Abstract
Objectives
A previously unidentified mecA homologue, mecALGA251, has recently been described in methicillin-resistant Staphylococcus aureus (MRSA) from humans and dairy cattle. The origin and epidemiology of this novel homologue are unclear. The objective of this study was to provide basic descriptive information of MRSA isolates harbouring mecALGA251 from a range of host animal species.
Methods
A number of S. aureus isolates from historical animal isolate collections were chosen for investigation based on their similarity to known mecALGA251 MRSA isolates. The presence of mecALGA251 was determined using a multiplex PCR and antimicrobial susceptibility testing performed by disc diffusion.
Results
MRSA harbouring mecALGA251 were found in isolates from a domestic dog, brown rats, a rabbit, a common seal, sheep and a chaffinch. All of the isolates were phenotypically MRSA, although this depended on which test was used; some isolates would be considered susceptible with certain assays. All isolates were susceptible to linezolid, rifampicin, kanamycin, norfloxacin, erythromycin, clindamycin, fusidic acid, tetracycline, trimethoprim/sulfamethoxazole and mupirocin. Five multilocus sequence types were represented (2273, 130, 425, 1764 and 1245) and six spa types (t208, t6293, t742, t6594, t7914 and t843).
Conclusions
The discovery of MRSA isolates possessing mecALGA251 from a diverse range of host species, including different taxonomic classes, has important implications for the diagnosis of MRSA in these species and our understanding of the epidemiology of this novel mecA homologue.
Keywords: animal infections, animal reservoirs, wildlife, MRSA
Introduction
Staphylococcus aureus causes a wide range of diseases in humans, from minor skin infections to severe illnesses such as septicaemia, toxic shock, endocarditis and pneumonia. The emergence and dissemination of methicillin-resistant S. aureus (MRSA) has posed a major challenge in the treatment of S. aureus infections. S. aureus, including MRSA, can colonize and infect a wide variety of other host species, including cats, dogs, pigs, cattle, poultry and horses. This is not only of veterinary significance, but has zoonotic importance, with animals acting as a potential source for the emergence of novel MRSA clones in human beings. Pig to human transmission of MRSA ST398 (where ST stands for sequence type) is suggested to explain the emergence and spread of this clone in humans.1
Methicillin resistance in S. aureus is conferred by the acquisition of one of several staphylococcal cassette chromosome mec (SCCmec) elements, which carry the mecA gene encoding a penicillin-binding protein homologue (PBP2a) with low affinity for β-lactam antibiotics.2 We have identified a novel mecA homologue, mecALGA251, encoded in a new SCCmec cassette designated type XI.3 This mecA homologue exhibits only 70% identity at the DNA level and 63% identity at the protein level to the previously described mecA gene and is not detectable by routine mecA-specific PCR approaches and PBP2a slide agglutination tests. While mecALGA251 is present in MRSA isolates from humans and dairy cattle, its origin and epidemiology are currently unclear, with some evidence to suggest it may have spread from cattle to humans.3 Here we describe mecALGA251-containing MRSA isolates from additional host species. This has important implications for the diagnosis of MRSA infections in these hosts, and for our understanding of the epidemiology and evolution of this mecA homologue and the MRSA lineages that carry it.
Materials and methods
Candidate isolates were identified through personal contacts, reports to the multilocus sequence typing (MLST) database (http://saureus.mlst.net) and scientific reports of phenotypically resistant but MRSA that were mecA negative or S. aureus isolates related by MLST to known mecALGA251-positive lineages [clonal complexes (CCs) 425, 130, 705 and 1943].3,4 Not all requested isolates were obtainable. A total of 52 candidate isolates were tested from samples collected between 1993 and 2011.
The isolates identified were tested for the presence of femB, mecA and mecALGA251 by multiplex PCR using the following primers: femB, 1I 5′-CATGGTTACGAGCATCATGG-3′ and 1J 5′-AACGCCAGAAGCAAGGTTTA-3′, yielding a 533 bp product; mecA, 2W 5′-TGGTATGTGGAAGTTAGATTGGGAT-3′ and 2X 5′-CTAATCTCATATGTGTTCCTGTATTGGC-3′, as used by Nakagawa et al.,5 yielding a 155 bp product; and mecALGA251, 1A 5′-CATTAAAATCAGAGCGAGGC-3′ and 1B 5′-TGGCTGAACCCATTT TTGAT-3′, yielding a 188 bp product. The specificity of each primer pair was confirmed in preliminary experiments, with product identity confirmed by sequencing. The presence of mecALGA251 in positive isolates was also confirmed by sequencing.
