Abstract
The proteasome is the primary site for protein degradation in mammalian cells, and proteasome inhibitors have been invaluable tools in clarifying its cellular functions. The anticancer agent bortezomib inhibits the major peptidase sites in the proteasome’s 20S core particle. It is a “blockbuster drug” that has led to dramatic improvements in the treatment of multiple myeloma, a cancer of plasma cells. The development of proteasome inhibitors illustrates the unpredictability, frustrations, and potential rewards of drug development but also emphasizes the dependence of medical advances on basic biological research.
Multiple myeloma, a cancer of the antibody-generating plasma cells, is the second most common hematological malignancy. In this disease, myeloma cells proliferate in the bone marrow, leading to decreased blood cell formation and bone resorption locally and causing systemic disease (especially renal failure) through their production of large amounts of abnormal immunoglobins. The proteasome inhibitor bortezomib is now part of the preferred treatment for multiple myeloma (Raab et al., 2009; Goldberg, 2011), and >400,000 patients worldwide have now received the drug, which has over two billion dollars in annual sales. Most importantly, this agent has led to major improvements in disease management and increased the lifespan of patients by years. Also, new combinations with other drugs are continually being introduced that are proving more effective and have fewer side effects. Recently, a second proteasome inhibitor, carfilzomib, has also received Food and Drug Administration (FDA) approval (Siegel et al., 2012), and three others are in clinical trials primarily for treating myeloma (Kisselev et al., 2012). Bortezomib is also approved for mantle cell lymphoma, and trials against other conditions are now in progress, including other cancers and inflammatory diseases, and for immunosuppression (Kisselev et al., 2012).
Why are myeloma cells particularly sensitive to proteasome inhibition? This special sensitivity was not anticipated and was only discovered during human trials of bortezomib. The primary reason is that most of the proteins expressed by myeloma cells are abnormal immunoglobins, and a key role of the ubiquitin–proteasome pathway is eliminating misfolded, potentially toxic proteins (Cenci et al., 2012). In this quality control process, termed ER-associated degradation, misfolded secretory proteins are extracted from the ER to the cytoplasm for degradation by the proteasome (Meusser et al., 2005). This process is also very important in the functioning of normal plasma cells because immunoglobins are large multisubunit molecules with multiple postsynthetic modifications, and many steps can go wrong in its synthesis (Cenci et al., 2012). Another reason for their special sensitivity is that myeloma cells rely on the transcription factor NF-κB (Nuclear Factor-κB), which inhibits apoptosis and promotes expression of growth factors and cytokines important for tumor pathogenesis (Hideshima et al., 2002). The proteasome activates NF-κB primarily by degrading its key inhibitor IκB. Therefore, treatment with the proteasome inhibitors prevents NF-κB activation and leads to toxic accumulation of misfolded proteins, which activates JNK and eventually apoptosis. These key functions of the proteasome that explain bortezomib’s efficacy in myeloma—NF-κB activation and its role in ER-associated degradation—were elucidated through many basic studies that used proteasome inhibitors as research tools. In other words, the medical progress and advances in understanding proteasome biology went hand in hand.
The historical background
The development of proteasome inhibitors for treatment of cancers has had a curious history that reflects the multiple strands of my own research career (Goldberg, 2011). When we initiated this research, we were not aiming to find new cancer therapies. Instead, our goal was based upon my long-standing interest (spanning almost 50 yr) to clarify the mechanisms of muscle atrophy, as occurs upon disuse, aging, or disease (e.g., cancer). These early experiments demonstrated unexpectedly that the rapid loss of muscle protein after denervation or fasting was caused primarily by an acceleration of overall protein breakdown rather than a reduction in protein synthesis (Goldberg, 1969), thereby providing the first evidence that overall rates of protein breakdown in mammalian cells are precisely regulated and help determine muscle size.
