Abstract
Identifying immunodominant CTL epitopes is essential for studying CD8+ T-cell responses in populations, but remains difficult, as peptides within antigens typically are too numerous for all to be synthesized and screened. Instead, to facilitate discovery, in silico scanning of proteins for sequences that match the motif, or binding preferences, of the restricting MHC class I allele – the largest determinant of immunodominance – can be used to predict likely candidates. The high false positive rate with this analysis ideally requires binding confirmation, which is obtained routinely by an assay using cell lines such as RMA-S that have defective transporter associated with antigen processing (TAP) machinery, and consequently, few surface class I molecules. The stabilization and resultant increased life-span of peptide-MHC complexes on the cell surface by the addition of true binders validates their identity. To determine whether a similar assay could be developed for dogs, we transfected a prevalent class I allele, DLA-88*50801, into RMA-S. In the BARC3 clone, the recombinant heavy chain was associated with murine β2-microglobulin, and importantly, could differentiate motif-matched and -mismatched peptides by surface MHC stabilization. This work demonstrates the potential to use RMA-S cells transfected with canine alleles as a tool for CTL epitope discovery in this species.
Keywords: Canine, CD8+ T Lymphocytes, MHC, Peptides/Epitopes
1. Introduction
CD8+ T cells chiefly function to eliminate virus-infected and malignant cells, which they detect by specifically recognizing short peptides – their cognate epitopes – bound by MHC class I molecules on the target surface. Remarkably, despite the vast diversity of TCR repertoires and large number of peptides within a given antigen, CTL responses across individuals sharing class I alleles are predictably directed against one or a few common epitopes. For example, of the 550 possible nonamer peptides within lymphocytic choriomeningitis virus (LCMV) nucleoprotein (NP), all C57BL/6 mice are dominated by reactivity against a single epitope, NP396 (Yanagi et al., 1992), while in BALB/c mice, the major response is directed against NP118 (Schulz et al., 1991). Multiple mechanisms underlie this immunodominance phenomenon, including antigen density and processing, TCR availability, peptide-MHC class I binding affinity, and competition between T cell specificities (reviewed in Yewdell, 2006). While the study of specific CTL responses is made possible by the restrictions on diversity imposed by immunodominance, identifying epitopes still remains a difficult, resource-intensive task.
Ultimately, epitopes are confirmed by demonstrating CTL effector activity, such as IFN-γ production or target killing, upon incubation with the peptide and the appropriate class I restriction element. Yet simply testing all possible peptides within an antigen is usually not a feasible strategy, because of the costs of peptide synthesis, and the low frequency of specific memory T cells (10−2 to 10−4), which limits the number of peptides that can be evaluated in a single assay. The preferred approach is therefore to generate a smaller list of likely candidates by attempting to predict the effects of immunodominance on peptide selection, usually through a combination of in silico analysis and empirical determination. Of course, not all effects are weighted equally: some factors are minimally limiting (TCR availability; transporter associated with antigen processing [TAP] specificity) or difficult to reproducibly measure or estimate (proteasomal cleavage; TCR binding) (Assarsson et al., 2007; Lundegaard et al., 2007), and consequently, are often not included in prediction analysis. Quantitatively, the most important parameter in epitope selection is the affinity for class I molecules, which are relatively poor at binding peptides (Yewdell, 2006). Importantly, in those peptides that do bind, common amino acid groups in certain positions (anchor residues) can be identified. These residues (and peptide length) collectively form a motif, which can be used to scan proteins by computer program to eliminate unlikely binders for that allele (Rammensee et al., 1999). While useful, such predictions typically generate many more false than true positive binders – in one study, of the 1657 predicted peptides from vaccinia virus, only 263 strongly bound HLA-A*0201 (Assarsson et al., 2007) – and therefore, require experimental confirmation. One of the simplest, most cost-effective means of testing peptide binding to class I alleles is the peptide-induced MHC stabilization assay, which uses TAP-deficient cell lines such as T2 (human) or RMA-S (mouse). Without efficient TAP-mediated transport of cytosolic peptides into the ER, assembled class I complexes are structurally unstable, and retained only transiently at the cell surface. However, when RMA-S or T2 are incubated with an exogenous peptide capable of binding class I, surface pMHC complexes are stabilized and easily detected by flow cytometry with an anti-class I mAb.
