Amphipathic helices in the A-kinase anchoring protein RSP3 bind to spoke proteins involved in the assembly and modulation of the flagellar radial spoke complex, expanding the repertoire of these versatile helical protein motifs.
Abstract
A-kinase anchoring proteins (AKAPs) contain an amphipathic helix (AH) that binds the dimerization and docking (D/D) domain, RIIa, in cAMP-dependent protein kinase A (PKA). Many AKAPs were discovered solely based on the AH–RIIa interaction in vitro. An RIIa or a similar Dpy-30 domain is also present in numerous diverged molecules that are implicated in critical processes as diverse as flagellar beating, membrane trafficking, histone methylation, and stem cell differentiation, yet these molecules remain poorly characterized. Here we demonstrate that an AKAP, RSP3, forms a dimeric structural scaffold in the flagellar radial spoke complex, anchoring through two distinct AHs, the RIIa and Dpy-30 domains, in four non-PKA spoke proteins involved in the assembly and modulation of the complex. Interestingly, one AH can bind both RIIa and Dpy-30 domains in vitro. Thus, AHs and D/D domains constitute a versatile yet potentially promiscuous system for localizing various effector mechanisms. These results greatly expand the current concept about anchoring mechanisms and AKAPs.
Introduction
Many molecules involved in signal transduction are localized to specific subcellular compartments. The best example is cAMP-dependent PKA, a key effector for the cAMP signaling pathway. PKA holoenzyme is a tetramer composed of two catalytic subunits and two regulatory subunits. It is anchored to specific subcellular locations by a diverse array of A-kinase anchoring proteins (AKAPs; Welch et al., 2010). Aside from PKA, most AKAPs also anchor additional molecules involved in other signal transduction pathways (Klauck et al., 1996; Scott and Pawson, 2009). The multitude of molecular switches anchored by AKAPs inspires a widely accepted theory that AKAPs serve as signal transduction scaffolds localizing PKA, a critical enzyme of broad substrate specificity, near intended substrates and other signaling pathways. As such, AKAPs integrate synergistic or antagonistic pathways and enhance the spatial and temporal precision of phosphoregulation. Extensive evidence has shown that AKAP-mediated anchoring is critical for PKA-regulated cellular reactions, organ functions, and longevity (Greengard et al., 1991; Kammerer et al., 2003; Mauban et al., 2009).
The anchoring of PKA is mediated by the interaction between a 14–18-aa amphipathic helix (AH) in AKAPs and the dimer of RIIa, a 40-aa dimerization and docking (D/D) domain in the regulatory subunit RI or RII of PKA. This AH–RIIa interaction has been commonly used to discover AKAPs (Carr et al., 1991). Many molecules are designated as AKAPs if they bind RII in a blot overlay and if the binding can be specifically blocked by a high affinity RIIa-binding AH (AHR), like Ht-31 from AKAP–lymphoid blast crisis oncogene (Carr et al., 1992). However, accumulated evidence indicates that the applications of the AH–RIIa interaction are broader than simply anchoring PKA. First, the RIIa domain is present not only in PKA regulatory subunits but also in more than 200 eukaryotic proteins with distinct molecular architectures. Some of them are enriched in cilia and flagella and bind AKAPs in vitro just as RII does (Fujita et al., 2000; Carr et al., 2001; Yang et al., 2006; Newell et al., 2008). This suggests that the RIIa domains from various proteins are functionally equivalent and the RII overlay assay can potentially reveal AKAPs that actually anchor non-PKA RIIa proteins. Second, the Dpy-30 domain that is present in over 200 proteins is similar in sequence and structure to RIIa (Roguev et al., 2001; Wang et al., 2009). Both Dpy-30 and RIIa are composed of an X-shaped bundle of two helix-loop-helix monomers (Fig. 1), but differ at the N terminus (Gold et al., 2006; Kinderman et al., 2006; Wang et al., 2009). Proteins with either a RIIa domain or a Dpy-30 domain are classified into two families within the RIIa clan in the Pfam database.
Figure 1.
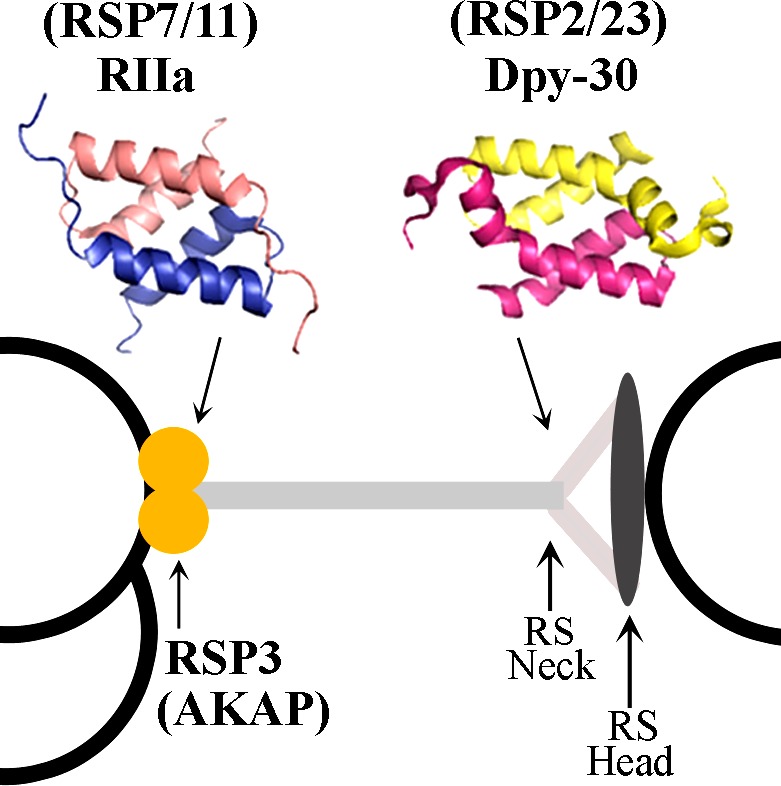
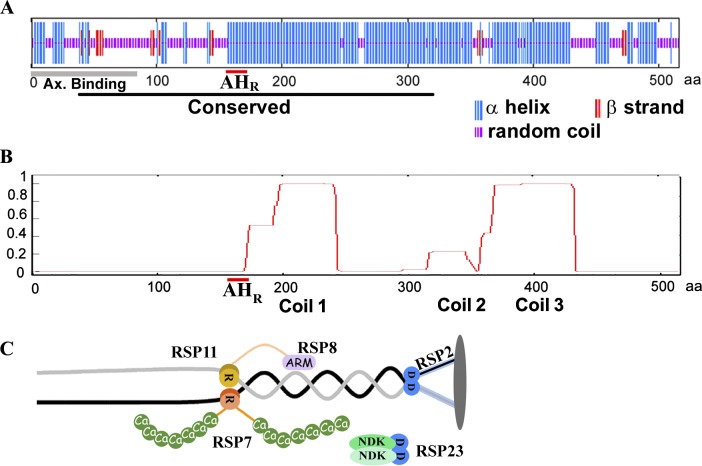
Predicted locations of RSP3 AKAP and the D/D domain–containing RSPs in the RS complex. RS is positioned between the microtubule outer doublet (left) and the CP (right). RSP3 dimer (orange circles) is thought to be the base of the RS. The RIIa domain–containing RSP7 and RSP11 are located toward the base, whereas the Dpy-30 domain–containing RSP2 and RSP23 are near the neck region underneath the spoke head. The prediction is based on RS deficiencies in RS mutants. Only a fraction of the 9 + 2 axoneme is depicted. The crystal structures of the RIIa and Dpy-30 domains were generated using PyMol (PDB accession numbers 1L6E [Morikis et al., 2002] and 3G36 [Wang et al., 2009]).
Emerging evidence shows that the non-PKA members in the RIIa clan are important for a wide range of functions distinct from PKA. For example, Dpy-30 protein, the namesake of the domain, is a small core subunit in Set1-like histone methyltransferase complexes in eukaryotic cells (Cho et al., 2007). It is dispensable for the enzymatic activity (Patel et al., 2009) but modulates H3K4 trimethylation (Jiang et al., 2011). A defective dpy-30 gene results in a dumpy body shape in Caenorhabditis elegans (Hsu and Meyer, 1994), whereas depletion of Dpy-30 transcripts blocks trans-Golgi trafficking (Xu et al., 2009) and neuronal differentiation of embryonic stem cells (Jiang et al., 2011). Despite the importance, the non-PKA RIIa clan members and their interacting partners remain poorly defined.
To elucidate the molecular interactions of these putative D/D domains and the functions that they may tether, we investigated the radial spoke (RS) complex in Chlamydomonas reinhardtii flagella that harbors an AKAP as revealed by the RII overlay (Gaillard et al., 2001) and four subunits with a RIIa domain or a Dpy-30 domain (Patel-King et al., 2004; Yang et al., 2006). The RS consists of a thin spoke stalk and an enlarged spoke head, nestled between outer doublet microtubules and the central pair (CP) apparatus in the 9 + 2 axoneme (Fig. 1). Diverse evidence suggests that the RS serves as a mechanical transducer, intermittently coupling the outer doublets and the CP to coordinate dynein motors that drive oscillatory beating (Warner and Satir, 1974; Omoto et al., 1999; Yang et al., 2008). Furthermore, the RS is involved in motility changes induced by second messengers and phosphorylation (Brokaw, 1987; Howard et al., 1994; Habermacher and Sale, 1996, 1997; Satir, 1999). The spoke AKAP RSP3 and the RS proteins (RSPs) with a putative D/D domain appear to be involved in these central functions and are exclusively located in the spoke stalk. RSP3 AKAP operates as a homodimer (Wirschell et al., 2008). It is essential to the assembly of the entire complex and considered to be the spoke base, targeting RSs to axonemes (Williams et al., 1989; Diener et al., 1993). RSP7 and RSP11 have a RIIa domain, whereas RSP2 and RSP23 have a Dpy-30 domain. Aside from the putative D/D domains, they all carry distinct additional sequences implicated in assembly, calcium signaling, motility modulation, or enzymatic activity (Patel-King et al., 2004; Yang and Yang, 2006; Yang et al., 2006). Most notably, RSP2 is critical for bridging the head module and stalk module together. The RSP deficiencies in RSP2 mutant pf24 (Huang et al., 1981; Patel-King et al., 2004) and chemical cross-linking (Kohno et al., 2011) strongly suggest that the two Dpy-30 domain–containing subunits are located underneath the spoke head, whereas the two RIIa domain–containing subunits are located near RSP3 at the base of the RS (Fig. 1; Yang et al., 2006). Intriguingly, mutations within the AHR of RSP3 abolished the interaction of the AHR with RII in the in vitro overlay assay, yet the same mutation in C. reinhardtii resulted in partially paralyzed flagella that still had the RIIa-containing RSP7 and RSP11 (Gaillard et al., 2006).
