Abstract
Taming a cyanobacterium in a pivitol event of endosymbiosis brought photosynthesis to eukaryotes, and gave rise to the plastids found in glaucophytes, red and green algae, and the descendants of the latter, the plants. Ultrastructural as well as molecular research over the last two decades has demonstrated that plastids have enjoyed surprising lateral mobility across the tree of life. Numerous independent secondary and tertiary endosymbiosis have led to a spread of plastids into a variety of, up to that point, non-photosynthetic lineages. Happily eating and subsequently domesticating one another protists conquered a wide variety of ecological niches. The elaborate evolution of secondary, or complex, plastids is reflected in the numerous membranes that bound them (three or four compared to the two membranes of the primary plastids). Gene transfer to the host nucleus is a hallmark of endosymbiosis and provides centralized cellular control. Here we review how these proteins find their way back into the stroma of the organelle and describe the advances in the understanding of the molecular mechanisms that allow protein translocation across four membranes.
The history of complex plastids
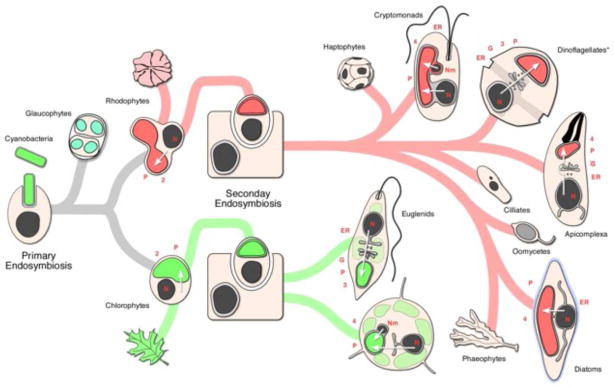
Endosymbiosis of a cyanobacterium and a heterotrophic eukaryote gave rise to primary plastids (Fig. 1). In much the same way the engulfment and subsequent domestication of an algal cell by another eukaryote resulted in the formation of complex plastids. There is now a broad consensus that primary plastids, found in the three lineages glaucophytes, red algae and green algae (from which land plants have developed), are monophyletic [1–4]. In contrast, multiple independent endosymbiotic events led to the formation of complex plastids. Chlorarachniophytes and euglenoids independently acquired green algal symbionts, which gave rise to their complex plastids (Fig 1) [5]. Chlorarachniophytes are single celled algae, some are entirely photosynthetic while others are mixotrophic and also ingest smaller organisms [6]. Euglenoids are mostly unicellular flagellated protists, with an even more divergent range of feeding habits: while many have plastid and are photosynthetic, others are phagotrophic, and some have no trace of plastids or have lost photosynthesis but retain a colourless plastid with a reduced genome [6].
Figure 1. Schematic flowchart representing the evolution of complex plastid.
A single endosymbiosis event gave raise to the primary plastids found in red and green algae and in glaucophytes. At least three separate events of secondary endosymbiosis gave raise to the complex plastids found in clorarachniophytes, euglenoids (green path) and in members of the chromalveolate superphylum (red path). Representative organisms for each of the resulting lineages are depicted as schemes and their corresponding cellular morphology is outlined. P; plastid, Number; number of plastid membranes, N; nucleaus, Nm; nucleomorph, G: Golgi apparatus, ER; endoplasmic reticulum. White arrows mark the pathway taken by nuclear/nucleomorph encoded proteins to the complex plastid in each organism. * Note that in dinoflagellates there is evidence for several independent secondary and tertiary endosymbioses that are not shown here. These involved green algal, haptophye, diatom and cryptomonad symbionts. See [88] for further reference to this fascinating complexity.
The complex plastid found in haptophytes, cryptomonads, heterokonts, dinoflagellates and apicomplexans, is of red algal origin [7]. While these algal taxa represent a wide diversity of nutritional modes and of cell types and structures, together they compose the eukaryotic superphylum chromalveolates [7]. The chromalveolate hypothesis suggests that a single endosymbiosis with a red alga gave rise to all these lineages [7]. Numerous studies have put this hypothesis to the test and, while there is significant support for its broad concept, some of the details remain the source of debate (reviewed in [8, 9]). The common origin of heterokonts, cryptophytes and haptophytes [5, 10, 11] is strongly supported by their plastid gene phylogenies. Such a relationship was initially not obvious for apicomplexans and dinoflagellates, as their plastid genomes are highly divergent. The substitution of apicomplexans cyanobacterial-type plastid glyceraldehyde-3-phosphate dehydrogenase (GAPDH) with a duplication of a eukaryotic GAPDH [12] argues in favor of the affiliation with chromalveolates. A similar duplication and takeover of the eukaryotic gene occurred in the case of fructose bisphosphate aldolase (FBA), this likely happened at a much earlier point, before the split of the red and green algal lineages [13]. However, chromalveolates share a distinct type of FBA that is now targeted to their plastids [13]. The recent discovery of a closely related yet still photosynthetic sister of Apicomplexa, Chromera velia [14], provided additional insights. The sequence of the plastid genome of Chromera enabled the demonstration of the phylogenetic link between the plastids of apicomplexans, dinoflagellates, and heterokonts [15]. New sequence data for the haptophyte Emiliana huxleyi prompted Felsner and coworkers to analyze the phylogeny of certain protein transport systems (more detail below). Their analysis is consistent with a monophyletic rhodophyte origin to the chromalveolate plastid. Surprizingly, other aspects of their study focused on a related host system localized to the endoplasmic reticulum linked cryptophytes and haptophytes with the green lineage [16]. The latter observation and other studies [17, 18] point to the possibility of host polyphyly among chromalveolates sub groups. One explanation may be that the recruitment of translocation machinery from rhodophytes occurred more than once, leading to a potentially more complex model of the origin of chromalveolates [16].
Complex plastids have numerous compartments of divergent origin
The evolution of plastids, both primary and secondary, goes hand in hand with the progressive loss of structural characters of the symbiont. These previously served functions that are now taken over by the host. In the case of the algal endosymbiont this loss includes organelles associated with secretion such as the endoplasmic reticulum (ER), the Golgi, and energy metabolism, like the mitochondrion and peroxisome. In most lineages the algal nucleus is also lost, however note that there are some highly informative exceptions that will be discussed below. In most cases, all that remains is the ancient primary plastid surrounded by its two membranes as well as one or two additional membranes around it.
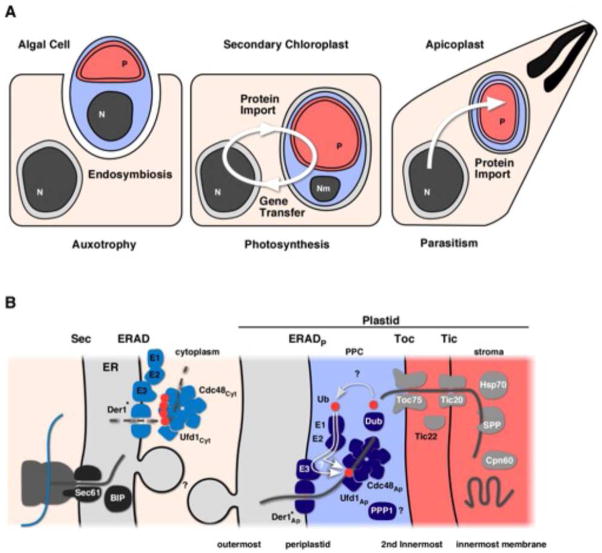
Uptake of the algal cell into a vacuole of the host’s endomembrane system likely represented the initial step of secondary endosymbiosis [19–21] (Fig. 2A). Therefore the outermost membrane (OMM) of complex plastids with four membranes is likely of host endomembrane origin. In fact, in haptohpytes, cryptomonads and heterokonts the outer membrane is continuous with the ER and the nuclear envelope (Fig. 1). The next membrane, the periplastid membrane (PPM), bounds the periplastid compartment (PPC). This compartment is the remainder of the algal cytoplasm and the PPM is likely a derivative of the algal plasma membrane. Finally, the two innermost membranes are thought to be equivalent to the two envelope membranes of the original algal chloroplast. Tight physical apposition between these two membranes, reminiscent of the original chloroplast, was observed in electron tomograms of secondary plastids [22].