While the ability to confirm the predictions of binding algorithms is critical for streamlining epitope discovery, there is unfortunately no corresponding cell line for evaluating putative CTL epitopes in dogs. The range of class I molecules of mice and humans that can be tested has been has been expanded beyond the endogenous alleles of RMA-S and T2 by production of transfectants; accordingly, we sought to determine whether peptide binding at the canine classical class I locus, Dog Leukocyte Antigen (DLA)-88 (Graumann et al., 1998), could be evaluated using this same strategy. An RMA-S clone expressing a prevalent allele, DLA-88*50801 (Ross et al., 2012), was therefore generated. Like the parent line, these cells could discriminate motif-matched and -mismatched peptides in a standard stabilization assay. This methodology should constitute a valuable immunologic tool for investigating and defining epitope-specific CD8+ T-cell responses in the dog.
2. Materials and methods
2.1 Cell culture and cloning of DLA-88-transfected RMA-S cells
The murine lymphoma line RMA-S was cultured in RPMI-1640 containing 10% FBS and 2 mM L-glutamine (R-10). A modified pcDNA3 expression plasmid encoding a DLA-88*50801 heavy chain (GenBank, JQ733514) with a FLAG epitope tag at the carboxyl terminus, previously generated in our lab (Ross P and Hess PR, manuscript submitted), was transfected into RMA-S using Lipofectamine 2000 (Invitrogen). Following G418 selection (1 mg/ml for 7 d, then 0.2 mg/ml for 8 d), individual clones were isolated by limiting dilution and screened for vector expression after permeabilization (Cytofix/Cytoperm; BD Biosciences) and intracellular staining with the anti-FLAG mAb M2 (Sigma-Aldrich) by flow cytometry. Clone number 3 (BARC3) was used throughout the study and maintained continuously under G418.
2.2 MHC class I surface stabilization assays
In order to accumulate class I molecules on the cell surface, RMA-S and BARC3 cells were cultured overnight at 27°C, as previously described (Ljunggren et al., 1990). In some experiments, accumulated surface class I molecules were peptide-loaded by adding K9 (KLFSGELTK), K11 (RFLDKDGFIDK) (both synthesized by Peptide 2.0), NP396 (FQPQNGQFI) or NP366 (ASNENMETM) (both synthesized by GenScript) peptides from DMSO stock solutions to overnight cultures, followed by an additional 5 hours of incubation at 37°C. Peptide loading of RMA-S and BARC3 cells (105 in 100 μl) was performed in R-10 or serum-free Opti-MEM I medium (Gibco/Life Technologies) in 96-well flat-bottom cell culture plates. To assess time-dependent stability of pMHC complexes, peptide-loaded BARC3 cells were washed with PBS and cultured with 5 μg/ml brefeldin A (BFA; BioLegend) for various lengths of time prior to collection.
2.3 Flow cytometry and data analysis
For staining, cells were washed in FACS buffer (PBS containing 2% FBS and 0.1% NaN3) and incubated with the relevant primary or secondary Ab for 15 min at 4°C in 96-well round-bottom polypropylene plates. The following unconjugated mAbs were used (clone names are listed parenthetically) at pre-determined optimal concentrations: anti-canine MHC class I (H58A, VMRD; 3F10, Ancell), anti-H2-Db (28-14-8), anti-Kb/d (34-1-2S) (both eBioscience), and anti-murine β2M (S19.8) (BD Pharmingen). In all experiments, Alexa Fluor 647-labeled donkey anti-mouse IgG (Jackson ImmunoResearch) was used as a secondary detecting Ab; background staining was established by omission of the primary mAb. Flow cytometric list mode data was collected using a FACSCalibur flow cytometer (BD Biosciences) and analyzed with FlowJo software (Tree Star). Viable cells were differentiated using forward and side scatter gating. Data were graphed and nonlinear regression analysis was performed using Prism 5 (GraphPad).
3. Results and discussion
To investigate whether a peptide stabilization assay could be developed for the dog, we generated a DLA-88-transgenic clone of RMA-S, and examined whether canine class I complexes were retained on the cell surface when synthetic peptides previously shown to bind this allele were added to the medium.
