SUMOylation of the small GTPase ARL13 is required for the proper ciliary targeting of sensory receptors and corresponding sensory functions.
Abstract
Primary cilia serve as cellular antenna for various sensory signaling pathways. However, how the sensory receptors are properly targeted to the ciliary surface remains poorly understood. Here, we show that UBC-9, the sole E2 small ubiquitin-like modifier (SUMO)-conjugating enzyme, physically interacts with and SUMOylates the C terminus of small GTPase ARL-13, the worm orthologue of ARL13B that mutated in ciliopathy Joubert syndrome. Mutations that totally abolish the SUMOylation of ARL-13 do not affect its established role in ciliogenesis, but fail to regulate the proper ciliary targeting of various sensory receptors and consequently compromise the corresponding sensory functions. Conversely, constitutively SUMOylated ARL-13 fully rescues all ciliary defects of arl-13–null animals. Furthermore, SUMOylation modification of human ARL13B is required for the ciliary entry of polycystin-2, the protein mutated in autosomal dominant polycystic kidney disease. Our data reveal a novel but conserved role for the SUMOylation modification of ciliary small GTPase ARL13B in specifically regulating the proper ciliary targeting of various sensory receptors.
Introduction
Primary cilia are built by intraflagellar transport (IFT) and act as sensory devices on the surface of most eukaryotic cells (Rosenbaum, 2002). Recently, mounting evidence has linked cilia dysfunction to a wide spectrum of human genetic disorders that have been collectively termed ciliopathies (Badano et al., 2006). Various signaling pathways that are crucial for either embryonic development or tissue pattern formation have been implicated in using cilia as a central cellular hub (Singla and Reiter, 2006; Eggenschwiler and Anderson, 2007). Proper ciliary targeting of the corresponding sensory receptors is critical for the signal transduction of a particular signaling pathway. However, little is known about how the ciliary entry of sensory receptors is regulated in vivo.
ARL13B was identified as one causal locus for the ciliopathy Joubert syndrome (JS; Cantagrel et al., 2008). Arl13b−/− mouse shows coupled defects in cilia structure and Sonic hedgehog (Shh) signaling (Caspary et al., 2007). Arl13b−/− zebrafish displays ciliogenesis defects in multiple ciliated organs (Sun et al., 2004; Duldulao et al., 2009). In Caenorhabditis elegans, ARL-13, the sole orthologue of ARL13B, is required for ciliogenesis and proper ciliary localization of ciliary membrane receptors (Blacque et al., 2005; Cevik et al., 2010; Li et al., 2010). 13 JS causal loci have been identified (Doherty, 2009; Juric-Sekhar et al., 2012). However, the connection between disease gene functions and pathologies remains largely elusive (Fliegauf et al., 2007; Parisi, 2009).
Modification of proteins by small ubiquitin-like modifier (SUMO) is a reversible posttranslational process that generates diverse molecular consequences (Johnson, 2004; Geiss-Friedlander and Melchior, 2007; Gareau and Lima, 2010). Although most SUMOylation substrates identified so far are nuclear proteins, the enzymes involved in SUMOylation are indeed present in the cytoplasm (Melchior et al., 2003; Johnson, 2004; Geiss-Friedlander and Melchior, 2007). Several pieces of evidence suggested distinct roles for SUMOylation in several cytoplasmic compartments, including mitochondria (Harder et al., 2004; Zunino et al., 2007) and ER (Dadke et al., 2007). SUMOylation is also implicated in regulating intermediate filament (Kaminsky et al., 2009), membrane receptors, and ion channels (Rajan et al., 2005; Benson et al., 2007; Martin et al., 2007).
The connections between SUMOylation and cilia or cilia-related structures are scarce. Several studies suggested that the nuclear localization of the centrosome protein ninein and centrin-2 depends on the SUMO system (Cheng et al., 2006; Klein and Nigg, 2009). One recent study indicated that TOPORS (topoisomerase I-binding arginine/serine rich), an E3 ligase for both SUMO and ubiquitin (Weger et al., 2003; Rajendra et al., 2004), localizes to the basal bodies of connecting cilia of photoreceptor cells and is implicated in retinal degeneration (Chakarova et al., 2011). However, it is unclear whether SUMOylation does play a role inside cilia and which ciliary proteins can be SUMOylated.
Here, we demonstrated that the E2 SUMO-conjugating enzyme UBC-9 physically interacts with and SUMOylates ARL-13/ARL13B in both C. elegans and mammalian cells. Remarkably, ARL-13/ARL13B SUMOylation does not affect the ciliary localization of ARL-13/ARL13B and its role in ciliogenesis, whereas it specifically regulates the ciliary targeting of various ciliary sensory receptors and corresponding downstream signaling pathways. These results provide the first evidence that SUMOylation machinery presents and functions in sensory organelle cilia. Furthermore, our data reveal the SUMOylated ARL-13/ARL13B as the key determinant in regulating the specific ciliary targeting of various sensory receptors.
Results and discussion
E2 SUMO-conjugating enzyme UBC-9 interacts with ARL-13 proline-rich domain (PRD) and SUMOylates its lysine 239 and 328
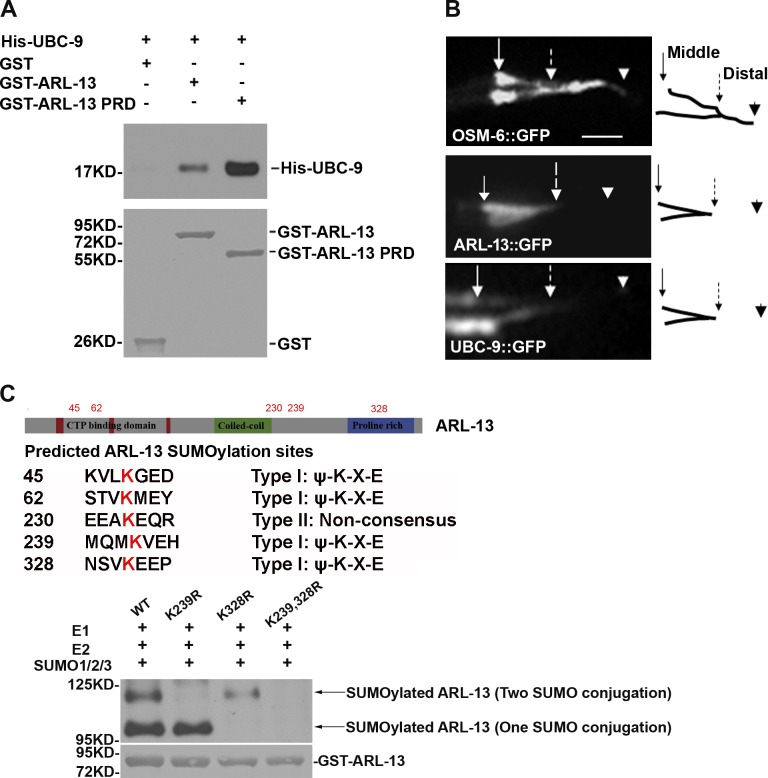
To identify potential effectors of ARL-13 in C. elegans, we used ARL-13 PRD, which is critical for ARL-13 ciliary function (Li et al., 2010), as bait to perform yeast two-hybrid screens. 107 cDNA clones were screened, and six plasmid-dependent genes were isolated. Among all interactors, UBC-9 was isolated 12 times (Table S2). An in vitro GST pull-down assay further confirmed that UBC-9 directly interacts with GST-tagged full-length or PRD domain of ARL-13, but not GST alone (Fig. 1 A).