These observations raise multiple questions. Does RSP3 in vivo bind PKA’s RII as demonstrated in the overlay assay (Gaillard et al., 2001) or, instead, bind its neighboring RSPs with their RIIa domains? Which spoke protein interacts with the Dpy-30 domain? How are these four proteins with similar D/D domains localized to distinct positions within the same complex? How do these molecules contribute to the mechanism of the RS? The answers to these questions will help to elucidate cellular reactions that use the RIIa clan members, including the RS-mediated motility control, and may broaden the impact from the discoveries of AKAPs beyond the realm of PKA. We postulate that the core of the spoke stalk is entirely composed of an RSP3 dimer that then directly anchors the RIIa and Dpy-30 domains in the four RIIa clan RSPs to form a rigid backbone for mechanical coupling. To test this, we generated a panel of RSP3 truncation mutants in conjunction with in vitro experiments. The combined approaches revealed two similar but discrete AHs in RSP3 for anchoring the RIIa and Dpy-30 domain. These D/D domains in turn tether the effector modules that mediate the assembly and modulation of the RS. Interestingly, in vitro, the Dpy-30 domain exhibited cross-reactivity for the AHR but the RIIa domain did not cross react with the Dpy-30–binding AH. These findings reveal that a single mechanism of unexpected versatility and promiscuity localizes distinct non-PKA effectors to specific microcompartments, bestowing the morphology and mechanisms central to this motility-regulating complex. This principle is expected to be applicable to the other RIIa clan members and their anchoring proteins.
Results
Prediction of the RIIa clan anchoring protein in the RS
Based on electron micrographs of beating cilia, it was predicted that RSs are rigid in order to mechanically couple the mobile CP and outer doublets intermittently at a high frequency (Warner and Satir, 1974). We reason that a RS with all four RIIa clan members docking to the same core protein extending throughout the complex may have higher rigidity than a RS made from a chain of consecutive individual RIIa clan members. The core molecule should be filamentous and long enough to span the distance from the outer doublets to the CP. Furthermore, it should be evolutionarily conserved, as the dimension of typical 9 + 2 axonemes is similar (Mastronarde et al., 1992; Pigino et al., 2011; Barber et al., 2012). Sequence analysis showed that among the 19 spoke components, the best candidate core protein is RSP3, the dimeric spoke AKAP (Gaillard et al., 2001; Wirschell et al., 2008; Diener et al., 2011), although experimental evidence positioned RSP3, in fact only its N-terminal ∼80-aa region, at the base of the RS, for binding the entire complex to the axoneme (Fig. 2; Diener et al., 1993).
Figure 2.
Sequence analysis and the new model of RSP3-centered RS. (A) The secondary structure of RSP3 appears to be divided into two distinct areas by the AHR (red underline). The N-terminal 160 aa contains random coils (purple) interspersed with short α helices (blue) and β strands (red), whereas the C terminus is primarily composed of α helices. The black bar corresponds to the RSP3 domain that is highly conserved among orthologues. The gray bar corresponds to the axoneme-binding region. (B) RSP3 contains three areas with different propensities to form coiled coils. The prediction was made by the COILS program using the window size of 28 aa. (C) A model depicting RSP3 dimer (black and gray lines) as the core in the RS, with two sites for anchoring RIIa (R) and Dpy-30 (D) domains in two pairs of RSPs. Each domain tethers distinct molecular moieties as effector mechanisms, like RSP8, an ARM repeat protein; calcium-binding EF-hands (Ca) in RSP7; and coiled coils (blue bars) and NDK in RSP2 and RSP23, respectively.
The 516-aa RSP3 could be viewed as two structurally distinct segments divided by the AHR at aa 161–178 (Fig. 2 A; Gaillard et al., 2001). The N-terminal 160-aa region that binds axonemes (Fig. 2 A, gray bar; Diener et al., 1993) and a stack of three to five LC8 dimers is equivalent to a 12–20-nm-long rod (Gupta et al., 2012). The remainder of RSP3 is largely helical (Fig. 2 A, blue lines) with a propensity to form three coiled coils (Fig. 2 B), structures known for protein–protein interactions. Only the first ∼320-aa region upstream to Coil 2 is highly conserved and is recognized as an RSP3 domain (Fig. 2 A, black bar) by the Pfam database. Coil 2, with a lower coiled coil propensity, is mildly conserved, whereas Coil 3 and the downstream sequence are unique to C. reinhardtii and are dispensable (Diener et al., 1993). The conserved 160-aa helix from AHR to Coil 2 will be ∼24 nm long, if each turn of the α helix contains 3.6 aa and is 0.54 nm long (Pauling et al., 1951). Together, the entire conserved 320-aa region, with 12–20-nm LC8–RSP31–160 complex and 24-nm RSP3160–320 dimeric coiled coil, is sufficient to extend throughout the stalk of the 38–41-nm RS (Yang et al., 2001; Nicastro et al., 2005; Pigino et al., 2011). Thus, we postulate that the dimer of the conserved 320-aa region in RSP3 (Fig. 2 C, black and gray wires) forms the core of the RS, with binding sites not only for the axoneme (Diener et al., 1993) and the RIIa domains (yellow and orange circles; Gaillard et al., 2001) but also for Dpy-30 domains (blue circles) and possibly the spoke head (gray oval disk). The RIIa and Dpy-30 domains tether to four types of effector modules—armadillo (ARM) repeats (Fig. 2 C, ARM in RSP8, which is reduced in RSP11 mutant), calcium-binding motifs (Ca in RSP7), nucleoside diphosphate kinase (NDK in RSP23) domain, and coiled coils (blue bar in RSP2).
RSP3 truncation mutagenesis reveals the binding region for the RIIa domain and its tethered moieties
This model explains the observation that although mutations in RSP3’s AHR abrogate the interaction of RSP3 with RII in an overlay assay (Gaillard et al., 2001), the same mutations did not perturb the assembly of the RIIa domain–containing proteins RSP7 and RSP11 into the RS (Gaillard et al., 2006). RSP7 and RSP11 may engage in multiple interactions, directly or indirectly, with RSP3 (Fig. 2 C) and hence a perturbation in the AHR–RIIa interaction alone is insufficient to prevent their assembly. If the model is correct, deletion of all involved sequences in RSP3 will result in an axoneme devoid of these two RIIa-containing molecules and possibly their associated molecules, such as RSP8 with ARM repeats that are known to promote protein–protein interactions. In contrast, retention of the AHR alone will be sufficient for the assembly of some, if not all, of these molecules.
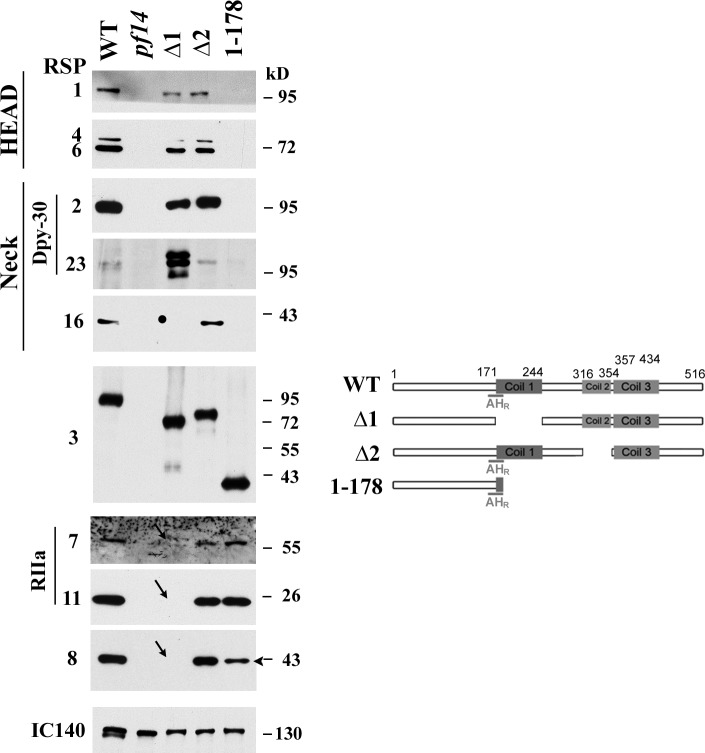
To test this, we first created two complementary deletion strains guided by the predicted molecular modules: the 1–178 strain that terminates at the end of the AHR and the Δ1 strain that lacks Coil 1 (aa 171–244) and, as a consequence, is missing part of the AHR (Fig. 2). The third strain, Δ2, which lacks the small Coil 2 (aa 316–354), and wild-type (WT) strain served as controls. All polypeptides retained the axonemal binding region at the first 80 aa so that they could be assembled into the axoneme. The C terminus of all RSP3 polypeptides was tagged with three HA epitopes and 12 His residues for detection, semiquantitative comparison, and protection of the truncated free end.
All of the deletion constructs were modified from a plasmid carrying the WT RSP3 genomic DNA. An antibiotic-resistant cassette was inserted into each plasmid to aid the selection of C. reinhardtii clones carrying the transgene. The intact and deletion constructs were transformed individually into the RSP3 mutant pf14 in which a premature stop codon results in diminished expression of the RSP3 polypeptide and the spokeless axonemes (Williams, et al., 1989; Diener et al., 1993). More than 40 antibiotic-resistant clones for each construct were screened microscopically. All clones in both the Δ1 and 1–178 groups were paralyzed. In contrast, among the 118 Δ2 clones screened, 50 clones contained motile cells (Table 1): a fraction of cells in the suspension were swimming. But unlike WT cells, they could not maintain helical trajectories. Their flagella beat with largely normal waveform but lost synchrony frequently. The flagella of immotile cells were paralyzed or twitching. The mixture of swimmers and immotile cells from a single clone resembles the phenotype of several mutants with mild RS deficiencies (Huang et al., 1981; Yang and Yang, 2006; Gaillard et al., 2006; Wei et al., 2010). This indicates that Δ2-RSP3 polypeptides were restored to the axoneme of the swimmers and were capable of rescuing paralyzed flagella, albeit partially. For those Δ2 clones that have paralyzed or twitching flagella only, the Δ2-RSP3 plasmid may not have inserted into the genome properly to restore the polypeptides at a sufficient quantity.
Table 1.
Screening of RSP3 truncation mutants
| Construct | Observed | Motilea | Flagella preparationb | HA+ |
| Δ1 | 160 | 0 | 23 | 5 |
| Δ2 | 118 | 50 | 16 | 7 |
| 1–178 | 40 | 0 | 10 | 2 |
| 1–244 | 50 | 0 | 10 | 3 |
| 1–269 | 92 | 0 | 10 | 2 |
| 1–316 | 114 | 47 | 10 | 7 |
Single colonies of antibiotic-resistant transformants were randomly picked and re-streaked on agar plates. A fraction of each colony was resuspended in water or media for light microscopy. The crude flagella preparation was made from a plate of clones randomly selected from the group of Δ1, 1–178, 1–244, and 1–269 that were 100% paralyzed; or from the clones with swimmers from the group of Δ2 and 1–316. The samples were then assessed by HA Western blots. Recorded in each column is the colony number.
The number of colonies that contained swimmers. The percentage of swimmers from each clone varied. The swimmers could not maintain linear trajectory and their flagella had a higher rate of asynchrony than WT cells.
The flagella quantity from each crude preparation for HA Western blots varied, affected by the population of flagellated cells and the deflagellation level.