3.1 Production and characterization of a DLA-88*50801-expressing murine cell line
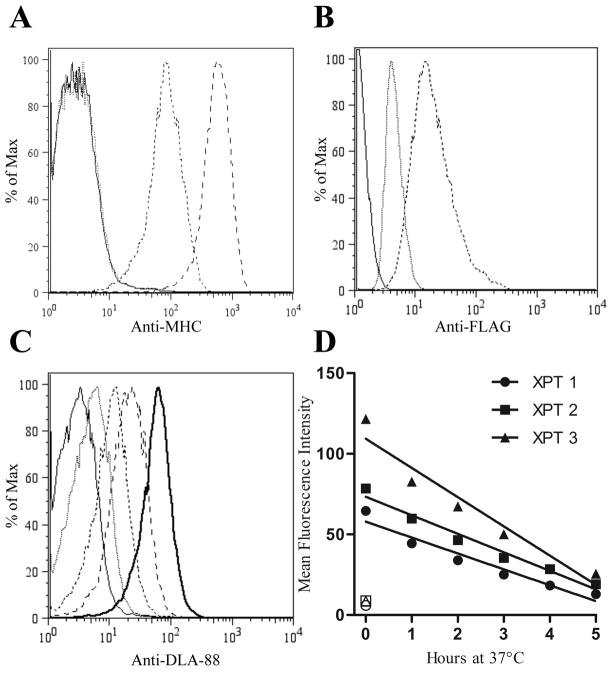
An important first step was to establish that the mAb that binds canine class I, H58A (Ladiges et al., 1988), which has a broad cross-species range (Davis et al., 1987), did not recognize the murine alleles. While not an insurmountable obstacle to our objective, such recognition would make the stabilization assay more laborious, as confirmation of any positive peptide as a true DLA-88 binder would require secondary assessment to exclude binding to endogenous class I molecules. We therefore cultured RMA-S cells overnight at 27°C, which increases surface expression of class I complexes non-specifically (Ljunggren et al., 1990). No staining with H58A was observed (Fig 1A), which was not due to the absence of murine complexes, as these were clearly detectable with H2-Db and -Kb-specific mAbs. Thus, H58A does not recognize endogenous class I complexes on RMA-S.
Fig. 1.
When cultured at 27°C, RMA-S cells stably transfected with a FLAG-tagged DLA-88*50801 construct (BARC3) express the canine class I molecule at the cell surface, which can be specifically detected with H58A. (A) The endogenous murine alleles H2-Db or -Kb are not recognized by H58A. RMA-S cells cultured overnight at 27°C were stained with H58A (dotted line), anti-H2-Kb/d (long dashed line), anti-H2-Db (dashed line), or no primary Ab (solid line). The lack of staining of cold-cultured RMA-S cells with H58A was confirmed in multiple experiments. (B) The BARC3 clone expresses the DLA-88*50801 vector gene products. Following overnight incubation at 27°C, RMA-S (dotted) and BARC3 (dashed) cells were assessed for FLAG expression by intracellular staining with the mAb M2. The solid line shows unstained BARC3 cells. Data are representative of three independent experiments. (C) Surface DLA-88*50801expression produced by incubation at cold temperatures is gradually lost upon return to standard culture conditions. BARC3 cells were cultured overnight at 27°C, followed by an additional 5 (dashed line), 3 (long dashed line), or no (bold solid line) h incubation at 37°C prior to harvesting; for clarity, data from 1, 2, and 4 h collection points are not shown. The dotted line represents cells that were maintained at 37°C for the assay duration, while the solid line indicates control cells maintained at 27°C that were stained with the secondary Ab only. (D) Graphical representation of data from three independent experiments (XPT) performed as described in (C). Open symbols indicate control samples that were cultured at 37°C for the complete assay duration.
To create cells with our desired phenotype, a plasmid encoding FLAG-tagged DLA-88*50801 was transfected into RMA-S cells. After selection, the FLAG-positive clone 3 (Bispecies Antigen Recognition Cells [BARC]3, Fig. 1B) was further characterized. We first asked whether DLA-88*50801 could be expressed on the cell surface, ideally seeking to make this determination in a manner independent of the our intended application (peptide binding). It was unclear, however, whether canine class I complexes would be stabilized by culturing at cold temperatures. The expression of human class I alleles transfected into RMA-S cells is not rescued by incubation at 26°C, even with co-transfection of human β2M (Anderson et al., 1993). Fortunately, unlike its HLA counterparts, DLA-88 was detectable on the surface of the transgenic RMA-S clone after 27°C overnight culture (Fig. 1C). Moreover, the differential expression of DLA-88 at 27 and 37°C encouraged the possibility of using BARC3s in a stabilization assay. Finally, while the staining of canine class I by H58A is well-known, the data here are the first to show that the classical (class Ia) gene product, DLA-88, is recognized by this mAb.