At the time, in 1969, virtually nothing was known about the pathways for protein catabolism in cells, and therefore, I decided to focus my research on the biochemical mechanisms of protein degradation in addition to exploring physiological regulation of this process in muscle (Goldberg and Dice, 1974; Goldberg and St John, 1976). Our physiological studies in the 1970s and 1980s showed that protein breakdown also increases and causes muscle wasting during cancer cachexia, sepsis, and renal or cardiac failure (Mitch and Goldberg, 1996; Lecker et al., 1999), whereas our biochemical study demonstrated the existence of a new, nonlysosomal proteolytic pathway in cells (later called the ubiquitin–proteasome system) that requires ATP and selectively eliminates misfolded proteins (Etlinger and Goldberg, 1977). A fundamental advance came with the Nobel prize winning discovery by Hershko, Ciechanover, and Rose of the involvement of ATP and the small protein ubiquitin in marking proteins for rapid hydrolysis (Hershko and Ciechanover, 1998; Glickman and Ciechanover, 2002). This knowledge enabled us, in the 1980s, to show that, in mammalian cells, ATP is also necessary for the degradation of ubiquitin-conjugated proteins (Tanaka et al., 1983), and in 1987, Rechsteiner’s (Hough et al., 1987) and our groups (Waxman et al., 1985) described the very large ATP-dependent protease complex that degraded ubiquitin-conjugated proteins, which we subsequently named the 26S proteasome.
The original rationale for generating proteasome inhibitors
Eventually, our two research interests in the physiological regulation of muscle protein breakdown and in the biochemical mechanism for proteolysis began to interconnect. In the late 1980s, we showed that the excessive proteolysis responsible for muscle wasting in many rodent disease models (e.g., cancers, renal failure, or denervation atrophy) was primarily caused by an activation of the ubiquitin–proteasome pathway (Mitch and Goldberg, 1996; Lecker et al., 1999), which was until then believed to degrade only misfolded or regulatory proteins (Hershko and Ciechanover, 1998; Glickman and Ciechanover, 2002; Goldberg, 2003). In fact, this system also digests long-lived proteins that comprise the bulk of cellular proteins. It is now clear that atrophying muscles undergo a series of transcriptional adaptations involving FoxO transcription factors that enhance their capacity for proteolysis (Lecker et al., 2004; Sandri et al., 2004), including increased expression of ubiquitin and key ubiquitination enzymes (Bodine et al., 2001; Gomes et al., 2001). These insights led me to propose that it could be beneficial to a large number of patients to pharmacologically inhibit this degradative process in muscle.
Starting a biotech company and my secret agenda
I decided to found a biotech company to inhibit proteasome function for two reasons: (1) There was no mechanism within the university to bring together a group of scientists with the expertise in chemistry, biochemistry, pharmacology, and medicine needed to develop a drug. (2) My experience consulting for biotechnology companies in the 1980s was a very positive one and had illustrated the satisfactions in seeing basic knowledge contribute to development of new therapies. Therefore, in the early 1990s, I convinced a group of Harvard colleagues to help found a small company, optimistically named MyoGenics, whose goal would be to try to block the debilitating loss of muscle in cancer and other diseases (Goldberg, 2011). Eventually, we found a venture capital group willing to gamble on this novel disease target (muscle wasting) and novel biochemical rationale.
In addition, I had a secret agenda. I realized that specific inhibitors of the proteasome could be very valuable tools to clarify the physiological functions of the ubiquitin–proteasome system in cells. However, this goal was kept secret because venture capitalists and business executives were not motivated to advance biological knowledge. Nevertheless, our success in this hidden agenda has proven to be a major legacy of the company as the lead compounds that led to bortezomib (e.g., MG132) have greatly advanced our understanding of many aspects of cell regulation, disease mechanisms, and immune surveillance (Rock and Goldberg, 1999). Unlike most companies, MyoGenics distributed our first proteasome inhibitors freely to academic investigators, whose efforts rapidly advanced our knowledge of the proteasome’s importance in cancer, apoptosis, and inflammation. In fact, MG132 has now been used as a research tool in over four thousand scientific studies because it is potent, inexpensive, and reversible.
Although MyoGenics (later renamed ProScript) was short lived as a separate entity, it was exceptionally successful in its scientific discoveries as well as in drug development. The company assembled a small, talented scientific team, including enzymologists led by Ross Stein, chemists led by Julian Adams, and cell biologists led by Vito Palombella. In addition, it had multiple close collaborations with us Harvard-based scientists. For example, when the first peptide aldehyde proteasomal inhibitors were available, their effects on muscle were analyzed (Tawa et al., 1997) in my laboratory and on antigen presentation in Kenneth Rock’s (Rock et al., 1994). Company scientists, in collaboration with Tom Maniatis’s laboratory, also made important findings about another key function of the proteasome, in the activation of NF-κB, the critical transcription factor in inflammation and cancer (Palombella et al., 1994; Silverman and Maniatis, 2001). This important role of the proteasome indicated that proteasome inhibitors could have dramatic effects in blocking inflammatory disease (e.g., arthritis) and cancer. Therefore, the company’s focus soon evolved to focus on these well-established disease targets and changed its name to ProScript (from proteasomes and transcription).
