Abstract
We describe the results of the first genome-wide association study of PTSD performed using trauma-exposed white non-Hispanic participants from a cohort of veterans and their intimate partners (295 cases and 196 controls). Several SNPs yielded evidence of association. One SNP (rs8042149), located in the retinoid-related orphan receptor alpha gene (RORA), reached genome-wide significance. Nominally significant associations were observed for other RORA SNPs in two African American replication samples—one from the veteran cohort (43 cases and 41 controls) and another independent cohort (100 cases and 421 controls). However, only the associated SNP from the veteran African American replication sample survived gene-level multiple testing correction. RORA has been implicated in prior GWAS studies of psychiatric disorders and is known to play an important role in neuroprotection and other behaviorally-relevant processes. This study represents an important step towards identifying the genetic underpinnings of PTSD.
Keywords: Posttraumatic stress disorder, Genome-wide association, RORA
Introduction
Posttraumatic Stress Disorder (PTSD) is a psychiatric disorder defined by profound disturbances in cognitive, emotional, behavioral and physiological functioning that occurs in response to a psychologically traumatic event. The DSM-IV1 specifies that the diagnosis applies to individuals who develop a constellation of symptoms after exposure to an event involving perceived or threatened loss of life, serious injury, or loss of physical integrity. The symptoms of PTSD in DSM-IV are organized under three clusters: 1) re-experiencing (e.g., intrusive thoughts, nightmares, flashbacks, and reactivity to reminders of the trauma); 2) avoidance and emotional numbing (e.g., avoiding stimuli associated with the trauma and anhedonia); and 3) hyperarousal (e.g., anger, hypervigilance, exaggerated startle response, and sleep disruption).
Epidemiological studies have shown that the majority of individuals in the general population experiences a traumatic event meeting the PTSD stressor criterion at some point during their lifetime though only a minority go on to develop PTSD2. After exposure, the probability of developing PTSD is estimated to be approximately 10% in the general population, though higher rates (i.e., closer to 25%) have been observed in samples of individuals exposed to severe traumas involving violence and/or life threat (e.g., combat or rape). The traumatic event is, by definition, necessary for the development of PTSD, but the fact that not everyone exposed to trauma goes on to develop PTSD indicates that individual difference factors, including genes, likely play a significant role in the etiology of the disorder3, 4.
Twin and heritability studies suggest that 30–70% of variation in PTSD risk is determined by genetic factors5–7 and recently there has been an increase in the number of studies examining the molecular genetics of PTSD (for review see 3). Most of these studies have been limited to candidate genes commonly examined in psychiatric disorders such as those involved with the regulation of dopamine, serotonin, and glucocorticoid metabolism. Results of those studies have been inconsistent and the heritability of PTSD remains largely unexplained.
In this article, we report the first positive results from a genome-wide scan for genetic risk factors for PTSD. We genotyped 2.5 million SNPs covering the entire genome and found an association at genome-wide levels of significance between PTSD and the Retinoic Acid Orphan Receptor A (RORA) gene in a U.S. Department of Veterans Affairs (VA) sample of trauma-exposed white non-Hispanic men and women. Nominally significant associations between RORA SNPs and PTSD were also observed in a replication sample of individuals of African American descent from the VA sample and in a cohort of trauma-exposed African Americans from the Detroit Neighborhood Health study8.
Materials and Methods
Participants
The VA sample was comprised of U.S. military veterans and a subset of their intimate partners (N=852) who were enrolled in one of two studies conducted at two U.S. Department of Veterans Affairs (VA) Healthcare facilities. Both studies were reviewed and approved by the appropriate human subjects and local institutional review boards. Data were not analyzed for 42 cases who did not complete the protocol, 25 cases who enrolled in both studies (in which case, only data from the first study was included) and DNA was not available for 24 participants due to problems with the blood draw. This yielded a subsample of (n=761) with complete data, of whom 729 had been exposed to a traumatic event meeting the DSM-IV PTSD Criterion A definition as determined by a clinician using the Clinician-Administered PTSD Scale (CAPS9). Of these events, the most common type was combat trauma, endorsed by 33.6% of the sample (54.7% of men; 2.4% of women). The trauma-exposed subsample was predominantly male (n=435, 59.7%) with a mean age of 51.5 (SD=10.9). 496 (68%) were veterans and the remaining 233 were their spouses or intimate partners. 409 participants (53.7%) met criteria for a lifetime diagnosis of PTSD based on the CAPS.
Ancestry was determined using 10,000 randomly chosen markers with MAF>.05 by the program STRUCTURE 10, 11 which performs a Bayesian clustering analysis to assign subjects to ancestry groups (see additional details below). From this analysis, 491 individuals with a history of trauma exposure and valid genotype data were identified as having primarily white non-Hispanic ancestry, 84 were identified as having African American ancestry, and 105 were of “other” ancestry (including individuals self-described as having Hispanic, Native American, or Asian ancestry). We found no evidence of PTSD-associated population substructure within the VA-sample ancestry groups (see supplementary materials for details).