On RMA-S cells, cold-stabilized class I complexes are thermolabile, with virtually all H2-Db and -Kb disappearing from the surface after return to 37°C by 4 h (Ljunggren et al., 1990). This phenomenon is likely due to the particular instability of empty complexes or the dissociation of low-affinity self-peptides (De Silva et al., 1999) at higher temperatures, as pMHC molecules on TAP-sufficient RMA cells have a half-life (t1/2) >6 h at 37°C (Ljunggren et al., 1990). To determine whether canine class I complexes were similarly temperature-sensitive, BARC3 cells were cultured overnight at 27°C, and then placed at 37°C. With progressively longer incubation times at the higher temperature, surface DLA-88*50801 decreased proportionately (Fig. 1C and D), returning to 85–90% of the 37°C baseline value by 5 h. It is likely that a greater percentage would have disappeared had we blocked the transport of new complexes, but our goal here was to simply determine whether most unstable surface class I molecules could be removed by this convenient manipulation, which would be useful in designing future experiments.
3.2 Binding of canine self-peptides stabilizes DLA-88*50801 on the BARC3 cell surface
When peptides capable of binding H2-Db or -Kb are added to RMA-S cells, surface class I expression is maintained by formation of stable pMHC complexes (Ljunggren et al., 1990; Schumacher et al., 1990). We hypothesized that the same effect would be observed with BARC3s, and hence, tested whether incubation with peptides known to bind DLA-88*50801 would cause retention of surface canine class I complexes. Using mass spectrometry, we have recently identified 36 canine self-peptides eluted from this allele, and the binding of two of these – K9 and K11 – was subsequently confirmed by demonstrating the relative thermostability of in vitro-folded K9- and K11-DLA-88*50801 complexes (Ross P and Hess P, manuscript submitted). To serve as a putative negative control, we used the LCMV-derived immunodominant peptide NP396; the amino acid sequence (FQPQNGQFI) does not match the peptide-binding motif that we had previously defined for DLA-88*50801, which prefers hydrophobic residues at position (P)2 and P3, and a K or R at P9. While an unlikely binder of the canine allele, the ability of NP396 to strongly bind H2-Db served simultaneously as internal control to confirm that peptide-loading conditions were correct.
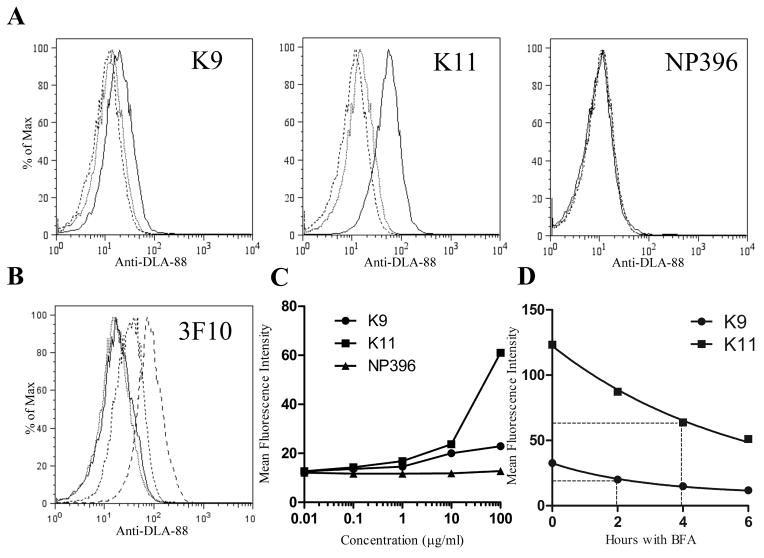
The surface class I complexes that accumulate on RMA-S during cold incubation are peptide-receptive – that is, exogenous high affinity peptides readily displace low affinity ones. Complexes emerging on the surface at 37°C are significantly less receptive (De Silva et al., 1999), and thus, RMA-S optimally are loaded with peptides during or immediately after cold incubation. Given that DLA-88*50801 behaved similarly to endogenous H2 class I molecules when exposed to low temperatures, it seemed reasonable to initially test the binding of canine peptides in the same fashion. Therefore, BARC3 cells were loaded at 27°C with K9, K11 or NP396 at various concentrations, and cultured for an additional 5 hours at 37°C to remove residual unstable complexes that could contribute background noise to the assay read-out. As shown in Figure 2A, the addition of K9 or K11 resulted in a dose-dependent increase in H58A staining, indicating stabilization of DLA88*50801 complexes on the BARC3 surface. This effect was consistent with specific binding of these peptides to the canine allele, as there was no staining of RMA-S cultured under the same conditions (not shown), nor staining of BARC3s after addition of the motif-mismatched peptide. The lack of H58A staining with NP396 was not the result of peptide degradation, suboptimal culture conditions, or other extraneous factors, as the nonamer did upregulate H2-Db expression on RMA-S cells (mean fluorescence intensity [MFI] of loaded cells was 217, vs. 14 for cells without peptide). Nor was the absence of H58A signal peculiar to NP396, as an influenza-derived peptide, NP366 (ASNENMETM) (Townsend et al., 1986), which also lacks the DLA88*50801 binding motif, similarly failed to increase expression of canine class I complexes (not shown). Finally, to corroborate that the increase in H58A staining observed with K9 and K11 peptide-pulsing truly reflected increased stabilization of surface DLA-88, we repeated the experiment using an alternative mAb that binds canine class I, 3F10 (Yang et al., 1987), and obtained identical results (Fig 2B). Collectively, these data demonstrate that BARC3 cells can be used in a standard stabilization assay to discriminate the binding of short peptides to the DLA88*50801 allele.