Creating proteasome inhibitors
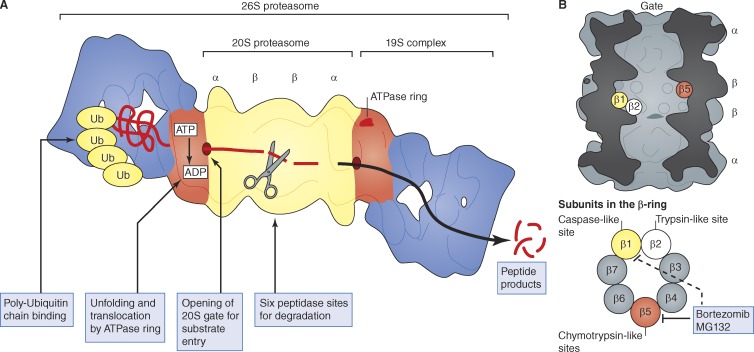
Most cell proteins are marked for degradation by attachment of a ubiquitin chain, which leads to their rapid degradation by the 26S proteasome, a 60-subunit particle composed of a 20S core and one or two 19S regulatory particles. Ubiquitinated proteins bind initially to the 19S particle, which contains enzymes to disassemble the ubiquitin chain and a ring of ATPases that unfold the protein and translocate it into the 20S proteasome (Pickart and Cohen, 2004; Finley, 2009; Peth et al., 2010). Degradation occurs within this hollow, cylindrical particle consisting of four stacked rings (Fig. 1). The outer rings contain seven distinct but homologous α subunits, and the inner rings contain seven homologous β subunits (Coux et al., 1996; Baumeister et al., 1998; Voges et al., 1999; Borissenko and Groll, 2007). Three β subunits contain the proteolytic sites, which face the inner chamber of the cylinder. In each β ring, there is a chymotrypsin-like, a trypsin-like, and a caspase-like site (Fig. 1), which act synergistically to cleave proteins to small peptides.
Figure 1.
Structure and function of the 26S proteasome. (A) Structure and components of the 26S proteasome. For more accurate images, see Lander et al. (2012) and Lasker et al. (2012). (B) Location of active sites in the 20S proteasome core. There are three types of proteolytic sites in the 20S proteasome’s central chamber, and each β ring contains three active sites. Bortezomib and MG132 act primarily on the chymotrypsin-like site in the β subunit but also inhibit the caspase-like site at high concentrations.
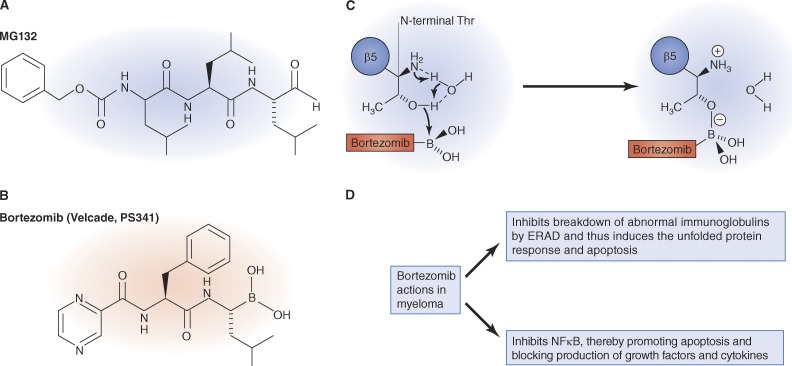
The first proteasome inhibitors synthesized were simple peptide aldehydes (Rock et al., 1994; Lee and Goldberg, 1998; Kisselev and Goldberg, 2001), which were analogues of the preferred substrates of the proteasome’s chymotrypsin-like active site. These inhibitors were not obtained through random screening of huge chemical libraries but instead were initially synthesized based on our knowledge of the substrate specificity of the proteasome’s active sites. Although the proteasome’s architecture and enzymatic mechanisms were unknown at the time, it was clear that the chymotrypsin-like site is the most important one in protein breakdown (Coux et al., 1996; Voges et al., 1999), and we knew that small hydrophobic peptides could often penetrate cell membranes. Therefore, the C termini of hydrophobic peptide substrates were derivatized to form peptide aldehydes, which were known to be effective inhibitors of serine and cysteine proteases. (Thus, MG132 is, in fact, simply carbobenzyl-Leu-Leu-Leu-aldehyde; Fig. 2). This compound was the lead molecule in medicinal chemistry efforts led by Julian Adams to enhance potency, selectivity, and stability. In place of the aldehyde (Fig. 2), he introduced the critical boronate “warhead,” which increased its potency 50–100-fold, and modifications in the peptide backbone then generated bortezomib within months.