Figure 2. The origin of plastid membranes and the translocons that allow proteins to cross them.
(A) Schematic outline of the acquisition and evolution of a complex plastid (using the apicoplast as an example). An algal cell (blue) carrying a chloroplast (red) is taken up into the endomembrane system (gray) of a protist host (pink). Co-adaptation lead to the establishment of complex protein import systems across each membrane allowing for a stable endosymbiotic relationship driven by massive gene transfer. P; Plastid, N; nucleus, Nm; nucleomorph. (B) Schematic outline of the protein import pathways of complex plastids of the red lineage (color coding for origin matches panel A, note that his only partially reflects systems in the green lineages). Question marks highlight elements that have not been experimentally validated or where the direct molecular function of a given protein remains to be established. *Note that Der1 is only one of several proteins that are considered candidates for the actual pore of the translocon, we only show one hypothesis for simplicity. Cargo proteins are shown as grey lines and proteins destined for degradation as dashed lines. PPC; the periplastid compartment.
It should be noted that there are complex plastids that are bound by only three membranes. This is the case for most photosynthetic dinoflagellates, and also for the green plastids of phototrophic euglenoids [7]. Some authors have suggested that this is a consequence of the route of uptake, in particular the use of myzocytosis, a specialized feeding strategy used by dinoflagellates and euglenoids. During myzocytosis the predator attaches to its prey cell without engulfing it and sucks its cellular contents into a digestive vacuole leaving behind the emptied wall and plasma membrane of the prey organism (reviewed in [23]). An alternative model suggests more conventional endocytosis followed by the loss of either the membrane of the food vacuole, or the algal plasma membrane [24].
Plastid proteins by and large are encoded in the nuclear genome
The transition from endosymbiont to organelle was accompanied by massive gene transfer from symbiont to host [25]. This is equally true for primary and secondary endosymbioses. This allows centralized control over metabolic function and inheritance. While in most cases the endosymbiont’s nuclear genome has been lost entirely, in cryptomonads and chlorarchniophytes, a remnant of the algal nucleus, the nucleomorph, was retained in the PPC [26, 27]. The nucleomorph genomes of both groups are highly reduced. Interestingly, despite their independent origin, the nucleomorph genomes of cryptomonads and chlorarachniophytes share a core of conserved genes suggesting a similar overall route of progressive reduction [28].
All plastids, primary and secondary, have maintained an organellar genome localized to the stroma (lumen) of the organelle, these genomes vary in size and complexity. The non-photosynthetic plastid of apicomplexan parasites, the apicoplast, is an interesting example in which all that remained from an ancestral genome of a photosynthetic plastid are 35Kb, essentially all of the genes are concerned with transcribing and translating the genome [29]. Regardless of size, all plastid organellar genomes encode a number of proteins that appears to be much smaller than the proteome size required for a fully functional organelle. In the extreme case of the apicoplast of Plasmodium falciparum, 32 protein encoding genes are found in the organelle genome, however the entire organellar proteome is predicted to include about 450 proteins [30]. As organellar proteins are encoded in the nucleus and translated on cytosolic ribosomes, there is an obvious need for significant protein import across the organelle’s numerous membranes. The developing picture describing this route is one of evolutionary spatial conservatism. The signals and machinery required for the journey through each of the compartments of the complex organelle largely originate from the very organism that gave rise to the compartment during the organelle’s evolution (Fig. 2B).
Signals for organellar targeting
Accurate trafficking of nuclear encoded proteins to the stroma of primary plastids is controlled by a characteristic N-terminal signal sequence named the transit peptide (TP). Many of proteins of the outer compartments insert into the membrane without such a targeting peptide. The transit peptide shows no strict consensus sequence, but is characterized by common features such as a positive net charge and a high frequency of hydroxylated amino acids (reviewed in [31]). Similarly, most nuclear encoded proteins destined to complex plastids studied to date possess an N-terminal signal, named the bipartite targeting signal [32]. In accordance with the need to traverse one or two additional membranes, these peptides contain an additional element, a sequence equivalent to the signal peptide (SP) of secretory proteins. The journey thus commences with entry to the secretory system, likely by the mechanism described for a large variety of secretory proteins: courtesy of the Sec61 translocon [33]. The SP element in the bipartite leader peptid has been studied in detail for proteins that target to the apicoplast (the non-photynthetic plastid of apicomplexan parasites). The signal was shown to be sufficient to guide reporter proteins to the secretory system [30, 34–36]. The sequence can be replaced with a canonical SP from a secretory protein, this chimera still confers plastid targeting of a reporter [37]. The SP is likely removed cotranslationally [38]. These first steps occur rapidly and may conclude before the synthesis of a full-length protein is concluded [38]. As a result of SP cleavage the TP is exposed and available to lead the rest of the way. Once the protein has reached the stroma the TP is proteolytically cleaved like the SP before [38]. Pulse-chase measurements in Apicomplexa showed that it takes 45–60 minutes from the moment of translation to TP removal [38]. A homolog of the plant chloroplast transit peptide peptidase was identified in the nuclear genome of the apicomplexans T. gondii and P. falciparum and proposed to execute this cleavage [38, 39]. Interestingly, the TP elements of complex plastid proteins from apicomplexans and cryptophytes were shown to be sufficient for targeting into isolated pea chloroplasts [34, 40] and cryptomonads peptide was substrate for the pea stromal processing peptidase [40].
The primary sequence of the apicoplast TP is not conserved, yet several features reminiscent of the plant chloroplast TP have been noted [30]. In apicomplexans it was demonstrated that an overall positive charge is essential for targeting to the apicoplast lumen, while the position of the positively charged amino acids in the TP can vary [30, 41, 42]. In addition, the compartmentalization of complex plastids imposes the need for specific signals to discriminate between proteins that continue all the way to the stroma from those that home to the outer compartments. The minute size of the apicoplast and the proximity of its four membranes have been a challenge to the study of these signals in Apicomplexa. However, in diatoms intermediates that are transported across only one or two of the four plastid membranes can be discriminated from those imported across all four, and studies in the model system Phaeodactylum tricornutum revealed a complex set of signals [43]. It was shown that the positive net charge of the TP is important to cross the two innermost membranes, and required to cross the second outermost membrane into the PPC [44]. Negative charges in the TP were shown to inhibit entry into the PPC of diatoms [44], but in contrast are required for PPC targeting in chlorarachniophytes [45]. This may represent a fundamental difference in these pathways originating from different algal lineages. In diatoms and in cryptophytes the presence of a specific residue, a highly conserved aromatic amino acid at position +1 of the TP, is crucial for import into the lumen, and the lack of this residue results in PPC residence [43, 46, 47]. Finally, it was shown that the sequences outside of the TP e.g. within the N-terminal part of the mature protein can contribute to the correct targeting of certain proteins [44].