Instruments
The Clinician Administered PTSD Scale (CAPS)9
The CAPS is a 30-item structured diagnostic interview that assesses the frequency and severity of the 17 DSM-IV PTSD symptoms, 5 associated features, and functional impairment. VA study participants were administered the CAPS to assess current and lifetime PTSD symptoms. All interviews were video-recorded and approximately one-quarter (n=197) of them were viewed and scored by an independent rater for purposes of quality control and evaluating inter-rater reliability. Inter-rater reliability for lifetime PTSD diagnosis, based on comparison of the primary interviewer and secondary raters scores was excellent (kappa=.87).
Traumatic Life Events Questionnaire (TLEQ)12
The TLEQ is a self-report measure that assesses exposure to 22 types of potentially traumatic events that meet the DSM-IV PTSD Criterion A1 definition for a traumatic event. For each traumatic event that is endorsed, the respondent is subsequently asked to provide (1) the number of times the event was experienced using a 7-point scale ranging from “never” to “more than five times” and (2) whether he or she experienced intense fear, helplessness, or horror in response to the event (i.e., to assess DSM-IV PTSD Criterion A2). The TLEQ has shown good test-retest reliability over a two-week interval (mean kappa=.63, mean percent agreement=86%), excellent convergent validity with interview-based measures of trauma exposure (mean percent agreement=92%), and good predictive validity for PTSD status12.
Genotyping
In the VA sample, DNA was isolated from peripheral blood samples on a Qiagen AutoPure instrument with Qiagen reagents and samples normalized using PicoGreen assays (Invitrogen). Each DNA sample was run on an Illumina OMNI 2.5–8 array and scanned using an Illumina HiScan System according to the manufactures protocol. All available DNA samples from the two studies were genotyped (n = 810). Seven samples had an overall call rate of < 95% and were eliminated from analysis. In the remaining 803 samples, the call rate was 99.34%. Of those, one was excluded due to a typographical error in the ID key, which prevented linking their data to the phenotypic database. There were 36 duplicates from subjects who participated in both studies (the 24 mentioned earlier, plus additional duplicates from subjects who did not complete one protocol or the other, but which provided genetic samples), in which case the second sample was removed from analysis. In addition, subjects were checked for consistency between their self-reported sex and the sex indicated by X-chromosome homozygosity using PLINK. Ten mismatches were detected. Seven of these were determined to be sample swaps that could be resolved based on prior genotyping. Two of the mismatches could not be resolved and were excluded from further analysis. One of the apparent mismatches was explained due to the subject’s transgendered status, this subject was also removed to prevent bias in the estimation of sex-specific effects. This left 763 genotyped cases whose genotype data were analyzed with STRUCTURE.
SNPs were removed from the analysis if they yielded greater than 5% missing genotypes or if they were rare (< 5% minor allele frequency). After these filters were applied, 1,199,491 autosomal SNPs remained for analysis in the non-Hispanic white group. None of the reported SNPs failed Hardy-Weinburg Equilibrium tests in Controls.
Statistical Methods
Because PTSD is conditional on trauma exposure, we restricted our analyses to subjects who reported experiencing a qualifying trauma as assessed by a clinician during the CAPS interview. Association between the autosomal SNPs and PTSD was evaluated using a allelic test of association and performed in PLINK (v 1.07, October 2009)13. Significance was determined using Fisher’s Exact Test. The white non-Hispanic group was analyzed separately as the discovery sample. The African American group was analyzed as a replication sample. (We will not be presenting the results for the third [Hispanic, Native American, and Asian] group due to the small number of controls [n=35] and admixed nature of the sample.) Association between all common SNPs (MAF>.05) was assessed in the white non-Hispanics. We used established levels of significance for genome-wide studies and considered p<10−5th as suggestive evidence of association and p<5×10−8th as genome-wide significant. In the VA African American replication sample, all SNPs passing quality control and MAF filters within 5 kb of implicated genes were examined. The max(T) permutation procedure in PLINK was used to correct for the testing of multiple SNPs within a gene in the replication sets. Haploview14 was used to compute LD information. Plots of association in genes of interest were created with LocusZoom15. Subsequent exploratory analysis examining the possibility of a SNP×trauma-exposure interaction were performed in R16.
Detroit Neighborhood Health Study (DNHS)
Subsequent to our analysis of the VA discovery and replication samples, we examined data from the Detroit neighborhood Health study (DNHS), a longitudinal cohort of predominately African American adults (18+) living in Detroit, Michigan and screened for lifetime trauma exposure. Data collection and genotyping procedures are described in detail in our supplementary material and additional information reported elsewhere8, 17. In particular, association was assessed for SNPs from the Illumina HumanOmniExpress BeadChip in and near implicated genes in a sample of trauma-exposed African Americans (n= 100 cases and 421 controls). As in the VA replication sample, PLINK was used to compute the allelic test of association for all SNPs passing QC with MAF>.05 within 5 kb of implicated genes, and gene-level multiple-testing significance was assessed using the PLINK max(T) permutation procedure.