Figure 2.
Structure and mechanism of action of proteasome inhibitors. (A) Structure of MG132, an inhibitor widely used in uncovering many cellular functions of the proteasome. (B and C) Structure of bortezomib (B), the inhibitor used to treat multiple myeloma, and its chemical reaction with the active site terminal threonine residue of the β5 subunit (C). (D) The primary mechanisms by which bortezomib causes death of myeloma cells.
The 20S proteasome was subsequently found to have a unique proteolytic mechanism, through the x-ray crystallographic studies of Huber and Baumeister (Voges et al., 1999; Kisselev and Goldberg, 2001; Borissenko and Groll, 2007; Kisselev et al., 2012), the active sites use the hydroxyl group of the N-terminal threonine residues to attack peptide bonds. Bortezomib and the peptide aldehyde inhibitors form adducts with this threonine that mimic the transition state intermediate during peptide cleavage (Fig. 2). A key early finding was that blocking the proteasome did not immediately kill cells or prevent normal function (Rock et al., 1994; Tawa et al., 1997). My prime concern had been that proteasome inhibition would be very toxic, causing accumulation of misfolded or regulatory proteins in ubiquitinated forms. In fact, compared with typical chemotherapeutic agents, proteasome inhibitors are not very toxic. The presence in cells of many enzymes that disassemble ubiquitin conjugates and recycle the ubiquitin meant that, upon proteasome inhibition, only a small fraction of cell proteins accumulate as ubiquitin conjugates form. Therefore, cells could function well for many hours or days with reduced proteasomal capacity. Furthermore, bortezomib and the other proteasome inhibitors block primarily the chymotrypsin-like sites, leaving the other sites functional. Thus, at therapeutic doses, bortezomib probably inhibits protein degradation for hours at most by only 20–30% (Kisselev et al., 2006), which does not perturb most cells significantly. However, myeloma cells are susceptible to this degree of inhibition because of their very high rates of breakdown of abnormal immunoglobulins, which are continually being cleared by the ubiquitin–proteasome system (Cenci et al., 2012). Consequently, even mild inhibition of the proteasome in these cells causes toxic accumulation of abnormal proteins, and triggers apoptosis, especially when they are weakened by NF-κB depletion. It is important to note that the 19S regulatory complex contains many other subunits and enzymatic activities, which comprise additional possible targets for drug development.
Bortezomib’s tribulations and surprising success
Only a few years have passed from our finding that proteasome inhibitors could reduce intracellular proteolysis (Rock et al., 1994), to the synthesis of bortezomib, to the acquisition of evidence for efficacy against cancer in mouse models (Adams et al., 1999), which came largely from screening at the National Cancer Institute. Despite this rapid progress, at multiple junctures, its development came close several times to termination for financial reasons (Goldberg, 2011). Our initial corporate partner decided not to pursue proteasome inhibitors in the clinic, and no other pharmaceutical company was interested in gambling on bortezomib becoming a drug, despite the impressive preclinical data. Because investment in the biotechnology industry had dried up at the time, our investors decided to sell ProScript to a larger company, Leukocyte, owned by the same group. Perhaps the best indication of how poorly bortezomib was valued by the “experts” is that the company’s assets (i.e., bortezomib and promising related research) were sold for less than three million dollars. In contrast, sales of bortezomib this year were almost 1,000 times the cost of the entire company. Leukocyte was soon purchased by Millennium Pharmaceuticals, a larger company that had failed to generate drug candidates. Initially, Millennium also failed to evaluate this program highly and even failed to announce its purchase of bortezomib, the one program that eventually led to its dramatic growth. (Instead, it heralded the purchase of seven drug candidates—all of which failed on the way to clinic.)