To identify those clones with tagged RSP3 present in the axoneme, HA Western blots were conducted on crude flagella preparations made from at least 10 clones randomly selected from the paralyzed Δ1 and 1–178 groups and from the motile clones in the Δ2 group. Axonemes, purified from the positive clones (Table 1), were then probed for HA and representative RSPs (Fig. 3). The amount of RSP3 variants and WT RSP3-HAHis appeared similar. The axonemes from the Δ1 strain that was missing Coil 1 (aa 171–244) and part of the AHR (aa 161–178) in RSP3 lacked both the RIIa-containing RSP7 and RSP11, as well as the ARM repeat protein RSP8 as expected (Fig. 3, arrows, compare blots on the left and prediction on the right), whereas the more distally located proteins in the spoke head and neck region, including the Dpy-30 domain–containing RSPs, were present. This result confirms that the RS is not composed of a string of consecutive RSPs. RSP16, the spoke HSP40, was drastically reduced in the axoneme of the RSP2 mutant (Huang et al., 1981; Yang et al., 2008) and was absent in the Δ1 axoneme (Fig. 3, dot) despite the presence of RSP2. The HSP40 deficiency in mutants either defective in RSP2 or RSP3’s Coil 1 suggests that HSP40 interacts with both RSP2 and RSP3. For the 1–178 axonemes, in which the RSP3 fragment was terminated immediately after the AHR (aa 161–178), both RSP7 and RSP11 were present, but the head proteins (RSP1, RSP4, and RSP6) and neck proteins (RSP2, RSP23, and RSP16) were absent (Fig. 3). Therefore, the region up to AHR is sufficient to anchor the two non-PKA RIIa proteins. Note that 1–178 axonemes (Fig. 3, arrowhead) contained less RSP8 than the WT control, like the axoneme of the RSP11 mutant pf25 (Yang et al., 2006). The RSP8 deficiency in strains defective either in RSP3 Coil 1 (1–178 or Δ1 strains) or in RSP11 supports the prediction that RSP11 and RSP8 form a trimolecular subcomplex with RSP3 Coil 1 (Fig. 2 C). Furthermore, the fact that Dpy-30 domain–containing RSP2 and RSP23 are absent in the 1–178 strain but present in the Δ1 and Δ2 strains suggests that the Dpy-30 domain binding site is located between Coil 1 and Coil 2. The band patterns of RSP23 varied among preparations because of RSP23’s unusual susceptibility to degradation (Patel-King et al., 2004).
Figure 3.
Deletions around the Coil 1 region resulted in the deficiencies in RIIa domain–dependent assembly. Representative Western blots of axonemes (left) from WT and the RSP3 strains defective in the region around coiled coils (right) were probed for relevant RSPs as indicated. RIIa proteins (RSP7 and RSP11) and the ARM protein (RSP8) were missing in the Δ1 strain (arrows) in which half of the AHR was truncated. RSP16, whose assembly required RSP2, was also absent (dot). These proteins appeared normal in Δ2 axoneme. The 1–178 axonemes that retained RSP3 sequence up to the AHR lacked all the proteins in the spoke head and spoke neck but contained RIIa proteins (RSP7 and RSP11). Note that RSP8 was less abundant (arrowhead). The spokeless pf14 was the negative control. IC140, an inner dynein arm subunit, indicated the protein load.
RSP3’s helical region associates with Dpy-30 domain–containing proteins
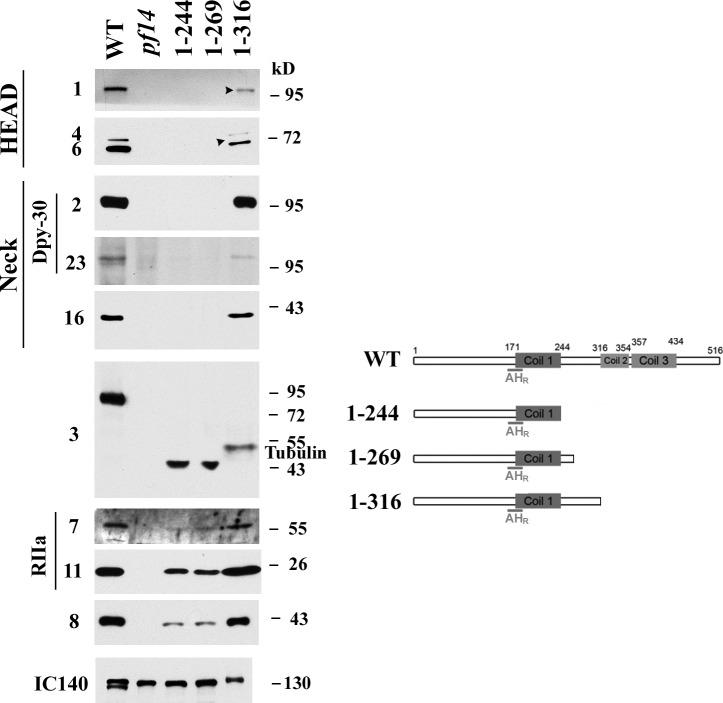
To identify the region that binds RSP2 and RSP23, we took the same strategy to generate three more strains in which RSP3 polypeptides terminate at different residues between Coil 1 and 2, i.e., aa 244, 269, and 316, respectively. As expected, all clones in the 1–244 and 1–269 groups were paralyzed (Table 1), whereas ∼40% of the clones in the 1–316 group that lacks Coil 2 and the downstream sequence contained swimmers, just like the Δ2 group that lacks Coil 2 only. This further confirms that the less conserved Coil 2 region is beneficial, albeit dispensable, to flagellar beating. Preliminary Western blots of flagella from selected clones identified RSP3 (HA)-positive clones. The axonemes from these clones were subjected to detailed Western blot analysis.
The axonemes from the paralyzed 1–244 and 1–269 strains resembled 1–178 axonemes, lacking the neck proteins (RSP2, RSP23, and RSP16) and the head proteins (RSP1, RSP4, and RSP6; Fig. 4). This suggests that the region involved in the head-neck assembly and in anchoring the Dpy-30 domain is within aa 269–316 in RSP3. For the 1–244 and 1–269 strains that retain both AHR and Coil 1, the axoneme had RIIa domain–containing RSP7 and RSP11 as well as the ARM repeat protein RSP8, consistent with the prediction that the mutually interacting RSP11 and RSP8 bind to AHR and Coil 1. The 1–316 axonemes, in which RSP3 lacks both Coil 2 and 3, were similar to Δ2 axonemes, in which all RSPs were present (compare Figs. 3 and 4) except that the head proteins (RSP1, RSP4, and RSP6) were drastically reduced (Fig. 4, arrowheads). Consistent with this, fewer cells expressing 1–316 swam in stationary phase cultures (axonemes were harvested from stationary phase cultures) than in the suspension from fresh plates or the log phase culture. This media effect on the motility level and assembly was noted previously in two spoke mutants (Yang and Yang, 2006; Wei et al., 2010). The head protein deficiency in this strain missing RSP3’s C terminus resembles the phenotype of the RSP2 mutant pf24 (Huang et al., 1981; Patel-King et al., 2004; Yang et al., 2006), suggesting that both RSP2’s and RSP3’s C termini are involved in, albeit not required for, the assembly of the spoke head, and thus head proteins are not entirely absent when only one of them is defective. Furthermore, the 47-aa RSP3269–316 may harbor the Dpy-30 domain binding site. The in vitro experiment for mapping the precise binding site will be described later.
Figure 4.
Deletions between the first two coils resulted in the deficiencies in Dpy-30 domain–dependent assembly. Western blots of axonemes (left) extracted from WT and three mutants with different truncations distal to Coil 1 (right) were probed for relevant RSPs as indicated. 1–244 strain and 1–269 strain lacked the spoke head and neck proteins including RSP16 and the two Dpy-30 domain–containing RSP2 and RSP23. The other spoke proteins appeared reduced because of less abundant 1–244 and 1–269 polypeptides. These proteins were present in 1–316 strain but the spoke head proteins were less abundant (arrowheads) than that in the WT control. The intensity of the 1–316 RSP3 band appeared lower than expected because of the interference of co-migrating tubulins in antibody binding.
Distinct dwarf RSs in RSP3 mutant axonemes
To assess the RSs with truncated RSP3 independently, we investigated axonemes using two additional approaches. First, RSs were extracted from axonemes and the dialyzed extract was fractionated by sucrose gradient velocity sedimentation. The fractions of the gradient were then assessed by Western blots (Fig. 5). The RS complex from the WT control sedimented as an intact particle at 20S (Fig. 5, arrow; Yang et al., 2001). For all other transgenic strains, RSs with truncated RSP3 sedimented near the middle of the gradient as smaller particles, similar to or smaller than RS stalk particles from the mutants lacking the head proteins (Yang et al., 2001). Although the gradients of 1–244, 1–269, and 1–316 strains contain a single RSP3 peak (Fig. 5, arrows), the gradients of the Δ1 and 1–178 strains that lack Coil 1 contained another minor peak (asterisk), suggesting that Coil 1, possibly through a coiled-coil interaction, is critical to the stability of the RS complex.
Figure 5.
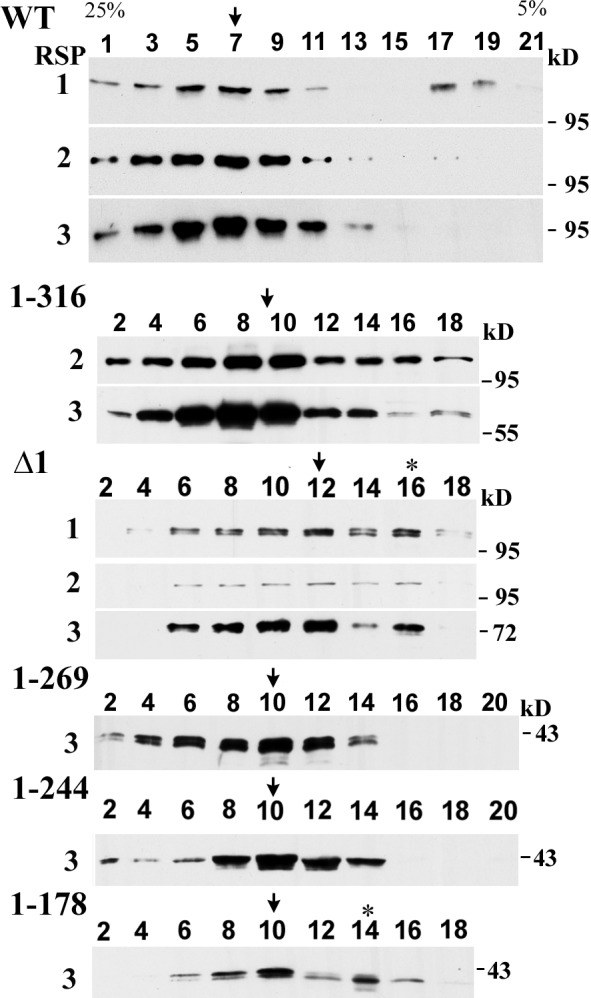
RSs extracted from RSP3 deletion mutants sedimented as smaller particles. The KI axonemal extracts from the indicated strains were centrifuged through a 5–25% sucrose density gradient and the fractions were assessed by Western blots probed for relevant RSPs as indicated. The major peaks are indicated by arrows. The RSPs from all truncation mutants largely sedimented in a single peak as intact particles smaller than WT RSs. A prominent second RS peak (asterisk) was present in the gradients from Δ1 and 1–178 strains that lacked Coil 1.
