Tail–tail interactions of desmoglein 2, promoted by its C-terminal unique region, inhibit its internalization, stabilizing it at the cell surface and promoting intercellular adhesion.
Abstract
Desmosomal cadherins, desmogleins (Dsgs) and desmocollins, make up the adhesive core of intercellular junctions called desmosomes. A critical determinant of epithelial adhesive strength is the level and organization of desmosomal cadherins on the cell surface. The Dsg subclass of desmosomal cadherins contains a C-terminal unique region (Dsg unique region [DUR]) with unknown function. In this paper, we show that the DUR of Dsg2 stabilized Dsg2 at the cell surface by inhibiting its internalization and promoted strong intercellular adhesion. DUR also facilitated Dsg tail–tail interactions. Forced dimerization of a Dsg2 tail lacking the DUR led to decreased internalization, supporting the conclusion that these two functions of the DUR are mechanistically linked. We also show that a Dsg2 mutant, V977fsX1006, identified in arrhythmogenic right ventricular cardiomyopathy patients, led to a loss of Dsg2 tail self-association and underwent rapid endocytosis in cardiac muscle cells. Our observations illustrate a new mechanism desmosomal cadherins use to control their surface levels, a key factor in determining their adhesion and signaling roles.
Introduction
Adherens junctions and desmosomes are essential for mediating intercellular adhesion in epithelial and cardiac tissues and, in addition, provide positional and signaling cues that regulate cell proliferation, polarity, migration, and differentiation (Schock and Perrimon, 2002; Green and Simpson, 2007; Niessen et al., 2011). The assembly and disassembly of cell–cell junctions is carefully choreographed during epithelial morphogenesis and remodeling (Niessen et al., 2011). Altering junction stability or assembly state through loss of function, mutation, or posttranslational modification can lead to inherited disorders, blistering diseases, and cancer (Holthöfer et al., 2007; Simpson and Green, 2007; Thomason et al., 2010; Amagai and Stanley, 2012; Brooke et al., 2012).
The adhesive core of adherens junctions and desmosomes comprises members of the cadherin superfamily—classical cadherins (e.g., E-cadherin) in adherens junctions and desmosomal cadherins (desmogleins [Dsgs] and desmocollins) in desmosomes (Green and Gaudry, 2000; Pokutta and Weis, 2007). In both cases, adhesive interactions are mediated by trans-interactions between the N-terminal cadherin ectodomains on the surface of neighboring cells. The C-terminal tails are embedded in a cytoplasmic plaque consisting of armadillo proteins, cytoskeletal adaptors, and their associated cytoskeletal connections. Although adherens junctions organize and regulate the assembly state of cortical actin, desmosomes provide integrity to tissues by anchoring intermediate filaments to sites of desmosomal adhesion.
The extracellular repeats that define the cadherin superfamily are fairly well conserved in classical and desmosomal cadherins, but the domain structure of the cytoplasmic tails exhibits unique features (Hulpiau and van Roy, 2009). The membrane proximal regions in both cases contain regions that associate with armadillo gene family members. More distally, Dsgs contain an extended C-terminal unique region (Dsg unique region [DUR]) with unknown function (Koch et al., 1990). This region can be divided into a linker region, a series of repeats each consisting of 29 ± 4 residues, and a terminal domain (Fig. 1 A). Electron microscopy showed that the predominant form of DUR is a monomer, consisting of a globular head attached to a thin tail. Dimers and oligomers were also observed but less frequently (Rutman et al., 1994). Another biophysical study demonstrated that the DUR is intrinsically disordered with an inducible structure (Kami et al., 2009). The potential modulatory roles conferred by the DUR on Dsg or desmosomes and how the DUR exerts these functions are unknown.
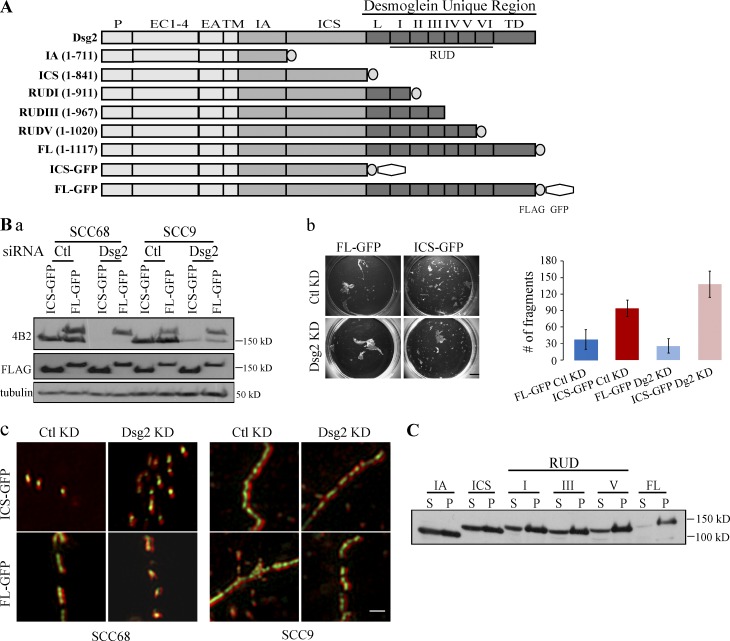
Figure 1.
DUR is required for strong cell–cell adhesion. (A) Schematic representation of Dsg2 and Dsg2 mutants. P, precursor sequence; EC, extracellular cadherin repeat; EA, extracellular anchoring domain; TM, transmembrane domain; IA, intracellular anchoring domain; ICS, intracellular cadherin-like sequence; L, linker domain; RUD, repeat unit domain; TD, terminal domain. (B, a) SCC68 or SCC9 cells expressing Dsg2.ICS-GFP or Dsg2.FL-GFP were transfected with 20 nM siRNA oligos targeting endogenous Dsg2 (Dsg2) or a nonspecific sequence (control [Ctl]). 48 h after transfection, cell lysates were collected and immunoblotted with the indicated antibodies. The epitope for 4B2 is in the C terminus of Dsg2 tail and therefore recognizes endogenous Dsg2 and Dsg2.FL-GFP but not Dsg2.ICS-GFP. (b) SCC68 cells stably expressing Dsg2.ICS-GFP or Dsg2.FL-GFP were transfected with 20 nM siRNA oligos targeting endogenous Dsg2 (Dsg2) or a nonspecific sequence (control KD). 24 h after transfection, cells were placed in medium containing 1.0 mM Ca2+. Another 24 h later, at which time the cells were confluent, a dispase assay was performed. Bar, 5 mm. The graph shows the number of fragments counted from one representative experiment with three replicates. This experiment was repeated twice. Error bars represent mean ± SEM. (c) SCC68 or SCC9 cells expressing Dsg2.ICS-GFP or Dsg2.FL-GFP were transfected with 20 nM siRNA oligos targeting endogenous Dsg2 (Dsg2) or nonspecific sequence (control KD). 24 h after transfection, cells were placed in medium containing 1.0 mM Ca2+. Another 24 h later, cells were stained for DP (red) and imaged using an N-SIM system. Bar, 1 µm. (C) Mutant expressing SCC68 cells grown in medium with 0.09 mM Ca2+ were lysed in 0.5% Triton X-100 buffer. Supernatants (S) were collected after centrifugation. To the remaining pellets (P), USB equal to the volume of the supernatant was added, and pellets were solubilized. Equal amount of supernatant and pellet samples from each mutant were blotted for FLAG. Similar results were obtained for cells in 1.0 mM Ca2+.
Cell–cell adhesion plays important roles in many cellular functions. Cell adhesive structures undergo dynamic changes during tissue remodeling (e.g., embryonic development, wound reepithelialization, and cell renewal in the epidermis), and endocytosis is a key regulator of this process. Some efforts have been made to determine how Dsg is endocytosed in the presence of various environmental stimuli: autoantibodies, kinase inhibitors, and calcium depletion (Holm et al., 1993; Delva et al., 2008; Klessner et al., 2009; Jolly et al., 2010; Jennings et al., 2011). Engagement of transmembrane receptors with internalization machinery is frequently dictated by the presence of specific sequences in their cytoplasmic tails (Bonifacino and Traub, 2003), but the contribution of Dsg cytoplasmic sequences to regulation of endocytosis is poorly understood. Among the four Dsg isoforms, Dsg2 is the first to be synthesized during development (Fleming et al., 1991), the most prevalently expressed in desmosomes of adult tissues, and has the longest DUR (Schäfer et al., 1994). Here, we address the contribution of the Dsg2 DUR sequence to intercellular adhesive strength and how DUR regulates Dsg2’s stability and endocytic behavior. Through analysis of a panel of Dsg2 mutants, we conclude that the DUR strengthens cell–cell adhesion and stabilizes Dsg2 at the cell surface by inhibiting its internalization. We also show that the DUR can mediate Dsg2 tail self-interactions and that forced dimerization of Dsg2 lacking the DUR is sufficient to attenuate internalization. Furthermore, we demonstrate that V977fsX1006, a Dsg2 DUR mutant identified in arrhythmogenic right ventricular cardiomyopathy (ARVC) patients, loses Dsg2 tail–tail interactions, and undergoes rapid endocytosis in cardiac muscle cells. These results reveal a new role for the DUR and a possible mechanism for regulating the level and organization of Dsg at the cell surface during development and homeostasis of adult tissues.
Results
Dsg2 DUR is required for strong cell–cell adhesion
To investigate the contribution of the DUR to cell–cell adhesion and Dsg2 dynamics, we generated a series of mutants with progressive deletions of previously designated domains within the cytoplasmic tail of Dsg2 (Fig. 1 A; Schäfer et al., 1994). Full-length (FL) Dsg2 and Dsg2 mutants were each tagged with a FLAG and/or a GFP epitope at the C terminus and designated by the name of the last remaining domain. Squamous cell carcinoma cell lines (SCC68 and SCC9) were retrovirally transduced to express each Dsg2 construct (Fig. S1). The GFP-tagged mutant-expressing cells were then sorted by flow cytometry before being used for further experiments. A dispase assay was performed to compare the mechanical integrity of epithelial cell sheets expressing Dsg2.ICS-GFP– and Dsg2.FL-GFP (Fig. 1 B, b). In this assay, cells were grown in high calcium medium until confluent. The cell sheet was then released from the substrate by specifically disrupting matrix adhesions by enzymatic digestion with dispase (Hudson et al., 2004). The lifted cell sheet was then subjected to mechanical sheer stress, and the number of cell sheet fragments generated was used as an inverse indicator of cell–cell adhesive strength. To rule out the possible contribution of endogenous Dsg2 in this system, we knocked down endogenous Dsg2 using siRNA oligonucleotides (oligos) targeting the 3′ UTR of Dsg2 and used nonspecific siRNA oligos as a control before assaying adhesive strength (Fig. 1 B, a). As shown in Fig. 1 B, regardless of the status of endogenous Dsg2, Dsg2.ICS-GFP–expressing cells produced two to three times more fragments under mechanical stress than Dsg2.FL-GFP–expressing cells, suggesting the DUR is required to facilitate strong cell–cell adhesion.