Fig. 2.
DLA-88*50801 is stabilized on the surface of BARC3 cells by the addition of DLA-88*50801 binding peptides to the culture medium. (A) The canine self peptides K9 and K11, but not the H2-Db-binding epitope NP396, increase surface expression of DLA-88*50801 in a dose-dependent manner. BARC 3 cells were cultured in the presence of 100 (solid line), 1 (dotted line), or 0.01 (dashed line) μg/ml of the indicated peptide; for clarity, data from 10 and 0.1 μg/ml peptide cultures are not shown. (B) Staining of K9- and K11-pulsed BARC3 cells with the anti-class I Ab 3F10 shows an increase in surface DLA-88*50801 expression that is similar to that observed using H58A. BARC3 cells were incubated with 100 μg/ml of NP396 (dotted line), K9 (dashed line), K11 (long dashed line), or no peptide (solid line). (C) Graphical representation of data shown in (A). (D) The K9 and K11 peptides dissociate from peptide-MHC complexes at different rates. BARC3 cells were cold-cultured overnight in the presence of 100 μg/ml of the indicated peptide, succeeded by an additional 5 hours at 37°C to remove unstable complexes. After washing to remove unbound peptide, cells were incubated at 37°C with BFA and harvested at the indicated time points. The dissociation half-life (t1/2 – shown by dotted lines) was calculated by non-linear regression, using a one phase exponential decay model for curve fitting. In the first experiment, the t1/2 for K9 was 1.9 h (95% CI 1.2 – 3.2 h), and 6.0 h for K11 (95% CI 3.6 – 18.9 h). In the second experiment (shown in graph), the t1/2 for K9 was 2.0 h (95% CI 1.7 – 2.4 h), and 4.0 h for K11 (95% CI 3.3 – 5.0 h). In this figure, data are representative of two [(B), (D)] or three [(A)] independent experiments.
The number of surface class I complexes on peptide-loaded RMA-S – measured as the MFI of the detecting mAb – is directly related to the peptide affinity for the MHC molecule (Andersen et al., 2003; Gairin and Oldstone, 1993), permitting the stabilization assay to be used semi-quantitatively. Our data with BARC3 cells show that, over a wide range of peptide concentrations, more DLA-88*50801 is retained on the surface with K11 than with K9 (Fig 2C), indicating a lower affinity of the nonamer. Interestingly, our previous study showed that complexes of K9-DLA-88*50801 and K11-DLA-88*50801 had roughly equivalent thermal denaturation profiles, which are measurements that also correlate with the strength of peptide-MHC binding (Morgan et al., 1997). The difference between these two findings suggests that the cell-based assay may be more sensitive, potentially related to the temperature at which complexes are assembled (10°C for in vitro folding vs. 27°C for cell loading). Such a discrepancy is not unprecedented: for example, wild-type and substituted HIV epitopes exhibited similar affinities for HLA-A*0201 in an in vitro assembly assay (4°C), but stabilized class I molecules on the surface of T2 cells (37°C) to differing degrees (Pogue et al., 1995). In particular, complexes with the substituted peptides had a three-fold increase in t1/2, the time point at which 50% of stabilized complexes are lost from the surface. To better characterize the relative binding strengths of K9 and K11, we therefore also compared their dissociation kinetics. After standard loading, BARC3s were washed extensively to remove unbound peptide and cultured with BFA to prevent surface transport of new complexes. Figure 2D shows that the t1/2 (dotted lines) of K9 is considerably shorter than that of K11. Importantly, these results demonstrate that BARC3 cells can be used to estimate the off-rate of DLA-88-binding peptides, which may be a better surrogate of immunogenicity than the relative degree of stabilization (van der Burg et al., 1996); in fact, t1/2 is used in some computer programs, such as BIMAS (http://www-bimas.cit.nih.gov/molbio/hla_bind/), to help predict CTL epitopes.
