 299/265 |
| OR(95% CI) | 2.16(1.64-2.86) | 2.21(1.69-2.89) | |
| P-value | 5.77×10−8 | 6.55×10−9 | |
| Q4 | n (cases/controls) |
447/266 | 387/268 |
| OR(95% CI) | 3.68(2.83-4.82) | 2.80(2.16-3.65) | |
| P-value | 1.17×10−21 | 1.10×10−14 |
Odds ratios (and 95% confidence intervals) adjusted for age and 1st 10 eigenvalues
Quartiles based on distribution in controls
Given the strongest associations at 8q24 noted in Japanese in stage 1, we also examined the effects of a risk score composed of only 11 variants at chromosome 8q24 (see Methods; Supplemental Table 5). The associations of the 8q24 risk score were greater in each population than the risk score comprised of all prostate cancer variants, highlighting the importance of this region in these populations. For Japanese, a 1.16-fold increased risk of prostate cancer was observed for each additional 8q24 risk allele (P=8.75×10−19); while for Latinos, a 1.10-fold increased risk of disease was seen (P=1.83×10−6). There was little evidence of heterogeneity in effects of the 8q24 risk score across race/ethnicity (Phet=0.15).
A risk score composed of risk variants outside of the 8q24 locus (SNPs=45) was associated with an 8% and 7% increased risk of disease, per additional risk allele, for Japanese (P=8.75×10−13) and Latinos (P=1.03×10−12), respectively (Phet for race/ethnicity=0.42).
DISCUSSION
In this GWAS of prostate cancer in Japanese and Latinos, two populations that experience the lowest incidence rates of prostate cancer in the United States, we did not identify novel risk variants that reached genome-wide significance. We did observe that the vast majority of the known prostate cancer risk variants were positively associated with risk, which extends our previous findings in these two populations (15). Specifically, effect estimates were >1 for 79% and 88% of the risk variants tested among Japanese and Latinos, respectively, suggesting that these markers are likely correlated with the biologically functional alleles in these populations. We also determined that, in aggregate, these variants significantly contribute to prostate cancer susceptibility in each population with each additional risk allele associated with a 10% and 7% increased risk of prostate cancer in Japanese and Latinos, respectively.
The inclusion of minorities in previous GWAS of prostate cancer has been notably absent. Of the 16 reports of GWAS of prostate cancer (2-8, 18, 19, 26, 33-37), only two studies have focused on minorities in the discovery stage, one of Japanese (19) and the other of African Americans (18), with the remaining reports limited to men of European ancestry (2-8, 26, 33-37). In the GWAS of prostate cancer in Japanese (cases/controls = 4,584/8,801) (19), five novel loci were identified (19). In the GWAS of prostate cancer in African Americans, a novel risk variant at 17q21-ZNF652 (18) was identified that is unique to men of African ancestry, suggesting that some prostate cancer risk variants may be population-specific. These findings from GWAS of prostate cancer in non-Europeans emphasize the importance of broadening GWAS to diverse populations to ensure the discovery of the complete spectrum of prostate cancer risk alleles. Whereas our GWAS of Japanese and Latinos did not identify novel loci for these two populations, we recognize that our sample size was smaller than contemporary GWAS; thus, limiting our ability to detect modest association signals. In addition, because of the lack of additional studies of prostate cancer in Latinos, we were unable to replicate our stage 1 findings in Latino populations and made use of available European data. The Latinos in the MEC are predominantly from Mexico and are highly admixed with Native American (38%), European (59%), and African (3%) ancestry (38). While replication testing of the most significant findings in Europeans allowed for discovery of alleles that are common in European groups, we may have missed alleles that may be important to Latinos. Additional large genetic studies of prostate cancer in Latinos will be needed to search for risk alleles that are more common in Native American populations.