One possible explanation for the observed decrease in intercellular adhesion is that without the DUR, Dsg2.ICS-GFP is unable to assemble into desmosomes. To test whether the DUR is required to target Dsg2 to junctions, we stained the GFP mutant-expressing cells for desmoplakin (DP), a component of the desmosome plaque. The immunofluorescence signals for DP and GFP were then captured and reconstructed using Nikon structured illumination microscopy (N-SIM), which produces two times the resolution of conventional optical microscopes. All desmosomes exhibit a bilaterally symmetrical structure comprising closely opposed plasma membranes containing Dsgs and desmocollins, sandwiched by two cytoplasmic plaques made of DP and its associated proteins. By N-SIM, we detected a DP-Dsg-DP arrangement along the plasma membrane only under conditions supporting desmosome assembly. Using this “railroad” pattern as an indicator, we demonstrate that both Dsg2.ICS-GFP and Dsg2.FL-GFP assemble into desmosomes regardless of the status of the endogenous Dsg2 (Fig. 1 B, c). These results suggested that the DUR is not required for Dsg2 to incorporate into desmosomes.
Even though both GFP mutants were observed to incorporate into desmosomes, the reduced adhesive strength exhibited by cell sheets expressing Dsg2.ICS-GFP suggested differences in the structure or stability of the Dsg2-containing adhesive complexes. To begin to test this idea, the Triton X-100 solubility of the Dsg2 mutants was tested (Fig. 1 C). Sequential deletion of distal regions of Dsg2 resulted in a progressive shift into the Triton X-100–insoluble pool, raising the possibility that the DUR promotes Dsg2 stabilization through strengthening association with detergent-insoluble cytoskeletal or membrane elements. The DUR decreased Dsg2 Triton X-100 insolubility even in low calcium medium, suggesting that association with the intermediate filament cytoskeleton is unlikely to completely account for these differences.
Dsg2 DUR inhibits Dsg2 internalization
The aforementioned observations suggested that Dsg2 mutants lacking the DUR form less stable structures than FL Dsg2 and prompted us to ask whether these mutants are also more likely to be subjected to endocytic turnover. To investigate whether the DUR regulates Dsg2 endocytosis, proteins on the surface of SCC68 cells were biotinylated, and the internalized pool of Dsg2 was tracked over time as described in the Materials and methods (Fig. 2 B). Among the Dsg2 mutants, Dsg2.ICS was internalized most efficiently. Dsg2.IA was the second best internalized construct, suggesting there may be a “signal” residing in the ICS domain that promotes internalization. Interestingly, mutants containing the DUR region (Dsg2.RUDI, Dsg2.RUDIII, Dsg2.RUDV, and Dsg2.FL) were also internalized less efficiently than the Dsg2.ICS (Fig. 2, B and C). These results suggest that the DUR negatively regulates Dsg2 internalization. The same effect of the DUR on Dsg2 internalization was also observed in SCC9 cells (Fig. 2 G), suggesting that the inhibitory function of the DUR is cell type independent.
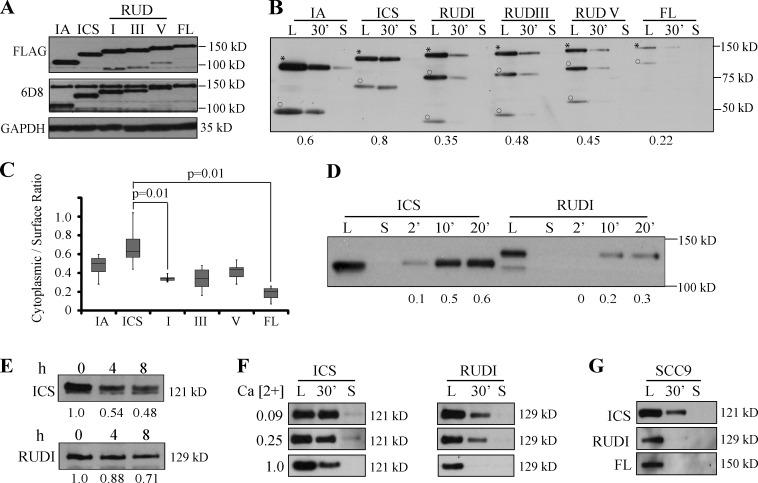
Figure 2.
DUR inhibits Dsg2 internalization. (A) Lysates from mutant-expressing SCC68 cells were immunoblotted with the indicated antibodies. The epitope for 6D8 is in the Dsg2 ectodomain. (B) Biotinylation assay was performed in mutant-expressing SCC68 cells grown in medium with 0.25 mM Ca2+. L, surface pool of the Dsg2 mutants. 30’, cytoplasmic pool of the Dsg2 mutants after 30 min of internalization. S, residual Dsg2 mutant protein left on the cell surface after stripping. Asterisks indicate FL proteins, and circles show cleavage products. Densitometric analysis was performed using ImageJ to determine the ratio of cytoplasmic/surface intensity. Numbers at the bottom indicate the internalization ratio for each mutant. (C) Collective results from seven independent biotinylation assays performed at 0.25 mM Ca2+. n ≥ 4 per mutant. Paired t test. The bottom and top of the box are the 25th and 75th percentile (the lower and upper quartiles), respectively. The horizontal line near the middle of the box is the 50th percentile (the median). The ends of the whisker are the minimum and maximum of all the data. (D) Biotinylation assay of SCC68 cells expressing Dsg2.ICS or Dsg2.RUDI grown in medium with 0.25 mM Ca2+ with varying internalization times: 2, 10, and 20 min. (E) SCC68 cells expressing Dsg2.ICS or Dsg2.RUDI were pretreated with 10 µg/ml cycloheximide for 30 min (time 0) and then kept in cycloheximide-containing medium for an additional 4 or 8 h before collection. Cell lysates were immunoblotted for FLAG. (F) SCC68 cells expressing Dsg2.ICS or Dsg2.RUDI were grown in medium containing 0.09, 0.25, or 1.0 mM Ca2+ and subjected to biotinylation assay with 30-min internalization. (G) Biotinylation assay conducted on SCC9 cells expressing the Dsg2 mutants.
Because the region consisting of the linker and the first repeat unit domain was sufficient to exhibit an inhibitory effect, we used Dsg2.ICS and Dsg2.RUDI to further investigate the impact of this region on Dsg2 stability. A biotinylation assay with higher resolution time course was conducted (Fig. 2 D). The difference in internalization efficiency between Dsg2.ICS and Dsg2.RUDI was observed as short at 2 min after the 37°C incubation. This suggests that the difference in the cytoplasmic/surface ratio is likely caused by inhibition of internalization rather than alterations in degradation and recycling. We also examined the turnover rate of these Dsg2 mutants. After cycloheximide treatment, Dsg2.ICS degraded more rapidly than Dsg2.RUDI (Fig. 2 E). This observation is consistent with the idea that more efficient internalization of Dsg2.ICS contributes to its faster turnover. Based on these observations, we conclude that the DUR of Dsg2 stabilizes Dsg2 at least in part by inhibiting its internalization.
Dsg2 DUR inhibits Dsg2 internalization independently of Dsg2 ectodomain engagement
Dsg ectodomains engage in homophilic and/or heterophilic interactions in the extracellular space (Chitaev and Troyanovsky, 1997; Nie et al., 2011). We used two approaches to examine the potential contribution of the Dsg2 ectodomain on DUR-dependent inhibition of Dsg2 internalization. Calcium binding is critical for ectodomain conformation and cadherin interactions (Watt et al., 1984; Syed et al., 2002), and cells are unable to assemble desmosomes in low extracellular calcium. Here in the first approach, we cultured and analyzed Dsg2 mutant-expressing SCC68 cells in medium containing 0.09 mM (low), 0.25 mM (medium), or 1.0 mM (normal) calcium (Fig. 2 F). Although the internalization rate of all the mutants tested decreased with increasing calcium concentration, the presence of the DUR sequence in Dsg2.RUDI conferred an inhibitory role on internalization compared with Dsg2.ICS independent of ectodomain engagement and under conditions that do not support desmosome assembly.
In the second approach, we fused variable lengths of the Dsg2 cytoplasmic tail with the nonadhesive IL2R (interleukin 2 receptor)-α chain ectodomain and transmembrane domain (Fig. 3 A). The resulting chimeras were stably expressed in SCC68 cells via retroviral transduction (Fig. 3 B), and their internalization efficiency was analyzed. Adding IA-ICS of Dsg2 to the IL2R ecto- and transmembrane domain did not alter the internalization rate (Fig. S2 A). However, addition of the region downstream of IA-ICS led to reduced internalization—IL2R:RUDI and IL2R:FL were internalized much less efficiently than IL2R:ICS, confirming the inhibitory role of DUR (Fig. 3 C).
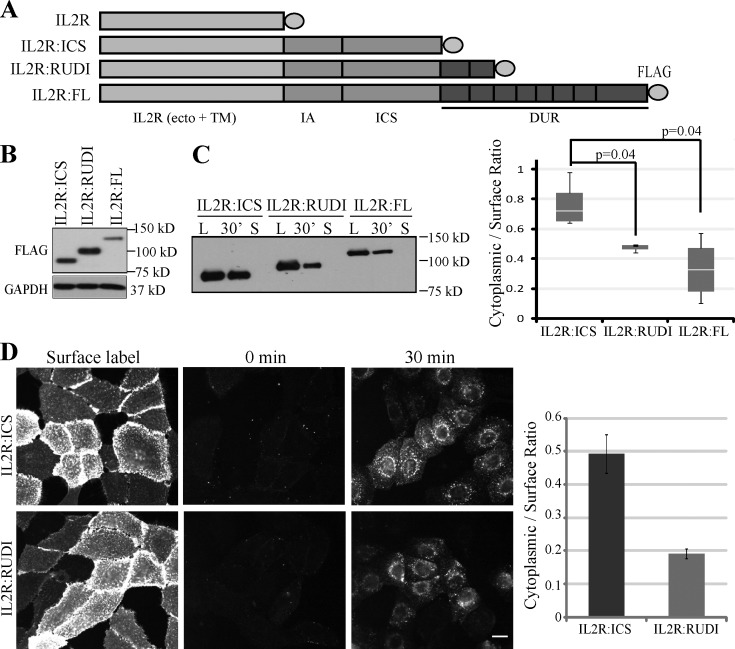
Figure 3.