Results
GWAS Results
The Manhattan Plot showing the results of the genome-wide analysis of lifetime PTSD within trauma-exposed individuals from the discovery sample (295 cases and 196 controls) is shown in Figure 1. Results for all SNPs with p<10−4 are available in supplementary Table 1. A total of 10 SNPs reached the “suggestive” level of significance (p<10−5). These are presented in Table 1 and could be categorized into 4 haplotype blocks: one each on chromosomes 1, 6, 15 and 17. The haplotype blocks on chromosomes 1 and 17 were at considerable distance from any well-characterized genes. SNPs on chromosome 6 and 15 were within haplotype blocks for the brefeldin A-inhibited guanine nucleotide-exchange protein 3 (BIG3) gene and the retinoid-related orphan receptor alpha (RORA) gene respectively.
Figure 1.
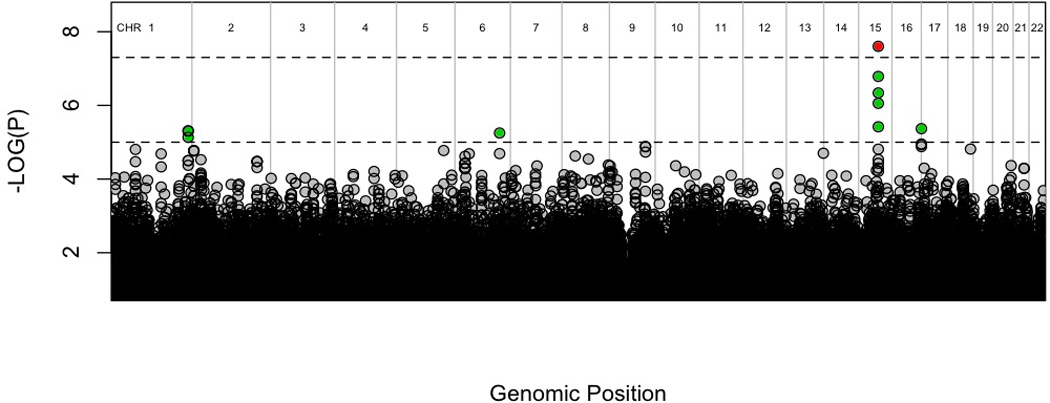
Manhattan plot of genome-wide association results in discovery group.
Dashed lines and colors represent suggestive and genome-wide significance respectively, with green=p<10−5 and red=p<5×10−8.
Table 1.
SNPs with P<10−5 from the discovery GWAS of PTSD in trauma-exposed white non-Hispanic subjects (n=295 cases and 196 controls).
| SNP | CHR | Position | Gene | Risk Allele |
AF Cases |
AF Controls |
OR | P |
|---|---|---|---|---|---|---|---|---|
| rs1021356 | 1 | 238,573,691 | -- | G | 76% | 62% | 1.9 | 4.9×10−6 |
| rs12118091 | 238,576,614 | C | 76% | 62% | 1.9 | 4.9×10−6 | ||
| rs113508478 | 238,576,715 | A | 75% | 61% | 1.9 | 7.4×10−6 | ||
| rs72986828 | 6 | 138,484,125 | BIG3 | C | 12% | 3.6% | 3.5 | 5.6×10−6 |
| rs8041061 | 15 | 61,124,838 | RORA | A | 58% | 41% | 2.0 | 1.6×10−7 |
| rs8042149 | 61,124,953 | C | 61% | 43% | 2.1 | 2.5×10−8 | ||
| rs4775301 | 61,126,859 | G | 60% | 43% | 2. | 4.6×10−7 | ||
| rs8024133 | 61,130,639 | A | 61% | 45% | 1.9 | 8.8×10−7 | ||
| rs11071561 | 61,131,683 | T | 64% | 49% | 1.8 | 3.8×10−6 | ||
| rs71355256 | 17 | 259,248 | C17orf97 | G | 97% | 90% | 3.7 | 4.3×10−6 |
One SNP within the RORA gene (rs8042149) met genome-wide significance (p<5×10−8) as well as Bonferroni-corrected (p<4.16×10−8 after correcting for 1,199,491 SNPs examined) significance thresholds. An additional 4 SNPs in RORA were suggestive of association (Figure 2A). The evidence of association was not specific to individuals of a particular sex. P-values were proportional to sample size for each of the sexes and OR estimates were similar; in n=319 males the pMale=1.35×10−5 and in n=172 females pFemale=.00050; ORMale=2.13 and ORFemale=2.19; p=.95 for a test of equality of rs8042149 effect by sex.
Figure 2.