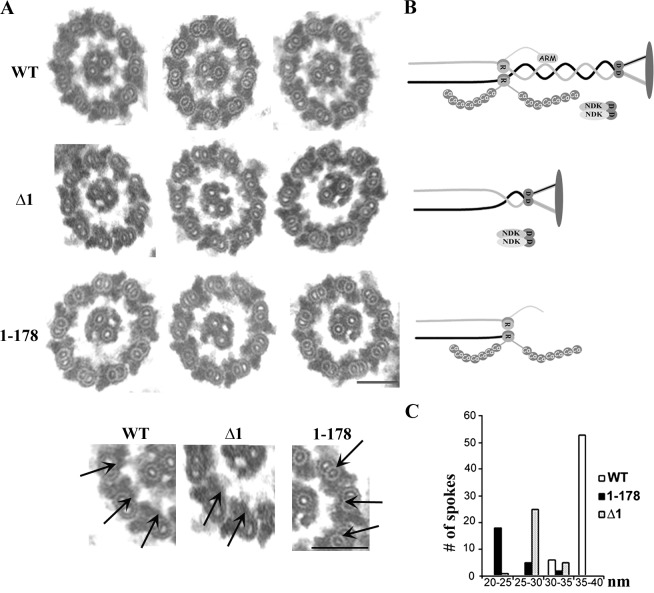
The morphology of the RS with truncated RSP3 was assessed by electron microscopy. Because of the resolution of electron microscopy and the similar sizes of extracted RS particles from mutant flagella, we only compared the axonemes of WT, Δ1, and 1–178 strains (Fig. 6). The electron microscopy images of axoneme cross sections revealed two main defects, stubby RSs (Fig. 6 A, arrows) and a lateral shift of the CP, a signature of RS deficiencies (Fig. 6 A and Table 2; Witman et al., 1978). Importantly, some RS stubs in Δ1 axonemes but not 1–178 axonemes exhibited an enlarged head (Fig. 6 A, bottom), consistent with the proposed model (Fig. 6 B) and the presence of head proteins only in the Δ1 axonemes (Fig. 4). The RSs are the longest in the WT and the shortest in 1–178, which has the shortest RSP3 polypeptide (Fig. 6 C). The CP shifted laterally in ∼92% of Δ1 images and ∼69% of 1–178 images (Table 2). The higher incidence of shifted CP in Δ1 axonemes despite their longer RSs than in 178 axonemes is consistent with the predicted interaction of the CP and the spoke head that is only present in Δ1 dwarf RSs (Warner and Satir, 1974) and may pull the CP away from the center.
Figure 6.
Distinct stubby spokes in the axonemes from the Δ1 and 1–178 mutants. (A) The representative transmission electron microscopic images of cross-sectioned axonemes from the WT, Δ1, and 1–178 strains. The bottom panel gives an enlarged view of the axoneme cross sections. The arrows highlight representative RSs in each strain. The enlarged spoke head is present in the RSs of WT and Δ1 axonemes. Bars, 100 nm. (B) Schematic pictures depicting the RSs in each strain. (C) The length distributions of RSs in cross-sectioned axonemes. The RSs with an identifiable morphology were measured from 13 WT sections, 24 Δ1 sections, and 22 1–178 sections. RSs were separated based on the spoke length, and the number in each group was plotted into a distribution histogram.
Table 2.
The incidence of lateral-shifted CP apparatus and the extent of deviation in electron micrograph of axonemes
| Strain | Percentage with shifted CPa | Number with measurable shiftb | Percentage of the shiftc |
| % | % | ||
| WT | 3.7 (n = 27) | 0 | NA |
| Δ1 | 92 (n = 38) | 1432 | 16.601520 |
| 1–178 | 69 (n = 39) | 95 | 16.6020 |
n, the total number of cross-sectioned axoneme images that were visually inspected.
The percentage of axoneme cross sections in which the CP appeared shifting from the center by visual inspection.
The number of axoneme cross sections in which the lateral shift of the CP was significant enough to be measured.
Percentage of the shift was derived from the distance between the centers of two CP microtubules and the axoneme divided by the radius of the axoneme.
Identification of the Dpy-30 domain binding site in RSP3
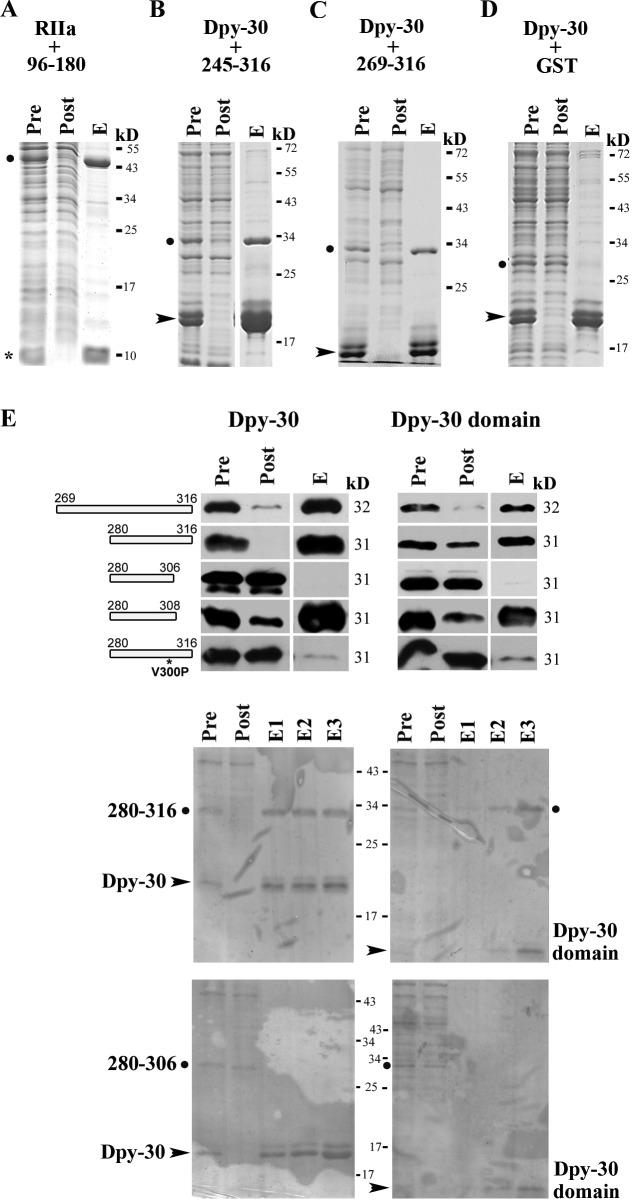
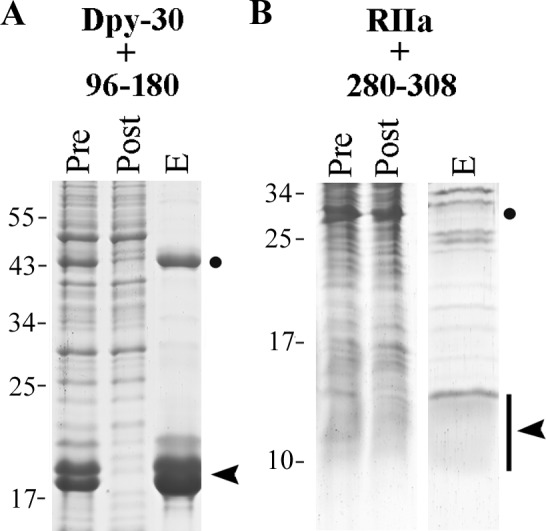
Based on the phenotypes of RSP3 transgenic strains (Fig. 4), an in vitro approach was taken to test that RSP3269–316 contains a Dpy-30 domain binding site. AHRs are 14–18 aa long. Some of AHs are clearly composed of ΦΦXX repeats in which the first two residues are hydrophobic residues (Φ) positioned to interact with complementary residues at the binding grooves of the RIIa domain (Burns-Hamuro et al., 2003; Kinderman et al., 2006; Sarma et al., 2010), whereas for the other AHRs the repeats are not as evident (Gaillard et al., 2001; Gold et al., 2006). Typical ΦΦXX repeats were noted within the Dpy-30 binding fragment in BIG1 at the trans-Golgi network but were not tested (Xia et al., 2010). For RSP3269–316, typical and degenerate ΦΦXX repeats are present throughout this region, thus the precise Dpy-30 binding site was determined objectively by nickel–nitrilotriacetic acid (Ni-NTA) pulldown. As a control, the His-tagged RIIa domain from RSP7 was coexpressed with the GST-tagged RSP396–180 that harbors AHR at aa 161–178 (Fig. 3; Gaillard et al., 2001) and both polypeptides were copurified by Ni-NTA (Fig. 7 A). Various Dpy-30 domain–containing RSP2 and RSP23 polypeptides were tested but were not compatible for this assay because of poor expression or precipitation propensity. Therefore we used the His-tagged human Dpy-30 protein for Ni-NTA pulldown instead. As expected, both GST-RSP3245–316 and GST-RSP3269–316 were copurified with His-Dpy-30 by Ni-NTA (Fig. 7, B and C). In contrast, the GST control was not copurified (Fig. 7 D).
Figure 7.
Two sites in RSP3 bind to the RIIa and the Dpy-30 domain. His-tagged RIIa from RSP7 (asterisk) or His-tagged Dpy-30 protein (arrowhead) were coexpressed with a GST-tagged RSP3 peptide (dot) in bacteria. The Ni-NTA pulldown from the extracts was analyzed by Coomassie-stained gel (A–D) or GST Western blots (E). (A) The positive control, the copurification of the RIIa domain, and RSP396–180 that contains the AHR (Gaillard et al., 2001). (B and C) The copurification of His-tagged human Dpy-30 protein and GST-tagged RSP3245–316 or RSP3269–316 (dot). (D) The negative control. GST alone did not interact with Dpy-30 protein. (E) The smallest region that binds the Dpy-30 domain is aa 280–308 in RSP3. The interaction is perturbed by V300P mutation. His-tagged full-length Dpy-30 protein or the Dpy-30 domain was only coexpressed with GST-tagged RSP3 peptides as indicated. The irregular patterns in the Ponceau-stained membranes for the demonstration of protein loads (middle and bottom) were from plastic wraps and staining solution. Pre, the bacterial extract; Post, the flow-through from Ni-NTA matrix; and E, one of the eluates that contained the most Dpy-30 (see Materials and methods). Dpy-30 often migrated as double bands because of the susceptibility of its C-terminal end to proteolysis.
The experiments were further conducted on smaller segments within aa 269–316. GST-RSP3280–316 and GST-RSP3280–308, but not GST-RSP3280–306, were also pulled down by His–Dpy-30 (Fig. 7 E, left). This suggests that the Dpy-30 binding site is near the end of RSP3280–308. To ensure that the copurification occurs through the Dpy-30 domain rather than its flanking sequence, the same experiment was conducted with the Dpy-30 domain only (Dpy-3045–99; Wang et al., 2009). The results from the experiments using Dpy-30 domain alone or full-length Dpy-30 protein were similar (Fig. 7 E, right). Based on the similar dimensions of the RIIa and Dpy-30 domains (Fig. 1), we tested if the Dpy-30 binding peptide is centered on the 18-aa helix at the aa 291–308 by mutating V300 in the middle of the region into P, a strategy used to study AHR (Carr et al., 1992; Gaillard et al., 2001). This V300P mutation abrogated the binding of RSP3280–316 to the full-length Dpy-30 and the Dpy-30 domain (Fig. 7 E, bottom). Together, these results strongly suggest that the Dpy-30 domain binding site is within the 18-aa RSP3291–308.
