Isolation of the intact TOB complex reveals a 1:1:1 stoichiometry of Tob55, Tob38, and Tob37 with a 140-kD molecular mass, providing new insight into complex structure and function.
Abstract
The TOB–SAM complex is an essential component of the mitochondrial outer membrane that mediates the insertion of β-barrel precursor proteins into the membrane. We report here its isolation and determine its size, composition, and structural organization. The complex from Neurospora crassa was composed of Tob55–Sam50, Tob38–Sam35, and Tob37–Sam37 in a stoichiometry of 1:1:1 and had a molecular mass of 140 kD. A very minor fraction of the purified complex was associated with one Mdm10 protein. Using molecular homology modeling for Tob55 and cryoelectron microscopy reconstructions of the TOB complex, we present a model of the TOB–SAM complex that integrates biochemical and structural data. We discuss our results and the structural model in the context of a possible mechanism of the TOB insertase.
Introduction
The evolution of mitochondria and chloroplasts by endosymbiosis of prokaryotes is reflected in the presence of membrane proteins with a β-barrel structure that are exclusively located in their outer membranes (Rapaport, 2002; Schleiff et al., 2003). It is also reflected in the relationship of the molecular machineries that mediate insertion of these classes of proteins into the outer membranes in prokaryotes and eukaryotes (Gray et al., 1999; Paschen et al., 2005a). In bacteria a key component is Omp85–BamA–YaeT, which is related to the eukaryotic mitochondrial Tob55–Sam50. These proteins are subunits of larger complexes, the bacterial β-barrel assembly machinery (BAM) and the mitochondrial TOB complex (topogenesis of the outer mitochondrial membrane β-barrel proteins; Paschen et al., 2003), also termed the SAM complex (sorting and assembly machinery; Kozjak et al., 2003). The latter complex has been demonstrated to be responsible for the insertion of β-barrel proteins into the outer mitochondrial membrane (Paschen et al., 2003; Wiedemann et al., 2003; Hoppins et al., 2007), but might also be involved in the assembly of the α-helical proteins Tom22, Tom5, Tom6, and Tom7 (Hoppins et al., 2007; Stojanovski et al., 2007; Thornton et al., 2010).
Tob55 (Sam50, Tom50) is the main subunit of the TOB complex (Neupert and Herrmann, 2007). Based on sequence analysis, secondary structure prediction, and circular dichroism measurements the C-terminal domain of Tob55 is able to form a β-barrel (Kozjak et al., 2003; Paschen et al., 2003; Gentle et al., 2004; Voulhoux and Tommassen, 2004; Jacob-Dubuisson et al., 2009). The hydrophilic domain at the N terminus is facing the intermembrane space and forms a characteristic structure, the polypeptide transport-associated (POTRA) domain. The POTRA domain is a module of ∼75 amino acid residues found in varying numbers of repeats at the N terminus of all members of the Omp85–TpsB transporter superfamily, including Tob55 (Sánchez-Pulido et al., 2003; Habib et al., 2007). To date, a high resolution structure of Tob55 is not available (Endo et al., 2011).
The other components of the TOB complex, Tob38 (Sam35, Tom38) and Tob37 (Mas37, Sam37, Tom37), are associated with Tob55, facing the cytosolic side of the mitochondrial outer membrane (Wiedemann et al., 2003; Ishikawa et al., 2004; Milenkovic et al., 2004; Waizenegger et al., 2004; Lackey et al., 2011). The structures of these subunits are unknown. Several membrane proteins such as Mdm10 (Meisinger et al., 2006, 2007; Yamano et al., 2010), Tom40 (Thornton et al., 2010), and Mim1 (Becker et al., 2008) were reported to associate with Tob55, Tob38, and Tob37. However, the nature and extent of these associations with the TOB complex, either as substrates, subunits, or temporary interaction partners, are unclear. The purification of the TOB complex is a prerequisite for the determination of its stoichiometric composition and organization. To date, the TOB complex, which is present in only very small quantities, has not been isolated.
We developed a procedure for the purification of the intact TOB complex from the filamentous fungus Neurospora crassa. Tob55, Tob38, and Tob37 were identified as stoichiometric components of the complex. Isotope dilution mass spectrometry (IDMS) analysis revealed a relative molar ratio between these subunits of 1:1:1. This, together with analysis by blue native gel electrophoresis (BNGE), showed that the TOB complex contains one copy of each subunit and has a molecular mass of 140 kD, which is much smaller than previously assumed. Homology modeling revealed a high similarity of Tob55 to the TpsB transporter FhaC. Three-dimensional reconstructions were obtained for the TOB complex by single-particle analysis of images from cryoelectron microscopy. These data led us to a structural model of the TOB complex that is in accordance with its biochemical characterization. Our results provide important initial steps toward understanding the complex mechanism of insertion of β-barrel proteins into the mitochondrial outer membrane.
Results
Tob38, Tob37, and Tob55 constitute the TOB complex
The TOB complex was isolated by Ni-NTA affinity purification from mitochondrial outer membrane vesicles (OMVs) of strains expressing Tob38 or Tob37 with an N-terminal His9 tag. The mitochondria were solubilized using either Triton X-100 (TX-100; Fig. 1, A and B) or digitonin (Fig.1, C and D). After affinity purification, the subunit proteins were separated by SDS-PAGE and detected by either immunodecoration (Fig. 1, A and C) or Coomassie staining (Fig. 1, B and D). Tob55 is expressed in three different isoforms of 54.7 kD (long), 54.1 kD (intermediate), and 50.7 kD (short) due to alternative mRNA splicing. Because the long and intermediate forms differ only by five amino acid residues in length, they could not be distinguished by the electrophoretic procedure used. Hence, only two bands representing the three Tob55 isoforms were visible upon SDS-PAGE (Hoppins et al., 2007). The Coomassie-stained bands of the eluted proteins were excised and the proteins identified by LC tandem mass spectrometry (LC-MS/MS).
Figure 1.
Tob38, Tob37, and Tob55 are the subunits of the TOB complex. (A–D) OMVs from N. crassa with His-tagged Tob37 or Tob38 were solubilized with TX-100 (A and B) or digitonin (C and D). Proteins were isolated by Ni-NTA affinity purification and analyzed by SDS-PAGE followed by immunodecoration (A and B) or Coomassie blue staining (C and D). The calculated molecular weights for the proteins decorated are shown in A and B. The positions of molecular weight standards are shown in C and D. Solubilized OMVs before (S) or after (L) a clarifying spin. FT, flow through (proteins not bound to the Ni-NTA column); E, eluate of bound proteins. 55lg+int, Tob55-long and intermediate isoform; 55sh, Tob55-short isoform; 37, Tob37; 38, Tob38.
Tob55, Tob38, and Tob37 were the only proteins that were detected in the eluate (Fig. 1). A very small amount of Mdm10 was consistently identified by immunodecoration. The quantity of this protein, however, was so low that it was not detected in our LC-MS/MS analysis. Very small amounts of porin, the most abundant protein of the mitochondrial outer membrane, were observed when digitonin, but not when TX-100 was used for solubilization. The proteins Tom40 and Mim1, which have been reported to interact with the TOB complex in yeast (Becker et al., 2008; Thornton et al., 2010), were not detected. This is in agreement with reports that depletion of Mim1 does not affect the assembly of the TOB complex (Waizenegger et al., 2005). These results therefore do not support models in which Tom40 and Mim1 are part of the TOB complex and suggest that Tob55, Tob38, and Tob37 form the TOB complex.
The purified complex was then analyzed by BNGE. Respiratory chain complexes from bovine heart mitochondria (BHM) with defined molecular masses were used as size markers because they have been reported to yield more reliable estimates of the apparent molecular mass of membrane complexes than soluble marker proteins (Wittig et al., 2010). The TOB complex was extracted from isolated OMVs containing either His-tagged Tob38 or His-tagged Tob37 using TX-100 (Fig. 2, A and B) or digitonin (Fig. 2, C and D). Isolation using both detergents yielded a major species of the TOB complex with an apparent molecular mass of 130–150 kD. The presence of all three subunits, Tob55, Tob38, and Tob37, was demonstrated by immunodecoration. The complex solubilized with TX-100 had the tendency to form higher molecular mass species (Fig. 2 B, asterisks).
Figure 2.
The native TOB complex. (A–D) The TOB complex was isolated from cells containing either His-tagged Tob38 or Tob37 using TX-100 (A and B) or digitonin (C and D). BHM were solubilized with TX-100 (A and B) or digitonin (C and D). The samples were analyzed by BNGE and immunodecoration (A and C) or Coomassie blue staining (B and D). Asterisks indicate possible oligomeric forms of the TOB complex. (E) The TOB complex remains intact during the isolation of OMVs from mitochondria. Mitochondria (M) and OMVs (O) from either wild-type (wt) N. crassa or strains bearing His-tagged Tob55 (55), Tob38 (38), or Tob37 (37) were solubilized with digitonin. They were then analyzed by BNGE and immunodecorated with Tob38 antiserum. (A–E) Membrane protein complexes from BHM were used as marker proteins for relative molecular weights.