DUR inhibits IL2R:Dsg2 chimera internalization. (A) Schematic representation of IL2R:Dsg2 chimeras. Ecto, ectodomain; TM, transmembrane domain. (B) Lysates from SCC68 cells expressing IL2R:Dsg2 chimeras were immunoblotted with the indicated antibodies. (C) Biotinylation assay of cells grown in medium with 0.25 mM Ca2+. The graph shows the results from five independent experiments. n ≥ 3 per mutant. Paired t test. L, surface pool of the Dsg2 mutants. 30’, cytoplasmic pool of the Dsg2 mutants after 30 min of internalization. S, residual Dsg2 mutant protein left on the cell surface after stripping. The bottom and top of the box are the 25th and 75th percentile (the lower and upper quartiles), respectively. The horizontal line near the middle of the box is the 50th percentile (the median). The ends of the whisker are the minimum and maximum of all the data. (D) Cells were labeled with anti-IL2R antibody at 4°C and then incubated at 37°C for 0 or 30 min. Residual surface antibodies were subsequently stripped. Internalized IL2R:ICS (top) and IL2R:RUDI (bottom) chimeras were visualized using indirect immunofluorescence. Bar, 20 µm. Graph, from one representative experiment, shows the ratio of mean cytoplasmic intensity per cell/mean surface label intensity per cell for each mutant. Number of cells quantified are as follows: IL2R:ICS (n = 63) and IL2R:RUDI (n = 66). Error bars are SEM.
Taking advantage of the absence of endogenous IL2R-α in SCC68 cells, we performed a fluorescence-based antibody internalization assay using the chimera-expressing cells. In this assay, the IL2R chimeras on the cell surface were labeled using an IL2R antibody at 4°C, and internalization was allowed to proceed by raising the temperature to 37°C (Xiao et al., 2005). Because the internalized pool is protected from biochemical removal of remaining surface-labeled antibody, this assay allows for specific detection of the internalized pool of Dsg2 constructs and differentiates between the exocytic and endocytic compartments. As shown in Fig. 3 D, ∼2.5-fold less internalized particles were observed in the cytoplasm of the IL2R:RUDI cells than in IL2R:ICS cells (Fig. 3 D). By placing the Dsg2 tail into a different molecular context (IL2R), these results support the idea that the DUR has an intrinsic ability to inhibit Dsg2 internalization in the absence of ectodomain engagement.
Dsg2 constructs with or without the DUR share endocytic pathways
The data presented so far demonstrate that the Dsg2 DUR controls the kinetics of Dsg2 internalization. It is possible that the difference in kinetics may be caused by the different internalization pathways taken by Dsg2 in the presence or absence of DUR. To address this, we compared the responses of Dsg2 constructs to various inhibitors of endocytosis. It is reported that Dsg2 resides in lipid rafts (Brennan et al., 2012), which are cholesterol-enriched hubs for signaling and endocytosis events (Simons and Gerl, 2010). Removal of cholesterol from SCC68 cells by brief methyl-β-cyclodextrin treatment led to reduced internalization of endogenous Dsg2 (Fig. S2 B), suggesting that the integrity of lipid raft structure plays a positive role in Dsg2 internalization. The sensitivity to cholesterol removal was also exhibited by Dsg2.ICS and Dsg2.RUDI (Fig. 4 A), which indicates that the internalization of both Dsg2.ICS and Dsg2.RUDI are cholesterol dependent.
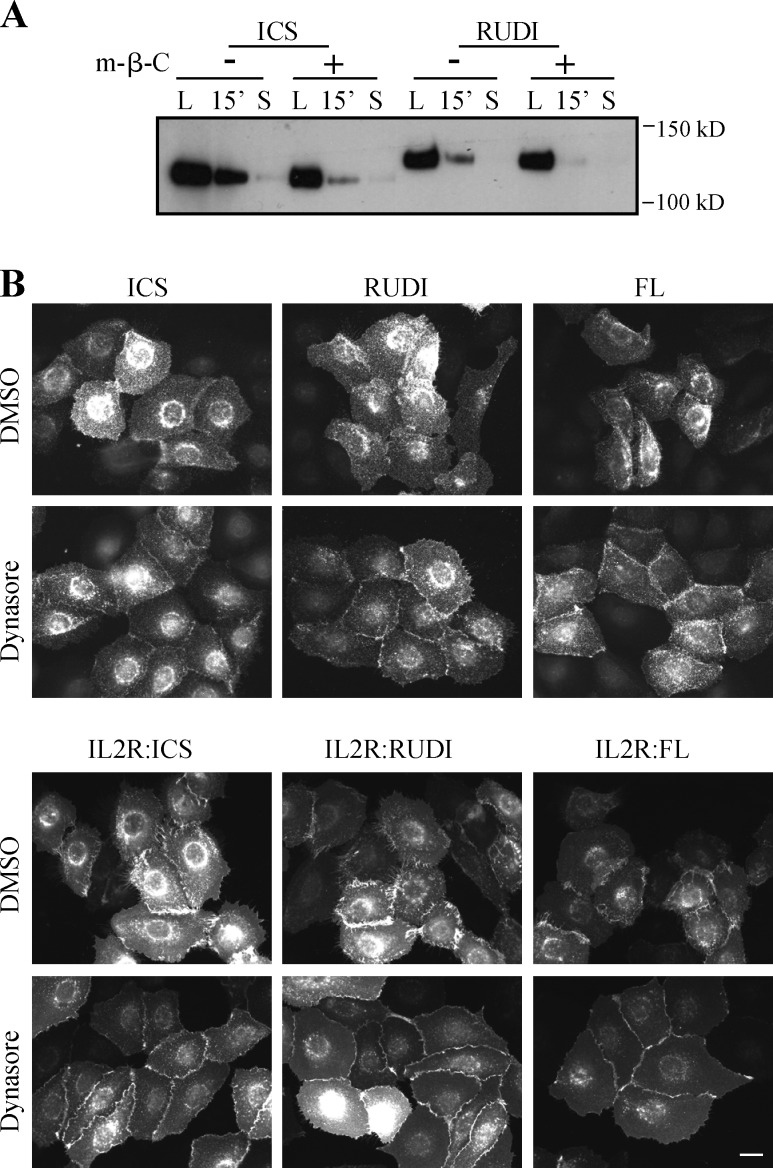
Figure 4.
Internalization of Dsg2 mutants depends on cholesterol and dynamin. (A) Biotinylation assay of cells that were pretreated with 5 mM methyl-β-cyclodextrin (m-β-C) for 30 min. L, surface pool of the Dsg2 mutants. S, residual Dsg2 mutant protein left on the cell surface after stripping. (B) Cells grown in medium with 0.25 mM Ca2+ were treated with 80 µM dynasore for 30 min and then stained for FLAG. Bar, 20 µm.
Lipid raft–mediated endocytosis encompasses various pathways characterized by their dependency on regulators including dynamin, caveolin-1 (Cav1), and flotillin (Doherty and McMahon, 2009). Dynamin is a GTPase involved in vesicle fission from the plasma membrane. Next, we tested whether Dsg2 internalization is dynamin dependent. Endogenous Dsg2 exhibited enhanced border localization in response to dynasore, a dynamin inhibitor (Fig. S2 C). Similar behaviors were observed for Dsg2.ICS, Dsg2.RUDI, Dsg2.FL, and their counterpart IL2R chimeras (Fig. 4 B), suggesting that Dsg2 internalization depends on dynamin regardless of the status of the DUR.
Cav1 is a scaffolding component of caveolae, a type of lipid raft. Dsg2 has been shown to bind to Cav1 (Brennan et al., 2012), but whether and how this interaction impacts Dsg2 internalization are unclear. We knocked down Cav1 with siRNA and tested the internalization of either endogenous Dsg2 or Dsg2 mutants (Dsg2.ICS, Dsg2.RUDI, IL2R:ICS, and IL2R:RUDI) by biotinylation or antibody internalization assays (Fig. S2, D–F). Knocking down Cav1 caused little change in the ratio of cytoplasmic/surface intensity, suggesting either Cav1 is not required for Dsg2 internalization or Dsg2 internalizes through multiple pathways that can compensate for the loss of Cav1. In either case, DUR does not play a determining role.
Other potential endocytosis regulators, flotillin-1 and clathrin, were tested using siRNA-mediated silencing, and neither stood out as a major sole determining factor in regulating Dsg2 internalization (Fig. S3). Collectively, these data support the idea that with or without the DUR, Dsg2 internalizes through a cholesterol-sensitive, dynamin-dependent pathway. Because of the complexity of the endocytic routing pathways, we cannot rule out the possibility that some fraction of Dsg2 is routed in a DUR-dependent manner by mechanisms not examined here.
DUR mediates intermolecular interactions of the Dsg2 tail
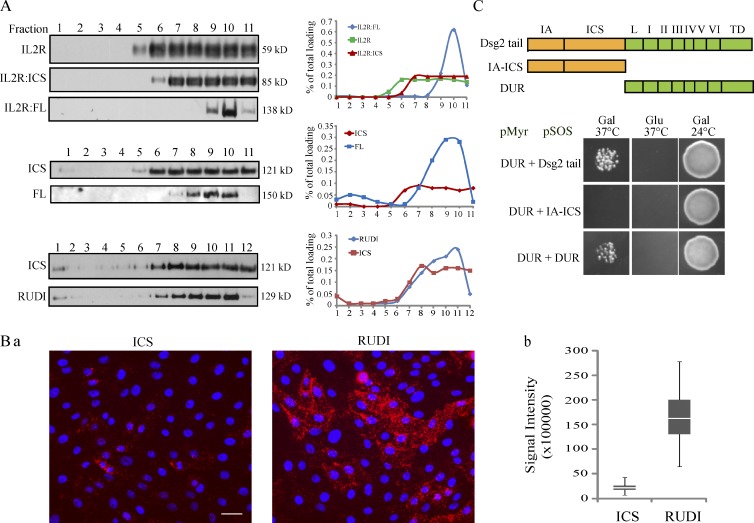
Another possible explanation for the DUR inhibitory effect is that the conformation of the Dsg2 tail depends on the DUR. DUR may provide or eliminate interaction sites for binding partners that are important for Dsg2 internalization. To examine the complex profile of Dsg2 mutants, sucrose gradient fractionation was performed. Because of the poor solubility of Dsg2 in conventional buffers, protein complexes in Dsg2 mutant-expressing cells were preserved by cross-linking, solubilized in urea buffer, and subjected to ultracentrifugation (Fig. 5 A). Both IL2R and IL2R:ICS were distributed evenly across the right half of the fraction spectrum (6–11). In contrast, IL2R:FL distribution was concentrated in fractions 9 and 10. The same difference in fraction distribution was observed between Dsg2.ICS and Dsg2.FL, consistent with the idea that DUR plays a role in controlling the distribution of Dsg2 in different subcellular complexes. The shift between Dsg2.ICS and Dsg2.RUDI was not as dramatic as the shift between Dsg2.ICS and Dsg2.FL, raising the possibility that the impact of multiple repeat unit domains is additive.
Figure 5.