Comparing the RIIa clan domains and their binding sequences
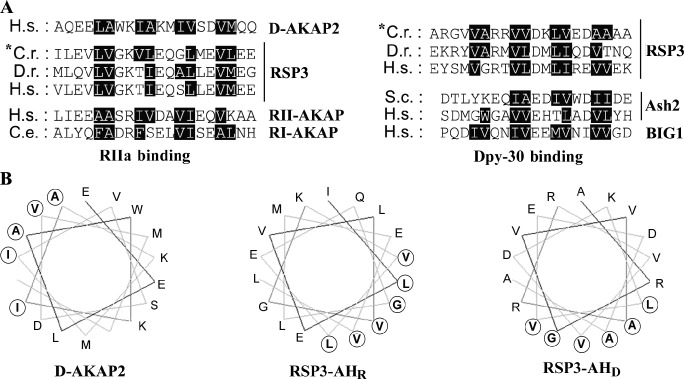
We compared RSP3291–308 with representative RIIa binding sequences (Fig. 8 A). As reported previously, typical and degenerate ΦΦXX repeats (Fig. 8 A, left) are noticeable in the AH of AKAPs that bind PKA’s RI (RI-AKAP), RII (RII-AKAP), or both (D-AKAP2) and in AHR at RSP3160–178 and the equivalent region in RSP3 orthologues. Likewise the loosely defined repeats are present at RSP3291–308, the counterparts in RSP3 orthologues, and a helix within the Dpy-30 binding fragments in BIG1 (Xia et al., 2010) and in Ash2 (Fig. 8 A, right; South et al., 2010). Like AHR in D-AKAP2 or at RSP3160–178, RSP3291–308 can be plotted into an AH by the Helical wheel program (Fig. 8 B) in which the hydrophobic residues (bold circled letters) are enriched at one side of the helix. Hence, the Dpy-30 binding helix in RSP3 is referred to here as AHD. Despite the differences between the Dpy-30 domain and RIIa domains and their distinct locations in the RS (Fig. 1), both associate with AHs of similar amino acid patterns.
Figure 8.
Similarity of the RIIa and Dpy-30 domain binding sequences. (A) Alignment of RIIa and Dpy-30 binding sequences in representative AKAPs and RSP3; and putative Dpy-30 binding sequence in Ash2 and BIG1. These regions contain three or four 4-aa repeats in which the first two residues are often, but not always, hydrophobic residues (black boxes). No particular conserved residues distinguish the two groups of sequences. D-AKAP2 is a dual-specific AKAP that binds the RIIa domain in RI and RII of PKA. The alignment for RSP3 sequences is generated by Multiple Sequence Alignment program and based on Gaillard et al. (2001). The alignment of AH sequences from AKAPs is based on Kinderman, et al. (2006) and Gold et al. (2006). The precise Dpy-30 binding sites in Ash2 and BIG1 remain to be tested. RII-AKAP, Q12802; RI-AKAP, AAC24507; C.r., C. reinhardtii; C.e., C. elegans; D.r., Danio rerio; H.s., Homo sapiens; S.c., Saccharomyces cerevisiae. (B) Helical wheel plots of RIIa and Dpy-30 binding sequences from D-AKAP2 and C. reinhardtii RSP3 (A, asterisks). One side of the RSP3’s Dpy-30 binding helix is enriched with hydrophobic residues (bold and circled) like AHD that binds RIIa and thus this region is designated as AHD.
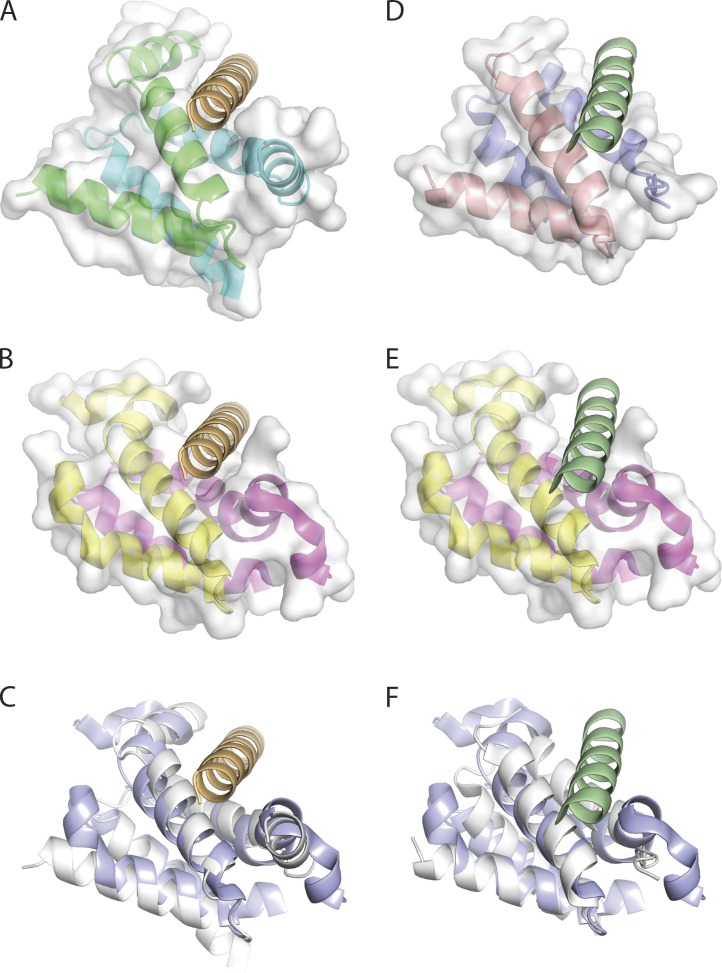
The similarity of AHR and AHD prompted us to align the crystal structures of the Dpy-30 domain and the complexes of AHR from the dual-specific D-AKAP2 and the RIIa domain from the RI and RII subunit (Fig. 9; Kinderman et al., 2006; Wang et al., 2009; Sarma et al., 2010). The N terminus of both RI’s RIIa and Dpy-30 forms an α helix that contributes to a deep pocket for the AH (Fig. 9, A and B). In contrast, the N terminus of RII’s RIIa is a β strand that contributes to a shallower binding cleft (Fig. 9 D). The AHR rests in the binding clefts from the two RIIa domains in a similar fashion except for a shift in helical register (Sarma et al., 2010). The binding pocket of the Dpy-30 domain bears close resemblance to both RIIa domains but more so to RI’s RIIa because of their common α helix at the N terminus. Nonetheless, all structures are highly similar (Fig. 9, C and F) and are able to accommodate AHR (Fig. 9, compare B and E). Thus, with regards to tertiary structure, the RIIa and Dpy-30 domain are not as distinctive as their classification.
Figure 9.
Similarity in x-ray structures of RIIa and Dpy-30 D/D domains with bound and superposed AHR peptide. (A) The PKA regulatory subunit RI D/D domain dimer in complex with the dual-specific D-AKAP2 AH (PDB accession number 3IM4 [Sarma et al., 2010]). The RI D/D dimer is represented in cartoon format and is colored by chain (cyan and green). The van der Waals surface for the RI D/D dimer is in semi-transparent white and the complexed D-AKAP2 AHR is in orange. (B) The human Dpy-30 C-terminal domain (PDB accession number 3G36 [Wang et al., 2009]) represented in cartoon format and colored by chain (pink and yellow). A structural overlay with the RI/D-AKAP2 structure (C) reveals that the dual-specificity AKAP2 AHR peptide can be accommodated in the Dpy-30 docking site. (C) Structural overlay of the D/D domains of human Dpy-30 (PDB accession number 3G36; blue) with the RI (PDB accession number 3IM4; white). The root mean squared deviation for equivalent polypeptide chains is 1.7 Å. (D) The PKA regulatory subunit RII D/D domain dimer in complex with the D-AKAP2 AH (PDB accession number 2HWN [Sarma et al., 2010]). The RII D/D dimer is represented in cartoon format and colored by chain (red and blue). The complexed D-AKAP2 AHR is colored in green. (E) A structural overlay of the human Dpy-30 C-terminal with the RII/D-AKAP2 structure (F) reveals that the dual-specificity AKAP2 AHR peptide can be accommodated in the Dpy-30 docking site. (F) Structural overlay of the D/D domains of human Dpy-30 (PDB accession number 3G36; blue) with the RII (PDB accession number 2HWN; white). The root mean squared deviation for equivalent polypeptide chains is 1.4 Å.
Dpy-30 binds AHR in RSP3 in vitro
Based on the structural similarity, we tested for the cross-recognition of His-Dpy-30 and AHR (GST-RSP396–180) and RSP7’s RIIa and AHD (GST-RSP3280–308) by the described Ni-NTA pulldown assay. His–Dpy-30 pulled down the mismatched AHR (Fig. 10 A), whereas His-RIIa did not significantly pull down the mismatched AHD (Fig. 10 B). Thus Dpy-30 that has a deep pocket from an additional α helix recognizes both AHR and AHD, whereas RSP7’s RIIa, as PKA’s RII, only binds RSP3’s AHR (Gaillard et al., 2001). The pocket depth in D/D domains does not seem related to the specificity. Consistent with this, the less diverse AH partners of RI compared with RII that has a shallow AH-binding groove is attributed to the RI-unique disulfide bonds that restrict the flexibility of the additional α helix (Banky et al., 2003; Sarma et al., 2010). From the perspective of AHs, RSP3’s AHR is analogous to a dual-specific AHR that binds both RI and RII, whereas RSP3’s AHD appears to be monospecific.
Figure 10.
Cross-recognition of the Dpy-30 domain and RSP3-AHR. His-tagged Dpy-30 protein (A, arrowhead) or His-tagged RIIa domain from RSP7 (B, line and arrowhead) was coexpressed with GST-tagged RSP3-AHR or AHD, respectively (dots). The extract was subjected to Ni-NTA affinity chromatography and the samples were analyzed by Coomassie-stained gel. RSP3-AHR in the bacterial extract (Pre) was mostly depleted after incubation with the Ni-NTA (see the flow-through; Post) and was enriched in the eluate (E). In contrast, RSP3-AHD did not bind to the His-tagged RIIa domain, appearing similar in the Pre and the Post and negligible in the eluate.
Discussion
A new RS model with dimeric RSP3 as a structural scaffold
The results from motility, biochemical, and morphological analyses consistently support the proposed model (Fig. 2 C). The two AHDs in a RSP3 dimer anchor the Dpy-30 domain present in RSP2 and RSP23. The nearby sequences in dimeric RSP3 and RSP2 further interact with each other and with the head components, leading to a Y-shaped spoke head, consistent with the Y-shaped images revealed by the recent cryotomographic studies (Pigino et al., 2011; Barber et al., 2012). As for RSP23, the Dpy-30 domain could tether the associated NDK to the RS complex for an unknown purpose. Based on the coassembly of RSP2 and RSP23 (Patel-King et al., 2004), we speculate that the two RIIa-containing proteins are also located in the same RS. Alternatively, the two RIIa-containing proteins may localize, respectively, in spoke 1 and 2 in each 96-nm axonemal unit, or they might form heterodimers. Regardless of the precise arrangement, RSP11’s short flanking sequence and the tethered ARM repeat protein (RSP8) interact with the Coil 1 region in RSP3 directly or indirectly, constitutively enhancing the rigidity of the dimeric RSP3 core to prevent paralysis in the exhausted media as exhibited by RSP11 mutants (Yang and Yang, 2006). In contrast, EF-hands in RSP7 may allosterically modulate the stalk when calcium increases, which is analogous to the cAMP-induced allosteric regulation of PKA. The collective evidence strongly suggests that dimeric RSP3 is a bona fide core scaffold of the entire RS complex. The various effectors anchored through AH–D/D domain interactions facilitate the assembly of the RS and possibly modulate the scaffold itself in a calcium-dependent and -independent manner.
The single AH–D/D system for anchoring various effector mechanisms
Such an anchoring system is not restricted to the RS, flagella, or C. reinhardtii. RSP3 and its AHR and AHD are conserved, as are Dpy-30 domain–containing NDK and potential RSP2 orthologues that were found to be partners in the human interactome (Rual et al., 2005). Neither will RSP3 be the only AKAP that binds non-PKA RIIa clan members. AKAP3 and a non-PKA RIIa clan member were pulled down together from testis extract, and RIIa clan members are not just located in cilia and flagella (Newell et al., 2008). As for the Dpy-30 protein, it may function primarily in the nucleus and trans-Golgi network (Xu et al., 2009). Thus AHs and D/D domains constitute a versatile system to anchor PKA and a wide spectrum of reactions in different cellular compartments.