Very minor amounts of a species with a higher apparent molecular mass of ∼200 kD were decorated by antibodies against all three TOB complex components and in addition by antibodies against Mdm10 (Fig. 2, A and C). We refer to this species as TOB–Mdm10 complex. Another minor species of ∼90 kD was observed that was decorated by antibodies against Tob55 and Tob38, but not Tob37 (Fig. 2 A). It is not clear whether this species occurs as such in the membrane or whether it arises from the TOB complex by the loss of Tob37 during isolation. The results obtained by immunodecoration were confirmed by Coomassie staining of the purified complexes (Fig. 2, B and D). To exclude a possible loss of subunits from the TOB complex during the preparation of OMVs, mitochondria and OMVs were solubilized with digitonin and subjected to BNGE. The TOB complex was identified by immunoblotting. The TOB complex preparations obtained from OMVs and mitochondria of wild-type cells and cells expressing His-tagged Tob proteins showed no difference in their electrophoretic mobility (Fig. 2 E). Taken together, these data show that Tob38, Tob37, and Tob55 constitute the TOB complex with an apparent molecular weight of 130–150 kD.
Characterization of the isolated TOB complex
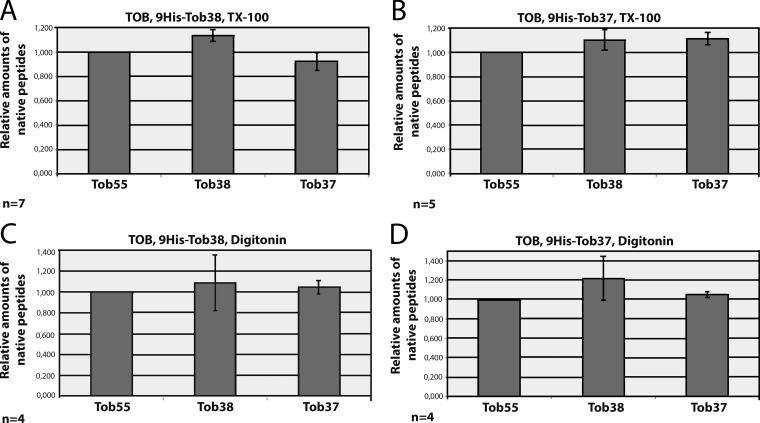
To determine the stoichiometry of the TOB complex, the purified protein complexes were excised from gels after BNGE and Coomassie staining and analyzed by IDMS. Unique, quantified isotope (13C/15N)–labeled internal peptides of Tob55, Tob38, and Tob37 were used as standards (Tob55, EDG FGV FIS DAR; Tob37, VYA DSQ AYK; Tob38, DPE YTD LLDR). TOB complex was purified from His9-Tob38– and His9-Tob37–expressing cells with TX-100 (Fig. 3, A and B) or digitonin (Fig. 3, C and D). The molar ratios of the components were found to be 1:1:1 between Tob55, Tob38, and Tob37 in the TOB complex (Fig. 3, A–D). LC-MS/MS did not detect any proteins other than Tob55, Tob38, and Tob37 in the TOB complex. A 1:1:1 ratio of Tob55, Tob38, and Tob37 with one of each subunit results in a calculated molecular mass of 140 kD. This is in agreement with the electrophoretic mobility of the complex relative to the standard membrane complexes from bovine heart mitochondria. The estimated mass of 200 kD of the TOB–Mdm10 complex determined by BNGE analysis suggests that one Mdm10 (52.7 kD) associates with the TOB complex.
Figure 3.
Tob55, Tob38, and Tob37 assemble in a 1:1:1 stoichiometry. (A–F) The TOB complex was isolated from cells expressing His9-Tob38 (A and C) or His9-Tob37 (B and D) using either TX-100 (A and B) or digitonin (C and D). The samples were subjected to BNGE, stained with Coomassie blue, and the bands were cut out from the gel and analyzed by IDMS. The amount of each subunit was determined by comparing the areas under the peaks for the labeled and native peptides. The relative amounts of the native peptides and thereby the native proteins were normalized relative to Tob55, which was set to 1. The error bars depict the standard deviation of the measurements from several (n) samples.
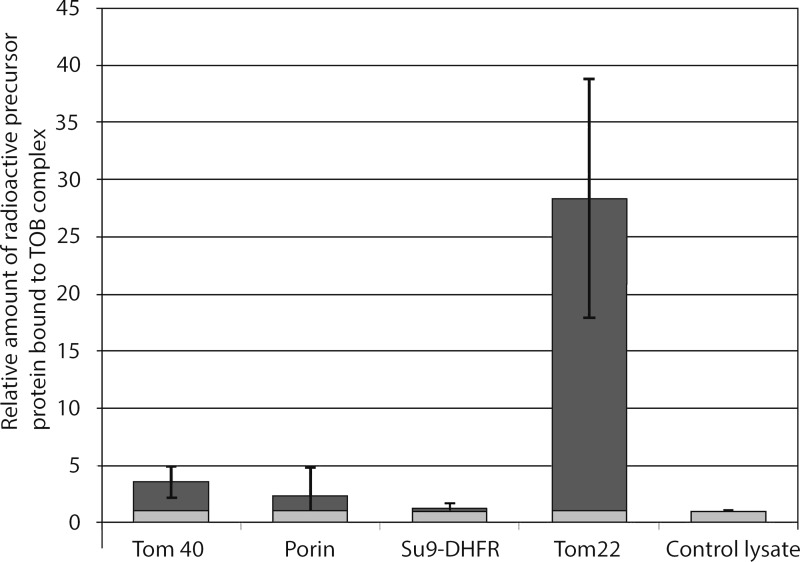
To assess the functionality of the isolated TOB complex, we analyzed its ability to interact with substrates. The TOB complex was immobilized on a Ni-NTA matrix, thoroughly washed, and then incubated with radioactive mitochondrial precursor proteins. Tom40 precursor showed a weak binding, slightly above background level. However, distinct binding of Tom22 to the TOB complex was observed (Fig. 4). The Tom22 precursor has previously been described as a substrate of the TOB complex, although it has no β-barrel structure (Stojanovski et al., 2007).
Figure 4.
Specific interaction of the Tom22 precursor with the isolated TOB complex. TOB complex was isolated from OMVs on a Ni-NTA matrix using TX-100. It was incubated while bound to the matrix with radioactive precursors of the indicated proteins (bars in dark gray). As a control and reference, mock isolation without OMVs was performed and the matrix was treated the same way with radioactive precursor proteins (bars in light gray). After washing, the bound precursor proteins were eluted and visualized by autoradiography. Binding in the mock control to the Ni-NTA matrix was set to 1. The amounts of precursor bound to the TOB complex immobilized on the Ni-NTA matrix were related to the mock control level. Error bars: standard deviation of five experiments. Radioactive precursor proteins: Tom40, porin, Su9-DHFR (matrix-targeting sequence of ATPase subunit 9 fused to mouse dihydrofolate reductase), Tom22. Control lysate (no plasmid was added to the transcription–translation reaction).
Antibody supershift assays were performed to further investigate the minor species of the TOB complex associated with Mdm10. OMVs from strains that expressed the His-tagged versions of Tob55, Tob38, or Tob37 were solubilized with digitonin. Subsequently, a monoclonal antibody against the His tag was added and BNGE was performed. Immunodecoration demonstrated the presence of Tob38, Tob37, and Tob55 in the TOB complex (Fig. 5, A–C). The TOB complex subunits were also detected in the far less abundant TOB–Mdm10 complex (Fig. 5, A–C). The vast majority of Mdm10, however, was present as a band with an apparent molecular mass of less than 100 kD, containing no subunits of the TOB complex (Fig. 5 D). No interaction of other β-barrel proteins, Tom40 and porin, with the TOB complex were detected in the antibody supershift assays (Fig. 5, E and F). These results confirm the composition of the TOB complex obtained by the analysis of the purified complex.
Figure 5.
A minor fraction of the TOB complex is associated with Mdm10 in a 200-kD complex, but not with Tom40 or porin. (A–F) OMVs from either wild-type (wt) or cells bearing a His-tagged Tob55 (55), Tob38 (38), or Tob37 (37) were lysed with digitonin. Where indicated, monoclonal His antibody was added to the lysates and the samples were subjected to BNGE. His antibody only was loaded as a control (c) to rule out unspecific interactions of the immunodecoration. Immunodecoration was performed with the indicated antibodies. Membrane protein complexes of BHM were used as relative molecular weight markers.
Analysis of OMVs from a strain expressing His-tagged Tob55 (Fig. S1) revealed not only the 140-kD TOB complex but also a species of ∼100 kD (Fig. 5 A). This latter species was detected likewise in OMVs from cells expressing His-tagged Tob38 or Tob37 as well as in the wild-type strain. Therefore, it is not a product of a dissociation of the TOB complex due to the attachment of the His tag to Tob55. This complex could only be shifted by the antibody against the His tag when the His tag was attached at the Tob55 (Fig. 5 A). Upon isolation of the TOB complex from this latter strain, a fraction of Tob55 was recovered in the 140-kD TOB complex containing also Tob38 and Tob37 in a molecular ratio of 1:1:1 (Fig. S2 A); another fraction of Tob55 was present as a 100-kD species containing only Tob55 (Fig. S2 B). These results suggest that a fraction of Tob55 was present in the form of dimers.
