The distinct morphological and dynamic properties of their respective filaments suggest that FtsZ1 facilitates Z-ring remodeling, whereas FtsZ2 forms the Z-ring backbone during chloroplast division.
Abstract
FtsZ, a cytoskeletal GTPase, forms a contractile ring for cell division in bacteria and chloroplast division in plants. Whereas bacterial Z rings are composed of a single FtsZ, those in chloroplasts contain two distinct FtsZ proteins, FtsZ1 and FtsZ2, whose functional relationship is poorly understood. We expressed fluorescently tagged FtsZ1 and FtsZ2 in fission yeast to investigate their intrinsic assembly and dynamic properties. FtsZ1 and FtsZ2 formed filaments with differing morphologies when expressed separately. FRAP showed that FtsZ2 filaments were less dynamic than FtsZ1 filaments and that GTPase activity was essential for FtsZ2 filament turnover but may not be solely responsible for FtsZ1 turnover. When coexpressed, the proteins colocalized, consistent with coassembly, but exhibited an FtsZ2-like morphology. However, FtsZ1 increased FtsZ2 exchange into coassembled filaments. Our findings suggest that FtsZ2 is the primary determinant of chloroplast Z-ring structure, whereas FtsZ1 facilitates Z-ring remodeling. We also demonstrate that ARC3, a regulator of chloroplast Z-ring positioning, functions as an FtsZ1 assembly inhibitor.
Introduction
FtsZ is a self-assembling GTPase related to tubulins that facilitates cell division in bacteria and chloroplast division in photosynthetic eukaryotes (Adams and Errington, 2009; Erickson et al., 2010; Miyagishima, 2011; Falconet, 2012). Bacterial FtsZ, a soluble protein, assembles at the midcell into a dynamic “Z ring,” which is tethered to the membrane at the division site by interaction with membrane proteins. The Z ring acts as a scaffold for recruitment of other cell division proteins to the division site and generates at least some contractile force for membrane constriction (Bi and Lutkenhaus, 1991; Löwe, 1998; Osawa et al., 2008; Adams and Errington, 2009).
In vitro, Escherichia coli FtsZ typically polymerizes into single-stranded protofilaments in a GTP-dependent manner, but also assembles into bundles, helices, and sheets under various assembly conditions (Erickson et al., 2010; Mingorance et al., 2010). Polymerization stimulates GTPase activity, which destabilizes protofilaments and promotes their fragmentation (Huecas et al., 2007). These activities do not require accessory proteins, though a number of such proteins regulate protofilament and Z-ring dynamics in vivo. Although the mechanism of Z-ring constriction remains uncertain, a current model suggests that tethered protofilaments generate a bending force on bacterial membranes as a consequence of their fixed direction of curvature (Osawa et al., 2009). Protofilament turnover, which may include fragmentation and dissociation of subunits from protofilament ends, facilitates nucleotide exchange and recycling of subunits back into the Z ring (Mukherjee and Lutkenhaus, 1998; Mingorance et al., 2005; Huecas et al., 2007; Chen and Erickson, 2009). Continuous turnover of protofilaments has recently been shown to be required for the sustained contractile activity of Z rings reconstituted on liposomes (Osawa and Erickson, 2011). The rates of Z-ring turnover in vivo and of protofilament turnover in vitro correlate with GTPase activity, which varies among FtsZs from different bacteria (Mukherjee and Lutkenhaus, 1998; Chen et al., 2007; Huecas et al., 2007; Srinivasan et al., 2008; Chen and Erickson, 2009).
In contrast to bacteria in which the Z ring is composed of only a single FtsZ protein, plants have two FtsZ families, FtsZ1 and FtsZ2, which both function in chloroplast division (Osteryoung et al., 1998; Strepp et al., 1998; Osteryoung and McAndrew, 2001). Both proteins are nuclear encoded and imported to the chloroplast stroma by N-terminal transit peptides that are cleaved upon import (Osteryoung and Vierling, 1995; Fujiwara and Yoshida, 2001; McAndrew et al., 2001; Mori et al., 2001). Inside the chloroplast, the mature FtsZ1 and FtsZ2 proteins colocalize to form the mid-plastid Z ring (McAndrew et al., 2001; Vitha et al., 2001). Overexpression or depletion of FtsZ1 or FtsZ2 in vivo results in fewer and larger chloroplasts per cell than in wild type, suggesting their stoichiometry may be critical for chloroplast division (Osteryoung et al., 1998; Stokes et al., 2000). Recent genetic analysis in Arabidopsis thaliana has established conclusively that FtsZ1 and FtsZ2 are not interchangeable, and therefore have distinct functions in vivo (Schmitz et al., 2009).
Except for their transit peptides, FtsZ1 and FtsZ2 are well conserved with their bacterial counterparts. They both bear a core region common to all FtsZs that is required for GTP binding and hydrolysis (Osteryoung and McAndrew, 2001; Vaughan et al., 2004; Margolin, 2005), and are each capable of GTP-dependent assembly into protofilaments in vitro and of assembly-stimulated GTP hydrolysis (El-Kafafi et al., 2005; Lohse et al., 2006; Olson et al., 2010; Smith et al., 2010). Importantly, however, they also coassemble and hydrolyze GTP as heteropolymers, apparently with variable stoichiometry (Olson et al., 2010). In the only two comparative in vitro studies, the GTPase activity of Arabidopsis FtsZ1 was slightly higher than that of FtsZ2, though both hydrolyze GTP more slowly than E. coli FtsZ (Olson et al., 2010; Smith et al., 2010). FtsZ1 and FtsZ2 differ primarily downstream of the core region in their C termini, within which only FtsZ2 has retained a short peptide conserved in most bacterial FtsZs (Ma and Margolin, 1999; Osteryoung and McAndrew, 2001). In chloroplasts, this C-terminal peptide mediates a specific interaction between FtsZ2 and a transmembrane protein localized at the chloroplast division site, presumably to tether the Z ring to the inner envelope membrane (Maple et al., 2005). However, the equivalent region in bacterial FtsZ is not required for protofilament assembly in vitro (Wang et al., 1997; Margolin, 2005), and other functional differences between FtsZ1 and FtsZ2 remain elusive.
Recently, Srinivasan et al. (2008) used the fission yeast Schizosaccharomyces pombe to study bacterial FtsZ in an in vivo–like environment. They showed that bacterial FtsZ expressed as a GFP fusion protein in S. pombe robustly reproduced behavior observed earlier in both in vivo and in vitro experiments, including the ability to self-assemble into filaments and rings without membrane tethering or other accessory proteins, and similar rates of subunit exchange. They also showed that coexpression of FtsZ with SulA, an inhibitor of FtsZ polymerization, disrupted FtsZ assembly in S. pombe (Srinivasan et al., 2007). Thus, S. pombe is a valuable system in which to analyze the intrinsic self-assembly behavior of FtsZ proteins and the effects of assembly regulators.
In this study, we exploit S. pombe to investigate the self-assembly and dynamic properties of fluorescently tagged Arabidopsis FtsZ1 and FtsZ2 expressed separately and together. We show that both proteins assemble into filaments in the S. pombe cytosol, but with different morphologies and subunit exchange dynamics. Coassembly experiments provide evidence that FtsZ2 controls filament morphology and FtsZ1 promotes protofilament turnover. The data suggest that, in vivo, FtsZ2 forms the chloroplast Z-ring backbone while FtsZ1 facilitates Z-ring remodeling. In addition, we show that the chloroplast Z-ring positioning factor ARC3 inhibits FtsZ1 assembly, consistent with its hypothesized role as a functional analogue of the bacterial Z-ring positioning factor MinC (Maple et al., 2007).
Results
Functionality of FtsZ1 and FtsZ2 C-terminal fusion proteins
In their experiments, Srinivasan et al. (2008) used C-terminal GFP fusions to study bacterial FtsZ in S. pombe. Although FtsZ-GFP cannot fully complement bacterial ftsZ mutants (Ma et al., 1996), at least partly because the tag blocks membrane tethering through the conserved C-terminal peptide (Ma and Margolin, 1999), they are nevertheless competent for assembly in bacteria, in vitro and in S. pombe (Ma et al., 1996; Srinivasan et al., 2008; Osawa et al., 2009; Fu et al., 2010). Further, as indicated in the Introduction, the behavior of the bacterial fusion proteins in S. pombe mirrors their behavior in vitro and in vivo (Chen et al., 2005, 2007; Srinivasan et al., 2008), indicating the tags do not interfere with their intrinsic cytoskeletal behavior. Therefore, we likewise generated C-terminal FtsZ1-eYFP and FtsZ2-eCFP fusion proteins for expression in S. pombe. A nearly identical FtsZ1-mCerulean fusion localized to the chloroplast division site and fully complemented the chloroplast division defect in an Arabidopsis ftsZ1 knockout mutant (Fig. S1 A–C), showing it is functional in vivo. In contrast, FtsZ2-GFP or FtsZ2-His fusions do not complement ftsZ2 mutants, probably because the C-terminal peptide involved in inner envelope tethering is blocked. However, FtsZ2 fluorescent fusions assemble into filaments in chloroplasts (Fig. S1 D; Suppanz et al., 2007) and in E. coli (Fig. S1 E). Further, FtsZ1 and FtsZ2 bearing C-terminal His tags assemble into protofilaments in vitro (Olson et al., 2010; Smith et al., 2010). These data indicate that C-terminally tagged chloroplast FtsZs, like bacterial FtsZs, are assembly competent and valuable for investigating their intrinsic behavior. Therefore, we expressed FtsZ1-eYFP and FtsZ2-eCFP in S. pombe and studied their assembly and dynamic properties. All experiments were performed 36–40 h after fusion protein induction.
FtsZ1 and FtsZ2 expressed separately in S. pombe assemble into filaments with distinct morphologies
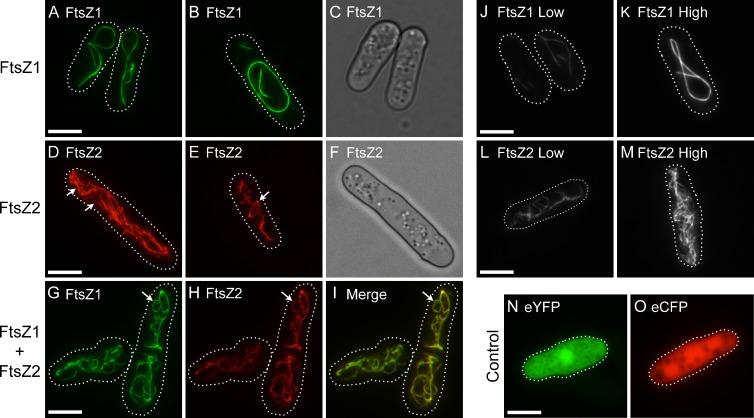
FtsZ1-eYFP and FtsZ2-eCFP (FtsZ1 and FtsZ2 hereafter) both formed filamentous structures (filaments) in the cytosol, as visualized by epifluorescence microscopy (Fig. 1). FtsZ1 typically formed long, gently curved cable-like filaments that looped around the cell, and also large closed rings (Fig. 1, A and B). Both structures appeared to follow the interior contours of the cell and the fluorescence distribution appeared even, suggesting uniform filament thickness. In contrast, FtsZ2 consistently formed elaborate networks (Fig. 1, D and E). Filaments within these networks were of variable thickness (fluorescence distribution; Fig. 1 D, arrows). Similar to FtsZ1, FtsZ2 formed closed rings (Fig. 1 E, arrow). This is the first evidence that FtsZ1 and FtsZ2 are capable of assembling into rings without the aid of accessory proteins or membrane attachment. However, FtsZ2 rings were observed less frequently than FtsZ1 rings, perhaps because the intricate FtsZ2 network obscured some of them. Similar FtsZ1 and FtsZ2 filament morphologies (i.e., FtsZ1 cables and FtsZ2 filamentous networks) were observed in cells with different levels of expression (Fig. 1, J–M; Fig. S2, A and B), indicating the distinct morphologies were not due to differences in protein level.
Figure 1.