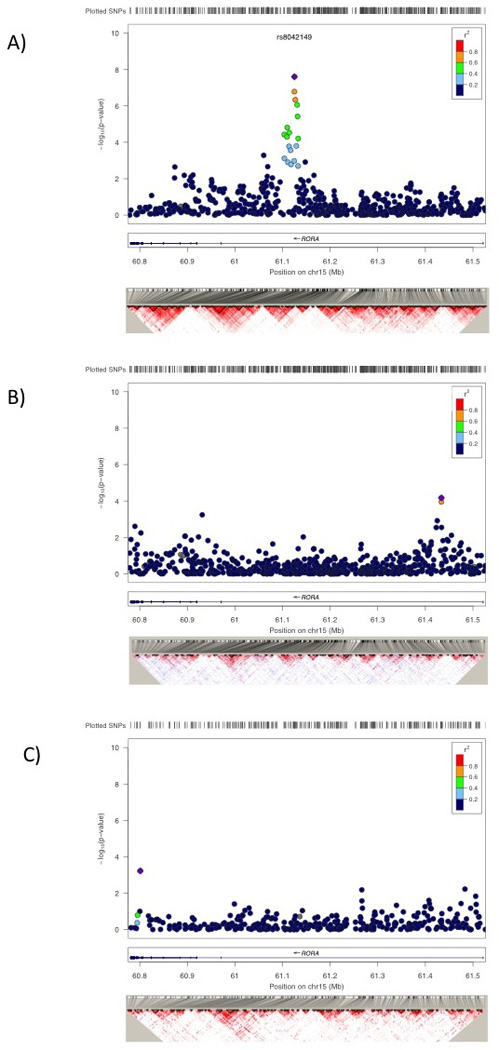
Association results in the RORA gene for VA Discovery sample of white non-Hispanics (A), the VA African American replication sample (B), and the Detroit Neighborhood Health Study African American replication sample (C).
To further explore the relationship between rs8042149 and PTSD, we next examined the relationship between its genotype, the probably of PTSD, and the total number of traumatic events (NTE) as measured by the TLEQ12 as a quantitative measure of environmental adversity. Table 3 lists the percentage of cases with PTSD for the entire sample as a function of Low/Medium/or High levels of traumatic exposure, with the cutoffs determined to approximate the terciles of the NTE distribution. Examining the probability of a PTSD diagnosis by genotype indicated that subjects with relatively low levels of exposure to trauma with the high-risk (GG) genotype had a similar likelihood of developing PTSD as those from the high-exposure group but with the low-risk (AA) genotype (63% and 61% respectively). A logistic regression model indicated that the NTE is strongly associated with the risk of PTSD (p=1.5×10−11). In a logistic regression model that included NTE and rs8042149, the effect of rs8042149 was significant (p=4.4×10−8). A further logistic model indicated that NTExSNP interaction was not significant (p=.4576).
Table 3.
Most significant RORA SNPs from case/control analysis of trauma-exposed subjects from the African American replication samples
| Group | SNP | Position | Risk Allele |
AF Cases |
AF Controls |
OR | Nominal P |
Adjusted P |
|---|---|---|---|---|---|---|---|---|
| VA-African Americans (n=43 cases and 41 Controls) |
rs11071587 | 61,433,543 | A | 45% | 17% | 4.03 | .00011 | .029 |
| rs11071588 | 61,433,578 | C | 53% | 23% | 3.81 | 6.77×10−5 | .018 | |
| African Americans from the DNHS(n= 100 cases and 421 controls) |
rs16942660 | 60,801,066 | G | 94% | 86% | 2.65 | .00090 | .202 |
| rs893290 | 61,483,157 | C | 18% | 11% | 1.85 | .0052 | .698 |
Replication Analyses
We next analyzed trauma-exposed cases and trauma-exposed controls in the African American subset of the VA cohort (43 cases/41 controls) for association to SNPs implicated in the discovery cohort. The peak SNP in white non-Hispanics, rs8042149, was not associated with PTSD in the African American sample (p=.87). However, when we expanded our search, other SNPs in RORA were found to be nominally significantly associated with PTSD (Figure 2B and Table 3). The most significant SNP, rs11071588, had a p=6.77×10−5 which remained significant after multiple testing correction (pADJUSTED=.018).
Subsequent to our analyses of the VA data, we requested association results for a set of SNPs in RORA from the DNHS. In trauma-exposed African Americans from the DNHS (100 cases/421 controls), the most significant SNP associated with lifetime PTSD within RORA was rs16942660 (p=.0009) 633 kb from the peak SNP in African American’s from the VA sample and 324 kb away from the peak SNP observed in the discovery sample of white non-Hispanic subjects. The most significant SNP in the VA African American sample, rs11071588, was not significant in the Detroit sample (p=.43). However, there was a cluster of nominally significant SNPs flanking this SNP; there were 5 SNPs with p<0.05 within 50 kb of rs11071588 compared to 7 nominally significant SNPs observed anywhere else in the gene region which spans 760 kb. The most significant of these was rs893290 (p=.0052) approximately 50 kb from rs11071588 followed by rs782928 (p=.018) which is 26 kb from rs11071588 in the opposite direction. While the number of nominally significant SNPs clustering around the peak African American SNP from the VA sample is interesting, none of the RORA SNPs in the DNHS survive correction for multiple testing at the gene-based level (Table 3).