Antimicrobial susceptibility testing was performed by disc diffusion (Oxoid, Basingtoke, UK) according to EUCAST methodology (www.eucast.org) for 12 antimicrobial agents: penicillin, cefoxitin, norfloxacin, erythromycin, clindamycin, kanamycin, tetracycline, linezolid, fusidic acid, rifampicin, trimethoprim/sulfamethoxazole and mupirocin. Growth on MRSA Brilliance 2 agar (Oxoid) was also assessed. All susceptibility results were interpreted according to EUCAST except for trimethoprim/sulfamethoxazole, for which interpretation was made according to CLSI guidelines. In addition, the MIC was determined for cefoxitin and oxacillin by microbroth dilution performed as described by EUCAST using Mueller–Hinton BBL II broth (Becton Dickinson, Heidelberg, Germany). An inoculum of 5 × 105 cfu of S. aureus ATCC 29213 was used for quality control.
Results
PCR testing identified mecALGA251 in MRSA isolated from four brown rats from Belgium (an identical strain from four different rats), one chaffinch from Scotland, one common seal from Scotland, three sheep from Denmark, one domestic dog from Scotland and one rabbit from Belgium (Table 1). The rat and sheep isolates were obtained from screening of apparently healthy individuals. The chaffinch and seal isolates were obtained from post mortem investigations of diseased animals, although it could not be determined if S. aureus was the primary cause of disease. The dog isolate was obtained from a clinical case, but further clinical details were not recorded. The rabbit isolate was obtained from a case of highly virulent staphylococcal disease.6 The MIC of oxacillin for all isolates ranged from 0.125 to 16 mg/L, and the MIC of cefoxitin ranged from 4 to 32 mg/L (Table 1). Eight of the 11 isolates were phenotypically MRSA as assessed by growth on MRSA indicator agar, disc diffusion (cefoxitin) and oxacillin and cefoxitin MICs (Table 1). Strains MRSA 1390 and PI 41/95 were susceptible to cefoxitin by disc diffusion, did not grow on the above MRSA indicator agar and had MICs of oxacillin beneath the breakpoint of 2 mg/L. However, they were resistant to cefoxitin as assessed by MIC (Table 1). Strain 07.7672.A had an oxacillin MIC below the breakpoint, but was phenotypically MRSA using the other assays (cefoxitin MIC, cefoxitin disc diffusion and growth on MRSA agar, although it produced small colonies). All isolates were susceptible to linezolid, rifampicin, kanamycin, norfloxacin, erythromycin, clindamycin, fusidic acid, tetracycline, trimethoprim/sulfamethoxazole and mupirocin. Five multilocus STs were represented (2273, 130, 425, 1764 and 1245) and six spa types (t208, t6293, t742, t6594, t7914 and t843) (Table 1 and Figure 1). DNA sequencing confirmed that the mecALGA251 in all these isolates was identical to that originally reported3 (data not shown).
Table 1.
Characteristics of mecALGA251-positive MRSA strains from this study
| Strain name | Host species | Country of isolation | Year of isolation | ST | spa type | Cefoxitin MIC (mg/L) | Oxacillin MIC (mg/L) | Resistance (disc diffusion)a | Additional notes |
|---|---|---|---|---|---|---|---|---|---|
| MRSA 1390 | brown rat (Rattus norvegicus) | Belgium | 2008–09 | 2273 (new) | t208 | 8 | 0.5 | penicillin | isolated from nasal mucosa of wild rats caught in River Demer basin |
| MRSA 1410 | brown rat (Rattus norvegicus) | Belgium | 2008–09 | 2273 (new) | t208 | 8 | 4 | cefoxitin and penicillin | isolated from nasal mucosa of wild rats caught in River Demer basin |
| MRSA 1421 | brown rat (Rattus norvegicus) | Belgium | 2008–09 | 2273 (new) | t208 | 8 | 4 | cefoxitin and penicillin | isolated from nasal mucosa of wild rats caught in River Demer basin |
| MRSA 1467 | brown rat (Rattus norvegicus) | Belgium | 2008–09 | 2273 (new) | t208 | 16 | 4 | cefoxitin and penicillin | isolated from nasal mucosa of wild rats caught in River Demer basin |
| B307063 | chaffinch (Fringilla coelebs) | Scotland | 2011 | 130 | t6293 | 16 | 4 | cefoxitin and penicillin | isolated at post mortem examination from the liver and intestines of a wild bird with severe necrotic esophagitis resulting from a Trichomonas gallinae infection |
| PI 41/95 | rabbit (Oryctolagus cuniculus) | Belgium | 1995 | 425 | t742 | 4 | 0.125 | penicillin | reported in a paper describing an isolate that caused a highly virulent infection in a rabbit6 |
| M1472/93/01 | common seal (Phoca vitulina) | Scotland | 1993 | 1764 | t6594 | 16 | 4 | cefoxitin and penicillin | isolated from a male seal pup with brain disease, Cromarty, Scottish Highlands |
| 07.7672.A | domestic dog (Canis lupus familiaris) | Scotland | 2007 | 1245 | t7914 | 8 | 1 | cefoxitin and penicillin | clinical isolate, but details not available |
| Får 2 | sheep (Ovis aries) | Denmark | 2011 | 130 | t843 | 16 | 16 | cefoxitin and penicillin | nasal swab from an apparently healthy animal |
| Får 7 | sheep (Ovis aries) | Denmark | 2011 | 130 | t843 | 16 | 8 | cefoxitin and penicillin | nasal swab from an apparently healthy animal |
| Får 9 | sheep (Ovis aries) | Denmark | 2011 | 130 | t843 | 32 | 8 | cefoxitin and penicillin | nasal swab from an apparently healthy animal |
aAntibiotics tested: penicillin, cefoxitin, linezolid, rifampicin, kanamycin, norfloxacin, erythromycin, clindamycin, fusidic acid, tetracycline, trimethoprim/sulfamethoxazole and mupirocin.