Figure 1.
UBC-9 interacts with and SUMOylates ARL-13. (A, top) GST pull-down assay with GST-tagged ARL-13 full-length or PRD and His-tagged UBC-9 protein. (A, bottom) Ponceau S staining. (B) ARL-13 and UBC-9 label the middle segments but not the distal segments of cilia. Arrows with solid lines, ciliary base; arrows with broken lines, middle-distal junction; arrowheads, ciliary tip. Bar, 3 µm. (C) Predicted ARL-13 SUMOylation sites (above) and in vitro SUMOylation assay results (below). (top) Western blot with anti-SUMO antibody. (bottom) Ponceau S staining.
UBC-9 is the sole E2 SUMO-conjugating enzyme and is predominantly a nuclear protein (Mingot et al., 2001). However, ARL-13 is exclusively expressed in cilia but not in nuclear or other cellular compartments (Li et al., 2010). To investigate if UBC-9 is a bona fide effector of ARL-13, we examined the cellular localization of GFP-tagged UBC-9 protein. Surprisingly, we found that, in addition to the expected nuclear enrichment, UBC-9 also presents in cilia (Fig. S1 A). Specifically, UBC-9 mainly localizes in the middle segments but not the distal segments of cilia (Fig. 1 B). In C. elegans, amphid and phasmid cilia are comprised of middle segments containing nine doublet microtubules and distal segments containing only singlet microtubules (Perkins et al., 1986). ARL-13 is one of the few proteins identified so far that shares this middle-segment–restrictive localization pattern, which further suggested the potential functional correlation between UBC-9 and ARL-13 (Fig. 1 B). The observation that UBC-9 still enters the truncated cilia in arl-13(gk513) mutant worms indicated that the ciliary entry of UBC-9 is not dependent on its association with ARL-13 (Fig. S1 B).
Unlike the ubiquitin system, which uses E3 ligases to ubiquitinate the substrates, the SUMO system uses the sole E2 SUMO-conjugating enzyme UBC-9 to recognize and SUMOylate the substrates (Kerscher et al., 2006). We then asked whether the binding of UBC-9 leads to the SUMOylation of ARL-13. By using anti-SUMO antibodies, we detected two slowly migrating bands in in vitro SUMOylation assays with full-length GST-tagged ARL-13 (Fig. 1 C), which represented the SUMOylated ARL-13. SUMOylation usually occurs in a highly conserved recognition motif (Sampson et al., 2001). Five strong candidate SUMO conjugation sites (K45, K62, K230, K239, and K328) were predicted in ARL-13 protein (SUMOsp algorithm). We then generated ARL-13 variants with lysine-to-arginine (K-to-R) mutation in each of the five lysines. Unlike ARL-13K45R, ARL-13K62R, and ARL-13K230R variants that were still SUMOylated at levels comparable to wild-type ARL-13, ARL-13K239R and ARL-13K328R were only partially SUMOylated, and ARL-13K239,328R completely lost SUMOylation (Fig. 1 C and Fig. S1 C). We confirmed that K-to-R mutation does not affect the binding between ARL-13 and UBC-9 (Fig. S1 D). Analyses of the size of SUMOylated ARL-13K239R or ARL-13K328R suggest that two SUMO molecules are conjugated on K239, and one SUMO molecular on K328 (Fig. 1 C). Because there is no other slowly migrating SUMOylated band being observed, we thus conclude that the SUMOylations on K239 and K328 are mutually exclusive (Fig. 1 C).
SUMOylation of ARL-13 is not required for its ciliary targeting and ciliogenesis
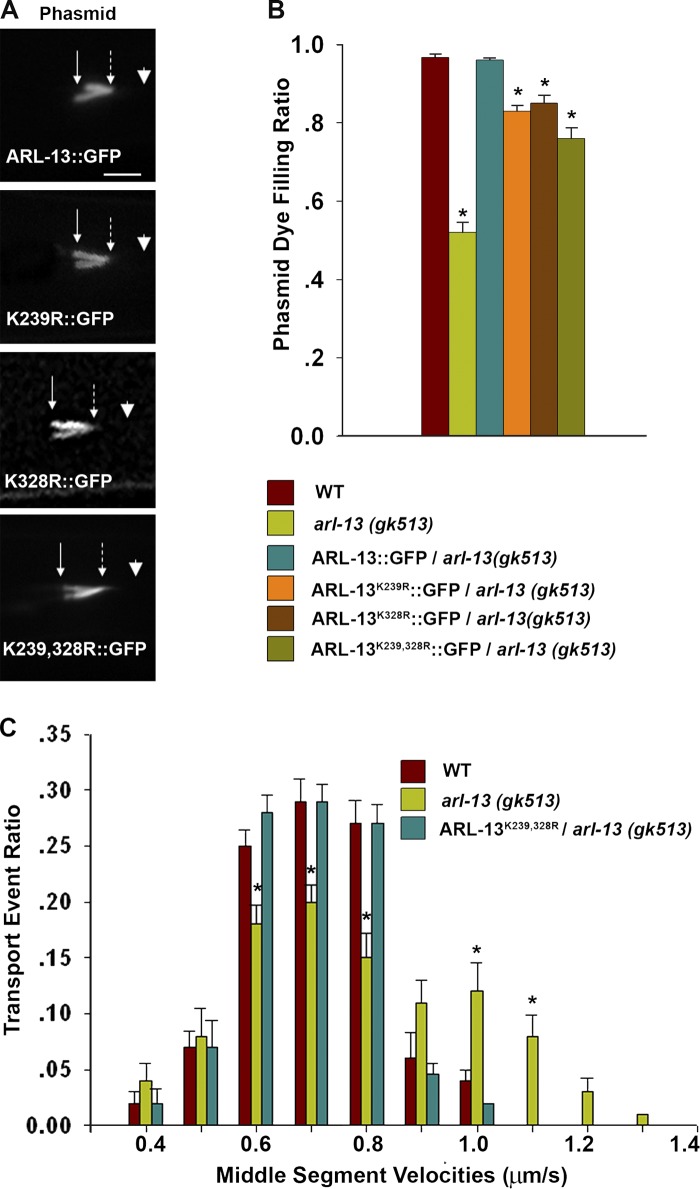
All GFP-tagged SUMOylation-deficient ARL-13 still show normal ciliary localization to the middle segments of cilia (Fig. 2 A), demonstrating that the SUMOylation status of ARL-13 is not required for maintaining its normal ciliary targeting. We next tested if SUMOylation plays a role in the reported function of ARL-13 in regulating ciliogenesis and IFT integrity in C. elegans (Cevik et al., 2010; Li et al., 2010). Interestingly, similar to GFP-tagged ARL-13, SUMOylation-deficient ARL-13 could fully rescue the ciliogenesis defect of arl-13(gk513) mutants (Fig. 2 B), which suggests a dispensable role for ARL-13 SUMOylation in cilia formation. We then checked the IFT integrity. In wild-type animals, slower motor protein kinesin II and faster motor protein OSM-3 cooperate in moving the same IFT particle along the middle segment at an intermediate rate of 0.7 µm/s (Ou et al., 2005). In arl-13(gk513) mutants, a significant amount of the IFT-A and IFT-B subcomplex dissociates, which leads to IFT-A–kinesin-II subcomplex moving at a slower rate of 0.5 µm/s, and the IFT-B–OSM-3 subcomplex moving faster than 1.0 µm/s (Li et al., 2010). As shown in Fig. 2 C, in arl-13(gk513) animals, a significant part of IFT-B component OSM-6 move around 1.1 µm/s, which indicates the breakage of the IFT integrity. However, the expression of SUMOylation-null ARL-13K239,328R can fully restore the compromised IFT integrity in arl-13(gk513) animals (Fig. 2 C). Collectively, we concluded that the SUMOylation of ARL-13 is not required for its normal ciliary targeting as well as its function in maintaining IFT integrity and regulating ciliogenesis.