N-terminal leader sequences have also been characterized for the proteins targeting to the three membrane bounded plastids of euglenoids and dinoflagellates. Bioinformatic analyses in euglenoids revealed two classes of plastid targeted proteins. In addition to those with bipartite signals there are those with tripartite leaders. In the latter, a third, usually hydrophobic, domain is present that acts as a stop transfer signal and allows the protein to be transported as an integral membrane protein [48]. This model is in agreement with earlier experimental evidence from Euglena [49, 50]. A recent study identified short introns within regions encoding the complex presequences of Euglena plastid proteins [51]. This likely indicates the involvement of introns and exon-shuffling in the acquisition of these targeting signals [51]. Interestingly, dinoflagellate plastid proteins feature leader sequences similar to those of Euglena [52, 53] indicating a potential link to the three-membrane topology of these plastids.
While most proteins destined to complex plastids are led by their N-terminal sequence, some proteins seem to possess a non-canonical targeting signal. Several examples have emerged recently from Apicomplexa. Among those are apicoplast proteins that lack a signal peptide at the immediate N- terminus but rather show a recessed hydrophobic patch [54–56]. Apicoplast proteins with multiple membrane spanning domains also often lack an obvious signal peptide [57–61].
The long way in – multiple translocons for multiple membranes
The two innermost membranes
In primary chloroplasts protein import is mediated by two protein complexes, the translocon of the outer chloroplast membrane (Toc) and the translocon of the inner chloroplast membrane (Tic) (Fig. 2B). These were defined in the plant chloroplast, and are composed of numerous proteins, some of which are likely derived from proteins already present in the cyanobacterial ancestor (see [31] for a thorough review). In agreement with their proposed algal chloroplast origin, the two inner membranes of complex plastids seem to rely on Tic and Toc derivates (reviewed in [62] and below). The identification of homologs of their components in the nuclear genomes of organisms with complex plastid, as well as in the nucleomorph genomes, support this notion: On the green side, the genome of the chlorarachniophyte alga Bigelowiella natans encodes homologs of at least six Tic components (TIC21/22/32/40/55/62, [63]) whose products join the Tic20 homolog encoded in its nucleomorph [64]. This model is supported by findings from the red lineage as well. Tic components were found to be encoded in the nucleomorph of the cryptophyte Guillardia theta [65] and in the nuclear genomes of several apicomplexans [62, 65, 66].
The most extensively characterized among these is a T. gondii homolog of Tic20. In the plant chloroplast Tic20 is an integral membrane protein involved in facilitating protein transport through the inner membrane [67, 68]. Van Dooren and coworkers, confirmed that TgTic20 is an integral membrane protein of the apicoplast, and demonstrated its specific localization in the inner most membrane using split-GFP reporters [69]. They further generated a conditional mutant of TgTic20, showing that this gene is essential for T. gondii survival, and demonstrated its involvement in protein import in vivo using pulse-chase import assays [69]. A second putative component of the apicomplexan Tic complex, the soluble protein Tic22, was localized to the apicoplast of P. falciparum [70]. In T. gondii TgTic22 also encodes apicoplast localized proteins and its loss results in a phenotype comparable to that observed for TgTic20 (Giel van Dooren, Swati Agrawal and Boris Striepen unpublished). Interestingly, a homolog of ClpC (Hsp93), a stromal chaperone component of the Tic complex, is encoded on the apicoplast genome both in P. falciparum and T. gondii [29, 71]. Addressing the potential role of ClpC in import (or as a peptidase) awaits the development of tools for genetic manipulation of the organellar genome.
Two critical components of the Toc complex in plants are the receptor protein Toc34 and the pore in the outer chloroplast membrane, Toc75 [31]. A Toc34 was identified in the genomes of green, red and secondary plastid containing algae of the red lineage [65]. In contrast, neither a Toc34 homolog, nor a homolog of the conserved cargo receptor, Toc159, were found in the genome of the chlorarachniophyte B. natans [63]. Interestingly, two proteins with similarity to Toc75 were found in this organism [63]. One is encoded on the nucleomorph genome [64], and a second, most similar to the cyanobacterial Omp85, is encoded in the nuclear genome [63]. The role of this second pore and its potential correlation with the absence of a receptor are unclear at this point. The search for Toc75 homologs in the genomes of organisms with complex plastids of the red lineage was more challenging, and an initial hypothesis was put forward that it might have been lost and replaced with another translocon [72][73]. Recently however, Bullmann and coworkers identified a homolog of Omp85 in the genome of the diatom P. tricornutum [74]. This gene is phylogenetically affiliated with the Toc75 group and encodes a protein with a bipartite signal [74]. Bullmann and coworkers demonstrated experimentally that PtOmp85 can act as a pore, and that it shares biochemical characteristics with plant the Toc75 [74]. Finally, this work has shown that both the N- and C- termini of PtOmp85 face the PPC [74]. This observation is consistent with a recent study showing that proteins of the chloroplast Omp85 family have changed their orientation as compared to their bacterial ancestors to present their polypeptide-transport–associated (POTRA) domain to the cytoplasm [75](or the PPC in the case of complex plastid). This way, the POTRA domain, whose affinity to precursor proteins is high, can take part in the perception of the targeting signal [75]. Homologs of the Phaeodactylum Opm85 are also found in the genomes of Plasmodium and Toxoplasma [62, 74] and our preliminary studies indicate that loss of this protein in Toxoplasma results in a pronounced apicoplast protein import defect (Agrawal, Brooks, Sheiner and Striepen unpublished).
Taken together, these observations make a compelling case for TIC and TOC derived translocons as mechanisms to cross the two innermost membranes of the secondary plastids.
The old algal plasma membrane
The PPM is thought to be derived from the plasma membrane of the algal endosymbiont, and establishment of protein import across this membrane was a critical early step in endosymbiosis [7]. The sequence of the cryptophyte G. theta nucleomorph genome revealed the crucial clue to the discovery of the machinery that breaches this barrier [76]. This genome encodes core elements of the endoplasmatic reticulum associated degradation (ERAD) system [76], this is despite the fact that an ER compartment was not reported in the PPC. Typically ERAD is responsible for retro-translocation of miss-folded proteins across the ER membrane into the cytoplasm, which is then followed by degradation by the proteasome [77]. Sommer and colleagues put forward the hypothesis that in cryptophytes the ERAD translocon had been retooled to import proteins into the PPC [76] (Fig. 2B). Consistent with this hypothesis it was found that other organisms with complex red plastids (but no nucleomorph) have duplicate sets of genes encoding ERAD components in their nuclear genome [16, 54, 70, 76, 78]. Several elements of the symbiont’s ERAD machinery have been characterized to date. These include the membrane protein Der-1, the AAA-ATPase Cdc48, and its cofactor, Ufd-1. In yeast and human cells ER-Der-1 is essential for the retro-translocation to occur, and it was hypothesized that it forms the translocation channel [79]. Recent data suggested it may aid the interaction with the E3 ubiquitin ligase (Hard1p) (reviewed in [77]).
The three ERAD core proteins localize to the periphery of complex plastids and more specifically to their PPMs and PPCs. This was shown using a variety of light and electron microscopy approaches in the apicomplexan T. gondii, the diatom P. tricornutum, and the haptophyte E. huxelyi [16, 54, 80]. Agrawal and colleagues generated a conditional mutant of the apicoplast Der1 [54]. Removal of Der1 results in ablation of apicoplast protein import, as measured using a variety of biochemical assays demonstrating a direct role of the ERAD system in import [54] and validating the initial hypothesis by Sommer and colleagues [76]. Importantly, the apicoplast ERAD machinery is derived from the ERAD system of the symbiont and does not represent a duplication of the host system [16, 54]. This observation is consistent with a symbiosis model under which the symbiont actively engaged the host by granting access to host synthesized proteins through modification of its membranes.