Only a small number of studies have investigated the known prostate cancer risk variants among Asians and Latinos (1, 13, 14, 18, 19, 39). For Asians, only two small Japanese studies have examined risk variants of prostate cancer (14, 40) separate from the MEC’s previous smaller reports (in sample size and number of SNPs) while a larger study of Chinese men has recently been conducted (15, 18, 41). Yamada et al. observed one variant at 3p12 (rs2660753) and 6 variants at chromosome 8q24 (rs13254738, rs6983561, rs16901979, rs1447295, rs10090154, and rs4430796) were associated with prostate cancer risk in 311 Japanese prostate cancer cases and 1,035 controls (14). Terada et al. reported an association between rs6983267 at 8q24 among 507 Japanese prostate cancer cases and 511 controls (40). Of the five novel risk loci (rs13385191, rs12653946, rs1983891, rs339331, and rs9600079) identified by the GWAS of prostate cancer in Japanese (19), we observed positive associations with all five variants and replicated significant associations with three of the risk variants (rs12653946, rs339331, and rs9600079). For the association at 3p12 (rs2660753) reported by Yamada et al. (14), we observed a non-significant positive association with the risk allele of rs2660753 among Japanese (OR=1.14; P=0.084) and a significant association in Latinos (OR=1.23; P=7.2×10−3). Of the previous 8q24 associations in these Japanese studies (14, 40), our findings in this larger MEC study confirm that there are multiple association signals at 8q24 (1, 42). Wang et al. in a study of Chinese men (41) examined the five prostate cancer risk variants identified in the Japanese GWAS of prostate cancer (19). Three of these risk variants (rs12653946, rs339331, and rs9600079) were associated with prostate cancer in Chinese men, providing evidence that some risk loci found in Japanese generalize to Chinese men (41). For Latinos, only one additional study outside of the MEC has reported the effects of prostate cancer risk variants (43). In this study of 196 Latino prostate cancer cases and 472 controls, 12 SNPs at 8q24 were associated with prostate cancer risk (43). Overall, aside from our reduced power to detect the originally reported effect estimates of small magnitude (Supplemental Table 6), our study was able to show positive associations for the majority of risk variants (> ~80%) among Japanese and Latinos. Moreover, our study not only corroborates previous reports (14, 19, 40, 43), but also provides the largest and most comprehensive evaluation to date of known prostate cancer risk variants and their cumulative genetic effect among Japanese and Latinos.
Given adequate statistical power, there are many questions directed towards understanding the reproducibility of risk variants across populations. There are three possible scenarios to consider. First, the disease locus identified by GWAS of European populations may not be relevant in other populations because the functional allele is limited to Europeans. Second, the locus is important in other populations, however a different variant (not the index SNP) is better in capturing risk in specific racial/ethnic populations as patterns of LD may vary between the index variant and functional allele across ancestral groups. Thus, fine-mapping of risk loci in different racial/ethnic groups could identify the most appropriate variant for a particular population. Lastly, the index risk variant identified in GWAS of Europeans is similarly associated with risk in other racial/ethnic groups. Directional consistency of an association for a given index signal across populations implies a shared functional common variant in each region and provides little support for the “synthetic association” model (44), which suggests that GWAS signals with common alleles are due to rare alleles, many of which are likely to be ethnically distinct. For the majority of the risk loci examined in this study, our observations support the existence of a common functional variant that is shared across populations.
As more prostate cancer risk variants are identified, the cumulative effects of these variants may have important clinical implications. With the 56 risk variants we examined, both Japanese and Latinos at the top quartile of the risk distribution had a highly significant ~3-fold increased risk of prostate cancer in comparison to those at the lowest quartile. In the absence of an established risk model of prostate cancer analogous to the Gail model for breast cancer (45), as more risk variants are identified, a SNP based risk model for prostate cancer may serve as a useful tool to define high-risk populations for targeted screening regimens and may better inform clinical decision making. Such models in development incorporate SNPs and family history in predicting prostate cancer risk (46). For individuals not at the high end of a genetic risk score, the clinical usefulness of such genetic information is unclear. Given the potential risks and costs associated with prostate cancer screening (47), these men may be less inclined to seek screening.