3.3 The DLA-88 heavy chain on the BARC3 cell surface is associated with murine β2M
The class I heavy chains on the surface of RMA-S, when stabilized by cold (Ljunggren et al., 1990) or peptide (Table 1, NP396-pulsed), are complexed with β2M. Hence, it seemed likely that peptide-loaded DLA-88*50801 molecules on BARC3 cells were also bound to β2M; however, the source was uncertain, as complexes potentially could form with murine β2M, or with bovine β2M contained in FBS (Bernabeu et al., 1984; Kefford et al., 1984). To distinguish these possibilities, we cultured BARC3 cells with K11 peptide in serum-containing and serum-free media, and found equivalent levels of surface DLA-88*50801, suggesting that the canine heavy chain could partner with the endogenous light chain. This hypothesis was confirmed by positive staining with an anti-mouse β2M mAb after pulsing with K11 in serum-free medium (Table 1). That DLA-88*50801 can form a functional heterotrimeric pMHC complex with endogenous β2M should not be entirely surprising, as the murine light chain can stabilize HLA class I molecules (Seong et al., 1988), albeit at reduced efficiency (Trymbulak and Zeff, 1997). While there is a ~30% difference in the sequence of human and murine β2M, many of the light chain residues at sites of contact between β2M and the α1, α2 and α3 domains of the heavy chain that are conserved between the two species are identical. An examination of these contact points in the first 66 amino acids of β2M – the region that is responsible for regulating the surface expression of class I molecules (Trymbulak and Zeff, 1997) – shows only one additional difference in canine β2M (I1V) from the murine protein, when compared to human β2M (Supplemental Table 1). Moreover, DLA-88*50801 contains 16 of the 19 conserved residues in the murine and human heavy chains that contact β2M (Ross et al., 2012).
Table 1.
Relative surface expression of murine β2m on untreated and peptide-pulsed RMA-S and BARC3 cells†
| Cell Line | Peptide (100 μg/ml)
|
||
|---|---|---|---|
| None | NP396 | K11 | |
| RMA-S | 80* | 240 | 91 |
| BARC3 | 161 | ND | 281 |
Data is representative of three independent experiments
Values indicate mean fluorescence intensity (MFI) of live cells;
ND – not done
In this study we showed that binding of 9- and 11-mer peptides to a recombinant canine class I molecule could be detected using a stably transfected clone of TAP-deficient RMA-S cells. This adaptation of the stabilization assay, developed originally with the murine parent line, is considerably simpler than alternate means for determining binding, such as in vitro competition experiments with radiolabeled peptides, or measurement of thermal denaturation by circular dichroism. As for mice and humans, binding confirmation should further reduce the pool of candidate epitopes predicted from large antigenic proteins to testable numbers. Additionally, the ability to compare relative affinities and dissociation kinetics with this cell-based method can likely provide a reasonable estimate of peptide immunogenicities, and therefore, a prioritized order for their evaluation in T-cell assays. In BARC3s, we found that the peptide-stabilized canine heavy chain was associated with endogenous β2M. Others have described the successful use of cell lines with such hybrid partnering, such as T2-Db, to measure peptide binding affinities (Gairin et al., 1995; Tian et al., 2007). Therefore, it may be fairly straightforward to construct a panel of lines by the transfection of prevalent canine class I alleles into RMA-S cells. In addition to screening putative epitopes, these BARC cells could serve, with peptide loading, as DLA-88-matched stimulators or targets to permit identification, for the first time, of immunodominant, epitope-specific CTL responses in the dog.
Supplementary Material
Acknowledgments
We thank Jeff Frelinger (University of Arizona) for the kind gifts of RMA-S cells and the NP366 and NP396 peptides. This work was supported by an NIH grant (K08 DK082264) and an NCSU Faculty Research and Development Fund award to P.R. Hess. The sponsors had no role in the design of the study, in the collection, analysis and interpretation of data, in the manuscript writing, or in the decision to submit the manuscript for publication.