In the ER, proteins retro-translocated by the ERAD system are marked for degradation by conjugation of ubiquitin resulting in poly-ubiquitin chains [81]. This appears to occur once the protein is on the cytoplasmic side of the ER membrane. Cdc48 then extracts these substrate proteins from the pore with the help of its cofactors, the Ufd-1–Npl4 complex [79]. This ERAD-associated ubiquitination is critical not only for the subsequent degradation of the protein but also appears critical to the translocation step across the ER membrane itself [77]. Consistent with this, deletion of the Ufd-1 amino terminus, known to bind polyubiquitin, results in disruption of protein translocation across the ER membrane in yeast [82, 83]. Moreover, for degradation of ER-lumen proteins, ubiquitination alone seemed sufficient to mark extraction by Cdc48, as overexpression of the yeast ubiquitin E3 ligase, Hard1p, can bypass the need of other ERAD components such as Der1 [84].
This ubiquitin dependent translocation model may apply also to complex plastids of the red lineage: a series of putative ubiquitination enzymes was shown to target to the complex plastid of Apicomplexa and diatoms or to contain presequences likely to lead to such targeting [78, 85–87]. Functional data linking the enzymatic activity of plastid ERAD components to protein import is emerging (Agrawal, van Dooren and Striepen unpublished). It is interesting to note in this context that plastid-specific deubiquitinases have also been reported in P. tricornitum [85, 87]. The precise role of ubiquitination in the scheme of import in complex plastid remains to be fully defined.
The current evidence strongly supports a model of protein import across the PPM of red lineage complex plastids based on the remodeling of the symbiont’s ERAD machinery. In stark contrast, data emerging from complex plastids of the green lineage chlorarachniophyte, so far fail to detect ERAD duplication and retooling in these organisms [63]. Genes for ERAD components are absent form the nucleomorph genome of the chlorarachniophyte B. natans [64]. Furthermore, Hirakawa and coworkers could only detect two homologs of Der1 in the nuclear genome of this alga (in contrast to four homologs found in the genomes of organisms with complex plastid of red origin), and both were shown to reside in the ER [63]. The machinery mediating import through this membrane in complex plastid of the green lineage remains an enigma.
Two recent studies have applied broad bioinformatics screens to identify additional PPC proteins [56, 85]. Recently we have identified two new T. gondii PPC proteins that are conserved among the red algal lineage and encoded in the nucleomorph genomes of cryptomonads [56]. One of those, PPP1 was also localized to the PPC of the diatom P. tricornutum [85]. Interestingly, like the symbiont ERAD components, PPP1 have no homolog in the nucleomorph of B. natans or in the nuclear genomes of organisms from the green lineage [56]. Mutation of TgPPP1 resulted in a loss of apicoplast protein import leading to apicoplast demise and ultimately to cell death. Overall the phenotype of these mutants is reminiscent of that found for the TgDer1 mutant [56]. Exactly how PPP1 integrates into the ERAD model of crossing the PPC awaits further clarification.
Moog and coworkers utilized the available data on PPC targeting to construct a search for PPC proteins encoded in the genome of P. tricornutum [85]. They then confirmed the localization of these hits as well as previously proposed PPC protein candidates using GFP fusions for in vivo localization [85]. This generated a map of PPC components indicative of its functions in P. tricornutum, and unraveled more candidate proteins likely involved in ERAD mediated import [85].
The outermost membrane – getting to the organelle
Finding and crossing the outermost membrane of complex plastids appears to occur via different mechanisms in different organisms. This may be a consequence of the initial uptake of the symbiont. A clear difference in the route to the organelle is found when comparing three and four membrane bounded plastids (Fig. 1 white arrows). In dinoflagellates and euglenids the third outer membrane represents the first barrier on the way into the plastid. Euglenid and dinoflagellate plastids are of independent origin [88], yet both groups appear to have evolved similar transport mechanisms. This might suggest that in this case the overall structure of the organelle dictates the import pathway. In both systems plastid proteins are cotranslationally inserted into the ER, from where they move to the Golgi, where they are packed into vesicles delivering them to the outermost plastid membrane upon fusion [50, 52, 89]. Interestingly in euglenids such vesicles were shown to fuse with the plastid in a manner that is independent of SNARE proteins [90].
In contrast in cryptophytes, haptophytes and heterokonts, the plastid resides within the ER and its outer membrane is decorated with 80S ribosomes. In this case import of proteins into the ER accomplishes the crossing the outermost plastid membrane [91]. Again this may be a consequence of the mechanism of plastid acquisition. This membrane may have evolved by fusion of the symbiont-containing compartment with host ER [92]. An alternative model could invoke a more immediate role for the ER in the endocytotic uptake of the symbiont [93].
Apicomplexa possess a four membrane bounded plastid for which no permanent connection with the ER have been detected thus far. Our understanding of the mechanisms involved in trafficking to the outermost membrane of the apicoplast is limited. The transport of apicoplast targeted GFP reporters was shown to be resistant to the action of the fungal toxin Brefeldin A, a potent disruptor of the Golgi apparatus [37, 94]. This may indicate direct ER to apicoplast transport, however the molecular details of this transport are not clear yet. Evidence from electron microscopy and tomographic studies suggested that the ER and apicoplast come into close contact, which may reflect functional interaction relevant for import [22, 95]. Alternative to the direct contact model, vesicles may shuttle apicoplast proteins from the ER while side stepping the Golgi. Several groups have shown vesicles around the apicoplast using light and electron microscopy [57, 59, 69, 96]. In T. gondii mutants with apicoplast import defects such vesicles become even more apparent, perhaps due to a “traffic jam”. Recent findings report the presence of phosphatidylinositol 3-monophosphate (PI3P) in the apicoplasts of both P. falciparum and T. gondii [97, 98]. Interference with PI3P in T. gondii through drug treatment or the overexpression of a heterologous PI3P binding proteins leads to profound and complex plastid biogenesis defects [98]. Interestingly, as in the case of protein import defect, these PI3P mutant parasites also show accumulation of vesicles around the apicoplast [98]. The role of PI3P in the regulation of endosomal trafficking is well established [99], and the observation of PI3P in the apicoplast suggests a potential connection between the apicoplast and the endosome. The precise mechanistic role of PI3P in this process remains to be defined.
Summary
Recent years have seen dramatic advances largely driven by genome analysis. There is now robust support for a model that invokes distinct translocon complexes that transport cargo proteins across subsequent membranes. There is also the understanding that both host and endosymbont adaptations contributed to the establishment of stable protein import. The mechanism used to traffic nuclear encoded proteins to complex plastids of similar structure appears conserved. While TIC and TOC still remain the unchallenged guardians of the two inner most membranes, evolution might have taken diverging routes through the other membranes, exploiting different mechanisms depending on the origin and structure of the specific plastid organelle.
While the data collected combines into a map of translocons marking the way of proteins into the plastid stroma, numerous mechanistic questions remain. Specifically, how these targeting machineries distinguish stromal proteins from those destined to the outer compartments. How the trafficking of membrane proteins differs from soluble proteins, also remains to be uncovered.
Highlights.
The evolutionary history of complex plastid includes at least three separate events of secondary endosymbiosis.
Complex plastids have numerous compartments that are derived from cellular components of host and symbiont.
A series of translocons enable the import of nuclear encoded proteins into the organelle.
Molecular details of these import are rapidly accumulating and a more and more detailed mechanistic model is emerging.