Through these troubled times, the core team of ProScript scientists continued working on bortezomib’s actions and pharmacology. Their efforts, led by Julian Adams (Adams et al., 1999), continued to generate evidence of bortezomib’s promise, and through his advocacy, it was given greater emphasis as Millennium’s other programs faltered. The promise of bortezomib against cancer received valuable support from screens against various tumor xenografts at the National Cancer Institute, and Millennium eventually initiated clinical trials against all human cancers.
Because of the financial challenges and unwarranted fears about its toxicity, bortezomib development was almost terminated several times before its success in the clinic could be established. The eventual dramatic success of bortezomib in the clinic has come as a real surprise to the industry and to the cancer community. I am certain that many other valuable treatments may have been terminated inappropriately for the lack of talented advocates, sufficient investment, or expert design of clinical trials, and their potential for helping patients were never realized.
Bortezomib’s clinical development is also a tale of serendipity. When it entered phase I trials against all cancers, one treated patient showed a complete remission. That patient had multiple myeloma, in which there had been no precedent for such dramatic improvement. Because some additional clear responses were evident in myeloma patients for whom there was no adequate therapy, phase II trials focused on this disease. They were performed by the team of Ken Anderson and Paul Richardson of the Dana-Farber Cancer Institute in an efficient and expert manner, which led to FDA approval after only phase II trials, as a result of the clear benefits found (Raab et al., 2009). Although initially approved for use only when other treatments failed (as “third-line therapy”), several subsequent clinical trials have led to it being approved now as “first-line” treatment (generally in combination with other drugs) and its wide acceptance throughout the world.
Proteasome inhibitors have advanced basic cell biology
Beyond their use in the clinic, proteasome inhibitors have contributed to dramatic advances in our understanding of cell regulation, immune responses, and disease mechanisms. Traditionally, the functions of the ubiquitin–proteasome pathway had to be studied by difficult approaches involving cell-free systems or genetic experiments in yeast, and many complex cellular processes could not be studied by these approaches. The availability of proteasome inhibitors has allowed rapid, simple analysis of the proteasome’s multiple cellular functions (Goldberg, 2011) and has led to fundamental insights about the cell cycle, metabolic regulation, immune surveillance, transcriptional responses, protein quality control, and disease mechanisms, especially cancer and neurodegenerative disease. The success of these inhibitors has also stimulated many ongoing efforts to find other ways to block the functioning of the ubiquitin–proteasome pathway (e.g., inhibitors of ubiquitination enzymes or deubiquitinases) and also to enhance proteasomal degradation of toxic proteins for treatment of neurodegenerative disease (Finley, 2009; Lee et al., 2010). Hopefully, these efforts will lead to other therapeutic advances in the near future.
Some lessons about drug development
The history of the development of proteasome inhibitors illustrates several key lessons about drug development that merit wide recognition:
Medical progress relies on advances in basic biochemistry and cell biology.
As bortezomib beautifully illustrates, the use of the proteasome inhibitors has led to tremendous advances in understanding cell regulation and disease mechanisms as well as to development of valuable drugs. Therefore, major credit for bortezomib, carfilzomib, and other inhibitors under development should go to the community of scientists who have advanced knowledge about protein degradation and to the National Institutes of Health and foundations that funded this work before its therapeutic importance was evident.
Major advances often emerge from outside established lines of research.
The proteasome was not viewed as a target for cancer drugs, and the novelty of this idea certainly slowed its acceptance. Also, at the time, there were no efforts in the pharmaceutical industry aimed at reducing muscle wasting or cachexia, although now these targets are being actively pursued in many companies. Thankfully, it is now widely recognized that many opportunities exist for drug development in the ubiquitin–proteasome pathway, especially against cancer, inflammation, and neurodegenerative disease.
Rapid drug development benefits from collaboration between basic and applied researchers.