To confirm that these dimers of Tob55, which can also be observed in the BNGE of mitochondria (Lackey et al., 2011), are not caused by a loss of Tob37 and Tob38 during the preparation of mitochondria, the post-mitochondrial supernatant (cytosolic fraction) was examined. No Tob37 or Tob38 could be detected in this fraction (Fig. S2 C). In addition, after the treatment of OMVs with cross-linker, Tob55 dimers could be detected after SDS-PAGE analysis by immunodecoration (Fig. S2 D). They had the same running behavior as the cross-linked Tob55 dimers in the eluate of the affinity preparation from OMVs bearing a His-tagged Tob55, but were not detected, as expected, when the preparation was performed with His-tagged Tob37 (Fig. S2 E). This demonstrates that the dimers are not an artifact of the solubilization of mitochondria or BNGE treatment. If the complex was dissociating during the isolation procedure, a relative excess of that subunit, which carried a His tag, would be expected upon affinity purification. However, in preparations of the TOB complex using His tags on Tob38 or Tob37, no such excess was observed. Furthermore, Tob38 and Tob37 were only observed in complex with a monomer of the Tob55, never with the dimer.
Cryoelectron microscopy and single-particle analysis of the TOB complex
For structural analysis, purified complexes were imaged in the frozen hydrated state by cryoelectron microscopy. Because complexes as small as the TOB complex (140 kD) are at or perhaps below the limit for which a correct alignment of cryoelectron microscopy images can be confidently obtained, we made efforts to maximize the signal-to-noise ratio. We performed imaging with an FEG (field emission gun) at 120 kV to increase image contrast and beam coherence and minimize the effect of the CCD MTF (charge-coupled device modulation transfer function or point spread function). The images were subjected to reference-free alignment. Classification of reference-free–aligned images should generate class averages representing specific projections of the complex. The individual class averages showed one to three additional masses occurring in different positions relative to the main mass (Fig. S4 A). None of these class averages appeared symmetric, nor did they have a visible channel. They appeared much like different projections of a three-dimensional complex containing four domains or subunits with a maximum diameter of ∼15 nm. Because Tob55 has two discrete domains, the β-barrel and the N-terminal domain, which contains the POTRA domain, the reference-free class averages is consistent with the determined stoichiometry. Still, it was not possible to determine relative orientations of the classes and thus generate a starting model. The reproducibility of the classification across all datasets and alignment methods, however, suggested that the real organization of the complex was being identified by the classification algorithm and that it was the low signal-to-noise ratio of the images and the small size of the complex which was limiting the reference-free alignment.
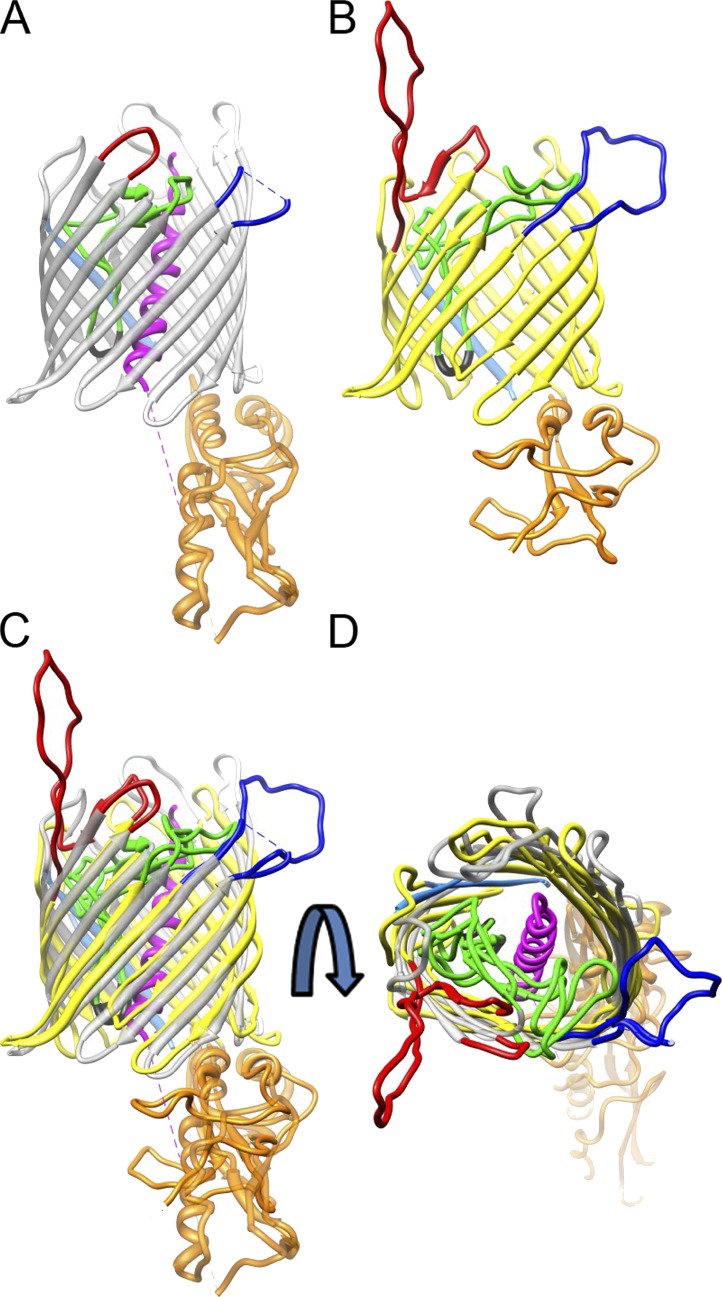
Because there was no structural information available with which to begin single-particle image alignment and reconstruction, we generated an initial reference model of the TOB complex. This reference was constructed based on a homology model of Tob55, and incorporated information about the previously determined orientations of the POTRA domain, Tob55, Tob37, and Tob38 relative to the membrane. Secondary structure prediction of Tob55 using HHpred (Söding et al., 2005) predicted a 16 β-strand barrel plus one POTRA domain. The only significant match for Tob55 with a known structure was to the TpsB transporter FhaC from Bordetella pertussis, also a 16-stranded β-barrel protein with two N-terminal POTRA domains (Clantin et al., 2007). Both proteins are members of the Omp85–TpsB transporter superfamily. Using MODELLER (Sali et al., 1995), which essentially maps the predicted Tob55 secondary structure on to the FhaC three-dimensional structure (Protein Data Bank accession no. 2QDZ), yielded a three-dimensional molecular model for Tob55. The resulting model follows the two-dimensional structure prediction well, and is in good agreement with the model of the human homologue Sam50 (Zeth, 2010). There are consistent features in both the structure of FhaC and the Tob55 model, for example the arrangements of the β-strands, the localization of the conserved VRGY/F tetrad at the tip of a loop spanning the interior of the β-barrel (Fig. 6, A and B, black; Fig. S3, framed in gray), or long loops at the cytoplasmic and extracellular side of Tob55 and FhaC, respectively (Figs. 6 and S3, blue and red loop). These similarities suggest that this model of Tob55 is realistic.
Figure 6.
Comparison of Tob55 and FhaC illustrates corresponding elements in the architecture of both transporters. (A and B) Ribbon representations of the crystal structure of FhaC (A, gray) and the predicted structure of Tob55 (B, yellow). The C termini of both proteins are colored in light blue and the N-terminal domains in orange. The loops that span the interior of both β-barrels are depicted in green. The conserved VRGY/F motif found in this loop is shown in black. The N-terminal helix in the interior of the FhaC β-barrel is shown in magenta. (C and D) Superposition of the two structures shown in A and B. The two loops of FhaC and Tob55 facing the extracellular and cytoplasmic side, respectively, are located at corresponding sites of the barrels (colored in red and dark blue). (C) Lateral view; (D) view from the exterior sides of the membrane.
An unstructured ring was used to account for the presence of Tob37 and Tob38 at the cytoplasmic side of the β-barrel. It was added to the homology model of Tob55 on the end opposite from the POTRA domain (Fig. S4 B, gray model). The transformation of the ring to a structure with two discrete densities is illustrated by five intermediate reconstructions (Fig. S4 B). These structures appeared to deteriorate in quality as more rounds of alignment were completed. Classification of reference-free–aligned images and the use of examples of “bad” images as references eliminated most incorrectly picked particles. After removal of bad images from the dataset the deterioration in quality was no longer observed.
We used the long-term (over many rounds of refinement) stability of alignment parameters for an individual image to screen the images for “goodness” to improve the reconstructions (Fig. 7, A–C; Yang et al., 2012). The most significant change was the appearance of domain-like features within the two subunits that should be Tob37 and Tob38 (Fig. 7, A–C). In addition, finer details such as the extra density found on the outside of the reconstructions of the complex isolated with His-tagged Tob55 became apparent (Fig. 7, B and C). The fit of the homology model to the Tob55 density was also improved. Projections of this final reconstruction are similar to the reference-free class averages, just more clearly defined (Fig. S4, A and C).