Figure 2.
ARL-13 SUMOylation is not required for its ciliary targeting or ciliogenesis. (A) SUMOylation-deficient ARL-13 variants show normal ciliary localization to the middle segments. Arrows with solid lines, ciliary base; arrows with broken lines, middle-distal junction; arrowheads, ciliary tip. Bar, 5 µm. (B) A dye-filling assay was used to examine the ciliogenesis of various transgenic animals. (C) IFT transport event ratios within the middle segments in various transgenic worms. n > 500 for total IFT events in each transgenic line. The data were analyzed using an unpaired Student’s t test and are presented as mean ± SEM (error bars); *, P < 0.01.
ARL-13 SUMOylation regulates the ciliary targeting of membrane sensory receptors
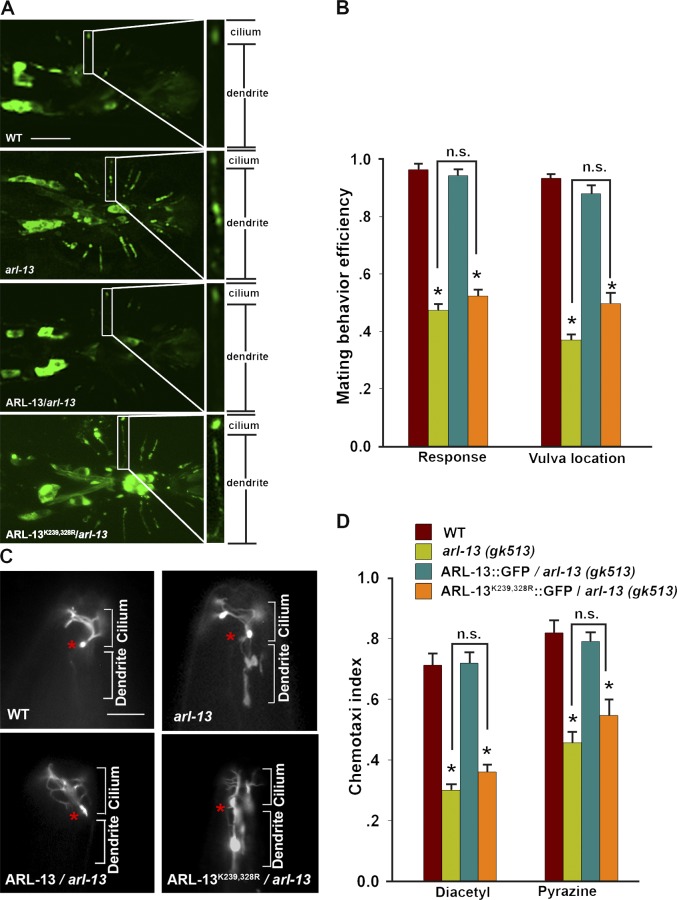
Other than the role in ciliogenesis, ARL-13 has also been implicated in the proper ciliary targeting of ciliary receptors (Cevik et al., 2010). We first examined PKD-2, which is the worm orthologue of human ciliopathy protein polycystin-2 and is required for the two aspects of mechanosensory behaviors in male worms, responding to hermaphrodite contact and vulva location (Barr and Sternberg, 1999; Barr et al., 2001). GFP-tagged PKD-2 localizes to the cilia of a subset of male-specific sensory neurons. As shown in Fig. 3 A, in wild-type animals, PKD-2 strongly enriches in the cilia, with little signal detected in the dendrites. In contrast, PKD-2 mislocalizes along the whole dendrites in arl-13(gk513), and this defect could be fully rescued by reintroducing a wild-type arl-13 gene. Notably, ARL-13K239,328R was unable to rescue PKD-2 mislocalization. Consistent with this, male mating behavior defects of arl-13(gk513) could not be rescued by expressing ARL-13K239,328R::GFP (Fig. 3 B). The fact that ARL-13K239R or ARL-13K328R can still rescue both PKD-2 mislocalization and male mating behavior defects of arl-13(gk513) animals suggests the functional redundancy for SUMOylation at either K239 or K328 (Fig. S2, A and B).
Figure 3.
ARL-13 SUMOylation is required for the proper ciliary targeting of sensory receptors and downstream sensory functions. (A) PKD-2 mislocalization in arl-13 can be rescued by the expression of wild-type ARL-13 but not ARL-13K239,328R variant. Bar, 10 µm. (B) ARL-13K239,328R fails to rescue the mating behavior defects of arl-13 animals. (C) ODR-10 mislocalization in fan-shape AWA cilia in arl-13 can be rescued by the expression of wild-type ARL-13 but not the ARL-13K239,328R variant. Asterisks indicate the base of AWA cilia. Note the strong accumulation of ODR-10 in dendrites in arl-13 and ARL-13K239,328R/arl-13 worms. Bar, 5 µm. (D) ARL-13K239,328R fails to rescue the chemotaxis defects of arl-13 animals. For each line, experiments were done five times to obtain statistical data. The data were analyzed using an unpaired Student’s t test and are presented as mean ± SEM (error bars); *, P < 0.01. n.s., not significant.