FtsZ1 and FtsZ2 filament morphologies in S. pombe single- and coexpression strains. (A and B, D and E, G–M) Epifluorescence micrographs of cells expressing FtsZ1-eYFP (A and B, J and K; green), FtsZ2-eCFP (D and E, L and M; red), FtsZ1-eYFP and FtsZ2-eCFP (G–I), eYFP (N), or eCFP (O). Because the eYFP signals in J and K or eCFP signals in L and M are from the same identically processed images, respectively, the differences in fluorescence intensity reflect differences in protein level. Dotted lines show cell outlines. (C and F) Differential interference contrast micrographs of cells in A and D, respectively. Bars, 5 µm.
FtsZ1 and FtsZ2 expressed together colocalize in FtsZ2-like filament networks
Arabidopsis FtsZ1 and FtsZ2 colocalize in vivo, not only to chloroplast Z rings in wild-type (WT) plants, but also to filaments with aberrant morphologies in various mutants and transgenic plants (McAndrew et al., 2001; Vitha et al., 2001, 2003). They also coassemble in bundled heteropolymers in vitro (Olson et al., 2010). To assess if these proteins colocalize in S. pombe, FtsZ1 and FtsZ2 were coexpressed and visualized by epifluorescence microscopy. Imaging was performed on a strain that displayed minimum cell-to-cell variability in coexpression of FtsZ1 and FtsZ2. Immunoblots of soluble culture extracts from this strain showed FtsZ1 and FtsZ2 to be at near-equal levels (Fig. S3).
In the coexpression strain, FtsZ1 and FtsZ2 colocalized to an intricate network of filaments (Fig. 1, G–I) that closely resembled the networks observed in the strain expressing only FtsZ2 (Fig. 1, D and E). They also colocalized to closed rings (Fig. 1, G–I; arrow). The extent of colocalization was quantified using the Pearson’s correlation coefficient (PCC), which gives a measure of both the overlap between the two fluorescence signals and of how closely the signal intensities are correlated (Bolte and Cordelières, 2006). FtsZ1 and FtsZ2 fluorescence signals had an average PCC of 0.77 ± 0.08 (n = 10 for all measurements), which indicates that FtsZ1 and FtsZ2 colocalize within filament networks. It also indicates that the FtsZ1 and FtsZ2 signals are directly proportional, i.e., as one signal increases, so does the other.
GTPase-deficient mutants exhibit altered filament morphology
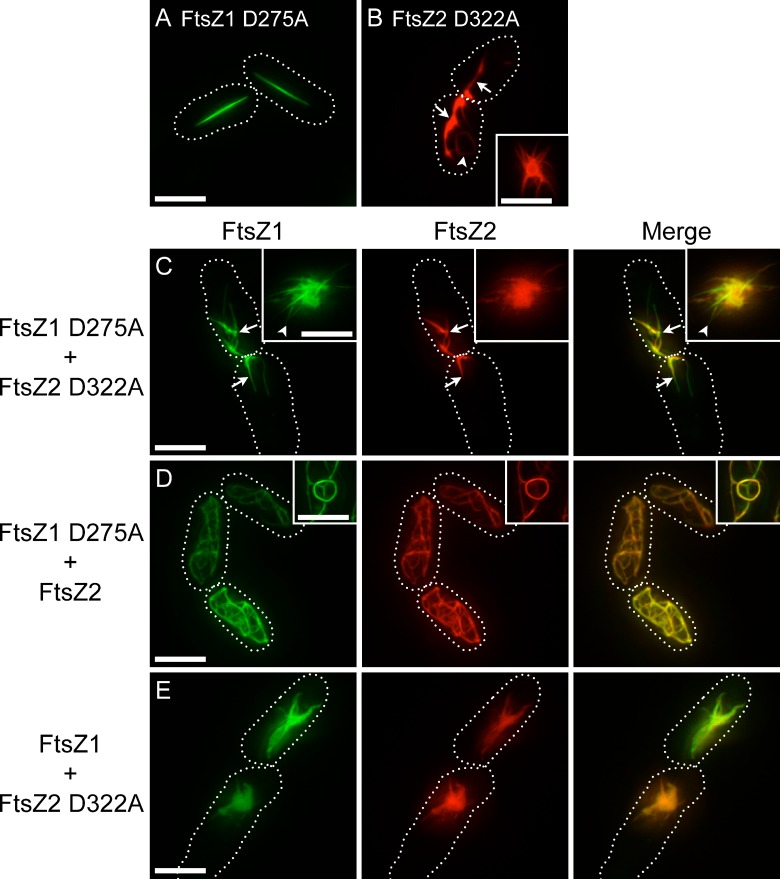
The effect of GTPase activity on chloroplast FtsZ filament morphology is not yet known. To test for this, the GTPase-deficient mutants FtsZ1 D275A and FtsZ2 D322A, which retain less than 10% of their WT activity (Olson et al., 2010), were expressed in S. pombe. The mutations alter a conserved aspartate required for GTP hydrolysis in presumably all FtsZ proteins, but do not prevent GTP binding (Scheffers et al., 2001; Oliva et al., 2004; Olson et al., 2010). Consistent with our previous findings that FtsZ1 D275A and FtsZ2 D322A are capable of assembling separately in vitro, the mutant proteins formed filaments when expressed individually in S. pombe. However, the filament morphologies differed from those of the WT proteins (Fig. 2). FtsZ1 D275A formed straight filaments of variable length (Fig. 2 A). Like the looping cables and rings formed by FtsZ1 (Fig. 1, A and B), the straight filaments formed by FtsZ1 D275A showed even fluorescence distribution. In contrast to FtsZ2 filament networks (Fig. 1, D and E), FtsZ2 D322A formed irregular filaments that appeared to be split and frayed (Fig. 2 B). However, similar to FtsZ2 filaments, FtsZ2 D322A filaments displayed regions of variable thickness (Fig. 2 B, arrows), and occasionally formed closed rings (Fig. 2 B, arrowhead). FtsZ2 D322A also formed aster-shaped structures and amorphous assemblies (Fig. 2 B, inset). Similar structures were observed in cells with different levels of expression (Fig. S2, C–F), indicating the distinct FtsZ1 D275A and FtsZ2 D322A morphologies were not due to differences in protein level. Although we cannot completely rule out that the structures formed by the mutant proteins result from misfolding in the S. pombe cytosol, we have shown that recombinant FtsZ1 D275A and FtsZ2 D322A, like the WT proteins, undergo GTP-dependent assembly into homopolymers and thick heteropolymer bundles in vitro (Olson et al., 2010), indicating they are capable of adopting their native structures. An FtsZ2 D322A-GFP fusion protein also assembles in E. coli (Olson, 2008) with the same localization pattern as FtsZ2-GFP (Fig. S1 E), suggesting proper folding in bacterial cells as well. Further, in the S. pombe coexpression strains described in the following two paragraphs, both FtsZ1 and FtsZ1 D275A colocalize tightly with FtsZ2 D322A and undergo active turnover, including in the amorphous assemblies. These latter observations suggest the mutant proteins coassemble in heteropolymers in S. pombe as in vitro, consistent with proper folding, and argue that the amorphous assemblies and other structures formed by the mutants represent thick filament bundles rather than protein aggregates. Finally, the equivalent E. coli FtsZ mutant (D212A) assembles on its own, coassembles with WT FtsZ, supports some degree of cell division in bacteria, and assembles reconstituted Z rings on liposomes (Stricker and Erickson, 2003; Redick et al., 2005; Osawa and Erickson, 2011). Collectively, these findings provide evidence that FtsZ1 D275A and FtsZ2 D322A fold similarly to the WT proteins in S. pombe.
Figure 2.
Filament morphologies in strains expressing GTPase-deficient FtsZ1 and FtsZ2. Epifluorescence micrographs of cells expressing FtsZ1 D275A (A), FtsZ2 D322A (B), FtsZ1 D275A and FtsZ2 D322A (C), FtsZ1 D275A and FtsZ2 (D), or FtsZ1 and FtsZ2 D322A (E). WT and mutant FtsZ1 and FtsZ2 proteins were fused to eYFP (green) and eCFP (red), respectively. Insets show regions in different cells from the same cultures as in larger panels. Dotted lines show cell outlines. Bars, 5 µm.
FtsZ1 D275A and FtsZ2 D322A were coexpressed to test how they would influence one another. In these strains and the mixed strains described in the following paragraph, coexpression levels were variable. For imaging, we chose cells that displayed fluorescence intensities equivalent to those in the WT coexpression strain described earlier in the Results in which FtsZ1 and FtsZ2 were at near-equal levels. FtsZ1 D275A and FtsZ2 D322A colocalized to irregular filaments, asters, and amorphous assemblies (Fig. 2 C). Similar to the FtsZ2 D322A filaments, irregular filaments in the coexpression strain had regions of variable thickness (Fig. 2 C, arrows). Rarely, amorphous assemblies displayed some variable composition. In the core of such structures, FtsZ1 D275A and FtsZ2 D322A were strongly colocalized whereas at the periphery, filaments more similar to those formed by FtsZ1 D275A were observed (Fig. 2 C, inset arrowhead). The latter regions were enriched in FtsZ1 D275A. However, overall FtsZ1 D275A and FtsZ2 D322A were highly colocalized (PCC 0.86 ± 0.07).
We also coexpressed WT FtsZ1 with GTPase-deficient FtsZ2 and vice versa. FtsZ1 D275A and FtsZ2 colocalized (PCC 0.85 ± 0.07) to an intricate filament network and occasionally to closed rings (Fig. 2 D). This morphology was visually indistinguishable from that observed when FtsZ2 was expressed alone (Fig. 1, D and E) or with WT FtsZ1 (Fig. 1, G–I). FtsZ1 coexpressed with FtsZ2 D322A colocalized (PCC 0.91 ± 0.06) in asters and amorphous assemblies (Fig. 2 E) closely resembling those formed when FtsZ2 D322A was expressed alone (Fig. 2 B) or with FtsZ1 D275A (Fig. 2 C).
A consistent result of the above experiments was that filament morphologies in all coexpression strains were very similar to those in the corresponding single FtsZ2 strain (FtsZ2 or FtsZ2 D322A), regardless of which form of FtsZ1 was present. These data suggest that FtsZ2 has structural dominance over FtsZ1 and controls filament morphology in the coexpression strains.
Dynamics of chloroplast FtsZ filaments
We studied the dynamics of subunit exchange within FtsZ1 and FtsZ2 filaments using FRAP. We measured the rate at which the proteins can diffuse into preexisting filaments, and also assessed the mobile and immobile fractions by calculating the extent of recovery after photobleaching. Cells selected for FRAP varied visibly in fluorescence intensity and hence in protein expression level. Photobleached regions were selected to represent the range of filament morphologies observed in a given strain.
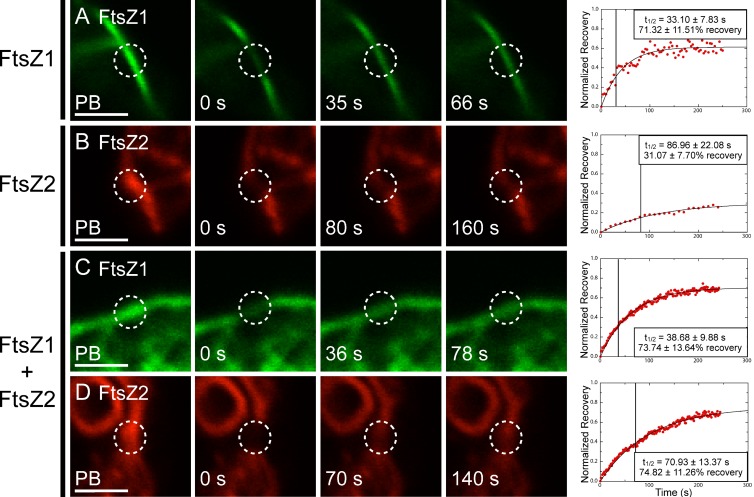
When expressed separately, FtsZ1 and FtsZ2 fluorescence signals showed recovery back into bleached regions (Fig. 3), indicating that filaments composed of either protein undergo subunit exchange. FtsZ1 recovered with a half-time (t1/2) of 33.10 ± 7.83 s (n = 10 for all FRAP experiments; Fig. 3 A and Video 1), whereas FtsZ2 had a slower t1/2 of 86.96 ± 22.08 s (Fig. 3 B and Video 2). FtsZ1 fluorescence recovered to 71.32 ± 11.51% of the prebleach intensity (Fig. 3 A), whereas FtsZ2 fluorescence only recovered to 31.07 ± 7.70% (Fig. 3 B). These differences were statistically significant (P < 0.01). No consistent correlation between the maximum fluorescence intensity in the photobleached cells and either t1/2 or percent recovery could be discerned (Fig. S4, A and B), suggesting that protein level did not influence FRAP measurements over the expression ranges represented in our experiments. FRAP data are summarized in Table S1.