The interactions of RSP3’s AHR with RII in vitro but with RSP7 and RSP11 in vivo firmly establish the functional equivalence of RIIa domains. This equivalence is also true for the Dpy-30 domains—Dpy-30 in the histone methyltransferase complex recognizes AHD in the RS. Furthermore, AHR and AHD share a similar pattern and AHR even recognizes both RIIa and Dpy-30. These findings strongly suggest that the interaction of diverged D/D domains and AHs uses an identical principle. This highlights the importance of using physiological evidence to interpret the effector mechanism while accentuating the question about specificity in vivo. How are the various effector mechanisms anchored to specific locations in the RS and in the cytoplasm? A dual-specific AHR/D, albeit conceivable from the structural perspective, could potentially misplace the spoke subunits in the RS or target Dpy-30 to AKAPs. There is no evidence that such mistargeting has occurred and thus mechanisms must be in place to ensure correct localization. One such cue could be differential affinities. In general, the affinity of the AHs for RI is lower than that for RII (Herberg et al., 2000; Sarma et al., 2010; Means et al., 2011) and those with a higher affinity may interact first. Conversely, the sequences flanking AHs and D/D domains may contribute to the specificity. The flanking sequences augment the recognition of RI and dual-specific AKAPs (Jarnaess et al., 2008) and may explain the assembly of RSP7 and RSP11 in the RS (Fig. 3) despite the abolished AHR (Gaillard et al., 2006). In such scenarios, flanking sequences may provide recognition, whereas the AH and D/D domain are more about docking than specific targeting as perceived currently. Although thought-provoking, dual-specific AHs are relatively rare. Most AH sequences, possibly including RSP3’s AHD, are monospecific because of distinct amino acid residues (Hirsch et al., 1992; Angelo and Rubin, 1998; Alto et al., 2003; Burns-Hamuro et al., 2003).
Although AKAPs are known as signal transduction scaffolds, the function of AH-containing proteins may be as functionally diverged as the RIIa clan family members they anchor. RSP3 not only anchors effectors related to signal transduction but also serves as the structural scaffold that anchors molecules involved in assembly. Similar dual roles may be applicable to BIG1 and BIG2, two large paralogous molecules forming a heterodimeric scaffold in the trans-Golgi network. Aside from binding to molecular switches that regulate membrane trafficking, they contain one to three AHs for anchoring RIIa- or Dpy-30 domain–containing proteins (Li et al., 2003; Xia et al., 2010), some of which may play a structural role. In contrast, monomeric Ash2 that interacts with Dpy-30 and multiple molecules that lack signaling moieties in the Set1-like histone methyltransferases (Patel et al., 2009; Cho et al., 2007; Cao et al., 2010; Chen et al., 2011) could be primarily a structural scaffold. This versatile AH–D/D system highlights the need of a new term to complement “AKAPs,” which implies exclusively PKA anchoring and signal transduction scaffolds. We propose to use D/D domain–anchoring proteins to encompass what appears to be a much broader spectrum of molecules that anchor various RIIa clan members.
Materials and methods
Cell strains, culture conditions, and biochemistry
C. reinhardtii WT strain and paralyzed RSP3 mutant strain (pf14) were used in this study. Cells were cultured in Tris-acetate-phosphate (TAP) medium with aeration over a 14-h/10-h light–dark cycle. Axoneme biochemistry was conducted at 4°C as described previously (Yang et al., 2008) with minor modifications. After sedimentation of cells at 1,800 g for 8 min and dibucaine deflagellation, flagella were centrifuged down at 11,000 g and demembranated with 0.5% NP-40. For velocity sedimentation, RSs were extracted from the axoneme pellets with 0.6 M KI at 5 mg/ml and the extract was sedimented through a 5–25% sucrose gradient at 220,000 g for 14 h.
Construct design
Genomic DNA constructs.
An NcoI fragment containing the RSP3 genomic sequence was released from a BAC clone and inserted into the same site in pGEM-T Easy vector (Promega). The SacI site and its downstream sequence in the 3′ flanking region were eliminated by limited restriction digest followed by treatment with T4 DNA polymerase (New England BioLabs, Inc.) and ligation. PCR with modified primers was performed to add the sequence for 6 His codons flanked by an XhoI site at one end and the endogenous stop codon followed by an Xba site at the other end. Subsequently, into the XhoI site a PCR product containing 3HA–6His coding sequence was inserted. This fragment was amplified using the p3HA plasmid (Silflow et al., 2001) as a template. This final RSP3 genomic construct, pRSP3-HAHis, expressed a polypeptide with a 3 HA and 12 His tag. This construct was used to create all the mutant constructs with a PCR-based approach. S and AS in primer names denote sense and antisense directions. To create the Δ1 construct, deleting the Coil 1–coding sequence, the sequences flanking Coil 1 (171–244 aa) were amplified from pRSP3-HAHis vector using the following primer pairs: XbaS (5′-TCCAACTCTACATCTAGAGCTCGCAGAGAGG-3′) and XhoAS (5′-TCTCCATCAGGCCCTGCTCGAGCACCTTGCCCAC-3′); and XhoS (5′-TCTCGAGCTGTCTGGCATTGTCAACACGGTG-3′) and AS (5′-TCTTGTCCGCCTCCCACTTGGCGTTG-3′). Underlined are built-in restriction site sequences. The PCR products Xba–Xho (1,055 bp) and Xho–AS (370 bp) were ligated into the pRSP3-HAHis construct digested with Spe and Not enzymes. Xba and Spe digestions generated identical adhesive ends compatible for ligation but the ligated mutant construct lost the Spe site, distinct from the parental clones. This mutated construct was further modified by adding the paromomycin (PMM) resistance cassette from pSI103 plasmid (Yang et al., 2008) into the AatII site in the vector to aid the selection of transgenic strains. The final plasmid is designated as pΔ1PMM. To create the Δ2 construct, sequences flanking Coil 2 coding region was amplified from pRSP3-HAHis using SpeS (5′-CCGCAAGCTCACTCGTTCACCATAAAC-3′) and NotAS (5′-AGCGGCCGCGCGATTGGCTGCCAGCGCCGCCGC-3′) primers. The amplified fragment was ligated into pΔ1PMM vector digested with Spe and Not.
To generate the remaining RSP3 constructs, the PMM cassette was first cloned into the AatII site in pGEM-T Easy to create pGEM-PMM. To create RSP31–178 construct, two fragments were amplified from the pRSP3-HAHis construct. The first fragment, which extended from the 5′UTR to the codon for aa 178, was generated using a sense primer with a built-in EcoRI site (EcoRIS: 5′-GGAATTCCCGCTCTGCTCTCCAGTCCGACTAGGG-3′) and an antisense primer with a built-in Xba site (XbaAS3: 5′-GCTCTAGACTCCTCCTCCTCCAGCACCTCCATCAG-3′). The second fragment, which extended from the HAHis tag to the 3′UTR, was generated using a sense primer with a built-in Xba site (XbaS: 5′-GCTCTAGACGCCAGGGTGCTGCGATTGGCTGCC-3′) and an antisense primer with a built-in EcoRI site (RIAS: 5′-GGAATTCTGTTGCCTGAGAGCTCCGCCTCGGCC-3′). To create the RSP31–244 construct, flanking sequences were amplified using the same set of primers for make RSP31–178 except XbaAS3 was replaced with the XbaAS2 primer (5′-GCTCTAGAGCCGCGCGCAAAGGCGCTGGCCGCC-3′). To create RSP31–316 construct, XbaAS2 primer was replaced by the XbaAS1 primer (5′-GCTCTAGACGCCAGGGTGCTGCGATTGGCTGCC-3′). Each paired EcoRI–Xba and Xba–EcoRI PCR fragments were ligated into the EcoRI site in pGEM-PMM. To create RSP31–269 construct, two fragments were amplified from the pRSP3-HAHis construct. The first fragment, which extended from the 5′UTR to the codon for aa 269, was generated using the EcoRIS primer and the ICAS primer with a built-in Xho site (5′-ACCTCGAGGGGGTCGTAGATGTAGCCGCT-3′). The second fragment, which extended from the HAHis tag to RSP3’s 3′UTR, was generated using the XhoS primer with a built-in Xho site (5′-ACCTCGAGCACCACCACCACCACCACTAAGCTAGAGGG-3′) and the RIAS primer. The two fragments (RI-Xho and Xho-RI) generated by PCR were also ligated into the EcoRI site in pGEM-PMM.
cDNA constructs.
The constructs expressing GST-tagged RSP396–180, RSP3245–316, and RSP3269–316 were generated by first PCR amplifying the corresponding sequences using a GST-RSP3 cDNA construct as a template (Diener et al.,1993). PCR products were then inserted into BamHI and EcoRI sites in pGEX-2T vector. The pGEX-RSP3245–316 was used as a template to amplify the coding sequence for GST-tagged RSP3280–316, RSP3280–306, and RSP3280–308. The PCR products were cloned into the NcoI and EcoRI site in pET-Duet vector (EMD Millipore). The V300 codon in pRSP3280–316 was replaced with the proline codon using QuikChange site-directed mutagenesis strategy (Agilent Technologies). The resulting plasmid was named pRSP3V300P. To create the HRSP3301–325 construct, the GST coding sequence was amplified from pGEX-2T and the RSP3 coding sequence was amplified from a human RSP3 cDNA clone. Both fragments were fused by PCR and the joint product was inserted between NcoI and EcoRI sites in pET-Duet vector. To create the His-tagged human Dpy-30, full length or 45–99 aa, the corresponding sequences were PCR amplified from a commercially available cDNA clone and inserted between the Nde and Xho sites in pET-28a vector. For expression of recombinant proteins, all constructs were transformed into BL21(DE3) cells. The cultures were induced with 1 mM IPTG overnight at 18°C.
Transformation of C. reinhardtii
All genomic constructs were transformed into the RSP3 mutant pf14 using the glass beads method (Kindle, 1990). In brief, autolysin-treated cells were washed with TAP medium and resuspended in the same solution to a final concentration of 108 cells/ml. 1–2 µg of plasmid, glass beads, and 100 µl of freshly prepared 20% polyethylene glycol was added to 500 µl of the cell suspension. This mixture was vortexed for 45 s followed by immediate suspension with 10 ml TAP media. The cells in the suspension were spun down and resuspended in fresh TAP media and recovered under light overnight. The following day, cells resuspended in TAP media were plated on TAP agar plates containing 10 µg/ml PMM. Single colonies that appeared after 4 to 5 d were transferred to fresh TAP plates. A fraction of each colony was resuspended in 200 µl TAP media in 96-well plates for observation using a compound microscope (BH-2; Olympus).
In vitro binding analysis
For Ni-NTA (QIAGEN) affinity copurification, GST-tagged RSP3 constructs and His-tagged HDpy-30 constructs were cotransformed into BL21(DE3) cells. The recombinant proteins were induced using 1mM IPTG overnight at 16°C. After induction, the cell pellet from a 5-ml culture was resuspended in 750 µl lysis buffer and sonicated using a Branson digital Sonifier (Emerson Industrial Automation). The sonicated mixtures were centrifuged at 4°C, 12,000 rpm for 25 min. The supernatant was incubated with 100 µl Ni-NTA for 1 h at room temperature. The matrix was subsequently washed thrice and elution was performed as instructed by the manufacturer, followed by a final elution with the SDS-PAGE sample buffer or directly with combined elution buffer and SDS- PAGE sample buffer for complete elution.