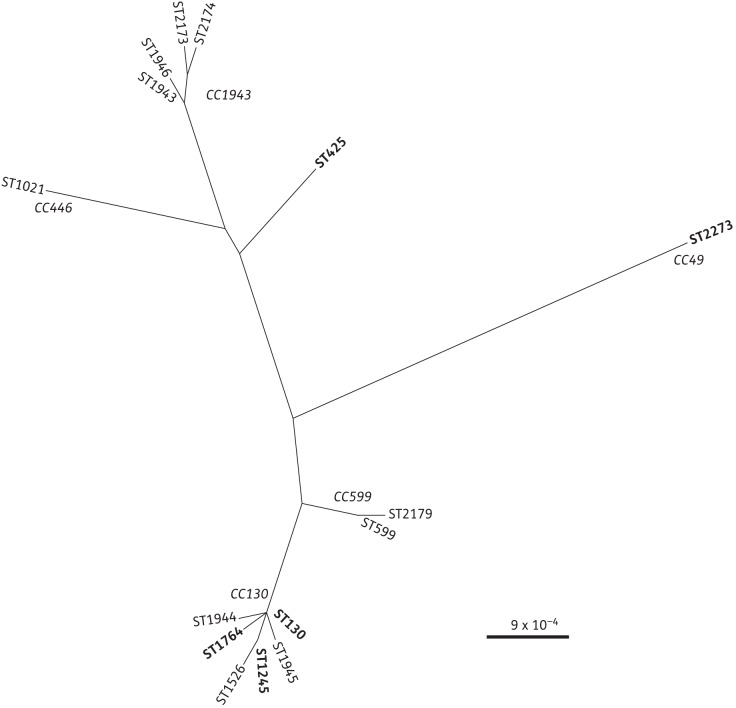
Figure 1.
Phylogenetic relationships between MRSA isolates positive for mecALGA251. An unrooted phylogenetic tree using the concatenated MLST allele sequences showing the relationship between all STs in which the divergent mecA gene has been found. The CCs of related STs are indicated in italic text. The STs of isolates that are described in this study are indicated in bold text. The tree was generated using Geneious v5.6 (www.geneious.com) using the Tamura-Nei distance model for genetic distance and the tree built using neighbour-joining with no outgroup.
Discussion
We have previously identified a divergent mecA homologue, mecALGA251, in MRSA strains from humans and dairy cattle.3 This homologue is not detected by routine PCR and PBP2a slide agglutination assays, which prevented its earlier detection, and its epidemiology and evolution are currently unclear. Initially mecALGA251 was only reported in MRSA from humans and/or dairy cattle from the UK, Denmark, Ireland and Germany.3,4,7,8 Using PCR, we have now identified this mecA homologue in MRSA isolates from several new host species (brown rat, rabbit, common seal, domestic dog, chaffinch and sheep) and from one new country, Belgium. Five STs were found among the 11 mecALGA251-positive isolates. Four of these STs (130, 425, 1764 and 1245) have previously been associated with mecALGA251. However, the four brown rat MRSA isolates from Belgium are of a new ST, ST2273, belonging to CC49, which has not previously been found among mecALGA251 MRSA. S. aureus belonging to CC49 have previously been isolated from Switzerland, the UK and Denmark from humans, pigs and red squirrels and include isolates of MRSA and methicillin-susceptible S. aureus (http://saureus.mlst.net, accessed May 2012). Of the six spa types identified in our isolates, mecALGA251 has been reported previously from three (t6293, t742 and t843),3 but not from the other three (t208, t6594 and t7914).