Figure 3.
Dynamics of FtsZ1 and FtsZ2 expressed singly or together. S. pombe cells expressing FtsZ1-eYFP (A, green; see also Video 1), FtsZ2-eCFP (B, red; see also Video 2), or FtsZ1-eYFP and FtsZ2-eCFP (C and D) were analyzed by FRAP. Images from left to right represent fluorescence signals in photobleached regions (circled) just before bleaching (PB), at the time of bleaching (0 s), at the time closest to t1/2, and at twice t1/2. Representative plots of fluorescence recovery vs. time are shown at right. Data in each plot were normalized to the PB fluorescence signal (1 on the y-axis) and the signal intensity at the time of bleaching (0 on the y-axis). Boxes in the plots show the average t1/2 (also indicated by vertical lines) and average percent recovery ± SD for 10 independent FRAP datasets obtained for each strain. Bars, 2 µm.
We also assessed how coexpression of FtsZ1 and FtsZ2 affected each of their dynamics. When coexpressed, FtsZ1 and FtsZ2 recovered with half-times of 38.68 ± 9.88 s and 70.93 ± 13.37 s, respectively (Fig. 3, C and D). These values were not statistically different from those measured when FtsZ1 and FtsZ2 were expressed alone (Fig. 3, A and B). FtsZ1 recovered to 73.74 ± 13.64% of the prebleach (Fig. 3 C), equivalent to the recovery when FtsZ1 was expressed by itself (Fig. 3 A). However, FtsZ2 recovered to 74.82 ± 11.26% (Fig. 3 D; Table S1), nearly 2.5-fold greater than when expressed on its own (Fig. 3 B), indicating an FtsZ1-dependent increase in FtsZ2 dissociation from filaments. This effect suggests that FtsZ1 destabilizes FtsZ2-containing protofilaments, consistent with the formation of heteropolymers.
GTPase-deficient proteins show altered turnover
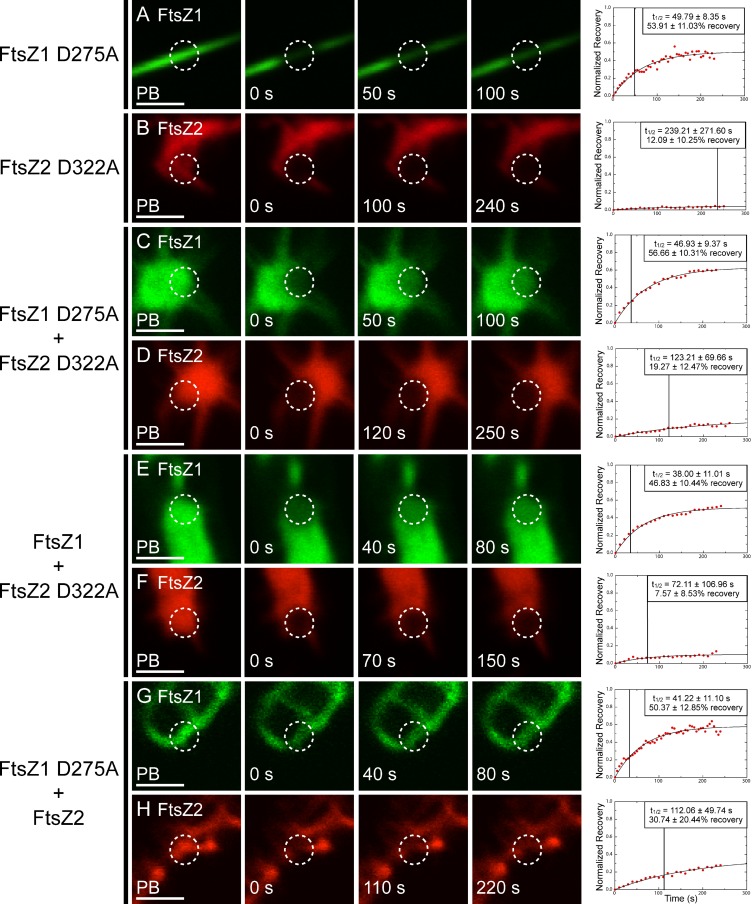
To assess whether GTPase activity affects FtsZ1 and FtsZ2 dynamics, FtsZ1 D275A and FtsZ2 D322A were analyzed for rate and extent of subunit turnover in S. pombe using FRAP (Fig. 4). The straight FtsZ1 D275A filaments recovered with a t1/2 of 49.79 ± 8.35 s and displayed a maximum recovery of 53.91 ± 11.03% (Fig. 4 A, Table S1, and Video 3). Both values were significantly reduced compared with those of FtsZ1 (Fig. 3 A). Surprisingly, however, even with its severely inhibited GTPase activity (Olson et al., 2010), FtsZ1 D275A filaments displayed a significant amount of subunit exchange. In contrast, turnover of FtsZ2 D322A was almost completely abolished. FtsZ2 D322A filaments had a t1/2 of 239.21 ± 271.60 s and recovered to a maximum of 12.09 ± 10.25% (Fig. 4 B and Video 4). As observed for FtsZ1 and FtsZ2, turnover of FtsZ1 D275A and FtsZ2 D322A filaments did not appear to be affected by protein expression level over the range of levels measured (Fig. S4, C and D). The reduced turnover of GTPase-deficient FtsZ filaments indicates that GTP hydrolysis is an important factor in promoting subunit exchange. However, in the case of FtsZ1, it may not be solely responsible, as filaments consisting of only FtsZ1 D275A still undergo significant turnover.
Figure 4.
Dynamics of GTPase-deficient FtsZ1 and FtsZ2 expressed in various combinations. S. pombe cells expressing FtsZ1 D275A (A; see also Video 3), FtsZ2 D322A (B, panel 3 is 100 s recovery, panel 4 is the image acquired closest to t1/2; see also Video 4), FtsZ1 D275A and FtsZ2 D322A (C and D), FtsZ1 and FtsZ2 D322A (E and F), and FtsZ1 D275A and FtsZ2 (G and H). WT and mutant FtsZ1 and FtsZ2 proteins were fused to eYFP (green) and eCFP (red), respectively. Images from left to right (except in B) represent fluorescence signals in photobleached regions (circled) just before bleaching (PB), at the time of bleaching (0 s), at the time closest to t1/2, and at twice t1/2. Representative plots of fluorescence recovery vs. time are shown at right. Data in each plot were normalized to the PB fluorescence signal (1 on the y-axis) and the signal intensity at the time of bleaching (0 on the y-axis). Boxes in the plots show the average t1/2 (also indicated by the vertical lines) and average percent recovery ± SD for 10 independent FRAP datasets obtained for each strain. Bars, 2 µm.
When FtsZ1 D275A and FtsZ2 D322A were coexpressed, half-times were 46.93 ± 9.37 s and 123.21 ± 69.66 s, and maximum recoveries were 56.66 ± 10.31% and 19.27 ± 12.47%, respectively (Fig. 4, C and D). These values were statistically similar to those observed when FtsZ1 D275A and FtsZ2 D322A were expressed alone.
To extend our analysis, mutant and WT FtsZ proteins were assayed in different combinations. When coexpressed, FtsZ1 and FtsZ2 D322A recovered with half-times of 38.00 ± 11.01 s and 72.11 ± 106.96 s, and had maximum recoveries of 46.83 ± 10.44% and 7.57 ± 8.53%, respectively (Fig. 4, E and F; Table S1). These were similar to values obtained when each protein was expressed individually, except that the percent recovery for FtsZ1 was statistically lower than when FtsZ1 was expressed alone (Fig. 3 A), indicating decreased FtsZ1 dissociation from the filaments. Conversely, when FtsZ1 D275A and FtsZ2 were coexpressed, half-times were 41.22 ± 11.10 s and 112.06 ± 49.74 s, and maximum recoveries were 50.37 ± 12.85% and 30.74 ± 20.44%, respectively (Fig. 4 H). In this combination, all values were similar to those observed when FtsZ1 D275A and FtsZ2 are expressed alone.
Based on the FRAP data, we conclude that FtsZ1 is more dynamic than FtsZ2 in S. pombe, that FtsZ2 but not FtsZ1 turnover is abolished by loss of GTPase activity, and that FtsZ1 enhances turnover of FtsZ2 in heteropolymers.
Effect of ARC3 on FtsZ1 assembly
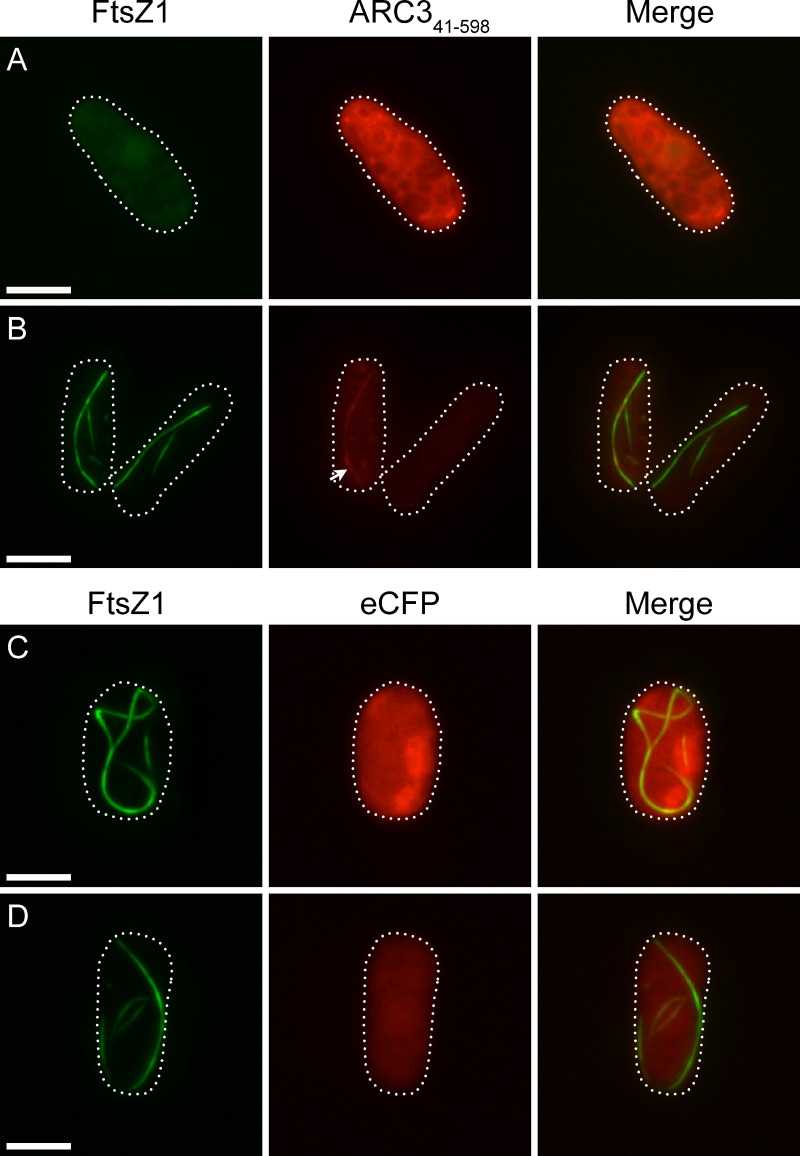
The chloroplast division protein ARC3 (Pyke and Leech, 1992; Shimada et al., 2004) regulates placement of the division site by restricting Z-ring assembly to the mid-plastid, as shown by the formation of multiple constrictions and Z rings in chloroplasts of Arabidopsis arc3 mutants (Glynn et al., 2007; Maple et al., 2007). ARC3 was reported to interact specifically with FtsZ1 in yeast two-hybrid assays and has been proposed to function similarly to bacterial MinC (Maple et al., 2007), suggesting it may be a direct inhibitor of FtsZ assembly (Hu et al., 1999; Lutkenhaus, 2007). To begin testing this hypothesis, we fused eCFP to a truncated form of ARC3, ARC341–598, which lacks the N-terminal chloroplast transit peptide and the C-terminal MORN domain. The MORN domain was shown to inhibit ARC3–FtsZ1 interaction in yeast (Maple et al., 2007), possibly because another Z-ring assembly regulator not present in yeast normally sequesters the MORN domain in vivo (Glynn et al., 2009).