Acknowledgments
BS is supported by grants from the National Institutes of Health to BS (AI084415 and AI64671), and is a GRA Distinguished Investigator. LS was supported by a postdoctoral fellowship form the Swiss National Science Foundation. We thank Giel van Dooren and Swati Agrawal for fruitful discussion over the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Lilach Sheiner, Email: lilash@uga.edu.
Boris Striepen, Email: striepen@uga.edu.
References
- 1.Adl SM, Simpson AG, Farmer MA, Andersen RA, Anderson OR, Barta JR, Bowser SS, Brugerolle G, Fensome RA, Fredericq S, James TY, Karpov S, Kugrens P, Krug J, Lane CE, Lewis LA, Lodge J, Lynn DH, Mann DG, McCourt RM, Mendoza L, Moestrup O, Mozley-Standridge SE, Nerad TA, Shearer CA, Smirnov AV, Spiegel FW, Taylor MF. The new higher level classification of eukaryotes with emphasis on the taxonomy of protists. J Eukaryot Microbiol. 2005;52:399–451. doi: 10.1111/j.1550-7408.2005.00053.x. [DOI] [PubMed] [Google Scholar]
- 2.Chan CX, Yang EC, Banerjee T, Yoon HS, Martone PT, Estevez JM, Bhattacharya D. Red and green algal monophyly and extensive gene sharing found in a rich repertoire of red algal genes. Curr Biol. 2011;21:328–333. doi: 10.1016/j.cub.2011.01.037. [DOI] [PubMed] [Google Scholar]
- 3.Price DC, Chan CX, Yoon HS, Yang EC, Qiu H, Weber AP, Schwacke R, Gross J, Blouin NA, Lane C, Reyes-Prieto A, Durnford DG, Neilson JA, Lang BF, Burger G, Steiner JM, Loffelhardt W, Meuser JE, Posewitz MC, Ball S, Arias MC, Henrissat B, Coutinho PM, Rensing SA, Symeonidi A, Doddapaneni H, Green BR, Rajah VD, Boore J, Bhattacharya D. Cyanophora paradoxa genome elucidates origin of photosynthesis in algae and plants. Science. 2012;335:843–847. doi: 10.1126/science.1213561. [DOI] [PubMed] [Google Scholar]
- 4.Rodriguez-Ezpeleta N, Brinkmann H, Burey SC, Roure B, Burger G, Loffelhardt W, Bohnert HJ, Philippe H, Lang BF. Monophyly of primary photosynthetic eukaryotes: green plants, red algae, and glaucophytes. Curr Biol. 2005;15:1325–1330. doi: 10.1016/j.cub.2005.06.040. [DOI] [PubMed] [Google Scholar]
- 5.Rogers MB, Gilson PR, Su V, McFadden GI, Keeling PJ. The complete chloroplast genome of the chlorarachniophyte Bigelowiella natans: evidence for independent origins of chlorarachniophyte and euglenid secondary endosymbionts. Mol Biol Evol. 2007;24:54–62. doi: 10.1093/molbev/msl129. [DOI] [PubMed] [Google Scholar]
- 6.Cavalier-Smith T. The phagotrophic origin of eukaryotes and phylogenetic classification of Protozoa. Int J Syst Evol Microbiol. 2002;52:297–354. doi: 10.1099/00207713-52-2-297. [DOI] [PubMed] [Google Scholar]
- 7.Cavalier-Smith T. Principles of protein and lipid targeting in secondary symbiogenesis: euglenoid, dinoflagellate, and sporozoan plastid origins and the eukaryote family tree. J Eukaryot Microbiol. 1999;46:347–366. doi: 10.1111/j.1550-7408.1999.tb04614.x. [DOI] [PubMed] [Google Scholar]
- 8.Cavalier-Smith T. Kingdoms Protozoa and Chromista and the eozoan root of the eukaryotic tree. Biol Lett. 2010;6:342–345. doi: 10.1098/rsbl.2009.0948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keeling PJ. Chromalveolates and the evolution of plastids by secondary endosymbiosis. J Eukaryot Microbiol. 2009;56:1–8. doi: 10.1111/j.1550-7408.2008.00371.x. [DOI] [PubMed] [Google Scholar]
- 10.Khan H, Parks N, Kozera C, Curtis BA, Parsons BJ, Bowman S, Archibald JM. Plastid genome sequence of the cryptophyte alga Rhodomonas salina CCMP1319: lateral transfer of putative DNA replication machinery and a test of chromist plastid phylogeny. Mol Biol Evol. 2007;24:1832–1842. doi: 10.1093/molbev/msm101. [DOI] [PubMed] [Google Scholar]
- 11.Yoon HS, Hackett JD, Pinto G, Bhattacharya D. The single, ancient origin of chromist plastids. Proc Natl Acad Sci U S A. 2002;99:15507–15512. doi: 10.1073/pnas.242379899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fast NM, Kissinger JC, Roos DS, Keeling PJ. Nuclear-encoded, plastid-targeted genes suggest a single common origin for apicomplexan and dinoflagellate plastids. Mol Biol Evol. 2001;18:418–426. doi: 10.1093/oxfordjournals.molbev.a003818. [DOI] [PubMed] [Google Scholar]
- 13.Gross W, Lenze D, Nowitzki U, Weiske J, Schnarrenberger C. Characterization, cloning, and evolutionary history of the chloroplast and cytosolic class I aldolases of the red alga Galdieria sulphuraria. Gene. 1999;230:7–14. doi: 10.1016/s0378-1119(99)00059-1. [DOI] [PubMed] [Google Scholar]
- 14.Moore RB, Obornik M, Janouskovec J, Chrudimsky T, Vancova M, Green DH, Wright SW, Davies NW, Bolch CJ, Heimann K, Slapeta J, Hoegh-Guldberg O, Logsdon JM, Carter DA. A photosynthetic alveolate closely related to apicomplexan parasites. Nature. 2008;451:959–963. doi: 10.1038/nature06635. [DOI] [PubMed] [Google Scholar]
- 15.Janouskovec J, Horak A, Obornik M, Lukes J, Keeling PJ. A common red algal origin of the apicomplexan, dinoflagellate, and heterokont plastids. Proc Natl Acad Sci U S A. 2010;107:10949–10954. doi: 10.1073/pnas.1003335107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Felsner G, Sommer MS, Gruenheit N, Hempel F, Moog D, Zauner S, Martin W, Maier UG. ERAD components in organisms with complex red plastids suggest recruitment of a preexisting protein transport pathway for the periplastid membrane. Genome Biol Evol. 2011;3:140–150. doi: 10.1093/gbe/evq074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hackett JD, Yoon HS, Li S, Reyes-Prieto A, Rummele SE, Bhattacharya D. Phylogenomic analysis supports the monophyly of cryptophytes and haptophytes and the association of rhizaria with chromalveolates. Mol Biol Evol. 2007;24:1702–1713. doi: 10.1093/molbev/msm089. [DOI] [PubMed] [Google Scholar]
- 18.Harper JT, Waanders E, Keeling PJ. On the monophyly of chromalveolates using a six-protein phylogeny of eukaryotes. Int J Syst Evol Microbiol. 2005;55:487–496. doi: 10.1099/ijs.0.63216-0. [DOI] [PubMed] [Google Scholar]
- 19.Archibald JM, Keeling PJ. Recycled plastids: a ‘green movement’ in eukaryotic evolution. Trends Genet. 2002;18:577–584. doi: 10.1016/s0168-9525(02)02777-4. [DOI] [PubMed] [Google Scholar]
- 20.Delwiche CF. Tracing the Thread of Plastid Diversity through the Tapestry of Life. Am Nat. 1999;154:S164–S177. doi: 10.1086/303291. [DOI] [PubMed] [Google Scholar]
- 21.Gould SB, Waller RF, McFadden GI. Plastid evolution. Annu Rev Plant Biol. 2008;59:491–517. doi: 10.1146/annurev.arplant.59.032607.092915. [DOI] [PubMed] [Google Scholar]
- 22.Tomova C, Geerts WJ, Muller-Reichert T, Entzeroth R, Humbel BM. New comprehension of the apicoplast of Sarcocystis by transmission electron tomography. Biol Cell. 2006;98:535–545. doi: 10.1042/BC20060028. [DOI] [PubMed] [Google Scholar]