ODR-10 is an olfactory G protein–coupled receptor specifically expressed on the fan-shape cilia of worm AWA olfactory neurons (Fig. 3 C; Sengupta et al., 1996). In arl-13(gk513), ODR-10 abnormally distributes along the dendrites and always shows strong accumulations in both cilia and dendrites (Fig. 3 C). Expression of wild-type ARL-13 but not SUMOylation-null ARL-13K239,328R can fully restore the ciliary targeting of ODR-10 (Fig. 3 C). AWA neurons utilize ODR-10 to sense the odorants diacetyl and pyrazine. As expected, arl-13(gk513) animals showed severe defects in sensing diacetyl or pyrazine. However, the expression of ARL-13K239,328R::GFP could not restore the chemosensory defects (Fig. 3 D). Collectively, our data suggested that ARL-13 SUMOylation is a key modification for regulating the normal ciliary targeting of different sensory receptors as well as their downstream signaling.
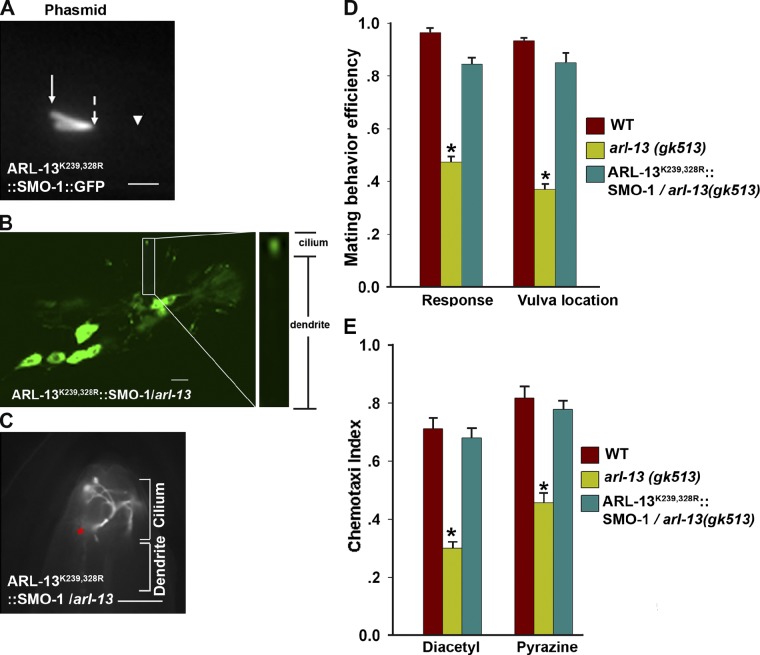
Besides SUMOylation, lysine can also be used for ubiquitination or acetylation (Deribe et al., 2010). We next asked whether the observations we made with ARL-13 mutants were solely due to the absence of SUMOylation and not other posttranslational alternations. Fusing a SUMO (or ubiquitin) to the substrate candidate has been favorably used to study the functional consequences of constitutive SUMOylation (or ubiquitination; Hu et al., 2007; Castillo-Lluva et al., 2010). Thus, we generated an ARL-13 chimera protein with SMO-1, the worm homologue of human SUMO-1, tagged at the C terminus of ARL-13K239,328R (ARL-13K239,328R::SMO-1), which potentially mimics the structure of constitutively SUMOylated ARL-13 at its C terminus. We found that GFP-tagged ARL-13K239,328R::SMO-1 show normal ciliary localization at the middle segments (Fig. 4 A) and can fully rescue the ciliogenesis defect of arl-13(gk513) (Fig. S2 C), which is indicative of no adverse effect of constitutively conjugating a SUMO molecule to the ARL-13 C terminus. As expected, this artificially SUMOylated ARL-13 can fully rescue the ciliary mislocalization of PKD-2 and ODR-10 (Fig. 4, B and C) as well as restore the defects in male mating and olfactory sensory behaviors, respectively (Fig. 4, D and E).
Figure 4.
Constitutively SUMOylated ARL-13 fulfills the function of wild-type ARL-13. (A) ARL-13K239,328R::SMO-1::GFP localizes normally. Arrow with solid line, ciliary base; arrow with broken line, middle-distal junction; arrowhead, ciliary tip. Bar, 5 µm. (B and C) ARL-13K239,328R::SMO-1 fusion protein completely rescues the mislocalization of PKD-2 and ODR-10 in arl-13(gk513) cilia. Bar, 5 µm. (D and E) Expression of ARL-13K239,328R::SMO-1 fully rescues the mating defects and chemotaxis defects of arl-13 animals. The data were analyzed using an unpaired Student’s t test and are presented as mean ± SEM (error bars); *, P < 0.01.
ARL13B SUMOylation and its functional consequences are conserved from worm to human
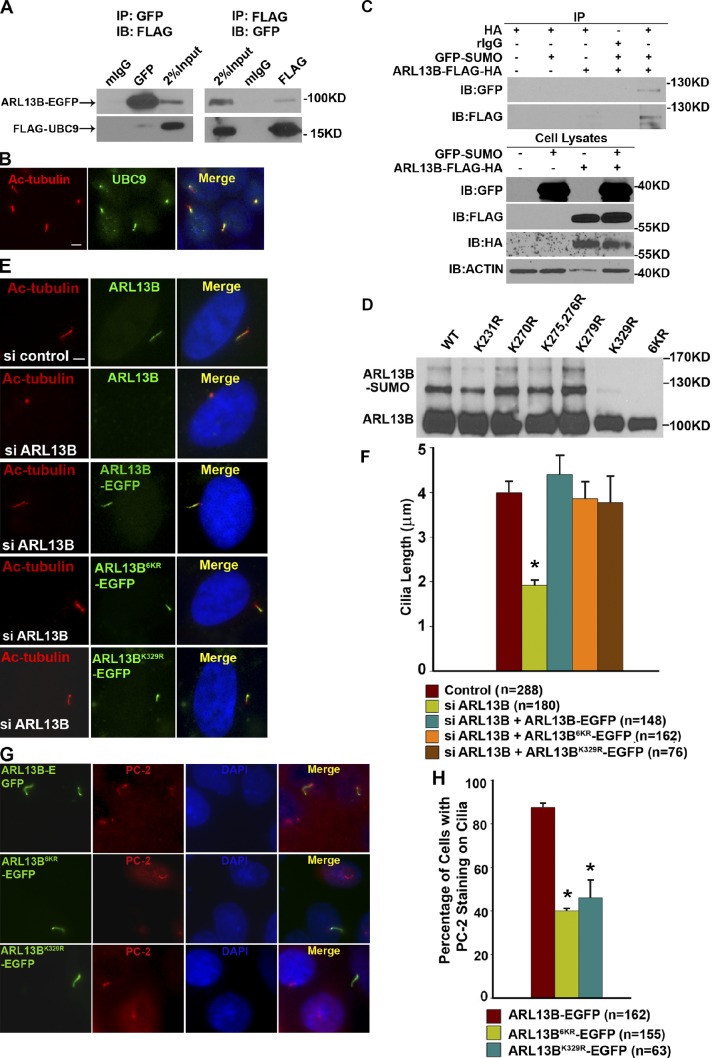
Cilia are sensory organelles that are highly conserved in both structure and function. Intriguingly, we found that GST-tagged human UBC-9 can also pull down FLAG-tagged human ARL13B (Fig. S3 A). Coimmunoprecipitations further confirmed that human UBC-9 associates with ARL13B in vivo in mammalian cells (Fig. 5 A). We mapped the ARL13B-interacting domain to the C terminus of UBC-9 (aa 70–158; Fig. S3 B). We further confirmed that endogenous human UBC-9 localizes to cilia of hTERT–immortalized retinal pigment epithelial cell line (hTERT-RPE1; Fig. 5 B).