ARC341–598 was coexpressed with FtsZ1 in S. pombe and FtsZ1 morphology was examined. In cells with strong ARC341–598 fluorescence, FtsZ1 adopted a diffuse localization pattern and did not produce any filaments (Fig. 5 A). In cells with weaker ARC341–598 fluorescence, some FtsZ1 filaments could be observed (Fig. 5 B). These filaments varied in length and displayed a straight or bent morphology, but were never as long as those observed when FtsZ1 was expressed alone (Fig. 1, A and B). ARC341–598 colocalized with FtsZ1 filaments in cells where filaments were visible (Fig. 5 B, arrow), consistent with the ARC341–598–FtsZ1 interaction in yeast (Maple et al., 2007). In contrast, when coexpressed with only eCFP as a control, FtsZ1 assembled into cable-like filaments and rings similar to those seen when FtsZ1 was expressed alone, regardless of eCFP protein level (Fig. 5, C and D). These results provide evidence that ARC341–598 functions as an inhibitor of FtsZ1 assembly, possibly in a dose-dependent manner, and support the hypothesis that ARC3 regulates division-site placement in vivo by interfering with protofilament formation away from the chloroplast midpoint.
Figure 5.
Effect of ARC341–598 on FtsZ1 assembly. Epifluorescence micrographs of S. pombe cells expressing FtsZ1-eYFP (green) and higher levels of ARC341–598–eCFP (A, red), lower levels of ARC341–598–eCFP (B), higher levels of eCFP (C), or lower levels of eCFP (D). Exposure times and image processing for cells in A and B were identical. Cells in C and D were taken from the same identically processed image; hence, differences in fluorescence intensity reflect differences in protein level. Dotted lines show cell outlines. Bars, 5 µm.
Discussion
Bacterial FtsZs, which function as homopolymers, have been studied extensively and, though questions remain, much has been learned about their assembly and dynamic properties (Mazouni et al., 2004; Adams and Errington, 2009; Erickson et al., 2010; Aylett et al., 2011). In contrast, the assembly properties of chloroplast FtsZ1 and FtsZ2 are poorly understood. Expression of fluorescent FtsZ1 and FtsZ2 fusions in fission yeast, which lacks the native assembly regulators present in chloroplasts, has allowed us to begin exploring their intrinsic self-assembly properties in an in vivo–like system. As observed by Srinivasan et al. (2008) for bacterial FtsZ, the behavior of FtsZ1 and FtsZ2 in S. pombe is consistent with findings from previous studies. For example, FtsZ1 and FtsZ2 are independently capable of forming filaments in S. pombe as they are in vitro and in plants (El-Kafafi et al., 2005; Lohse et al., 2006; Yoder et al., 2007; Schmitz et al., 2009; Olson et al., 2010; Smith et al., 2010). Further, the colocalization of FtsZ1 and FtsZ2 in S. pombe agrees with their tight colocalization in vivo and with recent work showing that they preferentially coassemble as heteropolymers in vitro (McAndrew et al., 2001; Vitha et al., 2001; Olson et al., 2010). Thus, S. pombe accurately reproduces key features of FtsZ1 and FtsZ2 behavior.
FtsZ filament morphology
We suggest that the distinct morphologies displayed by FtsZ1 and FtsZ2 in S. pombe represent protofilament bundles based on observations that E. coli FtsZ protofilaments, which are typically single stranded when polymerized in dilute solution in vitro, undergo bundling when assembled in crowding reagents more closely resembling the S. pombe cytosol (Mukherjee and Lutkenhaus, 1999; Popp et al., 2009; Erickson et al., 2010). The width and intensity of the fluorescence signals also suggest bundling, as assumed by Srinivasan et al. (2008) for the linear cables formed by bacterial FtsZ in S. pombe. Several studies suggest that bacterial FtsZ bundles and Z rings consist of loosely packed, overlapping protofilaments held together by weak lateral interactions, probably involving electrostatic forces (Li et al., 2007; Popp et al., 2009; Fu et al., 2010; Buske and Levin, 2012), though membrane tethering may enhance lateral interactions and packing between protofilaments (Milam et al., 2012). The split and frayed appearance and variable thickness of FtsZ2 and FtsZ2 D322A filaments (Fig. 1, D–E; Fig. 2 B) and more uniform appearance of FtsZ1 and FtsZ1 D275A filaments (Fig. 1, A and B; Fig. 2 A) could indicate that lateral interactions are stronger between FtsZ1 than FtsZ2 protofilaments. In vitro assembly experiments that mimic molecular crowding conditions in S. pombe and in chloroplasts will be important for understanding these differences.
Our finding that FtsZ1 and FtsZ2 are each capable of forming closed ring structures in S. pombe (Fig. 1, B and E), similar to bacterial FtsZ (Srinivasan et al., 2008), suggests that the ability to form closed rings, perhaps by annealing of protofilaments (Mingorance et al., 2005; Chen and Erickson, 2009; Erickson et al., 2010), is an inherent characteristic of FtsZ1 and FtsZ2 and does not require membrane tethering or accessory proteins. The fact that GTPase-deficient FtsZ2 D322A homopolymers could also form closed rings (Fig. 2 B) indicates that this property, at least for FtsZ2, does not depend on active protofilament turnover. This is consistent with recent reports showing that E. coli FtsZ forms closed rings on the surface of liposomes and on a mica surface even when GTPase activity and subunit exchange are severely inhibited (Osawa and Erickson, 2011; Mateos-Gil et al., 2012).
We presume that the consistent colocalization of FtsZ1 and FtsZ2 in S. pombe, also observed in chloroplasts (McAndrew et al., 2001; Vitha et al., 2001), represents coassembly in heteropolymers based on in vitro assembly experiments (Olson et al., 2010). The predominant structures in the S. pombe coexpression strains invariably resembled those assembled by whichever form of FtsZ2 was present (WT or FtsZ2 D322A), irrespective of the form of FtsZ1 present. This suggests that FtsZ2 exerts a significant degree of dominance over FtsZ1 in determining heteropolymer morphology, at least when unrestrained by membrane tethering or the action of assembly regulators. The reason for FtsZ2’s morphological dominance over FtsZ1 is not yet clear. A possibility is that the interface geometry between FtsZ1 and FtsZ2 subunits in heteropolymers is more similar to the geometry between subunits in FtsZ2 homopolymers. Structural approaches would be required to assess this. Whatever the explanation, morphological dominance by FtsZ2 may depend on the ratio between FtsZ1 and FtsZ2 in heteropolymers, as suggested by the enrichment of FtsZ1 D275A in straight filaments protruding from the amorphous assemblies of colocalized FtsZ1 D275A and FtsZ2 D322A (Fig. 2 C, inset arrowhead). We suggest these filaments arise from the ability of FtsZ1 D275A but not FtsZ2 D322A to dissociate from filaments, leading over time to the formation of the FtsZ1 D275A–enriched protrusions. Future studies in which the FtsZ1/FtsZ2 ratio in individual S. pombe cells is quantified and manipulated should provide insight into how the interplay between FtsZ1 and FtsZ2 influences filament morphology.
FtsZ filament dynamics
Studies on bacterial FtsZ have shown that GTPase activity correlates with the rate of subunit exchange from protofilaments and Z rings and is probably essential for Z-ring remodeling (Mukherjee and Lutkenhaus, 1998; Chen et al., 2007; Huecas et al., 2007; Chen and Erickson, 2009; Osawa and Erickson, 2011). Because FtsZ1 has higher GTPase activity than FtsZ2 in vitro (Olson et al., 2010; Smith et al., 2010), the higher turnover of FtsZ1 than FtsZ2 filaments in S. pombe (Fig. 3, A and B) may partly reflect this difference, though the difference in GTPase activity is fairly small. As these are the first comparative analyses of chloroplast FtsZ dynamics, it remains to be seen whether other methods yield similar data. However, turnover rates for E. coli and M. tuberculosis FtsZ in S. pombe were very close to those measured in bacterial cells and in vitro (Anderson et al., 2004; Chen et al., 2005, 2007; Srinivasan et al., 2008), suggesting that our measurements in this system reflect the intrinsic dynamics of the chloroplast proteins as well. Recovery half-times for FtsZ1 and FtsZ2 in homopolymers as well as in coassembled filaments were well below those reported for E. coli FtsZ, consistent with their lower GTPase activities (Lu et al., 1998; Redick et al., 2005; Olson et al., 2010; Smith et al., 2010). However, turnover of FtsZ1 in S. pombe was comparable to that of M. tuberculosis FtsZ (Srinivasan et al., 2008), which has a similar GTP hydrolysis rate (Chen et al., 2007).
The reduced recovery half-times and altered filament morphologies of GTPase-deficient FtsZ1 D275A and FtsZ2 D322A mutants demonstrate that GTPase activity is important for maintaining FtsZ1 and FtsZ2 protofilament structure and dynamics. However, in contrast with the static FtsZ2 D322A filaments, FtsZ1 D275A filaments still exhibited significant though reduced turnover compared with WT FtsZ1 filaments. Given the high degree of similarity between the conserved GTP-binding and hydrolysis domains of FtsZ1 and FtsZ2 (Osteryoung and McAndrew, 2001), it is not clear why equivalent mutations inhibit dynamics severely in FtsZ2 D322A and only moderately in FtsZ1 D275A, even though the GTPase activities in both cases are reduced below background (Olson et al., 2010). These findings suggest that some other factor facilitates turnover of FtsZ1 homopolymers and heteropolymers. One such factor could be the strength of their subunit interfaces. FtsZ1–FtsZ1 interfaces may be inherently weaker than FtsZ2–FtsZ2 interfaces, perhaps making FtsZ1 homopolymers, both WT and mutant, more prone to fragmentation and subunit dissociation than FtsZ2 homopolymers, resulting in higher turnover. As described in the following paragraph, this conjecture could also explain the behavior of heteropolymers.