- 23.Lukes J, Leander BS, Keeling PJ. Cascades of convergent evolution: the corresponding evolutionary histories of euglenozoans and dinoflagellates. Proc Natl Acad Sci U S A. 2009;106(Suppl 1):9963–9970. doi: 10.1073/pnas.0901004106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Whatley JM, John P, Whatley FR. From extracellular to intracellular: the establishment of mitochondria and chloroplasts. Proc R Soc Lond B Biol Sci. 1979;204:165–187. doi: 10.1098/rspb.1979.0020. [DOI] [PubMed] [Google Scholar]
- 25.Kleine T, Maier UG, Leister D. DNA transfer from organelles to the nucleus: the idiosyncratic genetics of endosymbiosis. Annu Rev Plant Biol. 2009;60:115–138. doi: 10.1146/annurev.arplant.043008.092119. [DOI] [PubMed] [Google Scholar]
- 26.Douglas S, Zauner S, Fraunholz M, Beaton M, Penny S, Deng LT, Wu X, Reith M, Cavalier-Smith T, Maier UG. The highly reduced genome of an enslaved algal nucleus. Nature. 2001;410:1091–1096. doi: 10.1038/35074092. [DOI] [PubMed] [Google Scholar]
- 27.Gilson PR, McFadden GI. The miniaturized nuclear genome of eukaryotic endosymbiont contains genes that overlap, genes that are cotranscribed, and the smallest known spliceosomal introns. Proc Natl Acad Sci U S A. 1996;93:7737–7742. doi: 10.1073/pnas.93.15.7737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tanifuji G, Onodera NT, Wheeler TJ, Dlutek M, Donaher N, Archibald JM. Complete nucleomorph genome sequence of the nonphotosynthetic alga Cryptomonas paramecium reveals a core nucleomorph gene set. Genome Biol Evol. 2011;3:44–54. doi: 10.1093/gbe/evq082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilson RJ, Denny PW, Preiser PR, Rangachari K, Roberts K, Roy A, Whyte A, Strath M, Moore DJ, Moore PW, Williamson DH. Complete gene map of the plastid-like DNA of the malaria parasite Plasmodium falciparum. J Mol Biol. 1996;261:155–172. doi: 10.1006/jmbi.1996.0449. [DOI] [PubMed] [Google Scholar]
- 30.Foth BJ, Ralph SA, Tonkin CJ, Struck NS, Fraunholz M, Roos DS, Cowman AF, McFadden GI. Dissecting apicoplast targeting in the malaria parasite Plasmodium falciparum. Science. 2003;299:705–708. doi: 10.1126/science.1078599. [DOI] [PubMed] [Google Scholar]
- 31.Soll J, Schleiff E. Protein import into chloroplasts. Nat Rev Mol Cell Biol. 2004;5:198–208. doi: 10.1038/nrm1333. [DOI] [PubMed] [Google Scholar]
- 32.Lang M, Apt KE, Kroth PG. Protein transport into “complex” diatom plastids utilizes two different targeting signals. J Biol Chem. 1998;273:30973–30978. doi: 10.1074/jbc.273.47.30973. [DOI] [PubMed] [Google Scholar]
- 33.Rapoport TA. Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature. 2007;450:663–669. doi: 10.1038/nature06384. [DOI] [PubMed] [Google Scholar]
- 34.DeRocher A, Hagen CB, Froehlich JE, Feagin JE, Parsons M. Analysis of targeting sequences demonstrates that trafficking to the Toxoplasma gondii plastid branches off the secretory system. J Cell Sci. 2000;113(Pt 22):3969–3977. doi: 10.1242/jcs.113.22.3969. [DOI] [PubMed] [Google Scholar]
- 35.Waller RF, Reed MB, Cowman AF, McFadden GI. Protein trafficking to the plastid of Plasmodium falciparum is via the secretory pathway. EMBO J. 2000;19:1794–1802. doi: 10.1093/emboj/19.8.1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Waller RF, Keeling PJ, Donald RG, Striepen B, Handman E, Lang-Unnasch N, Cowman AF, Besra GS, Roos DS, McFadden GI. Nuclear-encoded proteins target to the plastid in Toxoplasma gondii and Plasmodium falciparum. Proc Natl Acad Sci U S A. 1998;95:12352–12357. doi: 10.1073/pnas.95.21.12352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tonkin CJ, Struck NS, Mullin KA, Stimmler LM, McFadden GI. Evidence for Golgi-independent transport from the early secretory pathway to the plastid in malaria parasites. Mol Microbiol. 2006;61:614–630. doi: 10.1111/j.1365-2958.2006.05244.x. [DOI] [PubMed] [Google Scholar]
- 38.van Dooren GG, Su V, D’Ombrain MC, McFadden GI. Processing of an apicoplast leader sequence in Plasmodium falciparum and the identification of a putative leader cleavage enzyme. J Biol Chem. 2002;277:23612–23619. doi: 10.1074/jbc.M201748200. [DOI] [PubMed] [Google Scholar]
- 39.He CY, Striepen B, Pletcher CH, Murray JM, Roos DS. Targeting and processing of nuclear-encoded apicoplast proteins in plastid segregation mutants of Toxoplasma gondii. J Biol Chem. 2001;276:28436–28442. doi: 10.1074/jbc.M102000200. [DOI] [PubMed] [Google Scholar]
- 40.Wastl J, Maier UG. Transport of proteins into cryptomonads complex plastids. J Biol Chem. 2000;275:23194–23198. doi: 10.1074/jbc.M003125200. [DOI] [PubMed] [Google Scholar]
- 41.Harb OS, Chatterjee B, Fraunholz MJ, Crawford MJ, Nishi M, Roos DS. Multiple functionally redundant signals mediate targeting to the apicoplast in the apicomplexan parasite Toxoplasma gondii. Eukaryot Cell. 2004;3:663–674. doi: 10.1128/EC.3.3.663-674.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tonkin CJ, Roos DS, McFadden GI. N-terminal positively charged amino acids, but not their exact position, are important for apicoplast transit peptide fidelity in Toxoplasma gondii. Mol Biochem Parasitol. 2006;150:192–200. doi: 10.1016/j.molbiopara.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 43.Kilian O, Kroth PG. Identification and characterization of a new conserved motif within the presequence of proteins targeted into complex diatom plastids. Plant J. 2005;41:175–183. doi: 10.1111/j.1365-313X.2004.02294.x. [DOI] [PubMed] [Google Scholar]
- 44.Felsner G, Sommer MS, Maier UG. The physical and functional borders of transit peptide-like sequences in secondary endosymbionts. BMC Plant Biol. 2010;10:223. doi: 10.1186/1471-2229-10-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hirakawa Y, Gile GH, Ota S, Keeling PJ, Ishida K. Characterization of periplastidal compartment-targeting signals in chlorarachniophytes. Mol Biol Evol. 2010;27:1538–1545. doi: 10.1093/molbev/msq038. [DOI] [PubMed] [Google Scholar]
- 46.Gould SB, Sommer MS, Hadfi K, Zauner S, Kroth PG, Maier UG. Protein targeting into the complex plastid of cryptophytes. J Mol Evol. 2006;62:674–681. doi: 10.1007/s00239-005-0099-y. [DOI] [PubMed] [Google Scholar]
- 47.Gruber A, Vugrinec S, Hempel F, Gould SB, Maier UG, Kroth PG. Protein targeting into complex diatom plastids: functional characterisation of a specific targeting motif. Plant Mol Biol. 2007;64:519–530. doi: 10.1007/s11103-007-9171-x. [DOI] [PubMed] [Google Scholar]