Proteasome inhibitors were developed by a group of talented scientists in a small biotechnology company with extensive collaborations with academic experts and eventually clinicians. Academic investigators certainly do not need to compromise their ideals when working with profit-driven companies to develop agents that benefit suffering patients. Such collaborations should be fostered and can certainly be rewarding and fun!
The paths to scientific advances and the medical benefits of basic research are often unpredictable.
I never anticipated that our early observations on the mechanisms of muscle wasting might somehow contribute to therapies for multiple myeloma. In fact, had I ever suggested such outcomes in a grant proposal, the reviewers would have rejected such statements as ridiculous, fanciful, or naive and instead supported less innovative, more traditional lines of investigation.
Probably, the greatest rewards that a life in biological research can provide are seeing one’s work lead to both a greater understanding of living systems and to improvements in medical care.
Having focused for >40 yr on the mechanisms of intracellular protein breakdown and for 25 yr on understanding proteasome function, I have been fortunate to enjoy both rewards. It has been particularly gratifying to contribute to the development of the proteasome inhibitor bortezomib/velcade, which has had a major impact on the treatment of many patients. However, it has also been highly rewarding to witness the enormous advances in our knowledge about cell regulation, immune surveillance, and human disease that have been made using proteasome inhibitors as research tools.
Acknowledgments
The author is grateful to Lisa Bacis for her valuable assistance in the preparation of this manuscript.
The research from Dr. Goldberg’s laboratory reviewed here has been supported by grants from the National Institutes of Health (National Institute of General Medical Sciences and National Institute on Aging), the Muscular Dystrophy Association, Fidelity Biosciences Research Initiative, and the Multiple Myeloma Foundation. Illustrations were provided by Neil Smith, www.neilsmithillustration.co.uk.
References
- Adams J., Palombella V.J., Sausville E.A., Johnson J., Destree A., Lazarus D.D., Maas J., Pien C.S., Prakash S., Elliott P.J. 1999. Proteasome inhibitors: a novel class of potent and effective antitumor agents. Cancer Res. 59:2615–2622 [PubMed] [Google Scholar]
- Baumeister W., Walz J., Zühl F., Seemüller E. 1998. The proteasome: paradigm of a self-compartmentalizing protease. Cell. 92:367–380 10.1016/S0092-8674(00)80929-0 [DOI] [PubMed] [Google Scholar]
- Bodine S.C., Latres E., Baumhueter S., Lai V.K.-M., Nunez L., Clarke B.A., Poueymirou W.T., Panaro F.J., Na E., Dharmarajan K., et al. 2001. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science. 294:1704–1708 10.1126/science.1065874 [DOI] [PubMed] [Google Scholar]
- Borissenko L., Groll M. 2007. 20S proteasome and its inhibitors: crystallographic knowledge for drug development. Chem. Rev. 107:687–717 10.1021/cr0502504 [DOI] [PubMed] [Google Scholar]
- Cenci S., Oliva L., Cerruti F., Milan E., Bianchi G., Raule M., Mezghrani A., Pasqualetto E., Sitia R., Cascio P. 2012. Pivotal Advance: Protein synthesis modulates responsiveness of differentiating and malignant plasma cells to proteasome inhibitors. J. Leukoc. Biol. 10.1189/jlb.1011497 [DOI] [PubMed] [Google Scholar]
- Coux O., Tanaka K., Goldberg A.L. 1996. Structure and functions of the 20S and 26S proteasomes. Annu. Rev. Biochem. 65:801–847 10.1146/annurev.bi.65.070196.004101 [DOI] [PubMed] [Google Scholar]