In summary, we did not identify novel genome-wide significant prostate cancer loci for Japanese and Latino men. However, we established that known risk variants for prostate cancer contribute to prostate cancer risk susceptibility in these populations. The challenge remains to conduct large well-powered genome-wide scans and follow-up studies in diverse populations to further dissect the complete array of risk alleles that may contribute to prostate cancer across populations.
Supplementary Material
Acknowledgements
This work was supported by the National Institute of Health/National Human Genome Research Institute [U01 HG004726-01] and the Genome Coordinating Center was supported by U01 HG004446. We thank Lauren Hu for her contributions to this project.
This work was conducted as a part of the BioBank Japan Project that was supported by the Ministry of Education, Culture, Sports, Sciences and Technology of the Japanese government, and was supported in part by Research grant #22390306 (H. Nakagawa) from the Japan Society for the Promotion of Science, and by the Princess Takamatsu Cancer Research Fund (H. Nakagawa).
This work was also supported by Cancer Research UK Grants C5047/A7357, C1287/A10118, C5047/A3354, C5047/A10692, C16913/A6135, and C16913/A6835 and NIH grant U19 CA 148537-01.
We would also like to thank the following for funding support: The Institute of Cancer Research and The Everyman Campaign, The Prostate Cancer Research Foundation, Prostate Research Campaign UK (now Prostate Action), The National Cancer Research Network UK, The National Cancer Research Institute (NCRI) UK. We are grateful for support of NIHR funding to the NIHR Biomedical Research Centre at The Institute of Cancer Research and The Royal Marsden NHS Foundation Trust.
We are grateful to staff at the Welcome Trust Clinical Research Facility, Addenbrooke’s Clinical Research Centre, Cambridge, UK for their help in conducting the ProtecT study. We also acknowledge the support of the NIHR Cambridge Biomedical Research Centre, the DOH HTA (ProtecT grant) and the NCRI / MRC (ProMPT grant) for help with the bio-repository. The UK Department of Health funded the ProtecT study through the NIHR Health Technology Assessment Programme (projects 96/20/06, 96/20/99). The ProtecT trial and its linked ProMPT and CAP (Comparison Arm for ProtecT) studies are supported by Department of Health, England; Cancer Research UK grant number C522/A8649, Medical Research Council of England grant number G0500966, ID 75466 and The NCRI, UK. DNA extraction in ProtecT was supported by USA Dept of Defense award W81XWH-04-1-0280, Yorkshire Cancer Research and Cancer Research UK. The authors would like to acknowledge the contribution of all members of the ProtecT study research group. The views and opinions expressed therein are those of the authors and do not necessarily reflect those of the Department of Health of England.
We should like to acknowledge the NCRN nurses, data managers and Consultants for their work in the UKGPCS study. The bio-repository from ProtecT is supported by the NCRI (ProMPT) Prostate Cancer Collaborative and the Cambridge BMRC grant from NIHR.