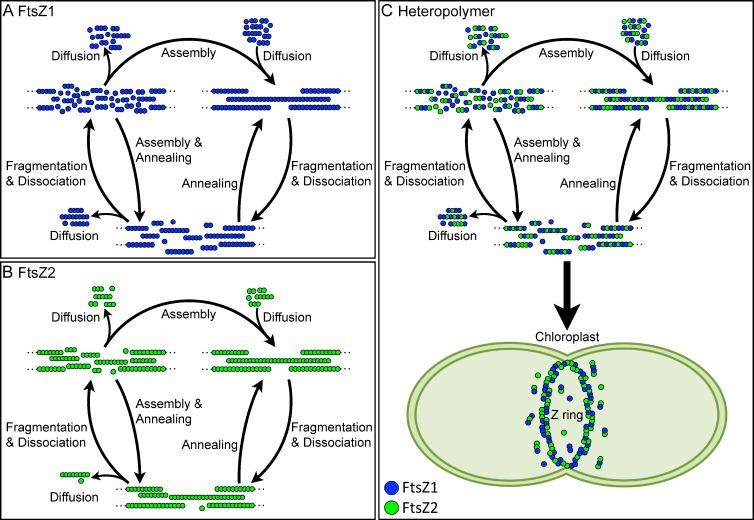
Our finding that FtsZ2 displays a 2.5-fold increase in fluorescence recovery when coexpressed with FtsZ1 suggests that more FtsZ2 dissociates from heteropolymers than from homopolymers (Fig. 3). A preliminary model consistent with these findings and with recent models of bacterial FtsZ behavior is depicted in Fig. 6. The model is meant to represent the intrinsic steady-state dynamic behavior of FtsZ1 and FtsZ2 in S pombe. In chloroplasts, self-assembly and turnover would be modulated by numerous assembly regulators (Yang et al., 2008; Maple and Møller, 2010; Pyke, 2010). We suggest that (1), in S. pombe, FtsZ1 and FtsZ2 homopolymers and heteropolymers assemble in loosely bundled filaments. This conjecture is based in part on in vivo data suggesting loose protofilament bundling through weak lateral interactions in bacteria (Li et al., 2007; Fu et al., 2010; Buske and Levin, 2012) and on our observation that changes in FtsZ1 and FtsZ2 expression levels do not appear to affect turnover dynamics under our experimental conditions (Fig. S4). If protofilaments were tightly packed, diffusion out of protofilament bundles might be reduced at higher expression levels, though additional experiments, perhaps coupled with new super-resolution imaging techniques (Li et al., 2007; Fu et al., 2010; Jennings et al., 2011), will be necessary to rigorously explore the arrangement of chloroplast FtsZ protofilaments. (2) The faster turnover and higher maximum recovery of FtsZ1 than FtsZ2 in the single-expression strains suggest that FtsZ1 homopolymers may be less stable and more likely to fragment and lose subunits from protofilament ends than FtsZ2 homopolymers (Fig. 6, A and B). Fragmentation of FtsZ1 filaments would therefore produce a larger pool of diffusible subunits and small oligomers that could be recycled back onto FtsZ1 protofilaments than fragmentation of FtsZ2 filaments, making FtsZ1 filaments more dynamic. Further disassembly of small FtsZ1 oligomers would contribute to FtsZ1 turnover. (3) Coassembly of FtsZ1 and FtsZ2 in heteropolymers would increase fragmentation and subunit dissociation (Fig. 6 C), producing a larger pool of diffusible FtsZ2-containing oligomers than would occur for FtsZ2 homopolymers (Fig. 6 B). Small oligomers containing FtsZ2 and/or FtsZ1 could further depolymerize, increasing the overall assembly-ready pool of FtsZ2 available for reassembly onto free protofilament ends. Additionally, as proposed for bacterial FtsZ (Osawa and Erickson, 2011), annealing of protofilament fragments could also contribute to recycling of both FtsZ1 and FtsZ2 subunits and short oligomers back into larger filaments, leading to the increased turnover of FtsZ2 observed in the coexpression strains (Fig. 3). Because FtsZ1 and FtsZ2 tightly colocalize to the chloroplast division site in vivo and form heteropolymers in vitro (McAndrew et al., 2001; Vitha et al., 2001; Olson et al., 2010), we suggest that this situation may be more representative of the dynamic behavior of FtsZ1 and FtsZ2 in dividing chloroplasts (Fig. 6 C, bottom). We also note that the assembly subunit is not yet known, but if it is a dimer or tetramer as proposed by Smith et al. (2011) for FtsZ1 and FtsZ2 homopolymers, or a hetero-oligomer of FtsZ1 and FtsZ2, then the minimal protofilament could be two or more subunits thick instead of a single subunit thick as depicted in Fig. 6. This would not necessarily alter the effect of FtsZ1 on heteropolymer dynamics, however.
Figure 6.
Working model of the intrinsic steady-state turnover of FtsZ1 and FtsZ2 homopolymers and heteropolymers in S. pombe based on FRAP analysis and models of bacterial FtsZ dynamics. (A and B) FtsZ1 homopolymers have higher rates of fragmentation and loss of subunits from protofilament ends than FtsZ2 homopolymers. The diffusible pool of small oligomers and subunits that can be recycled back into protofilaments is therefore larger for FtsZ1 than FtsZ2, resulting in higher FtsZ1 turnover. (C) FtsZ1 incorporation into heteropolymers enhances fragmentation and loss of subunits from protofilament ends. The diffusible pool of small FtsZ2-containing oligomers and FtsZ2 subunits that can be recycled back into protofilaments is therefore larger for heteropolymers than for FtsZ2 homopolymers, resulting in higher FtsZ2 turnover from heteropolymers. We hypothesize that heteropolymers represent the predominant protofilament form in the chloroplast Z ring in vivo (wide arrow). Annealing of protofilaments and addition of subunits onto protofilament ends may both contribute to turnover in all cases. Blue circles, FtsZ1; green circles, FtsZ2. Dotted lines represent continuation of FtsZ protofilaments. Important details are elaborated in the Discussion.
The mechanistic explanation for the increased turnover of FtsZ2 from heteropolymers is not yet clear. As FtsZ1 has somewhat higher GTPase activity than FtsZ2 and GTPase is correlated with turnover of bacterial FtsZ protofilaments, one potential explanation could be that heteropolymers hydrolyze GTP more rapidly than homopolymers, leading to increased turnover. However, coassembly of FtsZ1 and FtsZ2 in vitro only slightly increased (Olson et al., 2010) or decreased (Smith et al., 2010) GTPase activity, suggesting that GTP hydrolysis may not occur much more rapidly in FtsZ1–FtsZ2 than FtsZ1–FtsZ1 subunit interfaces. Another possibility, suggested by our finding that GTPase-deficient FtsZ1 D275A filaments are still dynamic (Fig. 4 A), is that weaker FtsZ1–FtsZ1 interfaces in heteropolymers stimulate fragmentation, enhancing turnover. Based on our previous in vitro work suggesting that FtsZ1 and FtsZ2 heteropolymers assemble with variable stoichiometry (Olson et al., 2010), in Fig. 6 C we have represented the arrangement of FtsZ1 and FtsZ2 subunits in heteropolymers as variable rather than as strictly alternating. Where FtsZ1–FtsZ1 interfaces are present, heteropolymers would fragment more readily if these interfaces are indeed weaker. This model would predict that increasing the FtsZ1-to-FtsZ2 ratio in heteropolymers would destabilize and promote turnover of heteropolymers. A combination of in vitro and in vivo approaches will be necessary to address these speculations. However, even if FtsZ1–FtsZ1 (or potentially FtsZ1–FtsZ2) interfaces are inherently weaker than FtsZ2–FtsZ2 interfaces, GTPase activity still appears to be necessary for the enhancement of FtsZ2 turnover by FtsZ1,as FtsZ1 D275A does not result in a statistically significant increase FtsZ2 turnover from heteropolymers as does FtsZ1 (Figs. 3 B and 4 H).
Function of ARC3
In E. coli, MinC antagonizes FtsZ polymerization by direct interaction near the cell poles. MinC is spatially regulated by MinD and MinE through a complex set of interactions, resulting in Z-ring formation and cell division only at the midcell (Lutkenhaus, 2007; de Boer, 2010). Green-lineage chloroplasts inherited homologues of MinD and MinE through endosymbiosis, and these proteins likewise function to restrict Z-ring formation to the mid-plastid (Colletti et al., 2000; Itoh et al., 2001; Maple et al., 2002; Vitha et al., 2003; Glynn et al., 2007). However, although MinC also occurs in the cyanobacterial relatives of chloroplasts (Mazouni et al., 2004; Miyagishima, 2005), higher plants lack MinC. Instead, the plant-specific FtsZ1-interacting protein ARC3 has been postulated as a functional replacement for MinC (Shimada et al., 2004; Maple et al., 2007). Our finding that ARC3 inhibits FtsZ1 assembly in S. pombe supports this hypothesis. Because ARC3 also interacts with MinD and MinE (Maple et al., 2007), these results also suggest that ARC3, like MinC, functions as a direct assembly inhibitor whose activity is controlled by MinD, MinE, and possibly several other plant-specific proteins (Miyagishima, 2011) to regulate Z-ring assembly and positioning. However, because ARC3 bears no obvious structural similarity to MinC, its mechanism of action may be different. MinC is thought to inhibit bacterial Z-ring formation by inhibiting bundling of protofilaments (Hu et al., 1999; Dajkovic et al., 2008; de Boer, 2010). ARC3 bears an FtsZ-like domain that interacts with FtsZ1, though it probably is not a functional GTPase (Shimada et al., 2004; Maple et al., 2007). We speculate that the FtsZ-like region of ARC3 might assemble directly into protofilaments and destabilize them, perhaps by promoting fragmentation. Further experimentation will be needed to understand the mechanism of ARC3 action.
Potential roles of FtsZ1 and FtsZ2 in chloroplast Z rings
Our analysis of the intrinsic assembly properties of FtsZ1 and FtsZ2 in S. pombe suggests potential specialized functions for these proteins. Our finding that coassembled filaments invariably adopt an FtsZ2-like morphology suggests that FtsZ2 may be the primary structural determinant of the chloroplast Z ring. This function may be enhanced in vivo by membrane tethering of FtsZ2 through its interaction with the transmembrane chloroplast division protein ARC6 (Vitha et al., 2003; Maple et al., 2005). The higher turnover of FtsZ1 than FtsZ2 homopolymers and the FtsZ1-dependent increase in FtsZ2 turnover from heteropolymers suggest that FtsZ1 may facilitate Z-ring constriction by enhancing Z-ring turnover. Consistent with these ideas, FtsZ2 occasionally forms mid-plastid Z rings and supports some degree of chloroplast division in an Arabidopsis ftsZ1-null mutant, but only in very small chloroplasts (Yoder et al., 2007). By promoting turnover, FtsZ1 may sustain Z-ring constriction during leaf growth, when chloroplasts are expanding and dividing. FtsZ1 and FtsZ2 maintain a constant 1:2 ratio in whole rosettes of Arabidopsis throughout plant development (McAndrew et al., 2008), but the ratio in individual chloroplasts, and more importantly in Z rings, remains unknown. The possibility that FtsZ1 and FtsZ2 assemble at variable ratios in vivo as observed in vitro (Olson et al., 2010) potentially represents a novel mechanism for regulating Z-ring constriction during plant growth and/or over a single chloroplast contractile cycle. Quantitative in vivo studies of FtsZ1 and FtsZ2 behavior and protein levels during plant development and in chloroplast Z rings will be important for further addressing the functional interplay between FtsZ1 and FtsZ2 and establishing how Z-ring assembly, positioning, and contractile activity are regulated in chloroplasts.
Materials and methods
Cloning and S. pombe transformation
Sequences encoding WT or GTPase-deficient Arabidopsis thaliana FtsZ1 (AtFtsZ1-1, At5g55280) and FtsZ2 (AtFtsZ2-1, At2g36250) lacking the predicted transit peptides (the first 57 and 48 amino acids, respectively) were amplified by PCR from the corresponding cDNA bacterial expression plasmids (Olson et al., 2010). The primers used were: 5′-TTTTTTCTCGAGACCATGAGGTCTAAGTCGATGCGATTGAGG-3′ (forward) and 5′-GCCCTTGCTCACCATCTGCATGAAGAAAAGTCTACGGGGAGAAGA-3′ (reverse) for FtsZ1, and 5′-TTTTTTCTCGAGACCATGGCCGCTCAGAAATCTGAATCTTCT-3′ (forward) and 5′-GCCCTTGCTCACCATCTGCATGACTCGGGGATAACGAGAGCT-3′ (reverse) for FtsZ2. The FtsZ1 and FtsZ2 PCR products were then fused to enhanced yellow fluorescent protein (eYFP) or enhanced cyan fluorescent protein (eCFP; Takara Bio Inc.), respectively, at their C termini by splicing by overlap extension (SOE) and asymmetric PCR (Warrens et al., 1997). The primers used to amplify eYFP and eCFP were 5′-ATGGTGAGCAAGGGCGAGGAGCTG-3′ (forward) and 5′-TTTTTTGGATCCTTACTTGTACAGCTCGTCCATGC-3′ (reverse). FtsZ1-eYFP and FtsZ2-eCFP fusion products were subcloned into pREP41X and pREP42X, under control of the medium-strength nmt1* promoter (Basi et al., 1993; Forsburg, 1993), using standard molecular biology techniques and Xho1 and BamH1 restriction sites.