- 48.Durnford DG, Gray MW. Analysis of Euglena gracilis plastid-targeted proteins reveals different classes of transit sequences. Eukaryot Cell. 2006;5:2079–2091. doi: 10.1128/EC.00222-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kishore R, Muchhal US, Schwartzbach SD. The presequence of Euglena LHCPII, a cytoplasmically synthesized chloroplast protein, contains a functional endoplasmic reticulum-targeting domain. Proc Natl Acad Sci U S A. 1993;90:11845–11849. doi: 10.1073/pnas.90.24.11845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sulli C, Fang Z, Muchhal U, Schwartzbach SD. Topology of Euglena chloroplast protein precursors within endoplasmic reticulum to Golgi to chloroplast transport vesicles. J Biol Chem. 1999;274:457–463. doi: 10.1074/jbc.274.1.457. [DOI] [PubMed] [Google Scholar]
- 51.Vesteg M, Vacula R, Steiner JM, Mateasikova B, Loffelhardt W, Brejova B, Krajcovic J. A possible role for short introns in the acquisition of stroma-targeting peptides in the flagellate Euglena gracilis. DNA Res. 2010;17:223–231. doi: 10.1093/dnares/dsq015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nassoury N, Cappadocia M, Morse D. Plastid ultrastructure defines the protein import pathway in dinoflagellates. J Cell Sci. 2003;116:2867–2874. doi: 10.1242/jcs.00517. [DOI] [PubMed] [Google Scholar]
- 53.Patron NJ, Waller RF, Archibald JM, Keeling PJ. Complex protein targeting to dinoflagellate plastids. J Mol Biol. 2005;348:1015–1024. doi: 10.1016/j.jmb.2005.03.030. [DOI] [PubMed] [Google Scholar]
- 54.Agrawal S, van Dooren GG, Beatty WL, Striepen B. Genetic evidence that an endosymbiont-derived ERAD system functions in import of apicoplast proteins. J Biol Chem. 2009 doi: 10.1074/jbc.M109.044024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nair SC, Brooks CF, Goodman CD, Strurm A, McFadden GI, Sundriyal S, Anglin JL, Song Y, Moreno SN, Striepen B. Apicoplast isoprenoid precursor synthesis and the molecular basis of fosmidomycin resistance in Toxoplasma gondii. J Exp Med. 2011;208:1547–1559. doi: 10.1084/jem.20110039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sheiner L, Demerly JL, Poulsen N, Beatty WL, Lucas O, Behnke MS, White MW, Striepen B. A systematic screen to discover and analyze apicoplast proteins identifies a conserved and essential protein import factor. PLoS Pathog. 2011;7:e1002392. doi: 10.1371/journal.ppat.1002392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.DeRocher AE, Coppens I, Karnataki A, Gilbert LA, Rome ME, Feagin JE, Bradley PJ, Parsons M. A thioredoxin family protein of the apicoplast periphery identifies abundant candidate transport vesicles in Toxoplasma gondii. Eukaryot Cell. 2008;7:1518–1529. doi: 10.1128/EC.00081-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fleige T, Fischer K, Ferguson DJ, Gross U, Bohne W. Carbohydrate metabolism in the Toxoplasma gondii apicoplast: localization of three glycolytic isoenzymes, the single pyruvate dehydrogenase complex, and a plastid phosphate translocator. Eukaryot Cell. 2007;6:984–996. doi: 10.1128/EC.00061-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Karnataki A, Derocher A, Coppens I, Nash C, Feagin JE, Parsons M. Cell cycle-regulated vesicular trafficking of Toxoplasma APT1, a protein localized to multiple apicoplast membranes. Mol Microbiol. 2007;63:1653–1668. doi: 10.1111/j.1365-2958.2007.05619.x. [DOI] [PubMed] [Google Scholar]
- 60.Karnataki A, Derocher AE, Coppens I, Feagin JE, Parsons M. A membrane protease is targeted to the relict plastid of toxoplasma via an internal signal sequence. Traffic. 2007;8:1543–1553. doi: 10.1111/j.1600-0854.2007.00637.x. [DOI] [PubMed] [Google Scholar]
- 61.Karnataki A, DeRocher AE, Feagin JE, Parsons M. Sequential processing of the Toxoplasma apicoplast membrane protein FtsH1 in topologically distinct domains during intracellular trafficking. Mol Biochem Parasitol. 2009;166:126–133. doi: 10.1016/j.molbiopara.2009.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Agrawal S, Striepen B. More membranes, more proteins: complex protein import mechanisms into secondary plastids. Protist. 2010;161:672–687. doi: 10.1016/j.protis.2010.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hirakawa Y, Burki F, Keeling PJ. Genome-based reconstruction of the protein import machinery in the secondary plastid of a chlorarachniophyte alga. Eukaryot Cell. 2012;11:324–333. doi: 10.1128/EC.05264-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gilson PR, Su V, Slamovits CH, Reith ME, Keeling PJ, McFadden GI. Complete nucleotide sequence of the chlorarachniophyte nucleomorph: nature’s smallest nucleus. Proc Natl Acad Sci U S A. 2006;103:9566–9571. doi: 10.1073/pnas.0600707103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.McFadden GI, van Dooren GG. Evolution: red algal genome affirms a common origin of all plastids. Curr Biol. 2004;14:R514–516. doi: 10.1016/j.cub.2004.06.041. [DOI] [PubMed] [Google Scholar]
- 66.Waller RF, McFadden GI. The apicoplast: a review of the derived plastid of apicomplexan parasites. Curr Issues Mol Biol. 2005;7:57–79. [PubMed] [Google Scholar]
- 67.Chen X, Smith MD, Fitzpatrick L, Schnell DJ. In vivo analysis of the role of atTic20 in protein import into chloroplasts. Plant Cell. 2002;14:641–654. doi: 10.1105/tpc.010336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kouranov A, Chen X, Fuks B, Schnell DJ. Tic20 and Tic22 are new components of the protein import apparatus at the chloroplast inner envelope membrane. J Cell Biol. 1998;143:991–1002. doi: 10.1083/jcb.143.4.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.van Dooren GG, Tomova C, Agrawal S, Humbel BM, Striepen B. Toxoplasma gondii Tic20 is essential for apicoplast protein import. Proc Natl Acad Sci U S A. 2008;105:13574–13579. doi: 10.1073/pnas.0803862105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kalanon M, Tonkin CJ, McFadden GI. Characterization of two putative protein translocation components in the apicoplast of Plasmodium falciparum. Eukaryot Cell. 2009;8:1146–1154. doi: 10.1128/EC.00061-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kohler S, Delwiche CF, Denny PW, Tilney LG, Webster P, Wilson RJ, Palmer JD, Roos DS. A plastid of probable green algal origin in Apicomplexan parasites. Science. 1997;275:1485–1489. doi: 10.1126/science.275.5305.1485. [DOI] [PubMed] [Google Scholar]