REFERENCES
- 1.Haiman CA, Patterson N, Freedman ML, Myers SR, Pike MC, Waliszewska A, et al. Multiple regions within 8q24 independently affect risk for prostate cancer. Nat Genet. 2007;39:638–44. doi: 10.1038/ng2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gudmundsson J, Sulem P, Gudbjartsson DF, Blondal T, Gylfason A, Agnarsson BA, et al. Genome-wide association and replication studies identify four variants associated with prostate cancer susceptibility. Nat Genet. 2009;41:1122–6. doi: 10.1038/ng.448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gudmundsson J, Sulem P, Manolescu A, Amundadottir LT, Gudbjartsson D, Helgason A, et al. Genome-wide association study identifies a second prostate cancer susceptibility variant at 8q24. Nat Genet. 2007;39:631–7. doi: 10.1038/ng1999. [DOI] [PubMed] [Google Scholar]
- 4.Gudmundsson J, Sulem P, Rafnar T, Bergthorsson JT, Manolescu A, Gudbjartsson D, et al. Common sequence variants on 2p15 and Xp11.22 confer susceptibility to prostate cancer. Nat Genet. 2008;40:281–3. doi: 10.1038/ng.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gudmundsson J, Sulem P, Steinthorsdottir V, Bergthorsson JT, Thorleifsson G, Manolescu A, et al. Two variants on chromosome 17 confer prostate cancer risk, and the one in TCF2 protects against type 2 diabetes. Nat Genet. 2007;39:977–83. doi: 10.1038/ng2062. [DOI] [PubMed] [Google Scholar]
- 6.Eeles RA, Kote-Jarai Z, Al Olama AA, Giles GG, Guy M, Severi G, et al. Identification of seven new prostate cancer susceptibility loci through a genome-wide association study. Nat Genet. 2009;41:1116–21. doi: 10.1038/ng.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eeles RA, Kote-Jarai Z, Giles GG, Olama AA, Guy M, Jugurnauth SK, et al. Multiple newly identified loci associated with prostate cancer susceptibility. Nat Genet. 2008;40:316–21. doi: 10.1038/ng.90. [DOI] [PubMed] [Google Scholar]
- 8.Thomas G, Jacobs KB, Yeager M, Kraft P, Wacholder S, Orr N, et al. Multiple loci identified in a genome-wide association study of prostate cancer. Nat Genet. 2008;40:310–5. doi: 10.1038/ng.91. [DOI] [PubMed] [Google Scholar]
- 9.Yeager M, Xiao N, Hayes RB, Bouffard P, Desany B, Burdett L, et al. Comprehensive resequence analysis of a 136 kb region of human chromosome 8q24 associated with prostate and colon cancers. Hum Genet. 2008;124:161–70. doi: 10.1007/s00439-008-0535-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Al Olama AA, Kote-Jarai Z, Giles GG, Guy M, Morrison J, Severi G, et al. Multiple loci on 8q24 associated with prostate cancer susceptibility. Nat Genet. 2009;41:1058–60. doi: 10.1038/ng.452. [DOI] [PubMed] [Google Scholar]
- 11.Lindstrom S, Schumacher FR, Campa D, Albanes D, Andriole G, Berndt SI, et al. Replication of Five Prostate Cancer Loci Identified in an Asian Population--Results from the NCI Breast and Prostate Cancer Cohort Consortium (BPC3) Cancer Epidemiol Biomarkers Prev. 2011 doi: 10.1158/1055-9965.EPI-11-0870-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haiman CA, Chen GK, Blot WJ, Strom SS, Berndt SI, Kittles RA, et al. Characterizing genetic risk at known prostate cancer susceptibility loci in African Americans. PLoS Genet. 2011;7:e1001387. doi: 10.1371/journal.pgen.1001387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang BL, Spangler E, Gallagher S, Haiman CA, Henderson B, Isaacs W, et al. Validation of genome-wide prostate cancer associations in men of African descent. Cancer Epidemiol Biomarkers Prev. 2011;20:23–32. doi: 10.1158/1055-9965.EPI-10-0698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamada H, Penney KL, Takahashi H, Katoh T, Yamano Y, Yamakado M, et al. Replication of prostate cancer risk loci in a Japanese case-control association study. J Natl Cancer Inst. 2009;101:1330–6. doi: 10.1093/jnci/djp287. [DOI] [PubMed] [Google Scholar]