Abbreviations
- β2M
Beta-2-microglobulin
- BARC
Bispecies Antigen Recognition Cells
- BFA
Brefeldin A
- DLA
Dog Leukocyte Antigen
- LCMV
Lymphocytic choriomeningitis virus
- MFI
Mean fluorescence intensity
- NP
Nucleoprotein
- TAP
Transporter associated with antigen processing
Footnotes
Conflict of Interest Statement
The authors declare no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andersen ML, Ruhwald M, Nissen MH, Buus S, Claesson MH. Self-peptides with intermediate capacity to bind and stabilize MHC class I molecules may be immunogenic. Scand J Immunol. 2003;57:21–27. doi: 10.1046/j.1365-3083.2003.01182.x. [DOI] [PubMed] [Google Scholar]
- Anderson KS, Alexander J, Wei M, Cresswell P. Intracellular transport of class I MHC molecules in antigen processing mutant cell lines. J Immunol. 1993;151:3407–3419. [PubMed] [Google Scholar]
- Assarsson E, Sidney J, Oseroff C, Pasquetto V, Bui HH, Frahm N, Brander C, Peters B, Grey H, Sette A. A quantitative analysis of the variables affecting the repertoire of T cell specificities recognized after vaccinia virus infection. J Immunol. 2007;178:7890–7901. doi: 10.4049/jimmunol.178.12.7890. [DOI] [PubMed] [Google Scholar]
- Bernabeu C, van de Rijn M, Lerch PG, Terhorst CP. Beta 2-microglobulin from serum associates with MHC class I antigens on the surface of cultured cells. Nature. 1984;308:642–645. doi: 10.1038/308642a0. [DOI] [PubMed] [Google Scholar]
- Davis WC, Marusic S, Lewin HA, Splitter GA, Perryman LE, McGuire TC, Gorham JR. The development and analysis of species specific and cross reactive monoclonal antibodies to leukocyte differentiation antigens and antigens of the major histocompatibility complex for use in the study of the immune system in cattle and other species. Vet Immunol Immunopathol. 1987;15:337–376. doi: 10.1016/0165-2427(87)90005-5. [DOI] [PubMed] [Google Scholar]
- De Silva AD, Boesteanu A, Song R, Nagy N, Harhaj E, Harding CV, Joyce S. Thermolabile H-2Kb molecules expressed by transporter associated with antigen processing-deficient RMA-S cells are occupied by low-affinity peptides. J Immunol. 1999;163:4413–4420. [PubMed] [Google Scholar]
- Gairin JE, Mazarguil H, Hudrisier D, Oldstone MB. Optimal lymphocytic choriomeningitis virus sequences restricted by H-2Db major histocompatibility complex class I molecules and presented to cytotoxic T lymphocytes. J Virol. 1995;69:2297–2305. doi: 10.1128/jvi.69.4.2297-2305.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gairin JE, Oldstone MB. Virus and cytotoxic T lymphocytes: crucial role of viral peptide secondary structure in major histocompatibility complex class I interactions. J Virol. 1993;67:2903–2907. doi: 10.1128/jvi.67.5.2903-2907.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graumann MB, DeRose SA, Ostrander EA, Storb R. Polymorphism analysis of four canine MHC class I genes. Tissue Antigens. 1998;51:374–381. doi: 10.1111/j.1399-0039.1998.tb02976.x. [DOI] [PubMed] [Google Scholar]
- Kefford RF, Calabi F, Fearnley IM, Burrone OR, Milstein C. Serum beta 2-microglobulin binds to a T-cell differentiation antigen and increases its expression. Nature. 1984;308:641–642. doi: 10.1038/308641a0. [DOI] [PubMed] [Google Scholar]
- Ladiges WC, Deeg HJ, Aprile JA, Raff RF, Schuening F, Storb R. Differentiation and function of lymphohemopoietic cells in the dog. In: Miyasaka M, Trnka Z, editors. Differentiation Antigens in Lymphohemopoietic Tissues. Marcel Dekker, Inc; New York: 1988. pp. 307–336. [Google Scholar]
- Ljunggren HG, Stam NJ, Ohlen C, Neefjes JJ, Hoglund P, Heemels MT, Bastin J, Schumacher TN, Townsend A, Karre K, et al. Empty MHC class I molecules come out in the cold. Nature. 1990;346:476–480. doi: 10.1038/346476a0. [DOI] [PubMed] [Google Scholar]