DUR controls Dsg2 complex profile and mediates Dsg2 tail–tail interactions. (A) Cells were crossed-linked with 20 µg/ml dithiobis(succinimidylpropionate) at 4°C for 10 min, lysed in 0.5× USB, and then subjected to sucrose gradient fractionation (13 to 4% discontinuous sucrose gradient in 0.5× USB; 150,000 g for 18 h at 20°C). Fractions were collected starting from the bottom of the tube and blotted with antibodies against IL2R (top) or FLAG (middle and bottom). Signal intensity of each fraction was measured by densitometric analysis. To calculate the percentage of total loading per fraction, the intensity of a fraction is divided by the total intensity of all the fractions. The data shown are from one representative experiment out of two independent repeats. (B, a) In situ PLA was conducted on Dsg2-ICS– or Dsg2-RUDI–expressing cells using 4B2 and anti-FLAG antibodies. Bar, 60 µm. (b) The integrated signal intensity per image was measured and plotted. Graph was from one representative experiment with number of images quantified: Dsg2-ICS (n = 77) and Dsg2-RUDI (n = 78). The bottom and top of the box are the 25th and 75th percentile (the lower and upper quartiles), respectively. The horizontal line near the middle of the box is the 50th percentile (the median). The ends of the whisker are the minimum and maximum of all the data. (C, top) Schematic representation of Dsg2 fragments. (bottom) Analysis of the Dsg2 fragments using a yeast CytoTrap two-hybrid system. The data shown here were from one representative experiment out of three independent repeats. Gal, galactose; Glu, glucose; TD, terminal domain; L, linker domain.
A previous study reported that sequences downstream of the ICS contribute to plakoglobin (Pg) association with the related Dsg1 molecule (Choi et al., 2009). To determine whether differences in Pg binding that could contribute to Dsg2 dynamic behavior are conferred by the DUR, we compared the amount of Pg that coimmunoprecipitated with FL and mutant Dsg2 molecules. As shown in Fig. S4 A, both the Dsg2 mutants and IL2R chimeras harboring the DUR sequences coprecipitated with more Pg than those containing just the IA or ICS domains. This correlated with a trend toward decreased total Pg levels in cells expressing the shorter mutants (Fig. S1 A). To test whether increased Pg binding contributes to DUR-mediated inhibition of internalization, we generated a triple alanine Dsg2 mutant (Pg binding deficient [PGde]; L828A, F832A, and L835A) on the Dsg2.FL (wild type [WT]) backbone, which largely abrogated Pg binding (Fig. S4 B). However, PGde did not exhibit any difference in its ability to internalize compared with WT (Fig. S4 B). Furthermore, RNAi-mediated silencing of Pg or silencing of the associated armadillo proteins PKP2 or PKP3 did not measurably alter the size of the internalized pool of Dsg2 using the cell surface biotinylation assay (unpublished data). Together, these results suggest that binding to Pg is not a determining factor in Dsg2 internalization.
Another potential factor controlling complex formation and protein stability is the oligomeric status of the core protein. Some evidence suggests that desmosomal cadherins form trans- and cis-interaction with each other through their ectodomains (Syed et al., 2002; He et al., 2003; Al-Amoudi et al., 2007). Little is known about the interaction of the cytoplasmic tail of cadherins within the desmosomal plaque. To test whether DUR assists in Dsg2 tail–tail interactions, we used the in situ proximity ligation assay (PLA) to test for evidence of DUR-dependent interactions of the Dsg2 tail (Fig. 5 B). This technique visualizes two proteins in close proximity (0–40 nm) by detecting an antibody-based fluorescence signal produced by the rolling cycle amplification reaction (Söderberg et al., 2006). Because an antibody for detecting homodimerization of Dsg2 mutants was not available, we asked whether endogenous Dsg2 dimerizes with Dsg2.ICS or Dsg2.RUDI (Fig. 5 B, a). Although Dsg2.ICS and Dsg2.RUDI were expressed at similar levels and in a similar percentage of cells in the population, the PLA signal produced in Dsg2.RUDI-expressing cells was significantly greater than that generated by the Dsg2.ICS-expressing cells (Fig. 5 B, b). This is consistent with the hypothesis that DUR facilitates a Dsg2 tail–tail interaction.
A yeast CytoTrap two-hybrid system was used to further confirm the DUR-mediated interaction. We observed that DUR, but not IA-ICS, interacted with DUR, suggesting there is no intramolecular interaction within the Dsg2 tail, but there is a DUR-mediated intermolecular interaction between Dsg2 tails (Fig. 5 C). Further definition of the minimal region required for this interaction was not possible because some of the smaller fragments of DUR were not compatible with the CytoTrap system and tended to grow as temperature-sensitive revertants. Nevertheless, collectively, the data support the idea that sequences in the DUR mediate intermolecular Dsg2 tail interactions.
Dsg2 tail–tail interaction negatively regulates internalization
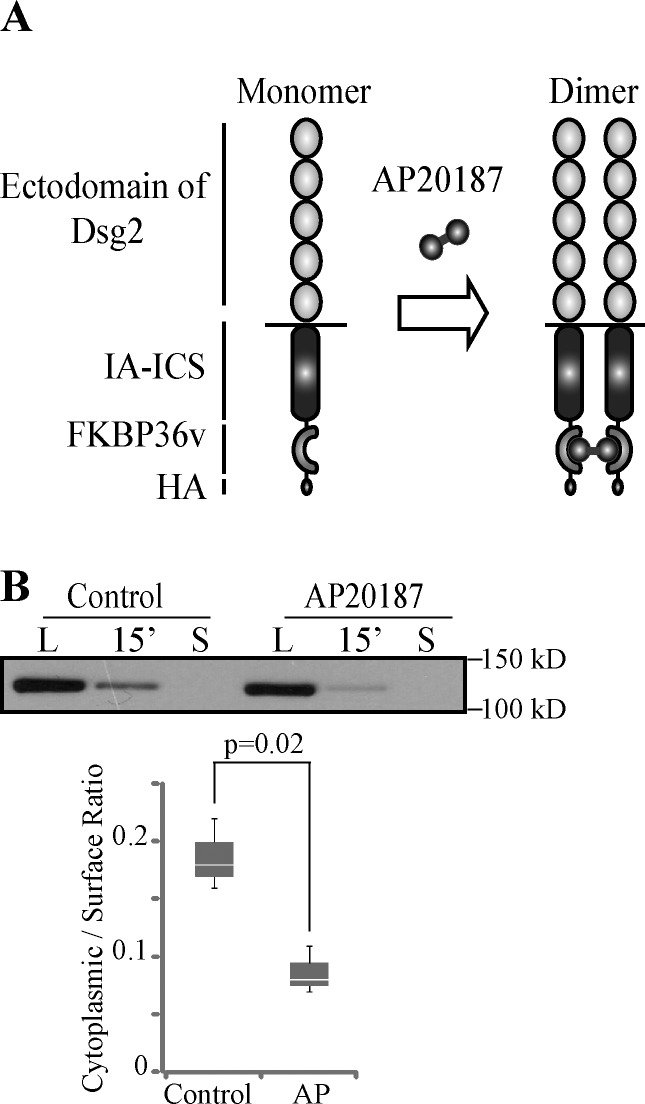
So far, we demonstrated two novel functions for the DUR: inhibition of Dsg2 internalization and mediation of Dsg2 tail–tail interactions. To investigate whether these two events are connected, we tested whether forced tail–tail interaction of Dsg2 inhibits its internalization. A regulated homodimerization system was designed based on the human FKBP (FK506-binding protein) 12 and its ligand (ARIAD Pharmaceuticals). One copy of a variant of FK506-binding protein (FKBP36v) followed by a HA epitope tag was fused to the C terminus of Dsg2.ICS. The fusion protein (Dsg2.ICS-Fv1E) was then expressed in SCC68 cells. Dimerization of Dsg2.ICS-Fv1E was induced by treating cells with AP20187, a cell-permeable bivalent compound (Fig. 6 A). The biotinylation assay was conducted to compare the internalization efficiency of Dsg2.ICS-Fv1E in the presence of AP20187 or ethanol (control). A decrease in internalization was observed when cells were treated with AP20187, supporting the idea that Dsg2 tail–tail interaction inhibits its endocytic internalization (Fig. 6 B).
Figure 6.
Dimerization of Dsg2.ICS-Fv1E leads to decreased internalization. (A) Schematic representation of the regulated homodimerization system. One copy of FKBP36v and HA epitope was placed in tandem with the C terminus of Dsg2-ICS. The dimerizer is AP20187. (B) SCC68 cells expressing Dsg2.ICS-Fv1E were pretreated with ethanol (control) or 10 nM AP20187 (AP) at 4°C for 30 min, and a biotinylation assay at 1.0 mM Ca2+ was then performed. Samples were immunoblotted with the anti-HA antibody. Graph presents collective results from three independent experiments. The bottom and top of the box are the 25th and 75th percentile (the lower and upper quartiles), respectively. The horizontal line near the middle of the box is the 50th percentile (the median). The ends of the whisker are the minimum and maximum of all the data. L, surface pool of the Dsg2 mutants. S, residual Dsg2 mutant protein left on the cell surface after stripping.
A mutation in DUR associated with human disease disrupts the Dsg2 tail–tail interaction and leads to rapid endocytosis in cardiac cells
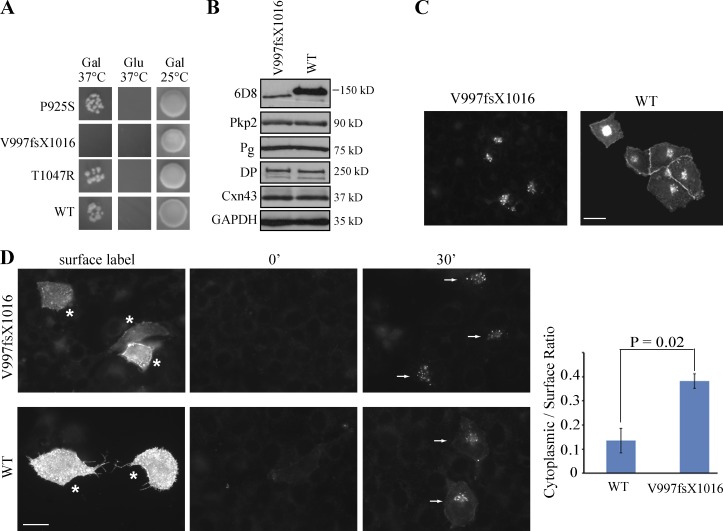
ARVC is an autosomal-dominant inherited disease, characterized by arrhythmias and progressive fibrofatty replacement of the myocardium. In ∼50% of cases, known mutations in desmosomal genes, including Dsg2, have been associated with ARVC, but the mechanisms by which these mutations contribute to disease pathogenesis are poorly understood (Syrris et al., 2007; Bhuiyan et al., 2009; Christensen et al., 2010; Tan et al., 2010; Bauce et al., 2011). Here, three pathogenic mutations in the Dsg2 DUR region (Arrhythmogenic Right Ventricular Dysplasia/Cardiomyopathy Genetic Variants Database) were generated and examined for their ability to mediate tail–tail interactions. One of these, V997fsX1016, which is caused by deletion of 2990G, abrogated DUR–DUR interactions based on yeast CytoTrap assay (Fig. 7 A). Human Dsg2.FL (WT) and the V997fsX1016 mutant were transiently expressed in mouse HL-1 cardiac cells (Claycomb et al., 1998). Although the expression level of the V977fsX1006 was lower than the Dsg2.FL (WT) control, other desmosomal protein levels were not affected (Fig. 7 B). By immunofluorescence, V977fsX1006 showed prominent perinuclear staining but little cell membrane staining, suggesting impaired delivery and/or increased internalization of the mutant protein (Fig. 7 C). To test whether alterations in internalization behavior contribute to this difference in distribution, we conducted an antibody internalization assay. The ratio of the internalized to cell surface fluorescence was over twofold greater for V977fsX1006, suggesting that it is more efficiently internalized from the cell surface (Fig. 7 D). These data raise the possibility that increased turnover of Dsg2, possibly leading to impaired stability and function of the cardiac intercalated disc, could contribute to ARVC disease pathogenesis.