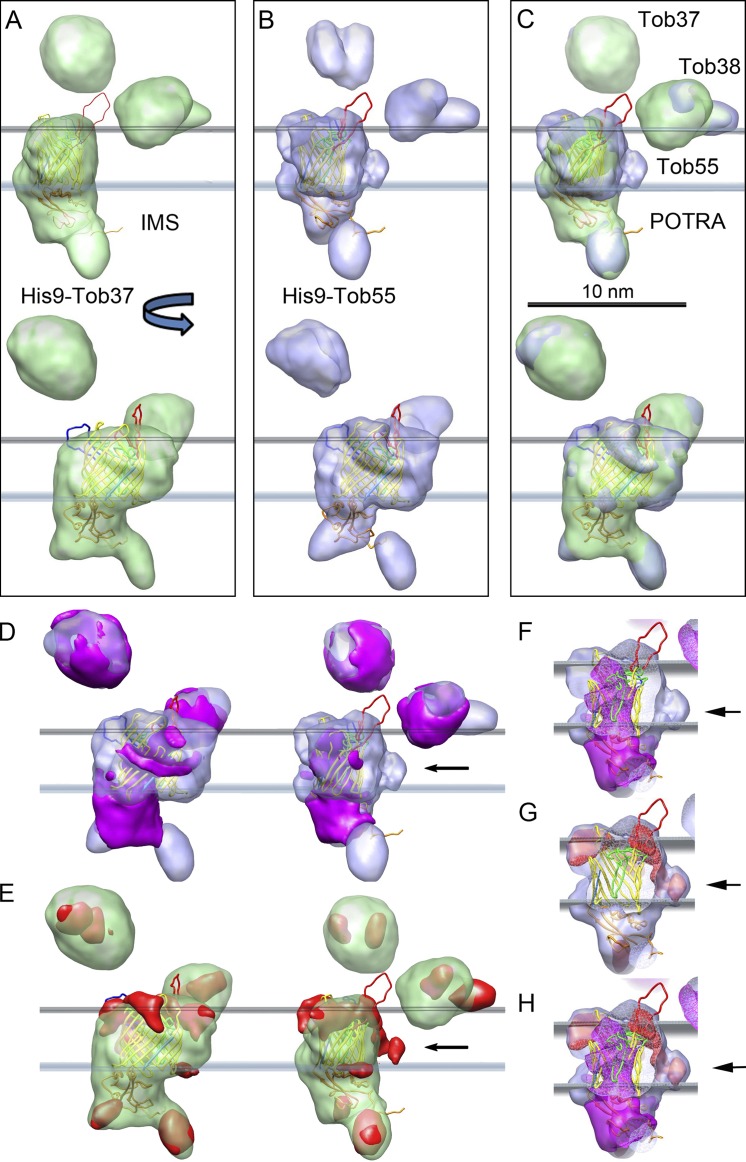
Figure 7.
Reconstruction of the isolated TOB complex. (A–C) Two views rotated by 90 degrees of the final reconstructions of the TOB complex from cells expressing His9-Tob37 (A) and His9-Tob55 (B). The presumed position of the outer membrane relative to Tob55 is indicated by planes defining the inner and outer surface of the outer mitochondrial membrane. The reconstructions are superimposed over the molecular homology model of Tob55. (C) Superposition of both reconstructions. Bar, 10 nm. The graphics and the positioning of the Tob55 homology model shown were generated using CHIMERA. (D) Two views of the difference map (magenta) resulting from the subtraction of the His9-Tob55 reconstruction from the His9-Tob37 reconstruction superimposed on the His9-Tob55 map. (E) The difference map (red) resulting from the subtraction of the His9-Tob37 reconstruction from the His9-Tob55 reconstruction superimposed on the His9-Tob37 map. (F–H) Side views with only the difference maps as shown in D (right) or E (right) superimposed on the Tob55 homology model and the His9-Tob55 map. The front of the His9-Tob55 map and the Tob55 homology model is cut away. The arrows in D–H all indicate the same region on one side of the β-barrel. (F and G) Difference maps of D and E, respectively. (H) Superposition of both difference maps from F and G.
The reconstructions of the His9-Tob37– and His9-Tob38–isolated complexes were very similar, whereas the maps of the His9-Tob55–isolated complexes differed from them in several ways (Fig. 7, A and B). A superposition of these reconstructions is shown in Fig. 7 C. Differences were calculated between the various maps that reveal a number of rearrangements (Fig. 7, D–H). There is significant density missing from the lumen of the β-barrel in all the reconstructions of the complexes isolated using His9-Tob55 relative to those isolated with His9-Tob37 and His9-Tob38 (Fig. 7, F–H). This missing density in the His9-Tob55 maps correlates with the appearance of density on the outside surface near β-strand 11 and 12 of the reconstructed complexes isolated with His9-Tob55, which is not present in the His9-Tob37 and His9-Tob38 maps.
A unique fit of the Tob55 homology model was obtained for all reconstructions using the FITMAP function of the visualization program CHIMERA (Pettersen et al., 2004) with all cross-correlation values exceeding 0.85. Fourier shell correlation (FSC) at 0.5 cutoff suggests a resolution of ∼15 Å (Table 1) for all reconstructions (Fig. 7, A–E). The two loops exposed at the cytoplasmic face of Tob55 (Fig. 6 B, red and blue) extend toward the densities that must be Tob37 and Tob38. Thus, these loops might form the connection sites between these proteins and Tob55 (Fig. 7). The assignment of density to Tob37 and Tob38 is not conclusive, but was chosen based on evidence that Tob38 is membrane associated in close proximity to Tob55 (Kutik et al., 2008). Also, the volume of the density assigned to Tob38 is smaller than the density assigned to Tob37, consistent with the fact that Tob38 has an 11.3-kD smaller molecular mass.
Table 1.
Representative samples of data used in the cryo-EM reconstructions with final resolution obtained
| Sample | No. of micrographs | No. of particles picked | Particles in final reconstruction | Resolution FSC 0.5 |
| His9-Tob37 | 546 | 175,000 | 76,700 | 14.7 |
| His9-Tob37 gold labeleda | 415 | 155,000 | 88,000 | 15.0 |
| His9-Tob38 | 318 | 104,000 | 43,500 | 15.0 |
| His9-Tob38 gold labeleda | 125 | 45,000 | 25,900 | 15.6 |
| His9-Tob55 | 215 | 80,000 | 40,600 | 14.7 |
| His9-Tob55 gold labeleda | 272 | 98,500 | 46,400 | 14.5 |
| Dimers from His9-Tob55 | N/A | 24,800 | 19,300 | 13.7 |
Although no gold label could be detected for His9-Tob37 and His9-Tob38, the structures are identical with or without added Ni-NTA nanogold.
Reference model bias is always a concern, especially in this case. 14 datasets were processed independently, four from His9-Tob55 preparations, five each from His9-Tob37 and His9-Tob38. Yet, two distinct structures were obtained from a common start; they correlate with the presence (His9-Tob37– and His9-Tob38–isolated complexes) or absence (His9-Tob55–isolated complexes) of the long and intermediate isoforms of Tob55 in expected places. When aligned to the same starting references using the same processing procedures, refinement of images of buffer did not produce the same structure.
We searched for and found the Tob55 dimer in the images collected from affinity purifications using His9-Tob55–expressing cells. A reference consisting of two cylinders simulating the Tob55 dimer was constructed. Approximately 10% of the images were classified as dimers, for which a structure was obtained (Fig. S5, A–E). The Tob55 dimer has two copies of the Tob55 β-barrel, which form a complex with twofold symmetry. The densities representing the POTRA/N-terminal domains appeared with opposing orientations, consistent with the complex having a twofold symmetry (Fig. S5, A–C). The final dimer structure was reconstructed using twofold symmetry with a resolution of 13.7 Å (FSC 0.5 cutoff; Fig. S5, F–H). The Tob55 homology model fits the Tob55 dimer density reasonably well with a cross-correlation value of 0.75. The lumen of the β-barrel is empty in this reconstruction, perhaps partially accounting for the lower cross-correlation value relative to the whole TOB complex. The Tob55 subunits are joined at the site of the loop shown in blue (see Figs. 6 and S5, F–H). When the structure of the Tob55 dimer was aligned with and superimposed on the TOB complex, it became clear that the Tob37–Tob55 interface and the dimer interface are in the same location (Fig. S5 H). Therefore, association of the subunit, tentatively identified as Tob37, would require dissociation of the dimer.
Discussion
We describe here the first isolation of a purified TOB complex. Tob55, Tob38, and Tob37 were identified as the only components of the complex. The stoichiometry of the TOB complex was determined to be 1:1:1 for the three components, resulting in a calculated molecular mass of the complex of 140 kD. This is consistent with that determined by BNGE analysis using mitochondrial respiratory complexes as standards. This molecular mass differs significantly from previous BNGE analyses, which suggested a molecular mass of 200–250 kD for the TOB complex (Paschen et al., 2003). This discrepancy is obviously due to the previous use of water-soluble proteins as molecular mass standards.
Does the TOB complex resulting from the isolation represent a functional entity? A functional assay in vitro is complicated by the rather complex assembly pathway of substrates of the TOB complex. After their synthesis in the cytosol, β-barrel precursor proteins pass through the TOM pore into the intermembrane space. Subsequently, they are transferred to the TOB complex with the help of the small Tim protein complexes and inserted into the outer membrane, presumably in a stepwise fashion. Distinct folding requirements may play a major role during this process. A highly complex reconstitution system would be needed to mimic the situation in vivo. Given this complexity, it is not surprising that the β-barrel precursor proteins showed only a weak binding to the isolated TOB complex in the assay we performed, measuring binding activity of TOB complexes in detergent micelles. However, using this assay we succeeded in demonstrating specific binding of the precursor of Tom22 to the isolated TOB complex. Tom22 was described as a substrate, which in contrast to a β-barrel precursor protein, is an α-helical transmembrane protein (Stojanovski et al., 2007). Tom22 does not pass through the intermembrane space, but needs the help of the TOB complex to be assembled with the TOM complex (Keil and Pfanner, 1993; Stojanovski et al., 2007).