- 15.Waters KM, Le Marchand L, Kolonel LN, Monroe KR, Stram DO, Henderson BE, et al. Generalizability of associations from prostate cancer genome-wide association studies in multiple populations. Cancer Epidemiol Biomarkers Prev. 2009;18:1285–9. doi: 10.1158/1055-9965.EPI-08-1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu J, Kibel AS, Hu JJ, Turner AR, Pruett K, Zheng SL, et al. Prostate cancer risk associated loci in African Americans. Cancer Epidemiol Biomarkers Prev. 2009;18:2145–9. doi: 10.1158/1055-9965.EPI-09-0091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheng I, Plummer SJ, Jorgenson E, Liu X, Rybicki BA, Casey G, et al. 8q24 and prostate cancer: association with advanced disease and meta-analysis. Eur J Hum Genet. 2008;16:496–505. doi: 10.1038/sj.ejhg.5201959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haiman CA, Chen GK, Blot WJ, Strom SS, Berndt SI, Kittles RA, et al. Genome-wide association study of prostate cancer in men of African ancestry identifies a susceptibility locus at 17q21. Nat Genet. 2011;43:570–3. doi: 10.1038/ng.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takata R, Akamatsu S, Kubo M, Takahashi A, Hosono N, Kawaguchi T, et al. Genome-wide association study identifies five new susceptibility loci for prostate cancer in the Japanese population. Nat Genet. 2010;42:751–4. doi: 10.1038/ng.635. [DOI] [PubMed] [Google Scholar]
- 20.Kolonel LN, Henderson BE, Hankin JH, Nomura AM, Wilkens LR, Pike MC, et al. A multiethnic cohort in Hawaii and Los Angeles: baseline characteristics. Am J Epidemiol. 2000;151:346–57. doi: 10.1093/oxfordjournals.aje.a010213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–9. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 22.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–1. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jia L, Landan G, Pomerantz M, Jaschek R, Herman P, Reich D, et al. Functional enhancers at the gene-poor 8q24 cancer-linked locus. PLoS Genet. 2009;5:e1000597. doi: 10.1371/journal.pgen.1000597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yeager M, Chatterjee N, Ciampa J, Jacobs KB, Gonzalez-Bosquet J, Hayes RB, et al. Identification of a new prostate cancer susceptibility locus on chromosome 8q24. Nat Genet. 2009;41:1055–7. doi: 10.1038/ng.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yeager M, Orr N, Hayes RB, Jacobs KB, Kraft P, Wacholder S, et al. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat Genet. 2007;39:645–9. doi: 10.1038/ng2022. [DOI] [PubMed] [Google Scholar]
- 27.Rafnar T, Sulem P, Stacey SN, Geller F, Gudmundsson J, Sigurdsson A, et al. Sequence variants at the TERT-CLPTM1L locus associate with many cancer types. Nat Genet. 2009;41:221–7. doi: 10.1038/ng.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zheng SL, Sun J, Cheng Y, Li G, Hsu FC, Zhu Y, et al. Association between two unlinked loci at 8q24 and prostate cancer risk among European Americans. J Natl Cancer Inst. 2007;99:1525–33. doi: 10.1093/jnci/djm169. [DOI] [PubMed] [Google Scholar]
- 29.Zheng SL, Stevens VL, Wiklund F, Isaacs SD, Sun J, Smith S, et al. Two independent prostate cancer risk-associated Loci at 11q13. Cancer Epidemiol Biomarkers Prev. 2009;18:1815–20. doi: 10.1158/1055-9965.EPI-08-0983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu J, Zheng SL, Isaacs SD, Wiley KE, Wiklund F, Sun J, et al. Inherited genetic variant predisposes to aggressive but not indolent prostate cancer. Proc Natl Acad Sci U S A. 2010;107:2136–40. doi: 10.1073/pnas.0914061107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun J, Zheng SL, Wiklund F, Isaacs SD, Purcell LD, Gao Z, et al. Evidence for two independent prostate cancer risk-associated loci in the HNF1B gene at 17q12. Nat Genet. 2008;40:1153–5. doi: 10.1038/ng.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takata R, Akamatsu S, Kubo M, Takahashi A, Hosono N, Kawaguchi T, et al. Genome-wide association study identifies five new susceptibility loci for prostate cancer in the Japanese population. Nat Genet. 42:751–4. doi: 10.1038/ng.635. [DOI] [PubMed] [Google Scholar]