Figure 5.
The SUMOylation of ARL13B and its role in cilia are conserved in mammalian cells. (A) Epitope-tagged proteins were expressed in HEK293 cells. FLAG and GFP immunoprecipitation were performed. Purified proteins were detected by Western blotting. (B) Endogenous UBC-9 was detected on hTERT-RPE1 cilia. (C) HEK293 cells were cotransfected with FLAG-HA double-tagged ARL13B and GFP-tagged SUMO1. ARL13B was immunoprecipitated with anti-HA antibody and the SUMOylation was visualized by Western blotting with anti-GFP and anti-FLAG antibodies. (D) HEK293 cells were transfected with EGFP-tagged ARL13B and different ARL13B variants. GFP antibody–precipitated proteins were then subjected to an in vitro SUMOylation assay. (E) The primary cilia in ARL13B knockdown hTERT-RPE1 cells were significant shorter than those in control cells. ARL13B-EGFP and ARL13BK329R-EGFP can both restore the cilia length to normal range. Bar, 2 µm. (F) Quantification of cilia length in different cells. (G) EGFP-tagged ARL13B, ARL13B6KR, and ARL13BK329R were transfected into IMCD3 cells. PC-2 localization was detected by antibody. Overexpression of SUMOylation-null mutant ARL13B but not wild-type ARL13B results in dramatically reduced ciliary staining of PC-2. Bar, 4 µm. (H) Quantification of the PC-2 ciliary signal. The data were analyzed using an unpaired Student’s t test and are presented as means ± SEM (error bars); *, P < 0.01.
Although we tried hard, we could not detect the SUMOylation of endogenous ARL13B protein in our experimental system. It is probable that our anti-ARL13B antibody is not sensitive enough to detect the SUMOylated ARL13B or that the amount of endogenous SUMOylated ARL13B at a certain time point is lower than the detectability of current methods. Thus, we decided to examine whether overexpressed human ARL13B can be SUMOylated or not, and we cotransfected HEK293 cells with FLAG-HA double-tagged ARL13B and GFP-tagged SUMO1. ARL13B was immunoprecipitated by anti-HA antibody, and the SUMOylation was visualized by anti-FLAG or anti-GFP antibody. As shown in Fig. 5 C, we confirmed that human ARL13B can be SUMOylated in vivo. Based on the size shifting, one SUMO molecule is conjugated on ARL13B in our assay. We noticed that, even for the overexpressed ARL13B, the SUMOylated band is rather weak (Fig. 5 C), which indicates that ARL13B SUMOylation only happens in a small portion of ARL13B protein and/or is a highly dynamic process in vivo. This observation also explains why it is extremely difficult to detect the SUMOylation of endogenous ARL13B.
Six strong candidate SUMO conjugation sites (K231, K270, K275, K276, K279, and K329) are predicted in human ARL13B protein. We found that mutation at K329 (ARL13BK329R) or all six putative SUMOylation lysines (ARL13B6KR) totally abolished the SUMOylation of ARL13B in an in vitro SUMOylation assay (Fig. 5 D).
We then generated a set of ARL13B mutations with the putative SUMOylation lysines altered to arginines. Similar to endogenous ARL13B (Fig. S3 C) or tagged ARL13B (Fig. S3 D), all ARL13B variants still localize to cilia normally (Fig. S3 G); this suggests that the SUMOylation is not required for the ciliary targeting of ARL13B, which is reminiscent of the observation obtained in the worm system (Fig. 2 A).
In hTERT-RPE1 cells, depletion of ARL13B by siRNAs led to significantly reduced cilia length as well as ciliated cells (Fig. S3, E and F). The defects can be fully rescued by reexpressing either an siRNA-insensitive ARL13B gene or an siRNA-insensitive ARL13BK329R variant (Fig. 5, E and F), which suggests that, just like in the worm system, the SUMOylation of ARL13B does not play a role in regulating ciliogenesis in mammalian cells.
We then examined PC-2 localization in mouse inner medullary collecting duct (IMCD3) cells. As shown in Fig. 5 G, the ciliary localization of PC-2 in IMCD3 cells is not affected when overexpressing wild-type ARL13B. However, overexpression of a SUMOylation-null ARL13B created a dramatic contrast with the ciliary staining of PC-2, which significantly decreases or even disappears (Fig. 5, G and H). These observations suggest that ARL13B SUMOylation plays a conserved role in regulating the proper ciliary targeting of sensory receptors.
The significance of ARL-13 SUMOylation
ARL-13 regulates cilia formation by maintaining the integrity of IFT particles (Cevik et al., 2010; Li et al., 2010). Because C. elegans polycystin-2 does not bind to the moving IFT particles (Qin et al., 2005), the reported role of ARL-13 in IFT assembly could not explain why PKD-2 mislocalizes in arl-13 mutant cilia. Here, we provide a novel mechanistic insight that ARL-13 SUMOylation specifically regulates the ciliary localization of sensory receptors in an IFT-independent manner.
SUMOylation modification on substrates usually facilitates protein–protein interaction by creating new binding surfaces or by affecting the conformational change of the target protein, which can result in altered subcellular localization, activity, or recruitment of regulatory factors to the modification site and multiprotein complex formation (Steinacher and Schär, 2005; Ulrich, 2005). Our data exclude the possibility that SUMO modification mediates the ciliary targeting of ARL-13/ARL13B. The ARL-13R83Q variant is proposed to have reduced GTPase activity (Cantagrel et al., 2008; Li et al., 2010). The observation that constitutively SUMOylated ARL-13R83Q does not show altered ability in rescuing the ciliogenesis defect of arl-13–null animals also suggests that SUMO conjugation may not affect the GTPase activity of ARL-13 either (Fig. S2 C). Therefore, it’s likely that the conjugated SUMO moiety on the ARL-13 surface serves as a mechanism for protein–protein interaction. The SUMO modification can be recognized by SUMO interaction motifs (SIMs) of downstream effector proteins. SIMs are very short motifs with a hydrophobic core (3–4 amino acids) followed (or preceded) by a stretch of 2–5 acidic amino acids (Minty et al., 2000). However, we did not find any SIM-like domain in the subset of sensory receptors regulated by ARL-13 SUMOylation. Thus, it is likely that there might be a SIM-containing protein that acts as a universal adaptor between SUMOylated ARL-13 and other transmembrane proteins.