Mdm10 in yeast has been suggested to be a subunit of the TOB–SAM complex. In addition, a regulatory function of Mdm10 in the assembly of the TOM complex was reported (Meisinger et al., 2004, 2006; Thornton et al., 2010; Yamano et al., 2010). Our experiments with Neurospora showed that only a very small amount of the total Mdm10 co-purified with the TOB complex; this TOB–Mdm10 complex was present at a very minor level compared with the TOB complex. Mdm10 is a β-barrel protein itself and, as such, a substrate of the TOB complex. Therefore, it might be co-isolated with the TOB complex. However, no other substrates such as the highly abundant β-barrel proteins porin or Tom40 were found in association with the isolated TOB complex. These observations strongly suggest that Mdm10 is not a subunit of the TOB complex, but rather agrees with a function as a regulatory interaction partner.
The TOB complex resulting from the three-dimensional reconstructions comprises the three subunits, one Tob55, Tob38, and Tob37 each. This is consistent with the results of the biochemical analysis of the TOB complex in the membrane and in the purified form as well as with all known topological characteristics of the TOB complex subunits. These experimental results assign Tob37 and Tob38 to the cytoplasmic face of the mitochondria and the POTRA domain to the intermembrane space (Habib et al., 2007; Lackey et al., 2011). Likewise, the volume obtained from the reconstruction agrees well with the calculated volume of the isolated complex. A preliminary two-dimensional structure (Paschen et al., 2003) suggested a contribution of several Tob55 subunits to the TOB complex. Although formation of higher order complexes of TOB cannot be excluded, we observed only a small percentage of larger species of the TOB complex in preparations extracted by the use of the detergent TX-100.
The role of the Tob55 dimers remains a matter of speculation. The structure obtained would disfavor an origin resulting from artificial dissociation of the TOB complex. The dimers might have a porin-like function for specific substrates or possibly be an assembly intermediate.
The organization of the TOB complex as an entity with only one Tob55 molecule and the conservation of structural and functional elements in Tob55 and FhaC allow us to speculate on the functional mechanism of the TOB complex. Tob55 and FhaC interact with their substrates in the intermembrane and periplasmic space, respectively. The POTRA domain of Tob55 and the POTRA domains of FhaC were proposed to serve as sites of substrate binding (Habib et al., 2007; Jacob-Dubuisson et al., 2009). However, the POTRA domain of Tob55 was recently shown to function also in the release of proteins from the TOB complex in the course of their insertion into the membrane (Stroud et al., 2011). In case of FhaC, the interaction of substrates was suggested to occur at the tip of the loop, which contains the VRGY/F tetrad (Jacob-Dubuisson et al., 2009; Delattre et al., 2010). This sequence is part of a conserved motif, motif 3, in the Omp85–TpsB transporter superfamily (Moslavac et al., 2005; Jacob-Dubuisson et al., 2009; Delattre et al., 2010). In view of its high conservation and its predicted localization to the intermembrane space in the Tob55 model, the VRGY/F tetrad might also serve as an interaction site for substrate proteins in Tob55. Binding of precursors to the VRGY/F tetrad of the loop seems to trigger a substantial rearrangement of the β-barrel and induce transport activity of FhaC (Delattre et al., 2010). This may also be the case with Tob55. Indeed, both Tob55 and FhaC exhibit channel activity that is responsive to substrate binding (Clantin et al., 2007; Kutik et al., 2008; Knowles et al., 2009).
Members of the Omp85 family mediate membrane insertion of β-barrel precursor proteins, in contrast to TpsB transporters, which mediate secretion of proteins. The lumen of a single β-barrel is too small to allow the folding of a new β-barrel substrate, therefore a lateral opening of the barrel of the translocase during the processing of the substrate protein seems to be required. Such a scenario was suggested to be unlikely in respect to thermodynamics (Wimley, 2003; Paschen et al., 2005b; Knowles et al., 2009). However, the junction between strand 1 (N-terminal) and strand 16 (C-terminal) is particularly short and not stabilized by a joining loop or turn. Significant rearrangements of the barrel during the processing of substrates by FhaC (Clantin et al., 2007) and after binding of precursor proteins to Tob55 (Kutik et al., 2008) have been reported. This tempts us to speculate that such a rearrangement might allow for a transient opening of the barrel in the TOB complex. A new barrel might be generated by sequential integration of antiparallel β-strands into the Tob55 structure to form a large intermediate β-barrel structure, which then would separate into two barrels and close. In this manner, both the outer, hydrophobic and the inner, hydrophilic sheath of the old and new barrel would stay in their respective environments. Tob37 and Tob38 might assist in such a mechanism. This would allow the TOB complex to act as a monomeric complex, the structure of which is supported by our analysis.
Materials and methods
Strains and growth of N. crassa
The N. crassa strains NCN251 (wild type), Tob55 short HT (Lackey et al., 2011), Tob38HT (9His-Tob38-3), and Tob37HT (9His-Tob37-2) were used (Table 2; Wideman et al., 2010). The Tob55 short HT strain expresses only the short isoform of Tob55 containing a 9× His-tag. In Tob38HT and Tob37HT all three Tob55 isoforms are expressed. Growth and handling of N. crassa were as described previously (Davis and de Serres, 1970). In brief, solid Vogel’s medium (1.5 g sodium citrate, 1 g KH2PO4, 0.1 g MgSO4, 0.05 g CaCl2, 12.5 µg biotin, 2% sucrose, 100 mg histidine, and 20 g agar per liter) in Erlenmeyer flasks was inoculated with granules from a silica stock of the strain of interest and grown in the dark at 30°C for 48 h. Subsequently, conidia were allowed to form under light for ∼190 h. Conidia were harvested and liquid cultures of Vogel’s minimal liquid medium supplemented with histidine (same as above without agar but containing 40 ug/L chloramphenicol) were inoculated and grown for 20 h. The mycelia were harvested by filtration.
Table 2.
Strains used in this paper
| Strain | Genotype | Origin or source and reference if applicable |
| NCN251 | A | FGSCa 2489 |
| Tob55 short HT | his-3 mtrR a Δ tob55:: hygR; Contains an ectopic copy of N-terminal 9x His-tagged tob55cDNA specific for the short form. Bleomycin resistant. | Nargang laboratory; Lackey et al., 2011 |
| Tob38HT (9His-Tob38-3) | his-3 mtrR a Δ tob38::hygR; Contains an ectopic copy of genomic tob38 with a C-terminal 9x His tag. Bleomycin resistant. | Nargang laboratory; Lackey et al., 2011 |
| Tob37HT (9His-Tob37-2) | his-3 mtrR a Δ tob37::hygR; Contains an ectopic copy of genomic tob37 with a C-terminal 9x His tag. Bleomycin resistant. | Nargang laboratory; Lackey et al., 2011 |
Fungal Genetics Stock Center, Kansas City, MO.
Isolation of mitochondria and mitochondrial OMVs
OMVs were isolated basically as described previously (Mayer et al., 1995) with some modifications. Mycelia were cultivated at 30°C, harvested by filtration, and ground in the presence of sand and 0.25 SET buffer (250 mM sucrose, 1 mM EDTA, 20 mM Tris-HCl, pH 8.5, and 1 mM PMSF). Sand was removed by two 5-min low-speed 2,600-g centrifugation steps. Organelles in the supernatant were pelleted by centrifugation at 11,000 g for 50 min at 4°C. Mitochondria were scraped off the surface of the pellet and resuspended in 0.25 M SET buffer. To generate OMVs, mitochondria were incubated with stirring in swelling buffer (1 mM EDTA, 20 mM Tris-HCl, pH 8.5, and 1 mM PMSF) at 4°C. Using an automated glass-Teflon homogenizer (40-min exposure) the outer membranes were sheared off the mitochondria. 20 ml of the homogenate were applied to a two-step gradient with 9 ml 0.9 M SET buffer on the bottom overlaid with 9-ml 0.25 M SET buffer. The samples were centrifuged for 1 h at 142,000 g at 4°C. The OMVs were collected from the boundary between the 0.25 M and 0.9 M SET buffers. This was mixed with one-half volume 2.0 M SET and overlaid with 0.7 M SET and an additional 3.0 ml Tris/EDTA buffer. The sample was centrifuged overnight at 142,000 g at 4°C. The OMVs were collected from the surface of the 0.7 M SET buffer. OMVs were diluted in OMV buffer (1 mM PMSF, 20 mM Tris, pH 8.5), then centrifuged for 1 h at 180,000 g to pellet the OMVs. The pellet was resuspended in OMV buffer.
Purification of the TOB complex
Mitochondria and OMVs from wild-type or strains expressing a His-tagged Tob55, Tob38, or Tob37 were solubilized with 0.2% (vol/vol) TX-100 or 0.64% (wt/vol) digitonin. After a clarifying spin, the supernatant was loaded on a matrix for Ni-NTA affinity purification. The proteins were bound in the presence of 15 mM imidazole. The column was rinsed stepwise with washing buffer containing 20 and 40 mM imidazole. Bound proteins were eluted with 200 mM imidazole. The eluted proteins were concentrated using an Amicon Ultra 30k device (EMD Millipore) with a cutoff of 30 kD. After separation by BNGE or SDS-PAGE, the proteins were analyzed by immunodecoration or Coomassie blue staining.