Control constructs were generated for the expression of eYFP and eCFP only. The primers used to amplify eYFP and eCFP were: 5′-TTTTTTCTCGAGACCATGGTGAGCAAGGGCGAGGAGCTG-3′ (forward) and the same reverse primer as described above for creation of the FtsZ fusion constructs. The eYFP and eCFP PCR products were subcloned into pREP41X and pREP42X as described above, but using the Xho1 and BamH1 HF restriction sites.
A truncated ARC3 (AtARC3, At1g75010) construct consisting of the FtsZ-like domain and the middle region (ARC341–598, amino acids 41–598; Shimada et al., 2004; Maple et al., 2007) was amplified from a cDNA clone by PCR. The primers used were: 5′-TTTTTTCATATGGCCAACTGTACATCTCGAAAGGCGCGTCG-3′ (forward) and 5′-GCCCTTGCTCACCATCTGCATATCTCCGGCGTCCACTTGTTTCC-3′ (reverse). eCFP was fused to the 3′ end of the ARC341–598 PCR product by SOE and asymmetric PCR (Warrens et al., 1997). The primers used to amplify eCFP were the same as described above. The ARC341–598–eCFP fusion product was subcloned into pREP42, under control of the nmt1* promoter (Basi et al., 1993; Forsburg, 1993), using the Nde1 and BamH1 restriction sites.
FtsZ constructs were transformed into S. pombe using a modified lithium acetate procedure (http://www.sanfordburnham.org/labs/wolf/Protocols/Protocols/Fission%20Yeast/Nurse%20Lab%20Manual.htm). S. pombe (strain MBY192 [h− leu1-32 ura4-D18]) cultures were grown in 50 ml of Pombe Glutamate medium (PMG) at 32°C with shaking at 250 rpm to an OD = 0.5 (107 cells/ml) and pelleted at 4,000 g at room temperature. The pellet was washed with 1/2 culture volume of TE (10 mM Tris-HCl, 1 mM EDTA, pH 7.5). The cells were pelleted again, resuspended in 1 ml of TE and LiAc (100 mM lithium acetate, pH 7.5) and allowed to incubate at 30°C for 30 min. 200 µl of cells were aliquoted into microfuge tubes containing 20 µl of 10 µg/µl carrier sperm DNA (Agilent Technologies) and 1 µg of plasmid DNA (∼2–3 µl in 2 mM Tris-HCl, pH 8.5) and mixed by vortexing. 1.2 ml of PEG solution (40% PEG, TE pH 7.5, and LiAc) was added and each tube was vortexed for 10 s to mix. Tubes were incubated at 30°C with 200 rpm shaking for 30 min and then heated for 15 min at 42°C. The cells were pelleted at 7,000 g for 30 s and the supernatant was discarded. The cells were resuspended in 300 µl of TE, plated on solid PMG with selection for the plasmids (−uracil for pREP41X or −leucine for pREP42 or pREP42X), and allowed to grow at 28°C until colonies formed. Coexpression lines were generated by taking a culture in which cells had approximately uniform fluorescence of FtsZ1-eYFP, transforming the FtsZ2-eCFP, ARC341–598–eCFP, or eCFP construct into that cell line by the same protocol described above, and plating on solid PMG with both selection markers (−uracil and −leucine). The FtsZ2-eCFP, ARC341–598–eCFP, and eCFP constructs were under the control of the same promoter but a different selection marker (ura4+) than the FtsZ1-eYFP construct (LEU2).
Growth and expression of transformed cell lines
The nmt1* promoter is repressible with 15 µM thiamine (Maundrell, 1990). Yeast strains were streaked for isolation and grown on solid PMG containing 15 µM thiamine in the absence of leucine and/or uracil to select for the FtsZ1-eYFP and/or FtsZ2-eCFP/ARC341–598–eCFP constructs, respectively, at 28°C until colonies formed. Colonies were used to inoculate liquid cultures without thiamine to activate expression of the fusion proteins, and allowed to grow at 32°C with 250 rpm shaking for 36–40 h.
A culture coexpressing FtsZ1-eYFP and FtsZ2-eCFP was identified in which cells displayed strong fluorescence signal for each protein and minimal cell-to-cell variation in expression. This strain was used to make a glycerol stock on which subsequent analyses were performed. Cultures grown from this stock were also used to determine the relative levels of FtsZ1-eYFP and FtsZ2-eCFP by immunoblotting using a monoclonal αGFP (Takara Bio Inc.). The resulting band intensities were quantified, FtsZ1-eYFP signal was normalized to 1, and FtsZ2-eCFP signal was normalized relative to FtsZ1-eYFP signal.
Fluorescence microscopy and FRAP analysis
Aliquots (2 µl) of the liquid culture were pipetted onto glass or poly-lysine–coated slides and covered with a coverslip. Samples were imaged by differential interference contrast and epifluorescence microscopy, using a microscope (model DMRA2; Leica) with an HCX PL Apochromat 63× (1.32 NA) oil-immersion objective (Leica) and a camera (Retiga Exi; QImaging) at room temperature. Z stacks were taken, 0.5-µm increments, and the images were de-blurred by performing nearest neighbor deconvolution with 70% haze removal using Image-Pro 7.0 software (Media Cybernetics). Further image manipulations were performed using ImageJ software (http://rsbweb.nih.gov/ij). Projections were made from Z stacks using the max-intensity algorithm and the images were falsely colored, green for FtsZ1-eYFP and red for FtsZ2-CFP and ARC341–598–eCFP. Coexpression overlays were generated with the merge channels option. Colocalization of FtsZ proteins in each coexpresion strain was quantified by averaging the PCCs calculated in 10 cells using the colocalization finder plug-in for ImageJ, ±SD.
FRAP was performed at room temperature on a laser-scanning confocal microscope (FluoView 1000; Olympus) with a Plan FLN 60× (1.42 NA) oil-immersion objective with a 3.4× zoom. Immediately before collecting FRAP data, the PMT voltage was adjusted so that the maximum fluorescence signal in each cell imaged was just below saturation. Data were collected with FV1000 ASW software (Olympus). 2 µl of cell culture was mounted on poly-lysine–coated slides. FtsZ1-eYFP filaments were photobleached for 20 ms with a 515-nm laser at 50%. FtsZ2-eCFP filaments were photobleached for 20 ms with a 458-nm laser at 50%. Fluorescence intensity measurements were taken over a time-course of 250 s after photobleaching for each photobleached region of interest, a background sample, and an area of fluorescence signal that was away from the bleached location. The FRAP raw data were processed to produce the normalized recovery curves (Rabut and Ellenberg, 2005). FRAP analysis was performed for yeast strains coexpressing all combinations of FtsZ1-eYFP and FtsZ2-eCFP (WT and GTPase-deficient mutant). Coexpressing cells chosen for FRAP analysis displayed strong fluorescence signals from both fluorescent proteins present. FRAP measurements for the strains coexpressing WT FtsZ1-eYFP and FtsZ2-eCFP were performed by taking 10 FtsZ1-eYFP datasets followed by 10 FtsZ2-eCFP datasets from different cells in the same culture. For the remainder of the coexpression strains, FRAP data for each FtsZ construct were obtained sequentially in the same cell. Recovery of FtsZ1-eYFP (WT or mutant) was measured first, as the eYFP emission spectrum does not overlap with the excitation spectrum of eCFP. Curve-fitting of FRAP data were performed using pro Fit software (QuantumSoft), where the data were fit to the single exponential equation f(t) = A(1−e−kt). The time for one-half recovery of the fluorescence signal (t1/2) was calculated as t1/2 = ln(1/2)/−k. Analysis of statistically significant differences between average recovery half-time and percent recovery in different strains were performed using a two-tailed Student’s t test (P < 0.01).
To assess the effect of protein expression level on filament dynamics, individual half-time and percent recovery values were plotted against the photomultiplier tube (PMT) voltage setting for each of the ten cells analyzed by FRAP in all four single-expression stains. PMT voltage inversely correlates with fluorescence intensity (http://www.olympusmicro.com/primer/techniques/confocal/pmtintro.html).
Functional analysis of FtsZ1 and FtsZ2 C-terminal fusion proteins
An FtsZ1-mCerulean construct was made as in Schmitz et al. (2009), except that the 3′ piece was fused to mCerulean (Addgene) at the 3′ end of FtsZ1 by splicing by overlap extension and asymmetric PCR (Warrens et al., 1997). The Multisite Gateway recombinations were performed using Gateway LR+ Clonase (Invitrogen), pMDC204 (http://botserv1.uzh.ch/home/grossnik/curtisvector/index_2.html) modified with a Gateway R4-R3 cassette, and 5′, middle, and 3′-mCerulean pENTR vectors to create the PFtsZ1::FtsZ1-mCerulean genomic clone.
ftsZ1 knockout plants were transformed using a standard floral dipping protocol (Clough and Bent, 1998) with Agrobacterium tumefaciens strain GV3101 containing PFtsZ1::FtsZ1-mCerulean. Transformants were selected on plates containing 20 µg/ml hygromycin B based on hypocotyl length after germination in the dark (Kadirjan-Kalbach et al., 2012). Positive transformants were transplanted to soil and grown in environmentally controlled growth chambers under white fluorescent light (100 μmol m−2 s−1, 16:8 h light/dark) at 21°C and relative humidity of 60%. T2 seeds harvested from T1 plants that showed partial complementation of the chloroplast division phenotype were grown and analyzed for chloroplast division complementation and fluorescence signal. Rosette leaf samples for phenotypic analysis were harvested on the same day from Col-0, ftsZ1 knockout, and T2 transgenic plants sown and grown together. Leaf samples were fixed with 3.5% glutaraldehyde for 3 h followed by 1.5 h at 50°C in 0.1 M Na2-EDTA (Pyke and Leech, 1991). Leaf samples were imaged by differential interference contrast microscopy using a microscope (model DMI 300B; Leica) with an HCX PL FLUOTAR 40× (0.75 NA) dry objective (Leica) at room temperature and Leica Application Suite. Images were acquired with a camera (DFC320; Leica). Mesophyll cell area was determined using ImageJ software and the chloroplast number per cell was manually counted for each cell. Various leaf types from a T2 transgenic plant that was fully complemented were analyzed for FtsZ1-mCerulean fluorescence signal by epifluorescence microscopy as described above, but with an HCX PL FLUOTAR 100× (1.30 NA) oil-immersion objective (Leica).
An FtsZ2-GFP fusion construct was generated by subcloning an FtsZ2-GFP fusion product (FtsZ2 beginning at residue 89, with a QGDIT linker) into pUC19. The FtsZ2-GFP construct was transformed into E. coli strain WM746 (Ma and Margolin, 1999), an ftsZ-null strain carrying a low copy number plasmid that displayed temperature-sensitive replication and contained an E. coli FtsZ gene. Bacterial cells were grown at 33°C until the exponential growth phase, then used to inoculate fresh medium. Fresh cultures were grown for 5 h at 42°C with 500 µM IPTG to induce expression of FtsZ2-GFP while depleting the bacterial FtsZ protein. Bright-field images were obtained with differential interference contrast optics and fluorescence microscopy using a microscope (model BH2; Olympus) with a 100× (1.25 NA) oil-immersion objective at room temperature; GFP epifluorescence images were captured with a color video camera (model DEI 750; Optronics) and Scion Image 1.62 software (Scion Corporation). Noise in the fluorescence images was reduced by applying a median filter (radius = 2). Images were assembled using Adobe Photoshop 5.0 (Adobe Systems Inc.) and Canvas 6.0 (Deneba Software) software.
An FtsZ2-eYFP fusion construct was generated by subcloning the FtsZ2-1 full-length coding sequence into a derivative of pCambia-1302 (Cambia) in which GFP was replaced by eYFP using standard molecular biology techniques. The FtsZ2-eYFP construct was transformed into an Arabidopsis ftsZ2-1 knockdown mutant (Schmitz et al., 2009) as described above. Positive transformants were selected for by growth on plates containing hygromycin, as described above. T1 plants were analyzed for FtsZ2-eYFP signal by epifluorescence microscopy.