- Lundegaard C, Lund O, Kesmir C, Brunak S, Nielsen M. Modeling the adaptive immune system: predictions and simulations. Bioinformatics. 2007;23:3265–3275. doi: 10.1093/bioinformatics/btm471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan CS, Holton JM, Olafson BD, Bjorkman PJ, Mayo SL. Circular dichroism determination of class I MHC-peptide equilibrium dissociation constants. Protein Sci. 1997;6:1771–1773. doi: 10.1002/pro.5560060819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogue RR, Eron J, Frelinger JA, Matsui M. Amino-terminal alteration of the HLA-A*0201-restricted human immunodeficiency virus pol peptide increases complex stability and in vitro immunogenicity. Proc Natl Acad Sci U S A. 1995;92:8166–8170. doi: 10.1073/pnas.92.18.8166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rammensee H, Bachmann J, Emmerich NP, Bachor OA, Stevanovic S. SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics. 1999;50:213–219. doi: 10.1007/s002510050595. [DOI] [PubMed] [Google Scholar]
- Ross P, Buntzman AS, Vincent BG, Grover EN, Gojanovich GS, Collins EJ, Frelinger JA, Hess PR. Allelic diversity at the DLA-88 locus in the Golden Retriever and Boxer breeds is limited. Tissue Antigens. 2012 doi: 10.1111/j.1399-0039.2012.01889.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz M, Aichele P, Schneider R, Hansen TH, Zinkernagel RM, Hengartner H. Major histocompatibility complex binding and T cell recognition of a viral nonapeptide containing a minimal tetrapeptide. Eur J Immunol. 1991;21:1181–1185. doi: 10.1002/eji.1830210513. [DOI] [PubMed] [Google Scholar]
- Schumacher TN, Heemels MT, Neefjes JJ, Kast WM, Melief CJ, Ploegh HL. Direct binding of peptide to empty MHC class I molecules on intact cells and in vitro. Cell. 1990;62:563–567. doi: 10.1016/0092-8674(90)90020-f. [DOI] [PubMed] [Google Scholar]
- Seong RH, Clayberger CA, Krensky AM, Parnes JR. Rescue of Daudi cell HLA expression by transfection of the mouse beta 2-microglobulin gene. J Exp Med. 1988;167:288–299. doi: 10.1084/jem.167.2.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian S, Maile R, Collins EJ, Frelinger JA. CD8+ T cell activation is governed by TCR− peptide/MHC affinity, not dissociation rate. J Immunol. 2007;179:2952–2960. doi: 10.4049/jimmunol.179.5.2952. [DOI] [PubMed] [Google Scholar]
- Townsend AR, Rothbard J, Gotch FM, Bahadur G, Wraith D, McMichael AJ. The epitopes of influenza nucleoprotein recognized by cytotoxic T lymphocytes can be defined with short synthetic peptides. Cell. 1986;44:959–968. doi: 10.1016/0092-8674(86)90019-x. [DOI] [PubMed] [Google Scholar]
- Trymbulak WP, Zeff RA. Expression of HLA class I molecules assembled with structural variants of human beta2-microglobulin. Immunogenetics. 1997;46:418–426. doi: 10.1007/s002510050296. [DOI] [PubMed] [Google Scholar]
- van der Burg SH, Visseren MJ, Brandt RM, Kast WM, Melief CJ. Immunogenicity of peptides bound to MHC class I molecules depends on the MHC-peptide complex stability. J Immunol. 1996;156:3308–3314. [PubMed] [Google Scholar]
- Yanagi Y, Tishon A, Lewicki H, Cubitt BA, Oldstone MB. Diversity of T-cell receptors in virus-specific cytotoxic T lymphocytes recognizing three distinct viral epitopes restricted by a single major histocompatibility complex molecule. J Virol. 1992;66:2527–2531. doi: 10.1128/jvi.66.4.2527-2531.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang TJ, Chandler JP, Dunne-Anway S. Growth stage dependent expression of MHC antigens on the canine transmissible venereal sarcoma. Br J Cancer. 1987;55:131–134. doi: 10.1038/bjc.1987.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yewdell JW. Confronting complexity: real-world immunodominance in antiviral CD8+ T cell responses. Immunity. 2006;25:533–543. doi: 10.1016/j.immuni.2006.09.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.