- Etlinger J.D., Goldberg A.L. 1977. A soluble ATP-dependent proteolytic system responsible for the degradation of abnormal proteins in reticulocytes. Proc. Natl. Acad. Sci. USA. 74:54–58 10.1073/pnas.74.1.54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley D. 2009. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu. Rev. Biochem. 78:477–513 10.1146/annurev.biochem.78.081507.101607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glickman M.H., Ciechanover A. 2002. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol. Rev. 82:373–428 [DOI] [PubMed] [Google Scholar]
- Goldberg A.L. 1969. Protein turnover in skeletal muscle. II. Effects of denervation and cortisone on protein catabolism in skeletal muscle. J. Biol. Chem. 244:3223–3229 [PubMed] [Google Scholar]
- Goldberg A.L. 2003. Protein degradation and protection against misfolded or damaged proteins. Nature. 426:895–899 10.1038/nature02263 [DOI] [PubMed] [Google Scholar]
- Goldberg A.L. 2011. Bortezomib’s scientific origins and its tortuous path to the clinic. In Bortezomib in the Treatment of Multiple Myeloma. Ghobrial I.M., Richardson P.G., Anderson K.C., editors. Springer Basel AG, Basel, Switzerland: 1–27 [Google Scholar]
- Goldberg A.L., Dice J.F. 1974. Intracellular protein degradation in mammalian and bacterial cells. Annu. Rev. Biochem. 43:835–869 10.1146/annurev.bi.43.070174.004155 [DOI] [PubMed] [Google Scholar]
- Goldberg A.L., St John A.C. 1976. Intracellular protein degradation in mammalian and bacterial cells: Part 2. Annu. Rev. Biochem. 45:747–803 10.1146/annurev.bi.45.070176.003531 [DOI] [PubMed] [Google Scholar]
- Gomes M.D., Lecker S.H., Jagoe R.T., Navon A., Goldberg A.L. 2001. Atrogin-1, a muscle-specific F-box protein highly expressed during muscle atrophy. Proc. Natl. Acad. Sci. USA. 98:14440–14445 10.1073/pnas.251541198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko A., Ciechanover A. 1998. The ubiquitin system. Annu. Rev. Biochem. 67:425–479 10.1146/annurev.biochem.67.1.425 [DOI] [PubMed] [Google Scholar]
- Hideshima T., Chauhan D., Richardson P., Mitsiades C., Mitsiades N., Hayashi T., Munshi N., Dang L., Castro A., Palombella V., et al. 2002. NF-kappa B as a therapeutic target in multiple myeloma. J. Biol. Chem. 277:16639–16647 10.1074/jbc.M200360200 [DOI] [PubMed] [Google Scholar]
- Hough R., Pratt G., Rechsteiner M. 1987. Purification of two high molecular weight proteases from rabbit reticulocyte lysate. J. Biol. Chem. 262:8303–8313 [PubMed] [Google Scholar]
- Kisselev A.F., Goldberg A.L. 2001. Proteasome inhibitors: from research tools to drug candidates. Chem. Biol. 8:739–758 10.1016/S1074-5521(01)00056-4 [DOI] [PubMed] [Google Scholar]
- Kisselev A.F., Callard A., Goldberg A.L. 2006. Importance of the different proteolytic sites of the proteasome and the efficacy of inhibitors varies with the protein substrate. J. Biol. Chem. 281:8582–8590 10.1074/jbc.M509043200 [DOI] [PubMed] [Google Scholar]
- Kisselev A.F., van der Linden W.A., Overkleeft H.S. 2012. Proteasome inhibitors: an expanding army attacking a unique target. Chem. Biol. 19:99–115 10.1016/j.chembiol.2012.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander G.C., Estrin E., Matyskiela M.E., Bashore C., Nogales E., Martin A. 2012. Complete subunit architecture of the proteasome regulatory particle. Nature. 482:186–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasker K., Förster F., Bohn S., Walzthoeni T., Villa E., Unverdorben P., Beck F., Aebersold R., Sali A., Baumeister W. 2012. Molecular architecture of the 26S proteasome holocomplex determined by an integrative approach. Proc. Natl. Acad. Sci. USA. 109:1380–1387 10.1073/pnas.1120559109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecker S.H., Solomon V., Mitch W.E., Goldberg A.L. 1999. Muscle protein breakdown and the critical role of the ubiquitin-proteasome pathway in normal and disease states. J. Nutr. 129(1S Suppl.):227S–237S [DOI] [PubMed] [Google Scholar]
- Lecker S.H., Jagoe R.T., Gilbert A., Gomes M., Baracos V., Bailey J., Price S.R., Mitch W.E., Goldberg A.L. 2004. Multiple types of skeletal muscle atrophy involve a common program of changes in gene expression. FASEB J. 18:39–51 10.1096/fj.03-0610com [DOI] [PubMed] [Google Scholar]