Figure 7.
ARVC mutation disrupts the Dsg2 tail–tail interaction and leads to rapid internalization. (A) ARVC mutations were introduced into pSOS-DUR vector individually. The interaction between the myristoylation-DUR and the SOS-DUR WT or SOS-DUR with mutation was tested in the CytoTrap two-hybrid system. Representative results from one experiment are shown. (B) HL-1 cells were transiently transfected with Dsg2.FL-WT or Dsg2.FL-V997fsX1016 with Lipofectamine. 48 h after transfection, cell lysates were collected and immunoblotted with the indicated antibodies. The 6D8 antibody exclusively recognizes the ectopic human Dsg2 constructs. (C) HL-1 cells expressing Dsg2.FL-WT or Dsg2.FL-V997fsX1016 were stained with the 6D8 antibody. Bar, 20 µm. (D) Mutant-expressing cells were labeled with the 6D8 antibody at 4°C (asterisks) and then incubated at 37°C for 0 or 30 min. Residual surface antibodies were subsequently stripped. Internalized molecules (arrows) were visualized using indirect immunofluorescence. Bar, 20 µm. Graph shows the ratio of mean cytoplasmic intensity per cell/mean surface label intensity per cell for each mutant. Error bars are SEM. n = 3. Paired t test. Gal, galactose; Glu, glucose.
Discussion
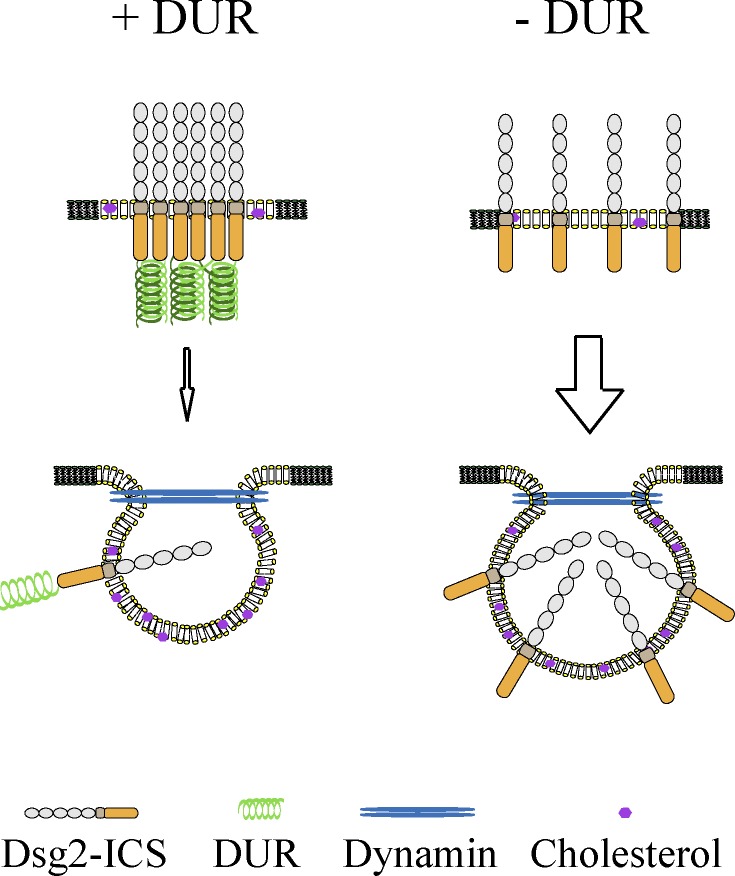
The desmosomal cadherins contain specialized structural features that set them apart from classical cadherins. How these motifs contribute to their adhesive functions or recently described roles in cell proliferation, apoptosis, differentiation, and virus infection (Eshkind et al., 2002; Brennan et al., 2007; Nava et al., 2007; Getsios et al., 2009; Wang et al., 2011) is not well understood. Here, we interrogate the function of the DUR, which was first identified in Dsg1 ∼20 yr ago (Koch et al., 1990). We show here that the DUR strengthens cell–cell adhesion, mediates cis-interactions between Dsg2 tails, and provides a brake on cadherin internalization (Fig. 8).
Figure 8.
Model of Dsg2 internalization. DUR inhibits Dsg2 internalization by promoting Dsg2 tail–tail interaction. DUR mediates Dsg2 tail–tail interaction, which stabilizes Dsg2 at the cell surface by inhibiting its internalization. When DUR–DUR interaction is disrupted (e.g., by mutation or deletion of the DUR), Dsg2 is more rapidly internalized from the cell surface. Dsg2 undergoes cholesterol- and dynamin-dependent endocytosis, independent of the status of DUR.
The total amount and organization of cell surface cadherins determines the adhesive strength they can provide. Regulation of cadherin trafficking through exo- and endocytosis is a critical determinant controlling cadherin functions, especially in dynamic, developing tissues and during tissue remodeling, including cancer invasion (Bryant and Stow, 2004; Green et al., 2010; Nekrasova et al., 2011). Here, we investigated the endocytosis of Dsg2 in epithelial cancer cell lines and demonstrate that the unique C-terminal region, or DUR, inhibits internalization. This inhibitory role is at least in part an intrinsic attribute of the DUR that does not depend on the presence of adhesive ectodomain interactions or desmosomes, as it occurs even in low calcium media that does not support assembly of intercellular junctions. Lipid raft and dynamin were reported to be involved in the endocytosis of some classical cadherins (Akhtar and Hotchin, 2001; Paterson et al., 2003; Taulet et al., 2009). Dsg2 shows the same dependence on these two factors. Interestingly, independent of DUR status, all Dsg2 constructs tested share the same endocytic features as endogenous Dsg2: they all internalize through a cholesterol- and dynamin-dependent pathway. This suggests that DUR is not rerouting Dsg2 but is in some way inhibiting entry into common pathways.
One way to achieve this inhibition would be to sequester Dsg2 in a complex that has less access to endocytic machinery. Consistent with this idea, the presence of the DUR region alters Dsg2 complex profiles. The DUR may control the formation of Dsg2 complexes by affecting its (a) posttranslational modification, (b) binding partners, or (c) oligomeric status. Although these outcomes are not mutually exclusive, we show here using a combination of yeast two-hybrid analysis and in situ proximity ligation that the DUR can mediate Dsg2 tail–tail interaction. It has been suggested that the disordered, flexible structure of DUR may provide an interaction surface for other binding partners or itself (Rutman et al., 1994; Kami et al., 2009). Our studies provide direct evidence that such interactions between Dsg2 tails occurs in cells, although we were unable to determine the exact oligomeric status (cis-dimer or higher order of oligomers) of Dsg2. Although the DUR was not absolutely required for targeting Dsg2 to desmosomes, DUR-dependent tail–tail interactions may promote clustering with other components of the membrane scaffolding and possibly strengthen association with the intermediate filament network. In the present work, we showed that induced dimerization of Dsg2 tails missing the DUR is sufficient to inhibit its internalization. However, it is likely that in the context of FL Dsg2, other DUR functions also contribute to Dsg2 dynamic behavior. Our work suggests that tail–tail interaction is one mechanism the DUR utilizes to inhibits Dsg2 internalization.
Our data raise the possibility that modulation or disruption of the DUR-mediated tail–tail interaction could target Dsg2 for internalization during normal tissue remodeling or disease pathogenesis. Consistent with this hypothesis, a pathogenic ARVC variant unable to mediate Dsg2 tail–tail interactions was observed to undergo rapid endocytosis in cardiac HL-1 cells. The abnormal behavior of this variant could destabilize cardiac intercalated discs and possibly contribute to the observed arrhythmias and sudden death that can occur in patients with this poorly understood disorder.
Another possible modulator of DUR function is proteolytic cleavage. Dsg1 is cleaved by caspase-3 at D888 in RUDIII in response to UV-induced apoptosis (Dusek et al., 2006). In addition, a prominent 130-kD Dsg2 form containing the ectodomain, but lacking the repeat unit domain or Dsg terminal domain was detected in human colonic mucosa (Kolegraff et al., 2011). In either case, a DUR-lacking Dsg may behave differently from FL Dsg because of altered clustering and internalization behavior.
Our observations demonstrate for the first time that the Dsg2 DUR mediates intermolecular interactions between the Dsg2 cytoplasmic domains and links this property to inhibition of Dsg2 internalization. This discovery could have important implications for both normal tissue remodeling and homeostasis as well as disease pathogenesis, including cancer and inherited diseases of the desmosome.
Materials and methods
Cell culture
The human-derived oral squamous cell carcinoma cell lines SCC68 and SCC9 were gifts from J. Rheinwald (Harvard Medical School, Boston, MA). SCC68 cells were maintained in keratinocyte serum-free media (Invitrogen) supplemented with 0.09 mM Ca2+, 0.2% bovine pituitary extract, and 0.3 ng/ml EGF. SCC9 cells were maintained in DMEM/F-12 media supplemented with 10% FBS. HL-1 cells were a gift from W. Claycomb (Louisiana State University School of Medicine, New Orleans, LA). HL-1 cells were grown in Claycomb medium containing 10% FBS, penicillin/streptomycin at 100 U/ml and 100 µg/ml, respectively, 0.1 mM norepinephrine, and 2 mM l-glutamine. For HL-1 cells, cell culture dishes/coverslips were precoated with a solution of 0.5% fibronectin and 0.2% gelatin from 3 h to overnight at 37°C.