Discussion
This report describes the results of a high-density GWAS of PTSD using diagnostic information based on clinical interview and genomic data from a 2.5 million SNP array. Analyses of data from a white non-Hispanic cohort of adult men and women with histories of exposure to trauma revealed a significant association between a lifetime diagnosis of PTSD and a SNP in the RORA gene (rs8042149) that met genome-wide and Bonferroni-corrected levels of significance. We observed a notably high (>60%) incidence of the disorder among those with limited histories of trauma exposure who carried the high-risk allele, though the prevalence of PTSD was elevated for individuals with the high-risk genotype across levels of exposure. In our analysis, which only included trauma-exposed individuals, no significant interaction between level of exposure and genotype was observed. Nominally significant associations between other RORA SNPs and PTSD were found in an African American subsample from the same study, and in a second much larger independent African American cohort, although only the most significant SNP from the VA-African American cohort survived gene-level multiple testing correction. To our knowledge, this is the first report of findings from a GWAS of PTSD and the first discovery of a significant risk marker for the disorder using this approach.
This is not, however, the first GWAS study to implicate RORA as a psychiatric risk factor. Prior studies have found RORA to be associated with attention-deficit hyperactivity disorder18, bipolar disorder19, and autism20, 21. In addition, perhaps the most extensive evidence for the role of RORA in psychiatric illness comes from research on the genetics of major depression. Depression is particularly relevant because of its high rates of comorbidity with PTSD and twin studies that have shown the two disorders to be influenced by a common genetic factor (e.g. 22, 23). Evidence for a significant association between RORA and depression comes from several sources including a GWAS of anti-depressant response in the Sequenced Treatment Alternatives to Relieve Depression (STAR*D) study that found the third most significant SNP to be located in RORA (p=8.19×10−6 at rs809736)24. This SNP (at 61.3 Mb) was also associated with depression remission (p=7.64×10−5). In a GWAS of trait depression, as indexed by a self-report measure of a facet of neuroticism, the single most significant SNP was also in RORA (rs12912233 p=6.3×10−7 at rs12912233 at 61.3 Mb)25. Similarly, a candidate gene study examining circadian rhythm genes involved in depression showed evidence of association with a SNP in Intron 2 of RORA (rs2028122 at 60.8 Mb multiple-testing corrected p=.03; Lavebratt et al. 2010)26. Taken together, prior GWAS studies suggest that polymorphisms of the RORA gene likely confer a general (i.e., non-disorder-specific) risk for the development of psychopathology, possibly via its relationship to a common vulnerability factor such as trait negative emotionality (i.e., neuroticism). This dimension of individual differences has long been conceptualized as the primary temperamental risk factor for disorders of the internalizing spectrum (i.e., anxiety and unipolar mood disorders27, 28) though more recent evidence suggests that it plays a more ubiquitous role in a broader array of psychopathologies including externalizing disorders and PTSD29, 30.
RORA belongs to the NR1 subfamily of nuclear hormone receptors. Other members of this family include the glucocorticoid, estrogen, progesterone, and androgen receptors, all of which have established roles in brain development and function. The function of the RORA gene is complex and the protein encoded by it involved in an intriguing variety of processes including brain development, neuroprotection, and the regulation of circadian rhythms and steroid hormones. Though research on the possible mechanism(s) by which polymorphisms of the RORA gene might confer risk for PTSD is in its infancy, we suggest a model in which alterations in RORA levels, due to inherited genetic variation, may reduce the capacity of neurons to respond to the biochemical stressors (oxidative stress, steroid hormone elevations, and inflammation) induced by traumatic stress. RORA is widely expressed in neurons of cortical and subcortical structures31 and serves several neuroprotective functions. It protects cortical neurons against oxidative stress-induced apoptosis by increasing the expression of the antioxidant proteins32. In glial cells, its expression is upregulated by the presence of destructive pro-inflammatory cytokines, and during hypoxia, its expression is upregulated in neurons and astrocytes33. Oxidative stress and inflammation have been implicated as possible mechanisms of the deleterious effects of traumatic stress on the brain34 and these neurodegenerative processes have been linked to functional and structural abnormalities in PTSD-relevant brain structures including the hippocampus35, dorsolateral and ventromedial prefrontal cortex36, 37, and anterior cingulate cortex38, 39 (for a review see 40). RORA’s role in this pathology is further supported by evidence that polymorphisms of the gene are linked to thickness of cortical gray matter and fractional anisotropy of cerebral white matter in humans41. In that study, RORA SNP rs341401, which was the 10th most strongly associated with PTSD in this study (p=6.20×10−5), was one of the strongest predictors of a composite index of structural brain imaging parameters.
In addition to RORA, we found other regions that showed suggestive evidence of association (Table 3). However, none of the SNPs were in or near genes that at this point would conceivably be related to PTSD pathogenesis (e.g., BIG-3 is involved in cartilage development, and the function of C17orf97 is unknown). We also failed to find significant or suggestive evidence of association with genes previously studied in PTSD including those involved in dopamine and serotonin metabolism. Thus, a limitation of this study, as in all GWAS studies, is the likelihood of false positive associations due to chance. At the same time, the stringent P-value thresholds applied to GWAS studies can limit the ability to detect true associations with modest levels of association.