It has been widely documented that most ciliopathies share the common feature of cystic kidney. Our finding that the deficiency of ARL-13/ARL13B SUMOylation could compromise the normal ciliary targeting of ADPKD candidate polycysitn-2 in both worm model and mammalian cells provides a new mechanistic insight into the pathogenesis of cystic kidney in JS patients. Considering the fact that more and more critical signaling pathways have been identified as using cilia as a central hub and that JS patients exhibit diverse phenotypes in different organs, it would be interesting to verify whether ARL13B SUMOylation also generally regulates cilia sensory functions in other affected organs by determining the proper ciliary targeting of the corresponding sensory receptors.
Materials and methods
C. elegans mutant alleles and strains
Nematodes were raised under standard conditions. N2 worms represented the wild-type animals in all assays. All strains used in this paper are listed in Table S1.
Yeast two-hybrid
The yeast strain AH109 (BD) was used for yeast two-hybrid experiments. Bait protein was expressed in the GAL4 DNA-binding domain vector pGBKT7. A cDNA library generated in the Barr laboratory was used (Hu and Barr, 2005). ARL-13 PRD domain (aa 250–367) was used as bait. Protein–protein interactions were accessed by growth rate on SD-Leu-Trp-His-Ade high-stringency plates and β-galactosidase filter assays.
Dye-filling assay
The stock DiI (2 mg/ml in dimethyl formamide; Molecular Probes) was diluted 1:200 in M9 buffer. Worms were incubated in diluted dye for 1 h at room temperature. After incubation, the animals were washed at least three times with M9 and observed using a fluorescence microscope (M2Bio; Carl Zeiss).
Microscopy
Animals were raised at 20°C and imaged by using standard C. elegans slide mounts and a Plan-Apochromat 60× 1.49 NA oil objective lens on an imaging microscope (TE 2000-U; all from Nikon).
IFT measurement
We performed all IFT analyses in phasmids for easier observation. IFT motility was observed by using a Plan-Apochromat 100× 1.49 NA oil total internal reflection fluorescence objective lens (Nikon). Motility stacks were recorded using a charge-coupled device camera (Photometrics QuantEM 512SC; Roper Scientific), and kymographs were produced with MetaMorph software. Worms were anesthetized in a drop of M9 containing 10 mM levamisole, transferred to an agarose mount slide, and imaged immediately.
Cell culture and RNAi
Human telomerase-immortalized retinal-pigmented epithelial cells (hTERT-RPE1) and IMCD3 cells were grown in DME/F12 with 10% FBS. Cells were grown to confluence and then starved for 24–48 h in media without serum to induce cilia. Ciliated cell numbers were quantified and cilia length was measured. Plasmids were transfected into cells using Lipofectamine 2000 (Invitrogen) or a Nucleofector kit V (Lonza). ARL13B were cloned in-frame in a pCDNA3-EGFP vector for expression in cells. siRNA sequences targeting human ARL13B (5′-GCUGCCACCUGAAACAUAAUU-3′) and luciferase (as a control) were transfected into cells with Lipofectamine RNAiMAX reagent (Invitrogen). Cells were fixed 48–72 h after transfection. The cells remaining on the plates were lysed for Western blot analysis using a rabbit anti-ARL13B antibody (Proteintech).
GST pull-down and immunoprecipitation
Purified His–UBC-9 protein was incubated with GST-ARL-13, GST-ARL-13PRD, and GST immobilized on glutathione Sepharose in the binding buffer (20 mM, Tris-HCl, pH 7.4, 150 mM KCl, 5 mM MgCl2, 0.5% Triton X-100, and 2 mM β-mercaptoethanol) for 4 h at 4°C. After four washes with binding buffer, the samples were subjected to SDS-PAGE and Western blotting with a monoclonal antibody against the Hisx6 epitope. FLAG-tagged UBC-9 and GFP-tagged ARL13B expressing HEK293 cells were lysed in buffer supplemented with protease inhibitors. For immunoprecipitation, whole cell lysates were precleared with protein G beads for at least 4 h at 4°C. FLAG antibody and 30-µl beads were added to the supernatant and incubated at 4°C overnight. Control immunoprecipitation with mIgG was also performed. After four washes, Western blotting was performed.
Immunofluorescence
For cilia staining, cells were fixed with 4% paraformaldehyde in PBS for 10 min followed by permeabilization with 0.2% Triton X-100 in PBS for 10 min. They were then blocked in PBS with 3% BSA, and sequentially blocked with primary and secondary antibodies. Ac-tubulin antibody was from Sigma-Aldrich.
In vitro SUMOylation
The in vitro SUMOylation kit reaction was purchased from Enzo Life Sciences. The reaction contains E1 and E2 enzymes, SUMO1/2/3, and GST-ARL-13 wild-type and mutant proteins in SUMOylation buffer. SUMOylation reactions were incubated at 30°C for 1 h. After termination with SDS-PAGE sample buffer, reaction products were subjected to SDS-PAGE. EGFP-tagged ARL13B and variants were expressed in HEK293 cells and precipitated by anti-GFP monoclonal antibody. The precipitations were incubated with SUMO reagents at 37°C for 3 h, and then separated by SDS-PAGE.
Online supplemental material
Fig. S1 show that the ciliary targeting of UBC-9 is independent of ARL-13. Fig. S2 shows the effects of SUMOylation on worm ARL-13. Fig. S3 shows that the SUMOylation of ARL13B is dispensable for its ciliary targeting in hTERT-RPE1 cells. Table S1 shows the strains used in this study. Table S2 shows the yeast two-hybrid candidates. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201203150/DC1.
Supplementary Material
Acknowledgments
We thank the Caenorhabditis Genetics Center and the Japanese Bioresource Project for strains.
J. Hu and coworkers were supported by the National Institutes of Health research grant 1R01DK090038 and P30 center grant P30DK90728, a Pilot and Feasibility Award from the Mayo Clinic Center for Cell Signaling in Gastroenterology (P30DK084567), and the PKD Foundation Young Investigator Award 04YI09a. K. Ling was supported by the National Cancer Institute (NCI; 1R01CA149039-01A1) and Susan G. Komen for the Cure (KG100902).