The MIC values for these isolates are similar to the range identified for cefoxitin in the original identification of mecALGA251 (4–64 mg/L).3 However, in the case of oxacillin, the MIC for strain PI 41/95, 0.125 mg/L is lower than previously described for mecALGA251-positive strains.3 No sequence diversity was discovered in the mecALGA251 gene, thus the basis for variation in the antimicrobial susceptibilities of these isolates is unclear (data not shown).
Our findings indicate that mecALGA251-carrying MRSA strains are present in diverse host species and can be responsible for clinical disease in species other than man and cattle. This has important implications for understanding the epidemiology and dissemination of mecALGA251. The ubiquitous status of the brown rat makes this species a strong candidate vector for the spread of mecALGA251, but it should be noted that to date only mecALGA251-positive CC49 strains of MRSA have been isolated from the brown rat, and CC49 mecALGA251-positive strains have not been found in other host species. The collection of isolates described in this report did not result from an exhaustive search of historical bacteriological collections or from a comprehensive survey of current clinical disease in animal species; however, mecALGA251-positive MRSA should be considered in the diagnosis of putative MRSA not only in the host species we highlight here, but also in additional hosts. Furthermore, our findings suggest that in addition to livestock and companion animal contact, wild animals and birds may pose a so far unregistered risk for transmission of MRSA between humans and animals.
To conclude, further MRSA surveillance in diverse host species including humans, companion animals, livestock and wildlife is required to fully understand mecALGA251 epidemiology and evolution, to evaluate its significance in disease and to implement control measures where necessary.
Funding
This work was supported by a Medical Research Council Partnership Grant (G1001787/1) held between the Department of Veterinary Medicine, University of Cambridge (M. A. H.), the School of Clinical Medicine, University of Cambridge (S. J. P.), the Moredun Research Institute (R. N. Z.) and the Wellcome Trust Sanger Institute (J. P. and S. J. P.).
Transparency declarations
None to declare.
References
- 1.Price LB, Stegger M, Hasman H, et al. Staphylococcus aureus CC398: host adaptation and emergence of methicillin resistance in livestock. mBio. 2012;3:e00305–11. doi: 10.1128/mBio.00305-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Utsui Y, Yokota T. Role of an altered penicillin-binding protein in methicillin-resistant and cephem-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 1985;28:397–403. doi: 10.1128/aac.28.3.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garcia-Alvarez L, Holden MTG, Lindsay H, et al. Meticillin-resistant Staphylococcus aureus with a novel mecA homologue in human and bovine populations in the UK and Denmark: a descriptive study. Lancet Infect Dis. 2011;11:595–603. doi: 10.1016/S1473-3099(11)70126-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stegger M, Andersen PS, Kearns A, et al. Rapid detection, differentiation and typing of methicillin-resistant Staphylococcus aureus harbouring either mecA or the new mecA homologue mecALGA251. Clin Microbiol Infect. 2012;18:395–400. doi: 10.1111/j.1469-0691.2011.03715.x. [DOI] [PubMed] [Google Scholar]
- 5.Nakagawa S, Taneike I, Mimura D, et al. Gene sequences and specific detection for Panton-Valentine leukocidin. Biochem Biophys Res Commun. 2005;328:995–1002. doi: 10.1016/j.bbrc.2005.01.054. [DOI] [PubMed] [Google Scholar]
- 6.Devriese LA, Hendrickx W, Godard C, et al. A new pathogenic Staphylococcus aureus type in commercial rabbits. Zentralbl Veterinarmed B. 1996;43:313–5. doi: 10.1111/j.1439-0450.1996.tb00319.x. [DOI] [PubMed] [Google Scholar]
- 7.Cuny C, Layer F, Strommenger B, et al. Rare occurrence of methicillin-resistant Staphylococcus aureus CC130 with a novel mecA homologue in humans in Germany. PLoS One. 2011;6:e24360. doi: 10.1371/journal.pone.0024360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shore AC, Deasy EC, Slickers P, et al. Detection of staphylococcal cassette chromosome mec type XI carrying highly divergent mecA, mecI, mecR1, blaZ, and ccr genes in human clinical isolates of clonal complex 130 methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2011;55:3765–73. doi: 10.1128/AAC.00187-11. [DOI] [PMC free article] [PubMed] [Google Scholar]