The primary strength of this study was the carefully assessed trauma-exposed discovery sample that was characterized by a high prevalence of lifetime PTSD. The primary weaknesses were the relatively modest sizes of the available samples. While the size of our discovery sample was not in the range of GWASs published by major consortia for disorders such as bipolar disorder or schizophrenia (e.g. 42, 43), it was within the range of samples used in other published GWAS studies of psychiatric disorders. For example, in a 2011 review of GWAS studies of schizophrenia and related phenotypes, there were 17 case/control studies identified, 5 of which had a smaller number of cases than in our discovery sample and 3 were published with a similar number or fewer controls than our discovery sample44. That said, given our modest sample sizes (especially in our replication samples) and lack of a white non-Hispanic replication sample, findings from this study should be considered provisional until further evidence of replication is obtained. As with any complex disease, much larger samples will be necessary to fully characterize the impact of RORA on PTSD susceptibility and to confirm our “suggestive” level associations on chromosomes 1, 6, and 17.
Based on these findings, we believe that it would be useful for future investigations to evaluate the hypothesis that RORA moderates the neurotoxic effects of traumatic stress though studies examining genetic associations with structural and functional neural imagining parameters in trauma exposed individuals with and without PTSD. In addition, research using methods such as whole genome sequencing and testing for association in other cohorts will be needed to identify the particular functional variant within RORA that is responsible for the association with PTSD. Our observation that different RORA SNPs are associated with PTSD in African Americans suggests that humans harbor multiple variants affecting protein function or gene expression of RORA. This variability may also be driven by differences in the pattern of linkage disequilibrium between the populations. In sum, the association of PTSD with RORA offers new insight into possible biological and neurological underpinnings of the psychopathological response to traumatic stress and we hope that these findings will stimulate the development of new models of gene-environment interactions in PTSD.
Supplementary Material
Table 2.
Percentage of participants with PTSD by trauma exposure and rs8042149 genotype in the white non-Hispanic discovery sample.
| Trauma Exposure |
Low (1 to 17) |
Medium (18 to 31) |
High (>31) |
|
|---|---|---|---|---|
| % PTSD for ALL Subjects: | 43% (n=159) | 59% (n=166) | 78% (n=166) | |
| % PTSD for subjects by genotype of rs8042149 |
AA | 22% (n=36) | 46% (n=37) | 61% (n=36) |
| AC | 40% (n=75) | 56% (n=84) | 77% (n=74) | |
| CC | 63% (n=48) | 76% (n=45) | 89% (n=56) | |
Acknowledgements
Funding for this study was provided by National Institute on Mental Health award RO1 MH079806 and a Department of Veterans Affairs Merit Review Grant awarded to Mark W. Miller. Mark W. Logue is funded by National Institute on Mental Health award K01 MH076100. The Detroit Neighborhood Health Study (PI: Allison Aiello) is funded by DA022720, DA 022720-S1, and MH 088283. Karestan C. Koenen is funded by MH093612, MH078928, P51RR000165 and RC4MH092707. Funding for genotyping of the VA sample was provided by the VA National Center for PTSD.
Footnotes
Supplementary information is available at Molecular Psychiatry's website
COI Statement: The authors report no financial conflicts of interest.
Authorship Statement:
Study concept and design: Miller, Baldwin, and Logue.
Sample collection and interpretation of VA data: Miller, Reardon, & Wolf.
Genotyping of VA sample: Baldwin and Melista.
Data cleaning and management of VA sample data: Logue, Reardon, & Wolf.
Analysis of VA genotype data: Logue.
Sample collection and interpretation of DNHS data: Galea, Aiello, Wildman, Uddin & Koenen
Genotyping of DNHS data: Uddin & Wildman
Data cleaning and management of DNHS data: Aiello, Guffanti & Koenen
Analysis of DNHS genotype data: Guffanti & Koenen
Drafting and critical revision of the Manuscript: Miller, Baldwin, Logue, Guffanti, and Koenen.
All authors gave approval to the final manuscript.