The authors of this paper declare that they have no conflicts of interest.
Footnotes
Abbreviations used in this paper:
- hTERT-RPE1
- hTERT–immortalized retinal pigment epithelial cell line
- IFT
- intraflagellar transport
- IMCD3
- murine inner medullary collecting duct
- JS
- Joubert syndrome
- PRD
- proline-rich domain
- SIM
- SUMO interaction motif
- SUMO
- small ubiquitin-like modifier
References
- Badano J.L., Mitsuma N., Beales P.L., Katsanis N. 2006. The ciliopathies: an emerging class of human genetic disorders. Annu. Rev. Genomics Hum. Genet. 7:125–148 10.1146/annurev.genom.7.080505.115610 [DOI] [PubMed] [Google Scholar]
- Barr M.M., Sternberg P.W. 1999. A polycystic kidney-disease gene homologue required for male mating behaviour in C. elegans. Nature. 401:386–389 [DOI] [PubMed] [Google Scholar]
- Barr M.M., DeModena J., Braun D., Nguyen C.Q., Hall D.H., Sternberg P.W. 2001. The Caenorhabditis elegans autosomal dominant polycystic kidney disease gene homologs lov-1 and pkd-2 act in the same pathway. Curr. Biol. 11:1341–1346 10.1016/S0960-9822(01)00423-7 [DOI] [PubMed] [Google Scholar]
- Benson M.D., Li Q.J., Kieckhafer K., Dudek D., Whorton M.R., Sunahara R.K., Iñiguez-Lluhí J.A., Martens J.R. 2007. SUMO modification regulates inactivation of the voltage-gated potassium channel Kv1.5. Proc. Natl. Acad. Sci. USA. 104:1805–1810 10.1073/pnas.0606702104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blacque O.E., Perens E.A., Boroevich K.A., Inglis P.N., Li C., Warner A., Khattra J., Holt R.A., Ou G., Mah A.K., et al. 2005. Functional genomics of the cilium, a sensory organelle. Curr. Biol. 15:935–941 10.1016/j.cub.2005.04.059 [DOI] [PubMed] [Google Scholar]
- Cantagrel V., Silhavy J.L., Bielas S.L., Swistun D., Marsh S.E., Bertrand J.Y., Audollent S., Attié-Bitach T., Holden K.R., Dobyns W.B., et al. 2008. Mutations in the cilia gene ARL13B lead to the classical form of Joubert syndrome. Am. J. Hum. Genet. 83:170–179 10.1016/j.ajhg.2008.06.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspary T., Larkins C.E., Anderson K.V. 2007. The graded response to Sonic Hedgehog depends on cilia architecture. Dev. Cell. 12:767–778 10.1016/j.devcel.2007.03.004 [DOI] [PubMed] [Google Scholar]
- Castillo-Lluva S., Tatham M.H., Jones R.C., Jaffray E.G., Edmondson R.D., Hay R.T., Malliri A. 2010. SUMOylation of the GTPase Rac1 is required for optimal cell migration. Nat. Cell Biol. 12:1078–1085 10.1038/ncb2112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cevik S., Hori Y., Kaplan O.I., Kida K., Toivenon T., Foley-Fisher C., Cottell D., Katada T., Kontani K., Blacque O.E. 2010. Joubert syndrome Arl13b functions at ciliary membranes and stabilizes protein transport in Caenorhabditis elegans. J. Cell Biol. 188:953–969 10.1083/jcb.200908133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakarova C.F., Khanna H., Shah A.Z., Patil S.B., Sedmak T., Murga-Zamalloa C.A., Papaioannou M.G., Nagel-Wolfrum K., Lopez I., Munro P., et al. 2011. TOPORS, implicated in retinal degeneration, is a cilia-centrosomal protein. Hum. Mol. Genet. 20:975–987 10.1093/hmg/ddq543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng T.S., Chang L.K., Howng S.L., Lu P.J., Lee C.I., Hong Y.R. 2006. SUMO-1 modification of centrosomal protein hNinein promotes hNinein nuclear localization. Life Sci. 78:1114–1120 10.1016/j.lfs.2005.06.021 [DOI] [PubMed] [Google Scholar]
- Dadke S., Cotteret S., Yip S.C., Jaffer Z.M., Haj F., Ivanov A., Rauscher F., III, Shuai K., Ng T., Neel B.G., Chernoff J. 2007. Regulation of protein tyrosine phosphatase 1B by sumoylation. Nat. Cell Biol. 9:80–85 10.1038/ncb1522 [DOI] [PubMed] [Google Scholar]
- Deribe Y.L., Pawson T., Dikic I. 2010. Post-translational modifications in signal integration. Nat. Struct. Mol. Biol. 17:666–672 10.1038/nsmb.1842 [DOI] [PubMed] [Google Scholar]
- Doherty D. 2009. Joubert syndrome: insights into brain development, cilium biology, and complex disease. Semin. Pediatr. Neurol. 16:143–154 10.1016/j.spen.2009.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duldulao N.A., Lee S., Sun Z. 2009. Cilia localization is essential for in vivo functions of the Joubert syndrome protein Arl13b/Scorpion. Development. 136:4033–4042 10.1242/dev.036350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggenschwiler J.T., Anderson K.V. 2007. Cilia and developmental signaling. Annu. Rev. Cell Dev. Biol. 23:345–373 10.1146/annurev.cellbio.23.090506.123249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fliegauf M., Benzing T., Omran H. 2007. When cilia go bad: cilia defects and ciliopathies. Nat. Rev. Mol. Cell Biol. 8:880–893 10.1038/nrm2278 [DOI] [PubMed] [Google Scholar]
- Gareau J.R., Lima C.D. 2010. The SUMO pathway: emerging mechanisms that shape specificity, conjugation and recognition. Nat. Rev. Mol. Cell Biol. 11:861–871 10.1038/nrm3011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiss-Friedlander R., Melchior F. 2007. Concepts in sumoylation: a decade on. Nat. Rev. Mol. Cell Biol. 8:947–956 10.1038/nrm2293 [DOI] [PubMed] [Google Scholar]
- Harder Z., Zunino R., McBride H. 2004. Sumo1 conjugates mitochondrial substrates and participates in mitochondrial fission. Curr. Biol. 14:340–345 [DOI] [PubMed] [Google Scholar]
- Hu J., Barr M.M. 2005. ATP-2 interacts with the PLAT domain of LOV-1 and is involved in Caenorhabditis elegans polycystin signaling. Mol. Biol. Cell. 16:458–469 10.1091/mbc.E04-09-0851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J., Wittekind S.G., Barr M.M. 2007. STAM and Hrs down-regulate ciliary TRP receptors. Mol. Biol. Cell. 18:3277–3289 10.1091/mbc.E07-03-0239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson E.S. 2004. Protein modification by SUMO. Annu. Rev. Biochem. 73:355–382 10.1146/annurev.biochem.73.011303.074118 [DOI] [PubMed] [Google Scholar]