Mass spectrometry analysis
After SDS-PAGE or BNGE and Coomassie blue staining, protein bands were excised and digested overnight with trypsin. For protein identification and quantification, the tryptic fragments were separated on a C18 reversed-phase column with a linear acetonitrile gradient and analyzed by nano-electrospray ionization-LC tandem MS (ESI-LC-MS/MS), recorded on an Orbitrap mass spectrometer. Analysis of spectra was performed with the MASCOT software (Matrix Science) on the complete N. crassa genome database (http://www.broad.mit.edu/annotation/fungi/neurospora). For IDMS, a defined amount of AQUA (AbsoluteQUAntification) peptides (Stemmann et al., 2001), quantified stable isotope (13C/15N)–labeled internal peptide standards, were mixed with the dehydrated gel pieces before tryptic digestion. The amounts of native peptides were calculated by comparing the areas under the peaks in the extracted ion chromatogram (EIC) of the AQUA peptide and its native counterpart. Several samples of each prepared protein complex were measured and the average was calculated. The error bars depict the standard deviations of measurements of (n) samples originating from the same preparation and demonstrates the reproducibility of the quantification by IDMS. Representative experiments for each measured complex are shown (Figs. 3, 5, and S2). A total of ten biological replicates of the TOB complex and five biological replicates of the Tob55–Tob38 complex isolated using TX-100 or digitonin, from strains differing in the His-tagged TOB complex subunit, were analyzed.
BN-PAGE antibody supershift assays
Mitochondria and OMVs were solubilized in 1x Native PAGE sample buffer (Invitrogen) with a detergent/protein ratio of 20:1 (wt/wt) for digitonin and 6:1 (wt/wt) for TX-100. Where indicated, 1 µg Penta-His antibody (QIAGEN) was added. After 1 h of incubation on ice, the samples were centrifuged for clarification at 36,000 g and the supernatant was mixed with Native PAGE 5% G-250 sample additive (Invitrogen) such that the final G-250 concentration in the sample was at least 25% of the detergent concentration. BNGE was performed as described previously (Schägger and von Jagow, 1991; Schägger et al., 1994) with Native PAGE Novex 4–16% Bis-Tris gel according to the manufacturer’s instructions (Invitrogen) using 1x Native PAGE anode and cathode buffers (Invitrogen) at 150 V for 60 min. The voltage was increased to 250 V and the electrophoresis continued for 4 h. For Western blotting, the proteins were transferred to a PVDF membrane at 80 mA for 80 min using 1× NuPAGE transfer buffer (Invitrogen).
Homology modeling
Secondary sequence prediction for Tob55 and searches for homology matches in the known databases were performed with HHpred (Söding et al., 2005). The results of HHpred were then used to generate a molecular homology model for Tob55 based on the FhaC structure using MODELLER (Sali et al., 1995); both programs are accessible for use online through the Bioinformatics toolkit, Gene Center, Ludwig Maximilians University Munich (http://toolkit.lmb.uni-muenchen.de).
Cryoelectron microscopy
The isolated TOB complexes were applied to a lacey carbon film on molybdenum EM grids and flash frozen in liquid ethane or an ethane–propane mixture cooled by liquid nitrogen. For labeling of His-tagged proteins, Ni-NTA Nanogold beads (1.8-nm gold particle; Nanoprobes) were mixed with the protein preparations in a 1:9 ratio (vol/vol). After incubation for 45 min on ice, the samples were applied to a grid and serially washed three times on droplets of aqueous buffer (50 mM Hepes, pH 8.5, 0.084% TX-100, and 1 mM PMSF) before they were blotted and flash frozen. Data were acquired on a microscope (model F20; FEI Tecnai) operating at 120 kV, using a cryo-holder (model 656; Gatan) with a specimen temperature of −180°C. Images were acquired on a 4K charge-coupled device camera (Eagle; FEI) at 84270× (1.78 Å/pixel) using the TOM_acquisition package (Korinek et al., 2011; available at http://www.biochem.mpg.de/baumeister/tom_e/get_tom/index.html). TOM_tool_box programs require MATLAB, a commercially available software package (for information see http://www.mathworks.com). The images were collected with a defocus range (ΔF) from −0.7 and −3.5 µm with an electron dose of 15–25 e−/Å2. The defocus contrast transfer function (CTF) for each good image was determined and the image phases were corrected using TOM_ctffindgui (Korinek et al., 2011; Scripts available in the online supplemental material).
Particles were selected from micrographs automatically by cross correlation (CC) with a reference using scripts (see online supplemental material) written for the SPIDER package (Shaikh et al., 2008; SPIDER is open software freely available with excellent documentation, tutorials and sample scripts at http://www.wadsworth.org/spider_doc/spider/docs/spider.html). The micrographs were filtered with a Gaussian band-pass filter, which emphasized the particles. A nearest-neighbor exclusion was applied to reject correlation peaks whose center-to-center distance was <1.5 particle diameters. After windowing into 140 × 140-pixel boxes, the images were masked and realigned to the selection reference. Particles whose center was not well determined, as indicated by a substantial in plane shift, were excluded from the dataset. Reference-free alignment followed by K means classification was used to identify and remove false positives from the dataset and an attempt to determine orientations from good class averages was made.
A maximum likelihood–based reference-free alignment and classification approach was also performed. Initial ML2D analysis (Sorzano et al., 2010) resulted in class averages that were very similar in appearance to the reference-free classes observed using SPIDER. Given the small size of the TOB complex and low signal-to-noise ratio, it is clear that both reference-free alignment methods were not good enough to allow the generation of an unbiased starting model.
A starting model was generated based on the stoichiometry results and the class averages obtained by reference-free alignment. The density map calculated from the Tob55 molecular homology model was the core of the starting model. Because Tob37 and Tob38 are present on the cytoplasmic side of the outer mitochondrial membrane, an unstructured ring of low density was placed above the cytoplasmic face of the Tob55 β-barrel (Fig. S4). The model was used only to start the reconstruction process. The final map shown in Fig. S4 was merged with the density map derived from the Tob55 homology model. This was the starting point for all subsequent alignments and reconstructions. The starting model for detecting and reconstructing the Tob55 dimer was a pair of somewhat hollow cylinders with a hemisphere of density added on one end to account for the POTRA domain of Tob55. This model was included in the initial multi-reference alignment of the images of His9-Tob55–isolated TOB complexes. Images were assigned to either TOB complex or dimer classes. The model and the progress of this initial approach are shown in Fig. S5. As control to ensure that the starting reference did not bias the progress of the reconstruction, images of vitreous ice (buffer only) were hand selected from micrographs lacking particles. The images were aligned to the same starting models as for both the TOB complex and the Tob55 dimers (shown in Figs. S4 B and S5 A–C), and the results were refined following the same processing steps as the real data. After completing the same initial 10 rounds of alignment, the aligned buffer only images were subjected to the same classification as the aligned data (Fig. S5 D). A subset of the resulting class averages is shown in Fig. S5 F.
Image processing was performed with the SPIDER software package (Shaikh et al., 2008). Iterative multi-reference alignment of the images to projections of a reference map was used to align the images and generate particle orientations. The images underwent many rounds of refinement with the current reconstruction becoming the new reference.
The stability of alignment parameters for each image was determined from the output of 4–5 successive rounds of alignment using a reference projection set with 900 equally distributed projections of the current map as references. The change in parameters over four iterations of alignment was calculated for each image. Only images that showed no change in which reference projection was matched and minimal changes for in-plane rotation and x and y shifts were included in further processing. Overall, 50–60% of the original images were eliminated from any given dataset. The remaining aligned images had a random in-plane rotation applied to them. Both the aligned and randomly rotated aligned images were reference-free aligned and classified. The class averages from classification were very similar to what was observed earlier, only cleaner with less background, yet it was still not possible to orient the class averages and generate a model. The alignment parameters revealed substantial x and y shifts away from the previously established image centers. These shifts make it unlikely that a common lines approach could work in this case.
The Fourier shell correlation was calculated to assess the resolution of the reconstructions. The SPIDER BP 32f Fourier-based reconstruction algorithm was used for reconstruction. The final reconstructions were normalized with respect to one another by setting all maps to a constant standard deviation. The maps were then Gaussian low-pass filtered to 12 Å for display and further analysis. Negative values were set to zero before calculating differences between maps. Maps are displayed using CHIMERA (Pettersen et al., 2004; free software available from http://www.cgl.ucsf.edu/chimera) with a threshold of 1 sigma and differences are displayed at a two-sigma threshold based on the original map statistics.
Examples of scripts used for the image processing are appended to the online supplemental material. These scripts are meant to be a representative sample of the scripts used.
Antibody production
Antibodies against N. crassa Tob37 were prepared by injecting peptides corresponding to Tob37 residues 165–184, 212–233, and 426–442 of the protein into rabbits or a peptide corresponding to Tob37 residues 305–319 into guinea pigs and mice. (Wideman et al., 2010).
Antibodies against N. crassa Tob38 were prepared by injecting guinea pigs with a fusion protein consisting of a hexahistidinyl tag, mouse dihydrofolate reductase, and N. crassa Tob38 (residues 1–185). The fusion protein was expressed in E. coli and was purified on a Ni-NTA column (QIAGEN) in 8 M Urea according to the manufacturer’s instructions. The protein was eluted in 0.1% SDS and 10 mM Tris-HCl, pH 7.4. The eluate was injected into guinea pigs and mice without further processing.