References
- 1.American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders 4th Edition. 4 edn. Washington, DC: 1994. [Google Scholar]
- 2.Kessler RC, Sonnega A, Bromet E, Hughes M, Nelson CB. Posttraumatic stress disorder in the National Comorbidity Survey. Archives of general psychiatry. 1995;52(12):1048–1060. doi: 10.1001/archpsyc.1995.03950240066012. [DOI] [PubMed] [Google Scholar]
- 3.Cornelis MC, Nugent NR, Amstadter AB, Koenen KC. Genetics of post-traumatic stress disorder: review and recommendations for genome-wide association studies. Curr Psychiatry Rep. 2010;12(4):313–326. doi: 10.1007/s11920-010-0126-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koenen KC, Amstadter AB, Nugent NR. Gene-environment interaction in posttraumatic stress disorder: an update. J Trauma Stress. 2009;22(5):416–426. doi: 10.1002/jts.20435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.True WR, Rice J, Eisen SA, Heath AC, Goldberg J, Lyons MJ, et al. A twin study of genetic and environmental contributions to liability for posttraumatic stress symptoms. Archives of general psychiatry. 1993;50(4):257–264. doi: 10.1001/archpsyc.1993.01820160019002. [DOI] [PubMed] [Google Scholar]
- 6.Stein MB, Jang KL, Taylor S, Vernon PA, Livesley WJ. Genetic and environmental influences on trauma exposure and posttraumatic stress disorder symptoms: a twin study. The American journal of psychiatry. 2002;159(10):1675–1681. doi: 10.1176/appi.ajp.159.10.1675. [DOI] [PubMed] [Google Scholar]
- 7.Sartor CE, McCutcheon VV, Pommer NE, Nelson EC, Grant JD, Duncan AE, et al. Common genetic and environmental contributions to post-traumatic stress disorder and alcohol dependence in young women. Psychological medicine. 2011;41(7):1497–1505. doi: 10.1017/S0033291710002072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Uddin M, Aiello AE, Wildman DE, Koenen KC, Pawelec G, de Los Santos R, et al. Epigenetic and immune function profiles associated with posttraumatic stress disorder. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(20):9470–9475. doi: 10.1073/pnas.0910794107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blake DD, Weathers FW, Nagy LM, Kaloupek DG, Gusman FD, Charney DS, et al. The development of a Clinician-Administered PTSD Scale. J Trauma Stress. 1995;8(1):75–90. doi: 10.1007/BF02105408. [DOI] [PubMed] [Google Scholar]
- 10.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155(2):945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164(4):1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kubany ES, Haynes SN, Leisen MB, Owens JA, Kaplan AS, Watson SB, et al. Development and preliminary validation of a brief broad-spectrum measure of trauma exposure: the Traumatic Life Events Questionnaire. Psychological assessment. 2000;12(2):210–224. doi: 10.1037//1040-3590.12.2.210. [DOI] [PubMed] [Google Scholar]
- 13.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. Epub 2007 Jul 2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 15.Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26(18):2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.R Development Core Team. R: A language and environment for statistical computing. 2008 [Google Scholar]
- 17.Goldmann E, Aiello A, Uddin M, Delva J, Koenen K, Gant LM, et al. Pervasive exposure to violence and posttraumatic stress disorder in a predominantly African American Urban Community: the Detroit Neighborhood Health Study. J Trauma Stress. 2011;24(6):747–751. doi: 10.1002/jts.20705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Neale BM, Lasky-Su J, Anney R, Franke B, Zhou K, Maller JB, et al. Genome-wide association scan of attention deficit hyperactivity disorder. American journal of medical genetics Part B, Neuropsychiatric genetics : the official publication of the International Society of Psychiatric Genetics. 2008;147B(8):1337–1344. doi: 10.1002/ajmg.b.30866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Le-Niculescu H, Patel SD, Bhat M, Kuczenski R, Faraone SV, Tsuang MT, et al. Convergent functional genomics of genome-wide association data for bipolar disorder: comprehensive identification of candidate genes, pathways and mechanisms. American journal of medical genetics Part B, Neuropsychiatric genetics : the official publication of the International Society of Psychiatric Genetics. 2009;150B(2):155–181. doi: 10.1002/ajmg.b.30887. [DOI] [PubMed] [Google Scholar]
- 20.Nguyen A, Rauch TA, Pfeifer GP, Hu VW. Global methylation profiling of lymphoblastoid cell lines reveals epigenetic contributions to autism spectrum disorders and a novel autism candidate gene, RORA, whose protein product is reduced in autistic brain. Faseb J. 2010;24(8):3036–3051. doi: 10.1096/fj.10-154484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sarachana T, Xu M, Wu RC, Hu VW. Sex hormones in autism: androgens and estrogens differentially and reciprocally regulate RORA, a novel candidate gene for autism. PloS one. 2011;6(2):e17116. doi: 10.1371/journal.pone.0017116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fu Q, Koenen KC, Miller MW, Heath AC, Bucholz KK, Lyons MJ, et al. Differential etiology of posttraumatic stress disorder with conduct disorder and major depression in male veterans. Biological psychiatry. 2007;62(10):1088–1094. doi: 10.1016/j.biopsych.2007.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wolf EJ, Miller MW, Krueger RF, Lyons MJ, Tsuang MT, Koenen KC. Posttraumatic stress disorder and the genetic structure of comorbidity. Journal of abnormal psychology. 