Generation of cDNA constructs
To generate the truncated Dsg2 constructs, PCR amplifications were performed on Dsg2-Myc (p685; available from GenBank under accession no. NM_001934) using corresponding primers. A FLAG tag was introduced at the C terminus of all the constructs, and the resulting Dsg2 fragments were then cloned into the LZRS vector (provided by M. Denning, Loyola University, Chicago, IL) through the BamHI (Dsg2.ICS, Dsg2.RUDI, Dsg2.RUDIII, and Dsg2.RUDV) or BamHI–NotI (Dsg2.FL) site. The Dsg2.ICS and Dsg2.FL were further amplified by PCR and cloned into the pEGFP-N2 vector. The resulting Dsg2.ICS-GFP and Dsg.FL-GFP fragments were then cloned into the LZRS vector through the XhoI and NotI sites. IL2R:Dsg2 chimeras were generated by fusing the cytoplasmic domain of Dsg2.ICS, Dsg2.RUDI, or Dsg2.FL with IL2R-α ectodomain and transmembrane domain (aa 1–259) with a 3-aa linker (TLV) between them. The chimera DNA fragments were first introduced into pBluescript II KS (+) through the SpeI and HindIII sites and then cloned into LZRS through the BamHI and HindIII sites. Dsg2.ICS-Fv1E was cloned using the regulated homodimerization kit (ARGENT; ARIAD Pharmaceuticals). Dsg2.ICS was PCR amplified with the SpeI site at both ends, digested with SpeI, and ligated into pC4M-Fv1E through XbaI sites. The Dsg2.ICS-Fv1E fragment was then cloned into LZRS through the BamHI and NotI sites. The following point mutations were individually introduced into the LZRS-Dsg2.FL backbone using a site-directed mutagenesis kit (QuikChange; Agilent Technologies): 2,773C→T (P925S), 3,140C→G (T1047R), and 2,990delG (V997fsX1016).
siRNA transfection, plasmid transfection, and retroviral transduction
SCC68 or SCC9 cells grown at 30% confluency were transfected with siRNA oligos targeting the 3′ UTR of human Dsg2, human GAPDH, clathrin heavy chain, Cav1, or a nonspecific at a final concentration of 20–100 nM using a transfection reagent (DharmaFECT 1; Thermo Fisher Scientific). HL-1 cells grown at 90% confluence were transfected with specific DNA plasmid using Lipofectamine LTX with Plus reagent (Invitrogen). Analysis was conducted 48 or 72 h later. Retroviral transduction was performed as previously described by Getsios et al. (2004). In brief, the LZRS vector was transfected into phoenix cells (gifted by G. Nolan, Stanford University, Palo Alto, CA) using Lipofectamine. 48 h later, the virus containing supernatant was collected, concentrated, and mixed with culture medium for SCC68 or SCC9 cells at a 1:1 ratio. SCC68 or SCC9 cells grown at 50% confluency were then incubated with the virus supernatant at 37°C for 2–3 h.
Dispase assay
Confluent cells were rinsed with PBS twice and then incubated with 2.4 U/ml dispase in PBS at 37°C until the cell sheets were released from the cell culture dishes (20–30 min). The released cell sheets were transferred into a 15-ml conical tube and washed with PBS twice. Tubes were inverted 10–20 times until the cell sheets broke apart into fragments. The fragments were then transferred into a 35-mm dish and imaged with a dissecting scope (MZ6; Leica) and MetaVue imaging software (Molecular Devices).
Biotinylation assay
Three P60 dishes were prepared for each construct to determine the total surface labeling, internalized pool of Dsg2 at 30 min, and a control for stripping efficiency. All three dishes were rinsed with ice-cold PBS and incubated with 2 mg/ml sulfo-NHS-SS-biotin (EZ-Link; Thermo Fisher Scientific) for 30 min at 4°C. Excess biotin was removed with PBS washing. Surface-labeling dish and stripping efficiency dishes were kept at 4°C, whereas the 30 min was transferred to 37°C and incubated with prewarmed culture medium for 30 min to allow internalization to proceed. The 30-min dish was then returned to 4°C and washed with cold PBS. To strip the residual biotin from the cell surface but keep the internalized biotin intact, the 30-min dish and stripping efficiency dish were washed 3 × 20 min at 4°C with washing solution (50 mM Tris-HCl, pH 8.6, 100 mM sodium, 2-mercaptoethanesulfonic acid [Sigma-Aldrich], 1 mM EDTA, and 0.2% BSA [Sigma-Aldrich]). After that, the 30-min dish and stripping efficiency dish were incubated with 120 mM iodoacetamide (Sigma-Aldrich) in PBS for 10 min at 4°C and then washed three times with PBS. Cell lysates were collected in radioimmunoprecipitation assay (RIPA) buffer supplemented with protease inhibitors. After centrifugation (14,000 rpm for 30 min at 4°C), the supernatants were collected and equalized using the amido black assay. 20 µl supernatant was saved to serve as the cell lysate input. The rest of the supernatant (∼230 µl) was added with 40 µl immobilized streptavidin (UltraLink; Thermo Fisher Scientific) beads plus 300 µl RIPA buffer supplemented with protease inhibitors. The mixture was tumbled end over end at 4°C overnight. After that, the beads were washed with RIPA buffer 4 × 15 min at 4°C. Biotinylated proteins were then released from the beads using Laemmli buffer at 95°C for 10 min and then analyzed by immunoblotting. ImageJ software (National Institutes of Health) was used to measure the band intensity. The intensity of the band from streptavidin pull-down was normalized to the intensity of tubulin/GAPDH/FLAG band from its counterpart cell lysate. Based on the normalized values, the internalization ratio was calculated by dividing the internalized pool signal minus the remaining stripped signal with the surface pool signal minus the remaining stripped signal.
Antibody internalization assay
Like the biotinylation assay, three coverslips of cells were prepared for each construct to determine total surface labeling, the internalized pool of Dsg2 at 30 min, and a control for stripping efficiency (0 min). All three coverslips were incubated at 4°C for 1 h with a specific antibody (anti-IL2R antibody at 1:100; 6D8 at 1:200) in either keratinocyte serum-free medium containing 3% BSA and 20 mM Hepes (for SCC68 cells) or Claycomb medium with 20 mM Hepes (for HL-1 cells). Unbound antibody was removed by washing with cold PBS three times. Surface-labeled and 0-min coverslips were kept at 4°C, whereas the 30-min coverslip was transferred to 37°C for 30 min in prewarmed culture media. After returning to 4°C, the 30-min coverslip was washed with PBS three times. To strip the residual from the cell surface, the 30- and 0-min coverslips were washed with acid solution (0.5 mM NaCl and 0.5 mM acetic acid) four times for 15 min and then with PBS three times for 10 min. All coverslips were then fixed in 4% paraformaldehyde, permeabilized with 0.2% Triton X-100, and processed using indirect immunofluorescence. To quantify internalization, the cytoplasmic fluorescence of 20–100 cells was measured from each coverslip using MetaMorph Imaging (Molecular Devices) or ImageJ software. Data are expressed as the ratio of mean cytoplasmic intensity/mean surface intensity.
Cross-linking and sucrose gradient fractionation
Cells grown in 10-cm dishes were rinsed in cold PBS three times and then incubated with cross-linking solution (20 µg/ml dithiobis(succinimidylpropionate) in PBS) on ice for 10 min. The cross-linking solution was then removed, and the remaining dithiobis(succinimidylpropionate) was quenched by a quick rinse in quenching buffer (50 mM Tris-HCL, pH 8.0, and 150 mM NaCl) followed by a 15-min incubation in quenching buffer on ice. Cells were solubilized in 300 µl urea sample buffer (USB; 8 M urea, 60 mM Tris-HCl, pH 6.8, 1% SDS, and 10% glycerol). The cell lysate was then passed through a 25-gauge needle 20 times and mixed with an equal volume of H2O. A discontinuous sucrose gradient was generated in a polyallomer tube (Beckman Coulter) by layering different concentrations of sucrose solutions (prepared in 0.5× USB): 1.5 ml of 15% sucrose solution was layered at the bottom, and 0.9 ml of 13, 11, 10, 9, 8, 7, 6, 5, and 4% sucrose solutions were successively layered upon one another with ∼800 µl cell lysate placed on the very top. The sucrose gradient was then centrifuged in a swinging bucket rotor (SW 40 Ti; Beckman Coulter) at 34,400 rpm (mean relative centrifugal force of 150,000 g; acceleration profile of 1 and deceleration profile of 0) for 18 h at 20°C. A total of 11 or 12 0.9–1.0-ml fractions were collected from the bottom of the tube. Fractions were reduced by incubating samples with 5% β-mercaptoethanol at 37°C for 1 h before performing immunoblotting.
In situ PLA
Cells were fixed with 4% paraformaldehyde, permeabilized with 0.2% Triton X-100, and blocked with 1% BSA and 10% goat serum in PBS. Samples were then incubated with anti-FLAG (anti-FLAG; rabbit; Sigma-Aldrich) and 4B2 (anti-Dsg2; mouse; Dusek et al., 2006) antibodies overnight. The epitope for 4B2 antibody is located in the distal region of the Dsg2 C terminus and therefore is not present in Dsg2.ICS or Dsg2.RUDI. Ligation and amplification were executed following the manufacturer’s manual (Duolink In Situ PLA; Olink Bioscience) using the following reagents contained in the kit: PLA probe anti–rabbit plus, PLA probe anti–mouse minus, and detection reagent red. Fluorescence signals were visualized, and images were taken using a microscope (DSM; Leica) fitted with a 40× oil objective (Plan Fluoratar, NA 1.0). Signal intensity was quantified using MetaMorph 7.0 imaging software.
CytoTrap two-hybrid system
The experiment was performed using a CytoTrap vector kit (Agilent Technologies), and procedures were executed following the manufacturer’s recommendation. The IA-ICS (aa 634–841), DUR (aa 842–1,117), and Dsg2 tail (aa 634–1,117) were cloned into the gateway-compatible pSOS vector. For ARVC mutants, 2,773C→T (P925S), 3,140C→G (T1047R), and 2,990delG (V997fsX1016) point mutations were individually introduced in the pSOS-DUR backbone using the site-directed mutagenesis kit (QuikChange). DUR and the Dsg2 tail were also cloned into the pMyr vector. Specific pMyr and pSOS vector pair (as indicated in Fig. 5 C) were cotransformed into the cdc25H yeast strain. The resulting cotransformants were each grown on glucose plates at 24°C for 5 d. Four to five colonies were picked from each plate and grown onto galactose plate at 37°C, glucose plate at 37°C (negative control), and galactose plate at 24°C (positive control) for 5 d. The colonies were imaged using MetaVue imaging software.
Immunoprecipitation and immunofluorescence
The FLAG immunoprecipitation was performed by lysing cells in RIPA buffer (10 mM Tris, pH 7.5, 140 mM NaCl, 1% Triton X-100, 0.1% SDS, 0.5% sodium deoxycholate, 5 mM EDTA, and 2 mM EGTA) supplemented with EDTA-free protease inhibitor (Roche). Lysates were centrifuged at 14,000 rpm at 4°C for 30 min. Supernatants were incubated with 30–40 µl anti-FLAG M2 affinity gel (Sigma-Aldrich) at 4°C for 2 h. The immune complexes were released by washing with reducing Laemmli buffer at 95°C and further analyzed by Western blotting.