- Lee B.H., Lee M.J., Park S., Oh D.C., Elsasser S., Chen P.C., Gartner C., Dimova N., Hanna J., Gygi S.P., et al. 2010. Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature. 467:179–184 10.1038/nature09299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D.H., Goldberg A.L. 1998. Proteasome inhibitors: valuable new tools for cell biologists. Trends Cell Biol. 8:397–403 10.1016/S0962-8924(98)01346-4 [DOI] [PubMed] [Google Scholar]
- Meusser B., Hirsch C., Jarosch E., Sommer T. 2005. ERAD: the long road to destruction. Nat. Cell Biol. 7:766–772 10.1038/ncb0805-766 [DOI] [PubMed] [Google Scholar]
- Mitch W.E., Goldberg A.L. 1996. Mechanisms of muscle wasting. The role of the ubiquitin-proteasome pathway. N. Engl. J. Med. 335:1897–1905 10.1056/NEJM199612193352507 [DOI] [PubMed] [Google Scholar]
- Palombella V.J., Rando O.J., Goldberg A.L., Maniatis T. 1994. The ubiquitin-proteasome pathway is required for processing the NF-kappa B1 precursor protein and the activation of NF-kappa B. Cell. 78:773–785 10.1016/S0092-8674(94)90482-0 [DOI] [PubMed] [Google Scholar]
- Peth A., Uchiki T., Goldberg A.L. 2010. ATP-dependent steps in the binding of ubiquitin conjugates to the 26S proteasome that commit to degradation. Mol. Cell. 40:671–681 10.1016/j.molcel.2010.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart C.M., Cohen R.E. 2004. Proteasomes and their kin: proteases in the machine age. Nat. Rev. Mol. Cell Biol. 5:177–187 10.1038/nrm1336 [DOI] [PubMed] [Google Scholar]
- Raab M.S., Podar K., Breitkreutz I., Richardson P.G., Anderson K.C. 2009. Multiple myeloma. Lancet. 374:324–339 10.1016/S0140-6736(09)60221-X [DOI] [PubMed] [Google Scholar]
- Rock K.L., Goldberg A.L. 1999. Degradation of cell proteins and the generation of MHC class I-presented peptides. Annu. Rev. Immunol. 17:739–779 10.1146/annurev.immunol.17.1.739 [DOI] [PubMed] [Google Scholar]
- Rock K.L., Gramm C., Rothstein L., Clark K., Stein R., Dick L., Hwang D., Goldberg A.L. 1994. Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell. 78:761–771 10.1016/S0092-8674(94)90462-6 [DOI] [PubMed] [Google Scholar]
- Sandri M., Sandri C., Gilbert A., Skurk C., Calabria E., Picard A., Walsh K., Schiaffino S., Lecker S.H., Goldberg A.L. 2004. Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell. 117:399–412 10.1016/S0092-8674(04)00400-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel D.S., Martin T., Wang M., Vij R., Jakubowiak A.J., Lonial S., Trudel S., Kukreti V., Bahlis N., Alsina M., et al. 2012. A phase 2 study of single-agent carfilzomib (PX-171-003-A1) in patients with relapsed and refractory multiple myeloma. Blood. 120:2817–2825 10.1182/blood-2012-05-425934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman N., Maniatis T. 2001. NF-kappaB signaling pathways in mammalian and insect innate immunity. Genes Dev. 15:2321–2342 10.1101/gad.909001 [DOI] [PubMed] [Google Scholar]
- Tanaka K., Waxman L., Goldberg A.L. 1983. ATP serves two distinct roles in protein degradation in reticulocytes, one requiring and one independent of ubiquitin. J. Cell Biol. 96:1580–1585 10.1083/jcb.96.6.1580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tawa N.E., Jr, Odessey R., Goldberg A.L. 1997. Inhibitors of the proteasome reduce the accelerated proteolysis in atrophying rat skeletal muscles. J. Clin. Invest. 100:197–203 10.1172/JCI119513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voges D., Zwickl P., Baumeister W. 1999. The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu. Rev. Biochem. 68:1015–1068 10.1146/annurev.biochem.68.1.1015 [DOI] [PubMed] [Google Scholar]
- Waxman L., Fagan J.M., Tanaka K., Goldberg A.L. 1985. A soluble ATP-dependent system for protein degradation from murine erythroleukemia cells. Evidence for a protease which requires ATP hydrolysis but not ubiquitin. J. Biol. Chem. 260:11994–12000 [PubMed] [Google Scholar]