- 72.Cavalier-Smith T. Genomic reduction and evolution of novel genetic membranes and protein-targeting machinery in eukaryote-eukaryote chimaeras (meta-algae) Philos Trans R Soc Lond B Biol Sci. 2003;358:109–133. doi: 10.1098/rstb.2002.1194. discussion 133–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tonkin CJ, Kalanon M, McFadden GI. Protein targeting to the malaria parasite plastid. Traffic. 2008;9:166–175. doi: 10.1111/j.1600-0854.2007.00660.x. [DOI] [PubMed] [Google Scholar]
- 74.Bullmann L, Haarmann R, Mirus O, Bredemeier R, Hempel F, Maier UG, Schleiff E. Filling the gap, evolutionarily conserved Omp85 in plastids of chromalveolates. J Biol Chem. 2010;285:6848–6856. doi: 10.1074/jbc.M109.074807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sommer MS, Daum B, Gross LE, Weis BL, Mirus O, Abram L, Maier UG, Kuhlbrandt W, Schleiff E. Chloroplast Omp85 proteins change orientation during evolution. Proc Natl Acad Sci U S A. 2011;108:13841–13846. doi: 10.1073/pnas.1108626108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sommer MS, Gould SB, Lehmann P, Gruber A, Przyborski JM, Maier UG. Der1-mediated preprotein import into the periplastid compartment of chromalveolates? Mol Biol Evol. 2007;24:918–928. doi: 10.1093/molbev/msm008. [DOI] [PubMed] [Google Scholar]
- 77.Smith MH, Ploegh HL, Weissman JS. Road to ruin: targeting proteins for degradation in the endoplasmic reticulum. Science. 2011;334:1086–1090. doi: 10.1126/science.1209235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Spork S, Hiss JA, Mandel K, Sommer M, Kooij TW, Chu T, Schneider G, Maier UG, Przyborski JM. An unusual ERAD-like complex is targeted to the apicoplast of Plasmodium falciparum. Eukaryot Cell. 2009 doi: 10.1128/EC.00083-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ye Y, Shibata Y, Yun C, Ron D, Rapoport TA. A membrane protein complex mediates retro-translocation from the ER lumen into the cytosol. Nature. 2004;429:841–847. doi: 10.1038/nature02656. [DOI] [PubMed] [Google Scholar]
- 80.Hempel F, Bullmann L, Lau J, Zauner S, Maier UG. ERAD-derived preprotein transport across the second outermost plastid membrane of diatoms. Mol Biol Evol. 2009;26:1781–1790. doi: 10.1093/molbev/msp079. [DOI] [PubMed] [Google Scholar]
- 81.Hampton RY. ER-associated degradation in protein quality control and cellular regulation. Curr Opin Cell Biol. 2002;14:476–482. doi: 10.1016/s0955-0674(02)00358-7. [DOI] [PubMed] [Google Scholar]
- 82.Park S, Isaacson R, Kim HT, Silver PA, Wagner G. Ufd1 exhibits the AAA-ATPase fold with two distinct ubiquitin interaction sites. Structure. 2005;13:995–1005. doi: 10.1016/j.str.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 83.Walters KJ. Ufd1 exhibits dual ubiquitin binding modes. Structure. 2005;13:943–944. doi: 10.1016/j.str.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 84.Carvalho P, Stanley AM, Rapoport TA. Retrotranslocation of a misfolded luminal ER protein by the ubiquitin-ligase Hrd1p. Cell. 2010;143:579–591. doi: 10.1016/j.cell.2010.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Moog D, Stork S, Zauner S, Maier UG. In silico and in vivo investigations of proteins of a minimized eukaryotic cytoplasm. Genome Biol Evol. 2011;3:375–382. doi: 10.1093/gbe/evr031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ponts N, Saraf A, Chung DW, Harris A, Prudhomme J, Washburn MP, Florens L, Le Roch KG. Unraveling the ubiquitome of the human malaria parasite. J Biol Chem. 2011;286:40320–40330. doi: 10.1074/jbc.M111.238790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hempel F, Felsner G, Maier UG. New mechanistic insights into pre-protein transport across the second outermost plastid membrane of diatoms. Mol Microbiol. 2010;76:793–801. doi: 10.1111/j.1365-2958.2010.07142.x. [DOI] [PubMed] [Google Scholar]
- 88.Keeling PJ. The endosymbiotic origin, diversification and fate of plastids. Philos Trans R Soc Lond B Biol Sci. 2010;365:729–748. doi: 10.1098/rstb.2009.0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Vacula R, Slavikova S, Schwartzbach SD. Protein trafficking to the complex chloroplasts of Euglena. Methods Mol Biol. 2007;390:219–237. doi: 10.1007/978-1-59745-466-7_15. [DOI] [PubMed] [Google Scholar]
- 90.Slavikova S, Vacula R, Fang Z, Ehara T, Osafune T, Schwartzbach SD. Homologous and heterologous reconstitution of Golgi to chloroplast transport and protein import into the complex chloroplasts of Euglena. J Cell Sci. 2005;118:1651–1661. doi: 10.1242/jcs.02277. [DOI] [PubMed] [Google Scholar]
- 91.Bhaya D, Grossman A. Targeting proteins to diatom plastids involves transport through an endoplasmic reticulum. Mol Gen Genet. 1991;229:400–404. doi: 10.1007/BF00267462. [DOI] [PubMed] [Google Scholar]
- 92.Cavalier-Smith T. Chloroplast evolution: secondary symbiogenesis and multiple losses. Curr Biol. 2002;12:R62–64. doi: 10.1016/s0960-9822(01)00675-3. [DOI] [PubMed] [Google Scholar]
- 93.Lewis JD, Lazarowitz SG. Arabidopsis synaptotagmin SYTA regulates endocytosis and virus movement protein cell-to-cell transport. Proc Natl Acad Sci U S A. 2010;107:2491–2496. doi: 10.1073/pnas.0909080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.DeRocher A, Gilbert B, Feagin JE, Parsons M. Dissection of brefeldin A-sensitive and -insensitive steps in apicoplast protein targeting. J Cell Sci. 2005;118:565–574. doi: 10.1242/jcs.01627. [DOI] [PubMed] [Google Scholar]
- 95.Tomova C, Humbel BM, Geerts WJ, Entzeroth R, Holthuis JC, Verkleij AJ. Membrane contact sites between apicoplast and ER in Toxoplasma gondii revealed by electron tomography. Traffic. 2009;10:1471–1480. doi: 10.1111/j.1600-0854.2009.00954.x. [DOI] [PubMed] [Google Scholar]
- 96.van Dooren GG, Reiff SB, Tomova C, Meissner M, Humbel BM, Striepen B. A novel dynamin-related protein has been recruited for apicoplast fission in Toxoplasma gondii. Curr Biol. 2009;19:267–276. doi: 10.1016/j.cub.2008.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tawk L, Chicanne G, Dubremetz JF, Richard V, Payrastre B, Vial HJ, Roy C, Wengelnik K. Phosphatidylinositol 3-phosphate, an essential lipid in Plasmodium, localizes to the food vacuole membrane and the apicoplast. Eukaryot Cell. 2010;9:1519–1530. doi: 10.1128/EC.00124-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tawk L, Dubremetz JF, Montcourrier P, Chicanne G, Merezegue F, Richard V, Payrastre B, Meissner M, Vial HJ, Roy C, Wengelnik K, Lebrun M. Phosphatidylinositol 3-monophosphate is involved in toxoplasma apicoplast biogenesis. PLoS Pathog. 2011;7:e1001286. doi: 10.1371/journal.ppat.1001286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lindmo K, Stenmark H. Regulation of membrane traffic by phosphoinositide 3-kinases. J Cell Sci. 2006;119:605–614. doi: 10.1242/jcs.02855. [DOI] [PubMed] [Google Scholar]