Immunofluorescence was performed as previously described (Klessner et al., 2009). Samples were mounted in Gelvatol medium and imaged at room temperature. Wide-field images were taken using a microscope (DMR; 40 or 63× objective lens, NA 1.0, Plan Fluotar and NA 1.32, Plan-Apochromat, respectively) and a digital camera (ORCA-100 model C4742-95; Hamamatsu Photonics). The images were then processed using MetaMorph software. Subdiffraction resolution images were taken using a structured illumination super-resolution microscope (N-SIM; Nikon; Gustafsson, 2000). For N-SIM analysis, the samples were illuminated with spatially high-frequency patterned excitation light (100× objective lens, NA 1.49; TiE N-SIM microscope [Nikon] and iXon X3 897 camera [Andor Technology]). The moiré patterns were produced and analytically processed (Elements version 4 software; Nikon) to reconstruct the subresolution structure of the samples.
Antibodies and inhibitors
The following antibodies were used in this study: anti-FLAG (anti-FLAG; Sigma-Aldrich), 6D8 (anti-Dsg2; gift from J. Wahl III, University of Nebraska Medical Center, Omaha, NE), anti–IL2R-α (anti–IL2R-α; Santa Cruz Biotechnology, Inc.), HECD1 (anti–E-cadherin; Abcam), C2206 (anti–β-catenin; C2206; Sigma-Aldrich), Ab12 (anti–EGF receptor; Thermo Fisher Scientific), E7 (anti–β-tubulin; University of Iowa Developmental Studies Hybridoma Bank), 1407 (anti-Pg; Aves Laboratories), NW6, anti-DP (Angst et al., 1990), GAPDH (anti-GAPDH; G9545; Sigma-Aldrich), 1905 (anti-Dsg3; gift from J. Stanley, University of Pennsylvania, Philadelphia, PA), 6013 (anti-Pkp2; Progen Biotechnik), Cav1 (anti-Cav1; BD), 3F10 (anti-HA; Santa Cruz Biotechnology, Inc.), HA.11 clone 16b12 (anti-HA; Covance), 7G6 and 23E3 (anti-Dsc2 and anti-Pkp3, respectively; provided by F. van Roy, University of Ghent, Ghent, Belgium), 4B2, and the anti-Dsg2 cytodomain (Dusek et al., 2006).
Peroxidase-conjugated secondary antibodies used for Western blot analysis included goat anti–mouse, goat anti–rabbit, and goat anti–chicken (Rockland). Fluorophore-conjugated secondary antibodies used for immunofluorescence included goat anti–mouse and goat anti–rabbit (Alexa Fluor; Invitrogen). Cycloheximide (EMD), dynasore (Sigma-Aldrich), methyl-β-cyclodextrin (Sigma-Aldrich), dithiobis(succinimidylpropionate), and AP20187 (ARIAD Pharmaceuticals) were used at concentrations of 10 µg/ml, 80 µM, 5 mM, 20 µg/ml, and 10 nM, respectively.
Online supplemental material
Fig. S1 shows that Dsg2 mutants can be delivered to cell–cell borders, and the expression level of most of the junctional proteins remains unchanged. Fig. S2 shows that cholesterol and dynamin promote the internalization of endogenous Dsg2, whereas Cav1 has no effect on the internalization of either endogenous Dsg2 or Dsg2 mutants. Fig. S3 shows that the internalization of endogenous Dsg2 remains unchanged when the function of flotillin-1 and clathrin heavy chain is disrupted. Fig. S4 shows that DUR promotes Pg association with Dsg2, but disruption of the Pg–Dsg2 interaction has no effect on Dsg2 internalization. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201202105/DC1.
Supplementary Material
Acknowledgments
We would like to thank those who generously contributed reagents and advice, including J. Wahl, F. van Roy, J.R. Stanley, M. Patterson, B. Harmon, S. Troyanovsky, V. Gelfand, and C. Gottardi. Thanks also go to Green laboratory members for useful discussions. We would also like to thank the Northwestern University Keck Biophysics Facility for their technical support. Superresolution imaging work was performed at the Northwestern University Cell Imaging Facility–Nikon Imaging Center generously supported by the National Cancer Institute Cancer Center Support Grant P30 CA060553 awarded to the Robert H. Lurie Comprehensive Cancer Center as well as Nikon.
This work was supported by National Institutes of Health grants CA122151 and AR041836 and a Transatlantic Network of Excellence grant from the Leducq Foundation as well as the J.L. Mayberry Endowment to K.J. Green.
Footnotes
Abbreviations used in this paper:
- ARVC
- arrhythmogenic right ventricular cardiomyopathy
- Cav1
- caveolin-1
- DP
- desmoplakin
- Dsg
- desmoglein
- DUR
- Dsg unique region
- FL
- full length
- IA
- intracellular anchoring
- ICS
- intracellular cadherin-like sequence
- N-SIM
- Nikon structured illumination microscopy
- Pg
- plakoglobin
- PGde
- Pg binding deficient
- PLA
- proximity ligation assay
- RIPA
- radioimmunoprecipitation assay
- USB
- urea sample buffer
- WT
- wild type
References
- Akhtar N., Hotchin N.A. 2001. RAC1 regulates adherens junctions through endocytosis of E-cadherin. Mol. Biol. Cell. 12:847–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Amoudi A., Díez D.C., Betts M.J., Frangakis A.S. 2007. The molecular architecture of cadherins in native epidermal desmosomes. Nature. 450:832–837 10.1038/nature05994 [DOI] [PubMed] [Google Scholar]
- Amagai M., Stanley J.R. 2012. Desmoglein as a target in skin disease and beyond. J. Invest. Dermatol. 132:776–784 10.1038/jid.2011.390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angst B.D., Nilles L.A., Green K.J. 1990. Desmoplakin II expression is not restricted to stratified epithelia. J. Cell Sci. 97:247–257 [DOI] [PubMed] [Google Scholar]
- Bauce B., Rampazzo A., Basso C., Mazzotti E., Rigato I., Steriotis A., Beffagna G., Lorenzon A., De Bortoli M., Pilichou K., et al. 2011. Clinical phenotype and diagnosis of arrhythmogenic right ventricular cardiomyopathy in pediatric patients carrying desmosomal gene mutations. Heart Rhythm. 8:1686–1695 10.1016/j.hrthm.2011.06.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhuiyan Z.A., Jongbloed J.D., van der Smagt J., Lombardi P.M., Wiesfeld A.C., Nelen M., Schouten M., Jongbloed R., Cox M.G., van Wolferen M., et al. 2009. Desmoglein-2 and desmocollin-2 mutations in dutch arrhythmogenic right ventricular dysplasia/cardiomyopathy patients: results from a multicenter study. Circ. Cardiovasc. Genet. 2:418–427 10.1161/CIRCGENETICS.108.839829 [DOI] [PubMed] [Google Scholar]
- Bonifacino J.S., Traub L.M. 2003. Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu. Rev. Biochem. 72:395–447 10.1146/annurev.biochem.72.121801.161800 [DOI] [PubMed] [Google Scholar]
- Brennan D., Hu Y., Joubeh S., Choi Y.W., Whitaker-Menezes D., O’Brien T., Uitto J., Rodeck U., Mahoney M.G. 2007. Suprabasal Dsg2 expression in transgenic mouse skin confers a hyperproliferative and apoptosis-resistant phenotype to keratinocytes. J. Cell Sci. 120:758–771 10.1242/jcs.03392 [DOI] [PubMed] [Google Scholar]
- Brennan D., Peltonen S., Dowling A., Medhat W., Green K.J., Wahl J.K., III, Del Galdo F., Mahoney M.G. 2012. A role for caveolin-1 in desmoglein binding and desmosome dynamics. Oncogene. 31:1636–1648 10.1038/onc.2011.346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooke M.A., Nitoiu D., Kelsell D.P. 2012. Cell-cell connectivity: desmosomes and disease. J. Pathol. 226:158–171 10.1002/path.3027 [DOI] [PubMed] [Google Scholar]
- Bryant D.M., Stow J.L. 2004. The ins and outs of E-cadherin trafficking. Trends Cell Biol. 14:427–434 10.1016/j.tcb.2004.07.007 [DOI] [PubMed] [Google Scholar]
- Chitaev N.A., Troyanovsky S.M. 1997. Direct Ca2+-dependent heterophilic interaction between desmosomal cadherins, desmoglein and desmocollin, contributes to cell–cell adhesion. J. Cell Biol. 138:193–201 10.1083/jcb.138.1.193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H.J., Gross J.C., Pokutta S., Weis W.I. 2009. Interactions of plakoglobin and beta-catenin with desmosomal cadherins: basis of selective exclusion of alpha- and beta-catenin from desmosomes. J. Biol. Chem. 284:31776–31788 10.1074/jbc.M109.047928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen A.H., Benn M., Bundgaard H., Tybjaerg-Hansen A., Haunso S., Svendsen J.H. 2010. Wide spectrum of desmosomal mutations in Danish patients with arrhythmogenic right ventricular cardiomyopathy. J. Med. Genet. 47:736–744 10.1136/jmg.2010.077891 [DOI] [PubMed] [Google Scholar]
- Claycomb W.C., Lanson N.A., Jr, Stallworth B.S., Egeland D.B., Delcarpio J.B., Bahinski A., Izzo N.J., Jr 1998. HL-1 cells: a cardiac muscle cell line that contracts and retains phenotypic characteristics of the adult cardiomyocyte. Proc. Natl. Acad. Sci. USA. 95:2979–2984 10.1073/pnas.95.6.2979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delva E., Jennings J.M., Calkins C.C., Kottke M.D., Faundez V., Kowalczyk A.P. 2008. Pemphigus vulgaris IgG-induced desmoglein-3 endocytosis and desmosomal disassembly are mediated by a clathrin- and dynamin-independent mechanism. J. Biol. Chem. 283:18303–18313 10.1074/jbc.M710046200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty G.J., McMahon H.T. 2009. Mechanisms of endocytosis. Annu. Rev. Biochem. 78:857–902 10.1146/annurev.biochem.78.081307.110540 [DOI] [PubMed] [Google Scholar]
- Dusek R.L., Getsios S., Chen F., Park J.K., Amargo E.V., Cryns V.L., Green K.J. 2006. The differentiation-dependent desmosomal cadherin desmoglein 1 is a novel caspase-3 target that regulates apoptosis in keratinocytes. J. Biol. Chem. 281:3614–3624 10.1074/jbc.M508258200 [DOI] [PubMed] [Google Scholar]
- Eshkind L., Tian Q., Schmidt A., Franke W.W., Windoffer R., Leube R.E. 2002. Loss of desmoglein 2 suggests essential functions for early embryonic development and proliferation of embryonal stem cells. Eur. J. Cell Biol. 81:592–598 10.1078/0171-9335-00278 [DOI] [PubMed] [Google Scholar]