- 33.Kote-Jarai Z, Olama AA, Giles GG, Severi G, Schleutker J, Weischer M, et al. Seven prostate cancer susceptibility loci identified by a multi-stage genome-wide association study. Nat Genet. 2011;43:785–91. doi: 10.1038/ng.882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schumacher FR, Berndt SI, Siddiq A, Jacobs KB, Wang Z, Lindstrom S, et al. Genome-wide association study identifies new prostate cancer susceptibility loci. Hum Mol Genet. 2011;20:3867–75. doi: 10.1093/hmg/ddr295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun J, Zheng SL, Wiklund F, Isaacs SD, Li G, Wiley KE, et al. Sequence variants at 22q13 are associated with prostate cancer risk. Cancer Res. 2009;69:10–5. doi: 10.1158/0008-5472.CAN-08-3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Duggan D, Zheng SL, Knowlton M, Benitez D, Dimitrov L, Wiklund F, et al. Two genome-wide association studies of aggressive prostate cancer implicate putative prostate tumor suppressor gene DAB2IP. J Natl Cancer Inst. 2007;99:1836–44. doi: 10.1093/jnci/djm250. [DOI] [PubMed] [Google Scholar]
- 37.Murabito JM, Rosenberg CL, Finger D, Kreger BE, Levy D, Splansky GL, et al. A genome-wide association study of breast and prostate cancer in the NHLBI’s Framingham Heart Study. BMC Med Genet. 2007;8(Suppl 1):S6. doi: 10.1186/1471-2350-8-S1-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang H, Haiman CA, Kolonel LN, Henderson BE, Wilkens LR, Le Marchand L, et al. Self-reported ethnicity, genetic structure and the impact of population stratification in a multiethnic study. Hum Genet. 2010;128:165–77. doi: 10.1007/s00439-010-0841-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou CH, Wang JY, Cao SY, Shi XH, Zhang YG, Liu M, et al. Association between single nucleotide polymorphisms on chromosome 17q and the risk of prostate cancer in a Chinese population. Chinese journal of cancer. 2011;30:721–30. doi: 10.5732/cjc.011.10070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Terada N, Tsuchiya N, Ma Z, Shimizu Y, Kobayashi T, Nakamura E, et al. Association of genetic polymorphisms at 8q24 with the risk of prostate cancer in a Japanese population. Prostate. 2008;68:1689–95. doi: 10.1002/pros.20831. [DOI] [PubMed] [Google Scholar]
- 41.Wang M, Liu F, Hsing AW, Wang X, Shao Q, Qi J, et al. Replication and cumulative effects of GWAS-identified genetic variations for prostate cancer in Asians: a case-control study in the ChinaPCa consortium. Carcinogenesis. 2012;33:356–60. doi: 10.1093/carcin/bgr279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Freedman ML, Haiman CA, Patterson N, McDonald GJ, Tandon A, Waliszewska A, et al. Admixture mapping identifies 8q24 as a prostate cancer risk locus in African-American men. Proc Natl Acad Sci U S A. 2006;103:14068–73. doi: 10.1073/pnas.0605832103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beuten J, Gelfond JA, Martinez-Fierro ML, Weldon KS, Crandall AC, Rojas-Martinez A, et al. Association of chromosome 8q variants with prostate cancer risk in Caucasian and Hispanic men. Carcinogenesis. 2009;30:1372–9. doi: 10.1093/carcin/bgp148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dickson SP, Wang K, Krantz I, Hakonarson H, Goldstein DB. Rare variants create synthetic genome-wide associations. PLoS Biol. 8:e1000294. doi: 10.1371/journal.pbio.1000294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gail MH, Greene MH. Gail model and breast cancer. Lancet. 2000;355:1017. doi: 10.1016/S0140-6736(05)74761-9. [DOI] [PubMed] [Google Scholar]
- 46.Macinnis RJ, Antoniou AC, Eeles RA, Severi G, Al Olama AA, McGuffog L, et al. A risk prediction algorithm based on family history and common genetic variants: application to prostate cancer with potential clinical impact. Genet Epidemiol. 35:549–56. doi: 10.1002/gepi.20605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brett AS, Ablin RJ. Prostate-cancer screening--what the U.S. Preventive Services Task Force left out. N Engl J Med. 365:1949–51. doi: 10.1056/NEJMp1112191. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.