Antibodies against N. crassa Mdm10 were prepared by injecting guinea pigs with a fusion protein composed of a hexahistidinyl tag, mouse dihydrofolate reductase, and residues 5–298 of the N. crassa Mdm10 protein. The fusion protein was purified as described above for the Tob38 fusion protein and injected into guinea pigs and mice. (Wideman et al., 2010).
Antibodies against N. crassa Tob55 were generated as for Tob38 above by injecting rabbits with a fusion protein containing a hexahistidinyl tag, mouse dihydrofolate reductase, and residues 1–108 of the short form of Tob55 (Hoppins et al., 2007).
Antibodies against N. crassa Tim8 were raised in rabbits against His6-tagged fusion proteins comprised of full-length mouse dihydrofolate reductase and the C terminus of Tim8 (residues 35–92; Hoppins and Nargang, 2004).
The antibodies against N. crassa Tom40 were generated against a C-terminal peptide of NcTom40 (QQEGAQSLNIPF) coupled to keyhole limpet hemocyanin and injected into rabbits using TiterMax Gold (Sigma-Aldrich) for the first injection and incomplete Freund’s adjuvant for subsequent injections.
Miscellaneous
Radiolabeled precursor proteins were synthesized in rabbit reticulocyte lysate in the presence of [35S]methionine (MP Biomedicals) after transcription by SP6 polymerase from vectors containing the gene of interest.
For chemical cross-linking experiments, OMVs and isolated TOB complex were incubated with DSG (disuccinimidyl glutarate) for 1.5 h at 4°C. Subsequently, glycine (0.1 M, pH 8.8) was added to quench excess of the cross-linker and the samples were analyzed by SDS-PAGE followed by immunodecoration.
EM reconstructions
Accession numbers for the EM reconstructions deposited at the Electron Microscopy Data Bank (http://www.emdatabank.org) are: EMD-2195, EMD-2196, EMD-2197, EMD-2200, EMD-2201, EMD-2202, and EMD-2203.
Online supplemental material
Fig. S1 shows TOB isolation via His-tagged Tob55. Fig. S2 shows that TOB isolation via His-tagged Tob55 leads to copurification of Tob55 dimers. Fig. S3 shows analysis of the secondary structure elements of FhaC and Tob55. Fig. S4 shows progress from model to structure and reference-free alignment of single-particle images. Fig. S5 shows reconstruction of Tob55 dimers. A text file featuring image-processing scripts is also available. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201207161/DC1.
Supplementary Material
Acknowledgments
We thank Doron Rapaport for advice and discussions. We also thank Carlos Oscar Sanchez Sorzano and Jose Maria-Carazo for help with applying their maximum-likelihood 2D alignment and classification to our data. Also, we thank Andreas Korinek for help with the automated data acquisition.
This work was supported by the Deutsche Forschungsgemeinschaft, Sonderforschungsbereich 594-B14, and a grant from the Canadian Institutes of Health Research (CIHR) to F.E. Nargang.
Footnotes
Abbreviations used in this paper:
- BAM
- β-barrel assembly machinery
- BHM
- bovine heart mitochondria
- BNGE
- blue native gel electrophoresis
- IDMS
- isotope dilution mass spectrometry
- LC-MS/MS
- LC tandem mass spectrometry
- NTA
- nitrilotriacetic acid
- OMV
- outer membrane vesicle
- POTRA
- polypeptide transport-associated domain
- SAM
- sorting and assembly machinery
- TOB
- topogenesis of the outer mitochondrial membrane β-barrel proteins
- TX-100
- Triton X-100
References
- Becker T., Pfannschmidt S., Guiard B., Stojanovski D., Milenkovic D., Kutik S., Pfanner N., Meisinger C., Wiedemann N. 2008. Biogenesis of the mitochondrial TOM complex: Mim1 promotes insertion and assembly of signal-anchored receptors. J. Biol. Chem. 283:120–127 10.1074/jbc.M706997200 [DOI] [PubMed] [Google Scholar]
- Clantin B., Delattre A.S., Rucktooa P., Saint N., Méli A.C., Locht C., Jacob-Dubuisson F., Villeret V. 2007. Structure of the membrane protein FhaC: a member of the Omp85-TpsB transporter superfamily. Science. 317:957–961 10.1126/science.1143860 [DOI] [PubMed] [Google Scholar]
- Davis R.H., de Serres F.J. 1970. [4] Genetic and microbiological research techniques for Neurospora crassa. Methods in Enzymology. Vol. Volume 17, Part 1 Herbert Tabor C.W.T., editor Academic Press; 79–143 [Google Scholar]
- Delattre A.S., Clantin B., Saint N., Locht C., Villeret V., Jacob-Dubuisson F. 2010. Functional importance of a conserved sequence motif in FhaC, a prototypic member of the TpsB/Omp85 superfamily. FEBS J. 277:4755–4765 10.1111/j.1742-4658.2010.07881.x [DOI] [PubMed] [Google Scholar]
- Endo T., Yamano K., Kawano S. 2011. Structural insight into the mitochondrial protein import system. Biochim. Biophys. Acta. 1808:955–970 10.1016/j.bbamem.2010.07.018 [DOI] [PubMed] [Google Scholar]
- Gentle I., Gabriel K., Beech P., Waller R., Lithgow T. 2004. The Omp85 family of proteins is essential for outer membrane biogenesis in mitochondria and bacteria. J. Cell Biol. 164:19–24 10.1083/jcb.200310092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray M.W., Burger G., Lang B.F. 1999. Mitochondrial evolution. Science. 283:1476–1481 10.1126/science.283.5407.1476 [DOI] [PubMed] [Google Scholar]
- Habib S.J., Waizenegger T., Niewienda A., Paschen S.A., Neupert W., Rapaport D. 2007. The N-terminal domain of Tob55 has a receptor-like function in the biogenesis of mitochondrial beta-barrel proteins. J. Cell Biol. 176:77–88 10.1083/jcb.200602050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppins S.C., Nargang F.E. 2004. The Tim8-Tim13 complex of Neurospora crassa functions in the assembly of proteins into both mitochondrial membranes. J. Biol. Chem. 279:12396–12405 10.1074/jbc.M313037200 [DOI] [PubMed] [Google Scholar]
- Hoppins S.C., Go N.E., Klein A., Schmitt S., Neupert W., Rapaport D., Nargang F.E. 2007. Alternative splicing gives rise to different isoforms of the Neurospora crassa Tob55 protein that vary in their ability to insert beta-barrel proteins into the outer mitochondrial membrane. Genetics. 177:137–149 10.1534/genetics.107.075051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa D., Yamamoto H., Tamura Y., Moritoh K., Endo T. 2004. Two novel proteins in the mitochondrial outer membrane mediate beta-barrel protein assembly. J. Cell Biol. 166:621–627 10.1083/jcb.200405138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob-Dubuisson F., Villeret V., Clantin B., Delattre A.S., Saint N. 2009. First structural insights into the TpsB/Omp85 superfamily. Biol. Chem. 390:675–684 10.1515/BC.2009.099 [DOI] [PubMed] [Google Scholar]
- Keil P., Pfanner N. 1993. Insertion of MOM22 into the mitochondrial outer membrane strictly depends on surface receptors. FEBS Lett. 321:197–200 10.1016/0014-5793(93)80107-6 [DOI] [PubMed] [Google Scholar]
- Knowles T.J., Scott-Tucker A., Overduin M., Henderson I.R. 2009. Membrane protein architects: the role of the BAM complex in outer membrane protein assembly. Nat. Rev. Microbiol. 7:206–214 10.1038/nrmicro2069 [DOI] [PubMed] [Google Scholar]
- Korinek A., Beck F., Baumeister W., Nickell S., Plitzko J.M. 2011. Computer controlled cryo-electron microscopy—TOM² a software package for high-throughput applications. J. Struct. Biol. 175:394–405 10.1016/j.jsb.2011.06.003 [DOI] [PubMed] [Google Scholar]
- Kozjak V., Wiedemann N., Milenkovic D., Lohaus C., Meyer H.E., Guiard B., Meisinger C., Pfanner N. 2003. An essential role of Sam50 in the protein sorting and assembly machinery of the mitochondrial outer membrane. J. Biol. Chem. 278:48520–48523 10.1074/jbc.C300442200 [DOI] [PubMed] [Google Scholar]
- Kutik S., Stojanovski D., Becker L., Becker T., Meinecke M., Krüger V., Prinz C., Meisinger C., Guiard B., Wagner R., et al. 2008. Dissecting membrane insertion of mitochondrial beta-barrel proteins. Cell. 132:1011–1024 10.1016/j.cell.2008.01.028 [DOI] [PubMed] [Google Scholar]
- Lackey S.W., Wideman J.G., Kennedy E.K., Go N.E., Nargang F.E. 2011. The Neurospora crassa TOB complex: analysis of the topology and function of Tob38 and Tob37. PLoS ONE. 6:e25650 10.1371/journal.pone.0025650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer A., Driessen A., Neupert W., Lill R. 1995. Purified and protein-loaded mitochondrial outer membrane vesicles for functional analysis of preprotein transport. Methods Enzymol. 260:252–263 10.1016/0076-6879(95)60143-0 [DOI] [PubMed] [Google Scholar]