- Juric-Sekhar G., Adkins J., Doherty D., Hevner R.F. 2012. Joubert syndrome: brain and spinal cord malformations in genotyped cases and implications for neurodevelopmental functions of primary cilia. Acta Neuropathol. 123:695–709 10.1007/s00401-012-0951-2 [DOI] [PubMed] [Google Scholar]
- Kaminsky R., Denison C., Bening-Abu-Shach U., Chisholm A.D., Gygi S.P., Broday L. 2009. SUMO regulates the assembly and function of a cytoplasmic intermediate filament protein in C. elegans. Dev. Cell. 17:724–735 10.1016/j.devcel.2009.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerscher O., Felberbaum R., Hochstrasser M. 2006. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu. Rev. Cell Dev. Biol. 22:159–180 10.1146/annurev.cellbio.22.010605.093503 [DOI] [PubMed] [Google Scholar]
- Klein U.R., Nigg E.A. 2009. SUMO-dependent regulation of centrin-2. J. Cell Sci. 122:3312–3321 10.1242/jcs.050245 [DOI] [PubMed] [Google Scholar]
- Li Y., Wei Q., Zhang Y., Ling K., Hu J. 2010. The small GTPases ARL-13 and ARL-3 coordinate intraflagellar transport and ciliogenesis. J. Cell Biol. 189:1039–1051 10.1083/jcb.200912001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin S., Nishimune A., Mellor J.R., Henley J.M. 2007. SUMOylation regulates kainate-receptor-mediated synaptic transmission. Nature. 447:321–325 10.1038/nature05736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melchior F., Schergaut M., Pichler A. 2003. SUMO: ligases, isopeptidases and nuclear pores. Trends Biochem. Sci. 28:612–618 10.1016/j.tibs.2003.09.002 [DOI] [PubMed] [Google Scholar]
- Mingot J.M., Kostka S., Kraft R., Hartmann E., Görlich D. 2001. Importin 13: a novel mediator of nuclear import and export. EMBO J. 20:3685–3694 10.1093/emboj/20.14.3685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minty A., Dumont X., Kaghad M., Caput D. 2000. Covalent modification of p73alpha by SUMO-1. Two-hybrid screening with p73 identifies novel SUMO-1-interacting proteins and a SUMO-1 interaction motif. J. Biol. Chem. 275:36316–36323 10.1074/jbc.M004293200 [DOI] [PubMed] [Google Scholar]
- Ou G., Blacque O.E., Snow J.J., Leroux M.R., Scholey J.M. 2005. Functional coordination of intraflagellar transport motors. Nature. 436:583–587 10.1038/nature03818 [DOI] [PubMed] [Google Scholar]
- Parisi M.A. 2009. Clinical and molecular features of Joubert syndrome and related disorders. Am. J. Med. Genet. C. Semin. Med. Genet. 151C:326–340 10.1002/ajmg.c.30229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins L.A., Hedgecock E.M., Thomson J.N., Culotti J.G. 1986. Mutant sensory cilia in the nematode Caenorhabditis elegans. Dev. Biol. 117:456–487 10.1016/0012-1606(86)90314-3 [DOI] [PubMed] [Google Scholar]
- Qin H., Burnette D.T., Bae Y.K., Forscher P., Barr M.M., Rosenbaum J.L. 2005. Intraflagellar transport is required for the vectorial movement of TRPV channels in the ciliary membrane. Curr. Biol. 15:1695–1699 10.1016/j.cub.2005.08.047 [DOI] [PubMed] [Google Scholar]
- Rajan S., Plant L.D., Rabin M.L., Butler M.H., Goldstein S.A. 2005. Sumoylation silences the plasma membrane leak K+ channel K2P1. Cell. 121:37–47 10.1016/j.cell.2005.01.019 [DOI] [PubMed] [Google Scholar]
- Rajendra R., Malegaonkar D., Pungaliya P., Marshall H., Rasheed Z., Brownell J., Liu L.F., Lutzker S., Saleem A., Rubin E.H. 2004. Topors functions as an E3 ubiquitin ligase with specific E2 enzymes and ubiquitinates p53. J. Biol. Chem. 279:36440–36444 10.1074/jbc.C400300200 [DOI] [PubMed] [Google Scholar]
- Rosenbaum J. 2002. Intraflagellar transport. Curr. Biol. 12:R125 10.1016/S0960-9822(02)00703-0 [DOI] [PubMed] [Google Scholar]
- Sampson D.A., Wang M., Matunis M.J. 2001. The small ubiquitin-like modifier-1 (SUMO-1) consensus sequence mediates Ubc9 binding and is essential for SUMO-1 modification. J. Biol. Chem. 276:21664–21669 10.1074/jbc.M100006200 [DOI] [PubMed] [Google Scholar]
- Sengupta P., Chou J.H., Bargmann C.I. 1996. odr-10 encodes a seven transmembrane domain olfactory receptor required for responses to the odorant diacetyl. Cell. 84:899–909 10.1016/S0092-8674(00)81068-5 [DOI] [PubMed] [Google Scholar]
- Singla V., Reiter J.F. 2006. The primary cilium as the cell’s antenna: signaling at a sensory organelle. Science. 313:629–633 10.1126/science.1124534 [DOI] [PubMed] [Google Scholar]
- Steinacher R., Schär P. 2005. Functionality of human thymine DNA glycosylase requires SUMO-regulated changes in protein conformation. Curr. Biol. 15:616–623 10.1016/j.cub.2005.02.054 [DOI] [PubMed] [Google Scholar]
- Sun Z., Amsterdam A., Pazour G.J., Cole D.G., Miller M.S., Hopkins N. 2004. A genetic screen in zebrafish identifies cilia genes as a principal cause of cystic kidney. Development. 131:4085–4093 10.1242/dev.01240 [DOI] [PubMed] [Google Scholar]
- Ulrich H.D. 2005. Mutual interactions between the SUMO and ubiquitin systems: a plea of no contest. Trends Cell Biol. 15:525–532 10.1016/j.tcb.2005.08.002 [DOI] [PubMed] [Google Scholar]
- Weger S., Hammer E., Engstler M. 2003. The DNA topoisomerase I binding protein topors as a novel cellular target for SUMO-1 modification: characterization of domains necessary for subcellular localization and sumolation. Exp. Cell Res. 290:13–27 10.1016/S0014-4827(03)00292-1 [DOI] [PubMed] [Google Scholar]
- Zunino R., Schauss A., Rippstein P., Andrade-Navarro M., McBride H.M. 2007. The SUMO protease SENP5 is required to maintain mitochondrial morphology and function. J. Cell Sci. 120:1178–1188 10.1242/jcs.03418 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.