Western blot
For SDS-PAGE, protein samples were mixed with 5× Laemmli sample buffer and boiled for 5 min. In general, samples from 10–20-µg axonemes or 10 µl of bacterial samples were loaded in each lane in acrylamide gels of different percentages varying from 7 to 14% based on the molecular mass of proteins to be examined. RSP7 that co-migrated with tubulins was resolved in 10% gels. After electrophoresis, proteins were transferred to nitrocellulose membranes and the blots were probed with the antibodies as indicated. Anti-RSP2 and -RSP23 were raised in rabbits with Ni-NTA–purified recombinant polypeptides of C. reinhardtii RSP27–119 and RSP231–201. Rabbit anti-RSP3 was raised against His-tagged recombinant human RSP3. The other rabbit polyclonal antibodies for the axonemal proteins were described previously (Yang et al., 2006). In brief, anti-RSP1, -RSP4, and -RSP6 antibodies were raised against proteins purified from two-dimensional gels. Anti-RSP11 and -RSP16 were raised against respective Hi-tagged fusion proteins. Anti-RSP8 was raised against conjugated RSP8’s C-terminal fragment. Anti-HA polyclonal antibody was purchased from Covance. Anti-GST monoclonal antibody was purchased from Genscript. Anti-RSP7 was raised in chicken against a purified 25-kD His-tagged RSP7 C-terminal fragment. The anti-RSP7 IgY was used at 1:1,000 dilution, whereas the other primary and second antibodies were used at 1:5,000 dilutions in 5% dry milk in Tris-buffer-saline, pH 7.4.
Electron microscopy
Axonemes from WT, RSP31–178, and Δ1 strains were prepared by two methods: (1) a standard electron microscopy procedure with a 2.5% glutaraldehyde and cacodylate buffer primary fixative, osmium secondary fixative, dehydrated in ethanol, en bloc stained, and embedded in PolyBed resin; (2) primary fixation with 1% tannic acid, 1% glutaraldehyde in cacodylate buffer (modification of Mitchell and Sale, 1999), osmium secondary fixative, and dehydrated in ethanol, en bloc stained, and embedded in PolyBed resin. Gold-silver sections were double stained with lead citrate and uranyl acetate and examined at a magnification of 80,000 with an electron microscope (T109; Carl Zeiss) with Gatan Digital Micrograph software operating at 80 kV. Focused axoneme cross sections (∼50 from each type of flagella and from each fixation) were analyzed further. The lengths of RSs were measured and plotted into a distribution histogram. The number of axonemes with a CP that was visibly deviated was divided by the number of total axonemes examined to generate the percentage of axonemes with a deviated CP. For axonemes in which the deviation was substantial enough to be measured, the maximum difference in the distance from the middle of the CP to the inner edge of the opposing outer doublets was determined as the deviated distance. This length divided by the mean radius was computed as percentage deviation.
Sequence analysis
All analyses were conducted using web-based programs. Protein secondary structure was analyzed using the HNN predict program in NPS@ (Network Protein Sequence Analysis). The COILS program was used to assess the propensity of coil formation. Amphipathic helices were plotted using the Helical wheel program.
Acknowledgments
We are grateful for the communications regarding RSP3 with Drs. Winfield Sale, Ann Gaillard, and Maureen Wirschell (Emory University, Atlanta, GA); to Oza McClain for excellent technical assistance in electron microscopy; and to Lin Yi in generating the expression constructs for human dpy-30 gene.
This work was supported by grants from the National Institutes of Health (GM090162 to P. Yang and GM070455 to M. St. Maurice).
Footnotes
Abbreviations used in this paper:
- AH
- amphipathic helix
- AKAP
- A-kinase anchoring protein
- ARM
- armadillo
- CP
- central pair
- D/D
- dimerization and docking
- NDK
- nucleoside diphosphate kinase
- Ni-NTA
- nickel-nitrilotriacetic acid
- PMM
- paromomycin
- RS
- radial spoke
- RSP
- RS protein
- TAP
- Tris-acetate-phosphate
- WT
- wild type
References
- Alto N.M., Soderling S.H., Hoshi N., Langeberg L.K., Fayos R., Jennings P.A., Scott J.D. 2003. Bioinformatic design of A-kinase anchoring protein-in silico: a potent and selective peptide antagonist of type II protein kinase A anchoring. Proc. Natl. Acad. Sci. USA. 100:4445–4450 10.1073/pnas.0330734100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelo R., Rubin C.S. 1998. Molecular characterization of an anchor protein (AKAPCE) that binds the RI subunit (RCE) of type I protein kinase A from Caenorhabditis elegans. J. Biol. Chem. 273:14633–14643 10.1074/jbc.273.23.14633 [DOI] [PubMed] [Google Scholar]
- Banky P., Roy M., Newlon M.G., Morikis D., Haste N.M., Taylor S.S., Jennings P.A. 2003. Related protein-protein interaction modules present drastically different surface topographies despite a conserved helical platform. J. Mol. Biol. 330:1117–1129 10.1016/S0022-2836(03)00552-7 [DOI] [PubMed] [Google Scholar]
- Barber C.F., Heuser T., Carbajal-González B.I., Botchkarev V.V., Jr, Nicastro D. 2012. Three-dimensional structure of the radial spokes reveals heterogeneity and interactions with dyneins in Chlamydomonas flagella. Mol. Biol. Cell. 23:111–120 10.1091/mbc.E11-08-0692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brokaw C.J. 1987. Regulation of sperm flagellar motility by calcium and cAMP-dependent phosphorylation. J. Cell. Biochem. 35:175–184 10.1002/jcb.240350302 [DOI] [PubMed] [Google Scholar]
- Burns-Hamuro L.L., Ma Y., Kammerer S., Reineke U., Self C., Cook C., Olson G.L., Cantor C.R., Braun A., Taylor S.S. 2003. Designing isoform-specific peptide disruptors of protein kinase A localization. Proc. Natl. Acad. Sci. USA. 100:4072–4077 10.1073/pnas.2628038100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao F., Chen Y., Cierpicki T., Liu Y., Basrur V., Lei M., Dou Y. 2010. An Ash2L/RbBP5 heterodimer stimulates the MLL1 methyltransferase activity through coordinated substrate interactions with the MLL1 SET domain. PLoS ONE. 5:e14102 10.1371/journal.pone.0014102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr D.W., Stofko-Hahn R.E., Fraser I.D., Bishop S.M., Acott T.S., Brennan R.G., Scott J.D. 1991. Interaction of the regulatory subunit (RII) of cAMP-dependent protein kinase with RII-anchoring proteins occurs through an amphipathic helix binding motif. J. Biol. Chem. 266:14188–14192 [PubMed] [Google Scholar]
- Carr D.W., Hausken Z.E., Fraser I.D., Stofko-Hahn R.E., Scott J.D. 1992. Association of the type II cAMP-dependent protein kinase with a human thyroid RII-anchoring protein. Cloning and characterization of the RII-binding domain. J. Biol. Chem. 267:13376–13382 [PubMed] [Google Scholar]
- Carr D.W., Fujita A., Stentz C.L., Liberty G.A., Olson G.E., Narumiya S. 2001. Identification of sperm-specific proteins that interact with A-kinase anchoring proteins in a manner similar to the type II regulatory subunit of PKA. J. Biol. Chem. 276:17332–17338 10.1074/jbc.M011252200 [DOI] [PubMed] [Google Scholar]
- Chen Y., Wan B., Wang K.C., Cao F., Yang Y., Protacio A., Dou Y., Chang H.Y., Lei M. 2011. Crystal structure of the N-terminal region of human Ash2L shows a winged-helix motif involved in DNA binding. EMBO Rep. 12:797–803 10.1038/embor.2011.101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho Y.W., Hong T., Hong S., Guo H., Yu H., Kim D., Guszczynski T., Dressler G.R., Copeland T.D., Kalkum M., Ge K. 2007. PTIP associates with MLL3- and MLL4-containing histone H3 lysine 4 methyltransferase complex. J. Biol. Chem. 282:20395–20406 10.1074/jbc.M701574200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diener D.R., Ang L.H., Rosenbaum J.L. 1993. Assembly of flagellar radial spoke proteins in Chlamydomonas: identification of the axoneme binding domain of radial spoke protein 3. J. Cell Biol. 123:183–190 10.1083/jcb.123.1.183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diener D.R., Yang P., Geimer S., Cole D.G., Sale W.S., Rosenbaum J.L. 2011. Sequential assembly of flagellar radial spokes. Cytoskeleton (Hoboken). 68:389–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita A., Nakamura K., Kato T., Watanabe N., Ishizaki T., Kimura K., Mizoguchi A., Narumiya S. 2000. Ropporin, a sperm-specific binding protein of rhophilin, that is localized in the fibrous sheath of sperm flagella. J. Cell Sci. 113:103–112 [DOI] [PubMed] [Google Scholar]
- Gaillard A.R., Diener D.R., Rosenbaum J.L., Sale W.S. 2001. Flagellar radial spoke protein 3 is an A-kinase anchoring protein (AKAP). J. Cell Biol. 153:443–448 10.1083/jcb.153.2.443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillard A.R., Fox L.A., Rhea J.M., Craige B., Sale W.S. 2006. Disruption of the A-kinase anchoring domain in flagellar radial spoke protein 3 results in unregulated axonemal cAMP-dependent protein kinase activity and abnormal flagellar motility. Mol. Biol. Cell. 17:2626–2635 10.1091/mbc.E06-02-0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold M.G., Lygren B., Dokurno P., Hoshi N., McConnachie G., Taskén K., Carlson C.R., Scott J.D., Barford D. 2006. Molecular basis of AKAP specificity for PKA regulatory subunits. Mol. Cell. 24:383–395 10.1016/j.molcel.2006.09.006 [DOI] [PubMed] [Google Scholar]
- Greengard P., Jen J., Nairn A.C., Stevens C.F. 1991. Enhancement of the glutamate response by cAMP-dependent protein kinase in hippocampal neurons. Science. 253:1135–1138 10.1126/science.1716001 [DOI] [PubMed] [Google Scholar]
- Gupta A., Diener D.R., Sivadas P., Rosenbaum J.L., Yang P. 2012. The versatile molecular complex component LC8 promotes several distinct steps of flagellar assembly. J. Cell Biol. 198:115–126 10.1083/jcb.201111041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habermacher G., Sale W.S. 1996. Regulation of flagellar dynein by an axonemal type-1 phosphatase in Chlamydomonas. J. Cell Sci. 109:1899–1907 [DOI] [PubMed] [Google Scholar]
- Habermacher G., Sale W.S. 1997. Regulation of flagellar dynein by phosphorylation of a 138-kD inner arm dynein intermediate chain. J. Cell Biol. 136:167–176 10.1083/jcb.136.1.167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herberg F.W., Maleszka A., Eide T., Vossebein L., Tasken K. 2000. Analysis of A-kinase anchoring protein (AKAP) interaction with protein kinase A (PKA) regulatory subunits: PKA isoform specificity in AKAP binding. J. Mol. Biol. 298:329–339 10.1006/jmbi.2000.3662 [DOI] [PubMed] [Google Scholar]
- Hirsch A.H., Glantz S.B., Li Y., You Y., Rubin C.S. 1992. Cloning and expression of an intron-less gene for AKAP 75, an anchor protein for the regulatory subunit of cAMP-dependent protein kinase II beta. J. Biol. Chem. 267:2131–2134 [PubMed] [Google Scholar]