- Meisinger C., Rissler M., Chacinska A., Szklarz L.K., Milenkovic D., Kozjak V., Schönfisch B., Lohaus C., Meyer H.E., Yaffe M.P., et al. 2004. The mitochondrial morphology protein Mdm10 functions in assembly of the preprotein translocase of the outer membrane. Dev. Cell. 7:61–71 10.1016/j.devcel.2004.06.003 [DOI] [PubMed] [Google Scholar]
- Meisinger C., Wiedemann N., Rissler M., Strub A., Milenkovic D., Schönfisch B., Müller H., Kozjak V., Pfanner N. 2006. Mitochondrial protein sorting: differentiation of beta-barrel assembly by Tom7-mediated segregation of Mdm10. J. Biol. Chem. 281:22819–22826 10.1074/jbc.M602679200 [DOI] [PubMed] [Google Scholar]
- Meisinger C., Pfannschmidt S., Rissler M., Milenkovic D., Becker T., Stojanovski D., Youngman M.J., Jensen R.E., Chacinska A., Guiard B., et al. 2007. The morphology proteins Mdm12/Mmm1 function in the major beta-barrel assembly pathway of mitochondria. EMBO J. 26:2229–2239 10.1038/sj.emboj.7601673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milenkovic D., Kozjak V., Wiedemann N., Lohaus C., Meyer H.E., Guiard B., Pfanner N., Meisinger C. 2004. Sam35 of the mitochondrial protein sorting and assembly machinery is a peripheral outer membrane protein essential for cell viability. J. Biol. Chem. 279:22781–22785 10.1074/jbc.C400120200 [DOI] [PubMed] [Google Scholar]
- Moslavac S., Mirus O., Bredemeier R., Soll J., von Haeseler A., Schleiff E. 2005. Conserved pore-forming regions in polypeptide-transporting proteins. FEBS J. 272:1367–1378 10.1111/j.1742-4658.2005.04569.x [DOI] [PubMed] [Google Scholar]
- Neupert W., Herrmann J.M. 2007. Translocation of proteins into mitochondria. Annu. Rev. Biochem. 76:723–749 10.1146/annurev.biochem.76.052705.163409 [DOI] [PubMed] [Google Scholar]
- Paschen S.A., Waizenegger T., Stan T., Preuss M., Cyrklaff M., Hell K., Rapaport D., Neupert W. 2003. Evolutionary conservation of biogenesis of beta-barrel membrane proteins. Nature. 426:862–866 10.1038/nature02208 [DOI] [PubMed] [Google Scholar]
- Paschen S.A., Neupert W., Rapaport D. 2005a. Biogenesis of beta-barrel membrane proteins of mitochondria. Trends Biochem. Sci. 30:575–582 10.1016/j.tibs.2005.08.009 [DOI] [PubMed] [Google Scholar]
- Paschen S.A., Neupert W., Rapaport D. 2005b. Biogenesis of beta-barrel membrane proteins of mitochondria. Trends Biochem. Sci. 30:575–582 10.1016/j.tibs.2005.08.009 [DOI] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605–1612 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- Rapaport D. 2002. Biogenesis of the mitochondrial TOM complex. Trends Biochem. Sci. 27:191–197 10.1016/S0968-0004(02)02065-0 [DOI] [PubMed] [Google Scholar]
- Sali A., Potterton L., Yuan F., van Vlijmen H., Karplus M. 1995. Evaluation of comparative protein modeling by MODELLER. Proteins. 23:318–326 10.1002/prot.340230306 [DOI] [PubMed] [Google Scholar]
- Sánchez-Pulido L., Devos D., Genevrois S., Vicente M., Valencia A. 2003. POTRA: a conserved domain in the FtsQ family and a class of beta-barrel outer membrane proteins. Trends Biochem. Sci. 28:523–526 10.1016/j.tibs.2003.08.003 [DOI] [PubMed] [Google Scholar]
- Schägger H., von Jagow G. 1991. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal. Biochem. 199:223–231 10.1016/0003-2697(91)90094-A [DOI] [PubMed] [Google Scholar]
- Schägger H., Cramer W.A., von Jagow G. 1994. Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional native electrophoresis. Anal. Biochem. 217:220–230 10.1006/abio.1994.1112 [DOI] [PubMed] [Google Scholar]
- Schleiff E., Eichacker L.A., Eckart K., Becker T., Mirus O., Stahl T., Soll J. 2003. Prediction of the plant beta-barrel proteome: a case study of the chloroplast outer envelope. Protein Sci. 12:748–759 10.1110/ps.0237503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaikh T.R., Gao H., Baxter W.T., Asturias F.J., Boisset N., Leith A., Frank J. 2008. SPIDER image processing for single-particle reconstruction of biological macromolecules from electron micrographs. Nat. Protoc. 3:1941–1974 10.1038/nprot.2008.156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söding J., Biegert A., Lupas A.N. 2005. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 33(Web Server issue):W244–W248 10.1093/nar/gki408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorzano C.O., Bilbao-Castro J.R., Shkolnisky Y., Alcorlo M., Melero R., Caffarena-Fernández G., Li M., Xu G., Marabini R., Carazo J.M. 2010. A clustering approach to multireference alignment of single-particle projections in electron microscopy. J. Struct. Biol. 171:197–206 10.1016/j.jsb.2010.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stemmann O., Zou H., Gerber S.A., Gygi S.P., Kirschner M.W. 2001. Dual inhibition of sister chromatid separation at metaphase. Cell. 107:715–726 10.1016/S0092-8674(01)00603-1 [DOI] [PubMed] [Google Scholar]
- Stojanovski D., Guiard B., Kozjak-Pavlovic V., Pfanner N., Meisinger C. 2007. Alternative function for the mitochondrial SAM complex in biogenesis of alpha-helical TOM proteins. J. Cell Biol. 179:881–893 10.1083/jcb.200706043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud D.A., Becker T., Qiu J., Stojanovski D., Pfannschmidt S., Wirth C., Hunte C., Guiard B., Meisinger C., Pfanner N., Wiedemann N. 2011. Biogenesis of mitochondrial β-barrel proteins: the POTRA domain is involved in precursor release from the SAM complex. Mol. Biol. Cell. 22:2823–2833 10.1091/mbc.E11-02-0148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton N., Stroud D.A., Milenkovic D., Guiard B., Pfanner N., Becker T. 2010. Two modular forms of the mitochondrial sorting and assembly machinery are involved in biogenesis of alpha-helical outer membrane proteins. J. Mol. Biol. 396:540–549 10.1016/j.jmb.2009.12.026 [DOI] [PubMed] [Google Scholar]
- Voulhoux R., Tommassen J. 2004. Omp85, an evolutionarily conserved bacterial protein involved in outer-membrane-protein assembly. Res. Microbiol. 155:129–135 10.1016/j.resmic.2003.11.007 [DOI] [PubMed] [Google Scholar]
- Waizenegger T., Habib S.J., Lech M., Mokranjac D., Paschen S.A., Hell K., Neupert W., Rapaport D. 2004. Tob38, a novel essential component in the biogenesis of beta-barrel proteins of mitochondria. EMBO Rep. 5:704–709 10.1038/sj.embor.7400183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waizenegger T., Schmitt S., Zivkovic J., Neupert W., Rapaport D. 2005. Mim1, a protein required for the assembly of the TOM complex of mitochondria. EMBO Rep. 6:57–62 10.1038/sj.embor.7400318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wideman J.G., Go N.E., Klein A., Redmond E., Lackey S.W., Tao T., Kalbacher H., Rapaport D., Neupert W., Nargang F.E. 2010. Roles of the Mdm10, Tom7, Mdm12, and Mmm1 proteins in the assembly of mitochondrial outer membrane proteins in Neurospora crassa. Mol. Biol. Cell. 21:1725–1736 10.1091/mbc.E09-10-0844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedemann N., Kozjak V., Chacinska A., Schönfisch B., Rospert S., Ryan M.T., Pfanner N., Meisinger C. 2003. Machinery for protein sorting and assembly in the mitochondrial outer membrane. Nature. 424:565–571 10.1038/nature01753 [DOI] [PubMed] [Google Scholar]
- Wimley W.C. 2003. The versatile beta-barrel membrane protein. Curr. Opin. Struct. Biol. 13:404–411 10.1016/S0959-440X(03)00099-X [DOI] [PubMed] [Google Scholar]
- Wittig I., Beckhaus T., Wumaier Z., Karas M., Schägger H. 2010. Mass estimation of native proteins by blue native electrophoresis: principles and practical hints. Mol. Cell. Proteomics. 9:2149–2161 10.1074/mcp.M900526-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamano K., Tanaka-Yamano S., Endo T. 2010. Mdm10 as a dynamic constituent of the TOB/SAM complex directs coordinated assembly of Tom40. EMBO Rep. 11:187–193 10.1038/embor.2009.283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Fang J., Chittuluru J., Asturias F.J., Penczek P.A. 2012. Iterative stable alignment and clustering of 2D transmission electron microscope images. Structure. 20:237–247 10.1016/j.str.2011.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeth K. 2010. Structure and evolution of mitochondrial outer membrane proteins of beta-barrel topology. Biochim. Biophys. Acta. 1797:1292–1299 10.1016/j.bbabio.2010.04.019 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.