- Fleming T.P., Garrod D.R., Elsmore A.J. 1991. Desmosome biogenesis in the mouse preimplantation embryo. Development. 112:527–539 [DOI] [PubMed] [Google Scholar]
- Getsios S., Amargo E.V., Dusek R.L., Ishii K., Sheu L., Godsel L.M., Green K.J. 2004. Coordinated expression of desmoglein 1 and desmocollin 1 regulates intercellular adhesion. Differentiation. 72:419–433 10.1111/j.1432-0436.2004.07208008.x [DOI] [PubMed] [Google Scholar]
- Getsios S., Simpson C.L., Kojima S., Harmon R., Sheu L.J., Dusek R.L., Cornwell M., Green K.J. 2009. Desmoglein 1–dependent suppression of EGFR signaling promotes epidermal differentiation and morphogenesis. J. Cell Biol. 185:1243–1258 10.1083/jcb.200809044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green K.J., Gaudry C.A. 2000. Are desmosomes more than tethers for intermediate filaments? Nat. Rev. Mol. Cell Biol. 1:208–216 10.1038/35043032 [DOI] [PubMed] [Google Scholar]
- Green K.J., Simpson C.L. 2007. Desmosomes: new perspectives on a classic. J. Invest. Dermatol. 127:2499–2515 10.1038/sj.jid.5701015 [DOI] [PubMed] [Google Scholar]
- Green K.J., Getsios S., Troyanovsky S., Godsel L.M. 2010. Intercellular junction assembly, dynamics, and homeostasis. Cold Spring Harb. Perspect. Biol. 2:a000125 10.1101/cshperspect.a000125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson M.G. 2000. Surpassing the lateral resolution limit by a factor of two using structured illumination microscopy. J. Microsc. 198:82–87 10.1046/j.1365-2818.2000.00710.x [DOI] [PubMed] [Google Scholar]
- He W., Cowin P., Stokes D.L. 2003. Untangling desmosomal knots with electron tomography. Science. 302:109–113 10.1126/science.1086957 [DOI] [PubMed] [Google Scholar]
- Holm P.K., Hansen S.H., Sandvig K., van Deurs B. 1993. Endocytosis of desmosomal plaques depends on intact actin filaments and leads to a nondegradative compartment. Eur. J. Cell Biol. 62:362–371 [PubMed] [Google Scholar]
- Holthöfer B., Windoffer R., Troyanovsky S., Leube R.E. 2007. Structure and function of desmosomes. Int. Rev. Cytol. 264:65–163 10.1016/S0074-7696(07)64003-0 [DOI] [PubMed] [Google Scholar]
- Hudson T.Y., Fontao L., Godsel L.M., Choi H.J., Huen A.C., Borradori L., Weis W.I., Green K.J. 2004. In vitro methods for investigating desmoplakin-intermediate filament interactions and their role in adhesive strength. Methods Cell Biol. 78:757–786 10.1016/S0091-679X(04)78026-7 [DOI] [PubMed] [Google Scholar]
- Hulpiau P., van Roy F. 2009. Molecular evolution of the cadherin superfamily. Int. J. Biochem. Cell Biol. 41:349–369 10.1016/j.biocel.2008.09.027 [DOI] [PubMed] [Google Scholar]
- Jennings J.M., Tucker D.K., Kottke M.D., Saito M., Delva E., Hanakawa Y., Amagai M., Kowalczyk A.P. 2011. Desmosome disassembly in response to pemphigus vulgaris IgG occurs in distinct phases and can be reversed by expression of exogenous Dsg3. J. Invest. Dermatol. 131:706–718 10.1038/jid.2010.389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly P.S., Berkowitz P., Bektas M., Lee H.E., Chua M., Diaz L.A., Rubenstein D.S. 2010. p38MAPK signaling and desmoglein-3 internalization are linked events in pemphigus acantholysis. J. Biol. Chem. 285:8936–8941 10.1074/jbc.M109.087999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kami K., Chidgey M., Dafforn T., Overduin M. 2009. The desmoglein-specific cytoplasmic region is intrinsically disordered in solution and interacts with multiple desmosomal protein partners. J. Mol. Biol. 386:531–543 10.1016/j.jmb.2008.12.054 [DOI] [PubMed] [Google Scholar]
- Klessner J.L., Desai B.V., Amargo E.V., Getsios S., Green K.J. 2009. EGFR and ADAMs cooperate to regulate shedding and endocytic trafficking of the desmosomal cadherin desmoglein 2. Mol. Biol. Cell. 20:328–337 10.1091/mbc.E08-04-0356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch P.J., Walsh M.J., Schmelz M., Goldschmidt M.D., Zimbelmann R., Franke W.W. 1990. Identification of desmoglein, a constitutive desmosomal glycoprotein, as a member of the cadherin family of cell adhesion molecules. Eur. J. Cell Biol. 53:1–12 [PubMed] [Google Scholar]
- Kolegraff K., Nava P., Laur O., Parkos C.A., Nusrat A. 2011. Characterization of full-length and proteolytic cleavage fragments of desmoglein-2 in native human colon and colonic epithelial cell lines. Cell Adh. Migr. 5:306–314 10.4161/cam.5.4.16911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nava P., Laukoetter M.G., Hopkins A.M., Laur O., Gerner-Smidt K., Green K.J., Parkos C.A., Nusrat A. 2007. Desmoglein-2: a novel regulator of apoptosis in the intestinal epithelium. Mol. Biol. Cell. 18:4565–4578 10.1091/mbc.E07-05-0426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekrasova O.E., Amargo E.V., Smith W.O., Chen J., Kreitzer G.E., Green K.J. 2011. Desmosomal cadherins utilize distinct kinesins for assembly into desmosomes. J. Cell Biol. 195:1185–1203 10.1083/jcb.201106057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie Z., Merritt A., Rouhi-Parkouhi M., Tabernero L., Garrod D. 2011. Membrane-impermeable cross-linking provides evidence for homophilic, isoform-specific binding of desmosomal cadherins in epithelial cells. J. Biol. Chem. 286:2143–2154 10.1074/jbc.M110.192245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niessen C.M., Leckband D., Yap A.S. 2011. Tissue organization by cadherin adhesion molecules: dynamic molecular and cellular mechanisms of morphogenetic regulation. Physiol. Rev. 91:691–731 10.1152/physrev.00004.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson A.D., Parton R.G., Ferguson C., Stow J.L., Yap A.S. 2003. Characterization of E-cadherin endocytosis in isolated MCF-7 and chinese hamster ovary cells: the initial fate of unbound E-cadherin. J. Biol. Chem. 278:21050–21057 10.1074/jbc.M300082200 [DOI] [PubMed] [Google Scholar]
- Pokutta S., Weis W.I. 2007. Structure and mechanism of cadherins and catenins in cell-cell contacts. Annu. Rev. Cell Dev. Biol. 23:237–261 10.1146/annurev.cellbio.22.010305.104241 [DOI] [PubMed] [Google Scholar]
- Rutman A.J., Buxton R.S., Burdett I.D. 1994. Visualisation by electron microscopy of the unique part of the cytoplasmic domain of a desmoglein, a cadherin-like protein of the desmosome type of cell junction. FEBS Lett. 353:194–196 10.1016/0014-5793(94)01049-8 [DOI] [PubMed] [Google Scholar]
- Schäfer S., Koch P.J., Franke W.W. 1994. Identification of the ubiquitous human desmoglein, Dsg2, and the expression catalogue of the desmoglein subfamily of desmosomal cadherins. Exp. Cell Res. 211:391–399 10.1006/excr.1994.1103 [DOI] [PubMed] [Google Scholar]
- Schock F., Perrimon N. 2002. Molecular mechanisms of epithelial morphogenesis. Annu. Rev. Cell Dev. Biol. 18:463–493 10.1146/annurev.cellbio.18.022602.131838 [DOI] [PubMed] [Google Scholar]
- Simons K., Gerl M.J. 2010. Revitalizing membrane rafts: new tools and insights. Nat. Rev. Mol. Cell Biol. 11:688–699 10.1038/nrm2977 [DOI] [PubMed] [Google Scholar]
- Simpson C.L., Green K.J. 2007. Identification of desmogleins as disease targets. J. Invest. Dermatol. 127(E1):E15–E16 [DOI] [PubMed] [Google Scholar]
- Söderberg O., Gullberg M., Jarvius M., Ridderstråle K., Leuchowius K.J., Jarvius J., Wester K., Hydbring P., Bahram F., Larsson L.G., Landegren U. 2006. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat. Methods. 3:995–1000 10.1038/nmeth947 [DOI] [PubMed] [Google Scholar]
- Syed S.E., Trinnaman B., Martin S., Major S., Hutchinson J., Magee A.I. 2002. Molecular interactions between desmosomal cadherins. Biochem. J. 362:317–327 10.1042/0264-6021:3620317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syrris P., Ward D., Asimaki A., Evans A., Sen-Chowdhry S., Hughes S.E., McKenna W.J. 2007. Desmoglein-2 mutations in arrhythmogenic right ventricular cardiomyopathy: a genotype-phenotype characterization of familial disease. Eur. Heart J. 28:581–588 10.1093/eurheartj/ehl380 [DOI] [PubMed] [Google Scholar]
- Tan B.Y., Jain R., den Haan A.D., Chen Y., Dalal D., Tandri H., Amat-Alarcon N., Daly A., Tichnell C., James C., et al. 2010. Shared desmosome gene findings in early and late onset arrhythmogenic right ventricular dysplasia/cardiomyopathy. J. Cardiovasc. Transl. Res. 3:663–673 10.1007/s12265-010-9224-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taulet N., Comunale F., Favard C., Charrasse S., Bodin S., Gauthier-Rouvière C. 2009. N-cadherin/p120 catenin association at cell-cell contacts occurs in cholesterol-rich membrane domains and is required for RhoA activation and myogenesis. J. Biol. Chem. 284:23137–23145 10.1074/jbc.M109.017665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomason H.A., Scothern A., McHarg S., Garrod D.R. 2010. Desmosomes: adhesive strength and signalling in health and disease. Biochem. J. 429:419–433 10.1042/BJ20100567 [DOI] [PubMed] [Google Scholar]
- Wang H., Li Z.Y., Liu Y., Persson J., Beyer I., Möller T., Koyuncu D., Drescher M.R., Strauss R., Zhang X.B., et al. 2011. Desmoglein 2 is a receptor for adenovirus serotypes 3, 7, 11 and 14. Nat. Med. 17:96–104 10.1038/nm.2270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt F.M., Mattey D.L., Garrod D.R. 1984. Calcium-induced reorganization of desmosomal components in cultured human keratinocytes. J. Cell Biol. 99:2211–2215 10.1083/jcb.99.6.2211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao K., Garner J., Buckley K.M., Vincent P.A., Chiasson C.M., Dejana E., Faundez V., Kowalczyk A.P. 2005. p120-Catenin regulates clathrin-dependent endocytosis of VE-cadherin. Mol. Biol. Cell. 16:5141–5151 10.1091/mbc.E05-05-0440 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.