- Howard D.R., Habermacher G., Glass D.B., Smith E.F., Sale W.S. 1994. Regulation of Chlamydomonas flagellar dynein by an axonemal protein kinase. J. Cell Biol. 127:1683–1692 10.1083/jcb.127.6.1683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu D.R., Meyer B.J. 1994. The dpy-30 gene encodes an essential component of the Caenorhabditis elegans dosage compensation machinery. Genetics. 137:999–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B., Piperno G., Ramanis Z., Luck D.J. 1981. Radial spokes of Chlamydomonas flagella: genetic analysis of assembly and function. J. Cell Biol. 88:80–88 10.1083/jcb.88.1.80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarnaess E., Ruppelt A., Stokka A.J., Lygren B., Scott J.D., Taskén K. 2008. Dual specificity A-kinase anchoring proteins (AKAPs) contain an additional binding region that enhances targeting of protein kinase A type I. J. Biol. Chem. 283:33708–33718 10.1074/jbc.M804807200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H., Shukla A., Wang X., Chen W.Y., Bernstein B.E., Roeder R.G. 2011. Role for Dpy-30 in ES cell-fate specification by regulation of H3K4 methylation within bivalent domains. Cell. 144:513–525 10.1016/j.cell.2011.01.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kammerer S., Burns-Hamuro L.L., Ma Y., Hamon S.C., Canaves J.M., Shi M.M., Nelson M.R., Sing C.F., Cantor C.R., Taylor S.S., Braun A. 2003. Amino acid variant in the kinase binding domain of dual-specific A kinase-anchoring protein 2: a disease susceptibility polymorphism. Proc. Natl. Acad. Sci. USA. 100:4066–4071 10.1073/pnas.2628028100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinderman F.S., Kim C., von Daake S., Ma Y., Pham B.Q., Spraggon G., Xuong N.H., Jennings P.A., Taylor S.S. 2006. A dynamic mechanism for AKAP binding to RII isoforms of cAMP-dependent protein kinase. Mol. Cell. 24:397–408 10.1016/j.molcel.2006.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindle K.L. 1990. High-frequency nuclear transformation of Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA. 87:1228–1232 10.1073/pnas.87.3.1228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klauck T.M., Faux M.C., Labudda K., Langeberg L.K., Jaken S., Scott J.D. 1996. Coordination of three signaling enzymes by AKAP79, a mammalian scaffold protein. Science. 271:1589–1592 10.1126/science.271.5255.1589 [DOI] [PubMed] [Google Scholar]
- Kohno T., Wakabayashi K., Diener D.R., Rosenbaum J.L., Kamiya R. 2011. Subunit interactions within the Chlamydomonas flagellar spokehead. Cytoskeleton (Hoboken). 68:237–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Adamik R., Pacheco-Rodriguez G., Moss J., Vaughan M. 2003. Protein kinase A-anchoring (AKAP) domains in brefeldin A-inhibited guanine nucleotide-exchange protein 2 (BIG2). Proc. Natl. Acad. Sci. USA. 100:1627–1632 10.1073/pnas.0337678100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde D.N., O’Toole E.T., McDonald K.L., McIntosh J.R., Porter M.E. 1992. Arrangement of inner dynein arms in wild-type and mutant flagella of Chlamydomonas. J. Cell Biol. 118:1145–1162 10.1083/jcb.118.5.1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauban J.R., O’Donnell M., Warrier S., Manni S., Bond M. 2009. AKAP-scaffolding proteins and regulation of cardiac physiology. Physiology (Bethesda). 24:78–87 10.1152/physiol.00041.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Means C.K., Lygren B., Langeberg L.K., Jain A., Dixon R.E., Vega A.L., Gold M.G., Petrosyan S., Taylor S.S., Murphy A.N., et al. 2011. An entirely specific type I A-kinase anchoring protein that can sequester two molecules of protein kinase A at mitochondria. Proc. Natl. Acad. Sci. USA. 108:E1227–E1235 10.1073/pnas.1107182108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell D.R., Sale W.S. 1999. Characterization of a Chlamydomonas insertional mutant that disrupts flagellar central pair microtubule-associated structures. J. Cell Biol. 144:293–304 10.1083/jcb.144.2.293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morikis D., Roy M., Newlon M.G., Scott J.D., Jennings P.A. 2002. Electrostatic properties of the structure of the docking and dimerization domain of protein kinase A IIalpha. Eur. J. Biochem. 269:2040–2051 10.1046/j.1432-1033.2002.02852.x [DOI] [PubMed] [Google Scholar]
- Newell A.E., Fiedler S.E., Ruan J.M., Pan J., Wang P.J., Deininger J., Corless C.L., Carr D.W. 2008. Protein kinase A RII-like (R2D2) proteins exhibit differential localization and AKAP interaction. Cell Motil. Cytoskeleton. 65:539–552 10.1002/cm.20279 [DOI] [PubMed] [Google Scholar]
- Nicastro D., McIntosh J.R., Baumeister W. 2005. 3D structure of eukaryotic flagella in a quiescent state revealed by cryo-electron tomography. Proc. Natl. Acad. Sci. USA. 102:15889–15894 10.1073/pnas.0508274102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omoto C.K., Gibbons I.R., Kamiya R., Shingyoji C., Takahashi K., Witman G.B. 1999. Rotation of the central pair microtubules in eukaryotic flagella. Mol. Biol. Cell. 10:1–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A., Dharmarajan V., Vought V.E., Cosgrove M.S. 2009. On the mechanism of multiple lysine methylation by the human mixed lineage leukemia protein-1 (MLL1) core complex. J. Biol. Chem. 284:24242–24256 10.1074/jbc.M109.014498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel-King R.S., Gorbatyuk O., Takebe S., King S.M. 2004. Flagellar radial spokes contain a Ca2+-stimulated nucleoside diphosphate kinase. Mol. Biol. Cell. 15:3891–3902 10.1091/mbc.E04-04-0352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauling L., Corey R.B., Branson H.R. 1951. The structure of proteins; two hydrogen-bonded helical configurations of the polypeptide chain. Proc. Natl. Acad. Sci. USA. 37:205–211 10.1073/pnas.37.4.205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pigino G., Bui K.H., Maheshwari A., Lupetti P., Diener D.R., Ishikawa T. 2011. Cryoelectron tomography of radial spokes in cilia and flagella. J. Cell Biol. 195:673–687 10.1083/jcb.201106125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roguev A., Schaft D., Shevchenko A., Pijnappel W.W., Wilm M., Aasland R., Stewart A.F. 2001. The Saccharomyces cerevisiae Set1 complex includes an Ash2 homologue and methylates histone 3 lysine 4. EMBO J. 20:7137–7148 10.1093/emboj/20.24.7137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rual J.F., Venkatesan K., Hao T., Hirozane-Kishikawa T., Dricot A., Li N., Berriz G.F., Gibbons F.D., Dreze M., Ayivi-Guedehoussou N., et al. 2005. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 437:1173–1178 10.1038/nature04209 [DOI] [PubMed] [Google Scholar]
- Sarma G.N., Kinderman F.S., Kim C., von Daake S., Chen L., Wang B.C., Taylor S.S. 2010. Structure of D-AKAP2:PKA RI complex: insights into AKAP specificity and selectivity. Structure. 18:155–166 10.1016/j.str.2009.12.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satir P. 1999. The cilium as a biological nanomachine. FASEB J. 13(Suppl 2):S235–S237 [DOI] [PubMed] [Google Scholar]
- Scott J.D., Pawson T. 2009. Cell signaling in space and time: where proteins come together and when they’re apart. Science. 326:1220–1224 10.1126/science.1175668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silflow C.D., LaVoie M., Tam L.W., Tousey S., Sanders M., Wu W., Borodovsky M., Lefebvre P.A. 2001. The Vfl1 Protein in Chlamydomonas localizes in a rotationally asymmetric pattern at the distal ends of the basal bodies. J. Cell Biol. 153:63–74 10.1083/jcb.153.1.63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- South P.F., Fingerman I.M., Mersman D.P., Du H.N., Briggs S.D. 2010. A conserved interaction between the SDI domain of Bre2 and the Dpy-30 domain of Sdc1 is required for histone methylation and gene expression. J. Biol. Chem. 285:595–607 10.1074/jbc.M109.042697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Lou Z., Dong X., Yang W., Peng Y., Yin B., Gong Y., Yuan J., Zhou W., Bartlam M., et al. 2009. Crystal structure of the C-terminal domain of human DPY-30-like protein: A component of the histone methyltransferase complex. J. Mol. Biol. 390:530–537 10.1016/j.jmb.2009.05.061 [DOI] [PubMed] [Google Scholar]
- Warner F.D., Satir P. 1974. The structural basis of ciliary bend formation. Radial spoke positional changes accompanying microtubule sliding. J. Cell Biol. 63:35–63 10.1083/jcb.63.1.35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei M., Sivadas P., Owen H.A., Mitchell D.R., Yang P. 2010. Chlamydomonas mutants display reversible deficiencies in flagellar beating and axonemal assembly. Cytoskeleton (Hoboken). 67:71–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch E.J., Jones B.W., Scott J.D. 2010. Networking with AKAPs: context-dependent regulation of anchored enzymes. Mol. Interv. 10:86–97 10.1124/mi.10.2.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams B.D., Velleca M.A., Curry A.M., Rosenbaum J.L. 1989. Molecular cloning and sequence analysis of the Chlamydomonas gene coding for radial spoke protein 3: flagellar mutation pf-14 is an ochre allele. J. Cell Biol. 109:235–245 10.1083/jcb.109.1.235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirschell M., Zhao F., Yang C., Yang P., Diener D., Gaillard A., Rosenbaum J.L., Sale W.S. 2008. Building a radial spoke: flagellar radial spoke protein 3 (RSP3) is a dimer. Cell Motil. Cytoskeleton. 65:238–248 10.1002/cm.20257 [DOI] [PubMed] [Google Scholar]
- Witman G.B., Plummer J., Sander G. 1978. Chlamydomonas flagellar mutants lacking radial spokes and central tubules. Structure, composition, and function of specific axonemal components. J. Cell Biol. 76:729–747 10.1083/jcb.76.3.729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia B., Joubert A., Groves B., Vo K., Ashraf D., Djavaherian D., Awe J., Xiong Y., Cherfils J., Ma D. 2010. Modulation of cell adhesion and migration by the histone methyltransferase subunit mDpy-30 and its interacting proteins. PLoS ONE. 5:e11771 10.1371/journal.pone.0011771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Gong Q., Xia B., Groves B., Zimmermann M., Mugler C., Mu D., Matsumoto B., Seaman M., Ma D. 2009. A role of histone H3 lysine 4 methyltransferase components in endosomal trafficking. J. Cell Biol. 186:343–353 10.1083/jcb.200902146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C., Yang P. 2006. The flagellar motility of Chlamydomonas pf25 mutant lacking an AKAP-binding protein is overtly sensitive to medium conditions. Mol. Biol. Cell. 17:227–238 10.1091/mbc.E05-07-0630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C., Owen H.A., Yang P. 2008. Dimeric heat shock protein 40 binds radial spokes for generating coupled power strokes and recovery strokes of 9 + 2 flagella. J. Cell Biol. 180:403–415 10.1083/jcb.200705069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P., Diener D.R., Rosenbaum J.L., Sale W.S. 2001. Localization of calmodulin and dynein light chain LC8 in flagellar radial spokes. J. Cell Biol. 153:1315–1326 10.1083/jcb.153.6.1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P., Diener D.R., Yang C., Kohno T., Pazour G.J., Dienes J.M., Agrin N.S., King S.M., Sale W.S., Kamiya R., et al. 2006. Radial spoke proteins of Chlamydomonas flagella. J. Cell Sci. 119:1165–1174 10.1242/jcs.02811 [DOI] [PMC free article] [PubMed] [Google Scholar]