Online supplemental material
Fig. S1 shows the functionality of FtsZ1 and FtsZ2 C-terminal fluorescent fusion proteins. Fig. S2 shows that the distinct morphologies of FtsZ1, FtsZ2, FtsZ1 D275A, and FtsZ2 D322A are maintained in cells with variable protein expression levels. Fig. S3 shows an immunoblot of soluble bulk culture extracts, indicating that FtsZ1 and FtsZ2 are at a near-equal ratio in that coexpression stain. Fig. S4 shows that there is no consistent variation in half-time or total percent recovery over the range of protein levels examined during FRAP analysis. Video 1 shows time-lapse images of FtsZ1 recovery during FRAP analysis that corresponds to Fig. 3 A. Video 2 shows time-lapse images of FtsZ2 recovery during FRAP analysis that corresponds to Fig. 3 B. Video 3 shows time-lapse images of FtsZ1 D275A recovery during FRAP analysis that corresponds to Fig. 4 A. Video 4 shows time-lapse images of FtsZ2 D322A recovery during FRAP analysis that corresponds to Fig. 4 B. Table S1 summarizes all the FRAP data and indicates statistically significant differences. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201205114/DC1.
Supplementary Material
Acknowledgments
We thank Susan Forsburg for providing plasmids, Mohan Balasubramanian for providing constructs and yeast strains, William Margolin for providing the E. coli strain in Fig. S1, Stanislav Vitha and Aaron Schmitz for data in Fig. S1, Melinda Frame for assistance with FRAP experiments, and Yamato Yoshida and John Froehlich for discussions and comments on the manuscript. Confocal microscopy was performed in the Michigan State University Center for Advanced Microscopy and sequence analysis was performed in the Michigan State University Research Technology Support Facility.
This work was supported by grant number 1121943 from the US National Science Foundation.
Footnotes
Abbreviations used in this paper:
- eCFP
- enhanced CFP
- eYFP
- enhanced YFP
- PCC
- Pearson’s correlation coefficient
- PMT
- photomultiplier tube
- t1/2
- half-time of fluorescence recovery
- WT
- wild type
References
- Adams D.W., Errington J. 2009. Bacterial cell division: assembly, maintenance and disassembly of the Z ring. Nat. Rev. Microbiol. 7:642–653 10.1038/nrmicro2198 [DOI] [PubMed] [Google Scholar]
- Anderson D.E., Gueiros-Filho F.J., Erickson H.P. 2004. Assembly dynamics of FtsZ rings in Bacillus subtilis and Escherichia coli and effects of FtsZ-regulating proteins. J. Bacteriol. 186:5775–5781 10.1128/JB.186.17.5775-5781.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aylett C.H.S., Löwe J., Amos L.A. 2011. New insights into the mechanisms of cytomotive actin and tubulin filaments. Int Rev Cell Mol Biol. 292:1–71 10.1016/B978-0-12-386033-0.00001-3 [DOI] [PubMed] [Google Scholar]
- Basi G., Schmid E., Maundrell K. 1993. TATA box mutations in the Schizosaccharomyces pombe nmt1 promoter affect transcription efficiency but not the transcription start point or thiamine repressibility. Gene. 123:131–136 10.1016/0378-1119(93)90552-E [DOI] [PubMed] [Google Scholar]
- Bi E.F., Lutkenhaus J. 1991. FtsZ ring structure associated with division in Escherichia coli. Nature. 354:161–164 10.1038/354161a0 [DOI] [PubMed] [Google Scholar]
- Bolte S., Cordelières F.P. 2006. A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 224:213–232 10.1111/j.1365-2818.2006.01706.x [DOI] [PubMed] [Google Scholar]
- Buske P.J., Levin P.A. 2012. Extreme C terminus of bacterial cytoskeletal protein FtsZ plays fundamental role in assembly independent of modulatory proteins. J. Biol. Chem. 287:10945–10957 10.1074/jbc.M111.330324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Erickson H.P. 2009. FtsZ filament dynamics at steady state: subunit exchange with and without nucleotide hydrolysis. Biochemistry. 48:6664–6673 10.1021/bi8022653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Bjornson K., Redick S.D., Erickson H.P. 2005. A rapid fluorescence assay for FtsZ assembly indicates cooperative assembly with a dimer nucleus. Biophys. J. 88:505–514 10.1529/biophysj.104.044149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Anderson D.E., Rajagopalan M., Erickson H.P. 2007. Assembly dynamics of Mycobacterium tuberculosis FtsZ. J. Biol. Chem. 282:27736–27743 10.1074/jbc.M703788200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16:735–743 10.1046/j.1365-313x.1998.00343.x [DOI] [PubMed] [Google Scholar]
- Colletti K.S., Tattersall E.A., Pyke K.A., Froelich J.E., Stokes K.D., Osteryoung K.W. 2000. A homologue of the bacterial cell division site-determining factor MinD mediates placement of the chloroplast division apparatus. Curr. Biol. 10:507–516 10.1016/S0960-9822(00)00466-8 [DOI] [PubMed] [Google Scholar]
- Dajkovic A., Lan G., Sun S.X., Wirtz D., Lutkenhaus J. 2008. MinC spatially controls bacterial cytokinesis by antagonizing the scaffolding function of FtsZ. Curr. Biol. 18:235–244 10.1016/j.cub.2008.01.042 [DOI] [PubMed] [Google Scholar]
- de Boer P.A. 2010. Advances in understanding E. coli cell fission. Curr. Opin. Microbiol. 13:730–737 10.1016/j.mib.2010.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Kafafi S., Mukherjee S., El-Shami M., Putaux J.-L., Block M.A., Pignot-Paintrand I., Lerbs-Mache S., Falconet D. 2005. The plastid division proteins, FtsZ1 and FtsZ2, differ in their biochemical properties and sub-plastidial localization. Biochem. J. 387:669–676 10.1042/BJ20041281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson H.P., Anderson D.E., Osawa M. 2010. FtsZ in bacterial cytokinesis: cytoskeleton and force generator all in one. Microbiol. Mol. Biol. Rev. 74:504–528 10.1128/MMBR.00021-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falconet D. 2012. Origin, evolution and division of plastids. Photosynthesis. Eaton-Rye J.J., Tripathy B.C., Sharkey T.D., 35–61 Springer [Google Scholar]
- Forsburg S.L. 1993. Comparison of Schizosaccharomyces pombe expression systems. Nucleic Acids Res. 21:2955–2956 10.1093/nar/21.12.2955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu G., Huang T., Buss J., Coltharp C., Hensel Z., Xiao J. 2010. In vivo structure of the E. coli FtsZ-ring revealed by photoactivated localization microscopy (PALM). PLoS ONE. 5:e12682 10.1371/journal.pone.0012682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara M., Yoshida S. 2001. Chloroplast targeting of chloroplast division FtsZ2 proteins in Arabidopsis. Biochem. Biophys. Res. Commun. 287:462–467 10.1006/bbrc.2001.5588 [DOI] [PubMed] [Google Scholar]
- Glynn J.M., Miyagishima S.-Y., Yoder D.W., Osteryoung K.W., Vitha S. 2007. Chloroplast division. Traffic. 8:451–461 10.1111/j.1600-0854.2007.00545.x [DOI] [PubMed] [Google Scholar]
- Glynn J.M., Yang Y., Vitha S., Schmitz A.J., Hemmes M., Miyagishima S.-Y., Osteryoung K.W. 2009. PARC6, a novel chloroplast division factor, influences FtsZ assembly and is required for recruitment of PDV1 during chloroplast division in Arabidopsis. Plant J. 59:700–711 10.1111/j.1365-313X.2009.03905.x [DOI] [PubMed] [Google Scholar]
- Hu Z., Mukherjee A., Pichoff S., Lutkenhaus J. 1999. The MinC component of the division site selection system in Escherichia coli interacts with FtsZ to prevent polymerization. Proc. Natl. Acad. Sci. USA. 96:14819–14824 10.1073/pnas.96.26.14819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huecas S., Schaffner-Barbero C., García W., Yébenes H., Palacios J.M., Díaz J.F., Menéndez M., Andreu J.M. 2007. The interactions of cell division protein FtsZ with guanine nucleotides. J. Biol. Chem. 282:37515–37528 10.1074/jbc.M706399200 [DOI] [PubMed] [Google Scholar]
- Itoh R., Fujiwara M., Nagata N., Yoshida S. 2001. A chloroplast protein homologous to the eubacterial topological specificity factor minE plays a role in chloroplast division. Plant Physiol. 127:1644–1655 10.1104/pp.010386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings P.C., Cox G.C., Monahan L.G., Harry E.J. 2011. Super-resolution imaging of the bacterial cytokinetic protein FtsZ. Micron. 42:336–341 10.1016/j.micron.2010.09.003 [DOI] [PubMed] [Google Scholar]
- Kadirjan-Kalbach D.K., Yoder D.W., Ruckle M.E., Larkin R.M., Osteryoung K.W. 2012. FtsHi1/ARC1 is an essential gene in Arabidopsis that links chloroplast biogenesis and division. Plant J. [DOI] [PubMed] [Google Scholar]
- Li Z., Trimble M.J., Brun Y.V., Jensen G.J. 2007. The structure of FtsZ filaments in vivo suggests a force-generating role in cell division. EMBO J. 26:4694–4708 10.1038/sj.emboj.7601895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse S., Hause B., Hause G., Fester T. 2006. FtsZ characterization and immunolocalization in the two phases of plastid reorganization in arbuscular mycorrhizal roots of Medicago truncatula. Plant Cell Physiol. 47:1124–1134 10.1093/pcp/pcj083 [DOI] [PubMed] [Google Scholar]
- Löwe J. 1998. Crystal structure determination of FtsZ from Methanococcus jannaschii. J. Struct. Biol. 124:235–243 10.1006/jsbi.1998.4041 [DOI] [PubMed] [Google Scholar]
- Lu C., Stricker J., Erickson H.P. 1998. FtsZ from Escherichia coli, Azotobacter vinelandii, and Thermotoga maritima—quantitation, GTP hydrolysis, and assembly. Cell Motil. Cytoskeleton. 40:71–86 [DOI] [PubMed] [Google Scholar]
- Lutkenhaus J. 2007. Assembly dynamics of the bacterial MinCDE system and spatial regulation of the Z ring. Annu. Rev. Biochem. 76:539–562 10.1146/annurev.biochem.75.103004.142652 [DOI] [PubMed] [Google Scholar]
- Ma X., Margolin W. 1999. Genetic and functional analyses of the conserved C-terminal core domain of Escherichia coli FtsZ. J. Bacteriol. 181:7531–7544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X., Ehrhardt D.W., Margolin W. 1996. Colocalization of cell division proteins FtsZ and FtsA to cytoskeletal structures in living Escherichia coli cells by using green fluorescent protein. Proc. Natl. Acad. Sci. USA. 93:12998–13003 10.1073/pnas.93.23.12998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maple J., Møller S.G. 2010. The complexity and evolution of the plastid-division machinery. Biochem. Soc. Trans. 38:783–788 10.1042/BST0380783 [DOI] [PubMed] [Google Scholar]
- Maple J., Chua N.-H., Møller S.G. 2002. The topological specificity factor AtMinE1 is essential for correct plastid division site placement in Arabidopsis. Plant J. 31:269–277 10.1046/j.1365-313X.2002.01358.x [DOI] [PubMed] [Google Scholar]
- Maple J., Aldridge C., Møller S.G. 2005. Plastid division is mediated by combinatorial assembly of plastid division proteins. Plant J. 43:811–823 10.1111/j.1365-313X.2005.02493.x [DOI] [PubMed] [Google Scholar]
- Maple J., Vojta L., Soll J., Møller S.G. 2007. ARC3 is a stromal Z-ring accessory protein essential for plastid division. EMBO Rep. 8:293–299 10.1038/sj.embor.7400902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolin W. 2005. FtsZ and the division of prokaryotic cells and organelles. Nat. Rev. Mol. Cell Biol. 6:862–871 10.1038/nrm1745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateos-Gil P., Paez A., Hörger I., Rivas G., Vicente M., Tarazona P., Vélez M. 2012. Depolymerization dynamics of individual filaments of bacterial cytoskeletal protein FtsZ. Proc. Natl. Acad. Sci. USA. 109:8133–8138 10.1073/pnas.1204844109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maundrell K. 1990. nmt1 of fission yeast. A highly transcribed gene completely repressed by thiamine. J. Biol. Chem. 265:10857–10864 [PubMed] [Google Scholar]