2010;119(2):320–330. doi: 10.1037/a0019035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garriock HA, Kraft JB, Shyn SI, Peters EJ, Yokoyama JS, Jenkins GD, et al. A genomewide association study of citalopram response in major depressive disorder. Biological psychiatry. 2010;67(2):133–138. doi: 10.1016/j.biopsych.2009.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Terracciano A, Tanaka T, Sutin AR, Sanna S, Deiana B, Lai S, et al. Genome-wide association scan of trait depression. Biological psychiatry. 2010;68(9):811–817. doi: 10.1016/j.biopsych.2010.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lavebratt C, Sjoholm LK, Partonen T, Schalling M, Forsell Y. PER2 variantion is associated with depression vulnerability. American journal of medical genetics Part B, Neuropsychiatric genetics : the official publication of the International Society of Psychiatric Genetics. 2010;153B(2):570–581. doi: 10.1002/ajmg.b.31021. [DOI] [PubMed] [Google Scholar]
- 27.Krueger RF. The structure of common mental disorders. Arch Gen Psychiatry. 1999;56(10):921–926. doi: 10.1001/archpsyc.56.10.921. [DOI] [PubMed] [Google Scholar]
- 28.Watson D, Clark LA. Negative affectivity: the disposition to experience aversive emotional states. Psychol Bull. 1984;96(3):465–490. [PubMed] [Google Scholar]
- 29.Miller MW. Personality and the etiology and expression of PTSD: a three-factor model perspective. Clinical Psychology: Science and Practice. 2003;(10):373–393. [Google Scholar]
- 30.Miller MW, Wolf EJ, Reardon AF, Greene A, Ofrat S, McInerney S. Personality and the latent structure of PTSD comorbidity. Journal of Anxiety Disorders. doi: 10.1016/j.janxdis.2012.02.016. (IN PRESS) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ino H. Immunohistochemical characterization of the orphan nuclear receptor ROR alpha in the mouse nervous system. J Histochem Cytochem. 2004;52(3):311–323. doi: 10.1177/002215540405200302. [DOI] [PubMed] [Google Scholar]
- 32.Boukhtouche F, Vodjdani G, Jarvis CI, Bakouche J, Staels B, Mallet J, et al. Human retinoic acid receptor-related orphan receptor alpha1 overexpression protects neurones against oxidative stress-induced apoptosis. Journal of neurochemistry. 2006;96(6):1778–1789. doi: 10.1111/j.1471-4159.2006.03708.x. [DOI] [PubMed] [Google Scholar]
- 33.Jolly S, Journiac N, Vernet-der Garabedian B, Mariani J. RORalpha, a Key to the Development and Functioning of the Brain. Cerebellum. 2012 doi: 10.1007/s12311-011-0339-1. [DOI] [PubMed] [Google Scholar]
- 34.Oosthuizen F, Wegener G, Harvey BH. Nitric oxide as inflammatory mediator in post-traumatic stress disorder (PTSD): evidence from an animal model. Neuropsychiatric disease and treatment. 2005;1(2):109–123. doi: 10.2147/nedt.1.2.109.61049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Woon FL, Sood S, Hedges DW. Hippocampal volume deficits associated with exposure to psychological trauma and posttraumatic stress disorder in adults: a meta-analysis. Progress in neuro-psychopharmacology & biological psychiatry. 2010;34(7):1181–1188. doi: 10.1016/j.pnpbp.2010.06.016. [DOI] [PubMed] [Google Scholar]
- 36.Etkin A, Wager TD. Functional neuroimaging of anxiety: a meta-analysis of emotional processing in PTSD, social anxiety disorder, and specific phobia. Am J Psychiatry. 2007;164(10):1476–1488. doi: 10.1176/appi.ajp.2007.07030504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lyoo IK, Kim JE, Yoon SJ, Hwang J, Bae S, Kim DJ. The neurobiological role of the dorsolateral prefrontal cortex in recovery from trauma. Longitudinal brain imaging study among survivors of the South Korean subway disaster. Archives of general psychiatry. 2011;68(7):701–713. doi: 10.1001/archgenpsychiatry.2011.70. [DOI] [PubMed] [Google Scholar]
- 38.Woodward SH, Kaloupek DG, Streeter CC, Martinez C, Schaer M, Eliez S. Decreased anterior cingulate volume in combat-related PTSD. Biological psychiatry. 2006;59(7):582–587. doi: 10.1016/j.biopsych.2005.07.033. [DOI] [PubMed] [Google Scholar]
- 39.Schuff N, Zhang Y, Zhan W, Lenoci M, Ching C, Boreta L, et al. Patterns of altered cortical perfusion and diminished subcortical integrity in posttraumatic stress disorder: an MRI study. NeuroImage. 2011;54(Suppl 1):S62–S68. doi: 10.1016/j.neuroimage.2010.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lanius RA, Bluhm R, Lanius U, Pain C. A review of neuroimaging studies in PTSD: heterogeneity of response to symptom provocation. Journal of psychiatric research. 2006;40(8):709–729. doi: 10.1016/j.jpsychires.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 41.Kochunov P, Glahn DC, Nichols TE, Winkler AM, Hong EL, Holcomb HH, et al. Genetic analysis of cortical thickness and fractional anisotropy of water diffusion in the brain. Front Neurosci. 2011;5:120. doi: 10.3389/fnins.2011.00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ripke S, Sanders AR, Kendler KS, Levinson DF, Sklar P, Holmans PA, et al. Genome-wide association study identifies five new schizophrenia loci. Nature genetics. 2011;43(10):969–976. doi: 10.1038/ng.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sklar P, Ripke S, Scott LJ, Andreassen OA, Cichon S, Craddock N, et al. Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nature genetics. 2011;43(10):977–983. doi: 10.1038/ng.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Valiente A, Lafuente A, Bernardo M. Systematic review of the Genomewide Association Studies (GWAS) in schizophrenia. Rev Psiquiatr Salud Ment (Barc) 2011;4(4):218–227. doi: 10.1016/j.rpsm.2011.09.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