- Mazouni K., Domain F., Cassier-Chauvat C., Chauvat F. 2004. Molecular analysis of the key cytokinetic components of cyanobacteria: FtsZ, ZipN and MinCDE. Mol. Microbiol. 52:1145–1158 10.1111/j.1365-2958.2004.04042.x [DOI] [PubMed] [Google Scholar]
- McAndrew R.S., Froehlich J.E., Vitha S., Stokes K.D., Osteryoung K.W. 2001. Colocalization of plastid division proteins in the chloroplast stromal compartment establishes a new functional relationship between FtsZ1 and FtsZ2 in higher plants. Plant Physiol. 127:1656–1666 10.1104/pp.010542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAndrew R.S., Olson B.J.S.C., Kadirjan-Kalbach D.K., Chi-Ham C.L., Vitha S., Froehlich J.E., Osteryoung K.W. 2008. In vivo quantitative relationship between plastid division proteins FtsZ1 and FtsZ2 and identification of ARC6 and ARC3 in a native FtsZ complex. Biochem. J. 412:367–378 10.1042/BJ20071354 [DOI] [PubMed] [Google Scholar]
- Milam S.L., Osawa M., Erickson H.P. 2012. Negative-stain electron microscopy of inside-out FtsZ rings reconstituted on artificial membrane tubules show ribbons of protofilaments. Biophys. J. 103:59–68 10.1016/j.bpj.2012.05.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mingorance J., Tadros M., Vicente M., González J.M., Rivas G., Vélez M. 2005. Visualization of single Escherichia coli FtsZ filament dynamics with atomic force microscopy. J. Biol. Chem. 280:20909–20914 10.1074/jbc.M503059200 [DOI] [PubMed] [Google Scholar]
- Mingorance J., Rivas G., Vélez M., Gómez-Puertas P., Vicente M. 2010. Strong FtsZ is with the force: mechanisms to constrict bacteria. Trends Microbiol. 18:348–356 10.1016/j.tim.2010.06.001 [DOI] [PubMed] [Google Scholar]
- Miyagishima S.-Y. 2005. Origin and evolution of the chloroplast division machinery. J. Plant Res. 118:295–306 10.1007/s10265-005-0226-2 [DOI] [PubMed] [Google Scholar]
- Miyagishima S.-Y. 2011. Mechanism of plastid division: from a bacterium to an organelle. Plant Physiol. 155:1533–1544 10.1104/pp.110.170688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori T., Kuroiwa H., Takahara M., Miyagishima S.Y., Kuroiwa T. 2001. Visualization of an FtsZ ring in chloroplasts of Lilium longiflorum leaves. Plant Cell Physiol. 42:555–559 10.1093/pcp/pce095 [DOI] [PubMed] [Google Scholar]
- Mukherjee A., Lutkenhaus J. 1998. Dynamic assembly of FtsZ regulated by GTP hydrolysis. EMBO J. 17:462–469 10.1093/emboj/17.2.462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee A., Lutkenhaus J. 1999. Analysis of FtsZ assembly by light scattering and determination of the role of divalent metal cations. J. Bacteriol. 181:823–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliva M.A., Cordell S.C., Löwe J. 2004. Structural insights into FtsZ protofilament formation. Nat. Struct. Mol. Biol. 11:1243–1250 10.1038/nsmb855 [DOI] [PubMed] [Google Scholar]
- Olson B.J.S.C. 2008. Biochemical analysis of the chloroplast division proteins FtsZ1 and FtsZ2. Biocehmistry and Molecular Biology. Vol. Ph.D Michigan State University, East Lansing, MI: 259 [Google Scholar]
- Olson B.J.S.C., Wang Q., Osteryoung K.W. 2010. GTP-dependent heteropolymer formation and bundling of chloroplast FtsZ1 and FtsZ2. J. Biol. Chem. 285:20634–20643 10.1074/jbc.M110.122614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa M., Erickson H.P. 2011. Inside-out Z rings—constriction with and without GTP hydrolysis. Mol. Microbiol. 81:571–579 10.1111/j.1365-2958.2011.07716.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa M., Anderson D.E., Erickson H.P. 2008. Reconstitution of contractile FtsZ rings in liposomes. Science. 320:792–794 10.1126/science.1154520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa M., Anderson D.E., Erickson H.P. 2009. Curved FtsZ protofilaments generate bending forces on liposome membranes. EMBO J. 28:3476–3484 10.1038/emboj.2009.277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osteryoung K.W., McAndrew R.S. 2001. The plastid division machine. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52:315–333 10.1146/annurev.arplant.52.1.315 [DOI] [PubMed] [Google Scholar]
- Osteryoung K.W., Vierling E. 1995. Conserved cell and organelle division. Nature. 376:473–474 10.1038/376473b0 [DOI] [PubMed] [Google Scholar]
- Osteryoung K.W., Stokes K.D., Rutherford S.M., Percival A.L., Lee W.Y. 1998. Chloroplast division in higher plants requires members of two functionally divergent gene families with homology to bacterial ftsZ. Plant Cell. 10:1991–2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popp D., Iwasa M., Narita A., Erickson H.P., Maéda Y. 2009. FtsZ condensates: an in vitro electron microscopy study. Biopolymers. 91:340–350 10.1002/bip.21136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyke K.A. 2010. Plastid division. AoB Plants. 2010:plq016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyke K.A., Leech R.M. 1991. Rapid image analysis screening procedure for identifying chloroplast number mutants in mesophyll cells of Arabidopsis thaliana (L.) Heynh. Plant Physiol. 96:1193–1195 10.1104/pp.96.4.1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyke K.A., Leech R.M. 1992. Chloroplast division and expansion is radically altered by nuclear mutations in Arabidopsis thaliana. Plant Physiol. 99:1005–1008 10.1104/pp.99.3.1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabut G., Ellenberg J. 2005. Photobleaching techniques to study mobility and molecular dynamics of proteins in live cells: FRAP, iFRAP, and FLIP. Live Cell Imaging: A Laboratory Manual. R.D.a.S Goldman D.L., editor Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY: 101–126 [DOI] [PubMed] [Google Scholar]
- Redick S.D., Stricker J., Briscoe G., Erickson H.P. 2005. Mutants of FtsZ targeting the protofilament interface: effects on cell division and GTPase activity. J. Bacteriol. 187:2727–2736 10.1128/JB.187.8.2727-2736.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffers D.J., de Wit J.G., den Blaauwen T., Driessen A.J. 2001. Substitution of a conserved aspartate allows cation-induced polymerization of FtsZ. FEBS Lett. 494:34–37 10.1016/S0014-5793(01)02310-9 [DOI] [PubMed] [Google Scholar]
- Schmitz A.J., Glynn J.M., Olson B.J.S.C., Stokes K.D., Osteryoung K.W. 2009. Arabidopsis FtsZ2-1 and FtsZ2-2 are functionally redundant, but FtsZ-based plastid division is not essential for chloroplast partitioning or plant growth and development. Mol Plant. 2:1211–1222 10.1093/mp/ssp077 [DOI] [PubMed] [Google Scholar]
- Shimada H., Koizumi M., Kuroki K., Mochizuki M., Fujimoto H., Ohta H., Masuda T., Takamiya K.-i. 2004. ARC3, a chloroplast division factor, is a chimera of prokaryotic FtsZ and part of eukaryotic phosphatidylinositol-4-phosphate 5-kinase. Plant Cell Physiol. 45:960–967 10.1093/pcp/pch130 [DOI] [PubMed] [Google Scholar]
- Smith A.G., Johnson C.B., Vitha S., Holzenburg A. 2010. Plant FtsZ1 and FtsZ2 expressed in a eukaryotic host: GTPase activity and self-assembly. FEBS Lett. 584:166–172 10.1016/j.febslet.2009.11.044 [DOI] [PubMed] [Google Scholar]
- Smith A.G., Johnson C.B., Vitha S., Holzenburg A. 2011. Oligomerization of plant FtsZ1 and FtsZ2 plastid division proteins. Arch. Biochem. Biophys. 513:94–101 10.1016/j.abb.2011.07.001 [DOI] [PubMed] [Google Scholar]
- Srinivasan R., Mishra M., Murata-Hori M., Balasubramanian M.K. 2007. Filament formation of the Escherichia coli actin-related protein, MreB, in fission yeast. Curr. Biol. 17:266–272 10.1016/j.cub.2006.11.069 [DOI] [PubMed] [Google Scholar]
- Srinivasan R., Mishra M., Wu L., Yin Z., Balasubramanian M.K. 2008. The bacterial cell division protein FtsZ assembles into cytoplasmic rings in fission yeast. Genes Dev. 22:1741–1746 10.1101/gad.1660908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes K.D., McAndrew R.S., Figueroa R., Vitha S., Osteryoung K.W. 2000. Chloroplast division and morphology are differentially affected by overexpression of FtsZ1 and FtsZ2 genes in Arabidopsis. Plant Physiol. 124:1668–1677 10.1104/pp.124.4.1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strepp R., Scholz S., Kruse S., Speth V., Reski R. 1998. Plant nuclear gene knockout reveals a role in plastid division for the homolog of the bacterial cell division protein FtsZ, an ancestral tubulin. Proc. Natl. Acad. Sci. USA. 95:4368–4373 10.1073/pnas.95.8.4368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stricker J., Erickson H.P. 2003. In vivo characterization of Escherichia coli ftsZ mutants: effects on Z-ring structure and function. J. Bacteriol. 185:4796–4805 10.1128/JB.185.16.4796-4805.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suppanz I., Sarnighausen E., Reski R. 2007. An integrated physiological and genetic approach to the dynamics of FtsZ targeting and organisation in a moss, Physcomitrella patens. Protoplasma. 232:1–9 10.1007/s00709-007-0284-5 [DOI] [PubMed] [Google Scholar]
- Vaughan S., Wickstead B., Gull K., Addinall S.G. 2004. Molecular evolution of FtsZ protein sequences encoded within the genomes of archaea, bacteria, and eukaryota. J. Mol. Evol. 58:19–29 10.1007/s00239-003-2523-5 [DOI] [PubMed] [Google Scholar]
- Vitha S., McAndrew R.S., Osteryoung K.W. 2001. FtsZ ring formation at the chloroplast division site in plants. J. Cell Biol. 153:111–120 10.1083/jcb.153.1.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitha S., Froehlich J.E., Koksharova O., Pyke K.A., van Erp H., Osteryoung K.W. 2003. ARC6 is a J-domain plastid division protein and an evolutionary descendant of the cyanobacterial cell division protein Ftn2. Plant Cell. 15:1918–1933 10.1105/tpc.013292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Huang J., Mukherjee A., Cao C., Lutkenhaus J. 1997. Analysis of the interaction of FtsZ with itself, GTP, and FtsA. J. Bacteriol. 179:5551–5559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warrens A.N., Jones M.D., Lechler R.I. 1997. Splicing by overlap extension by PCR using asymmetric amplification: an improved technique for the generation of hybrid proteins of immunological interest. Gene. 186:29–35 10.1016/S0378-1119(96)00674-9 [DOI] [PubMed] [Google Scholar]
- Yang Y., Glynn J.M., Olson B.J.S.C., Schmitz A.J., Osteryoung K.W. 2008. Plastid division: across time and space. Curr. Opin. Plant Biol. 11:577–584 10.1016/j.pbi.2008.10.001 [DOI] [PubMed] [Google Scholar]
- Yoder D.W., Kadirjan-Kalbach D., Olson B.J.S.C., Miyagishima S.-Y., Deblasio S.L., Hangarter R.P., Osteryoung K.W. 2007. Effects of mutations in Arabidopsis FtsZ1 on plastid division, FtsZ ring formation and positioning, and FtsZ filament morphology in vivo. Plant Cell Physiol. 48:775–791 10.1093/pcp/pcm049 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.