Abstract
Despite the importance of Mg2+ for numerous cellular activities, the mechanisms underlying its import and homeostasis are poorly understood. The CorA family is ubiquitous and is primarily responsible for Mg2+ transport. However, the key questions—such as, the ion selectivity, the transport pathway, and the gating mechanism—have remained unanswered for this protein family. We present a 3.2 Å resolution structure of the archaeal CorA from Methanocaldococcus jannaschii, which is a unique complete structure of a CorA protein and reveals the organization of the selectivity filter, which is composed of the signature motif of this family. The structure reveals that polar residues facing the channel coordinate a partially hydrated Mg2+ during the transport. Based on these findings, we propose a unique gating mechanism involving a helical turn upon the binding of Mg2+ to the regulatory intracellular binding sites, and thus converting a polar ion passage into a narrow hydrophobic pore. Because the amino acids involved in the uptake, transport, and gating are all conserved within the entire CorA family, we believe this mechanism is general for the whole family including the eukaryotic homologs.
Keywords: channel gating, membrane protein, X-ray structure
The magnesium ion (Mg2+) is the most abundant divalent cation in living cells and mediates in numerous cellular activities. The uptake of this ion in most prokaryotes is through the action of the CorA family, which is also one of the most studied families of divalent cation transporters (1, 2). Functional homologs of the CorA family have been found in eukaryotes, which would suggest conserved functional and regulatory mechanisms (3). In addition, the virulence of some human and plant pathogens is directly linked with the function of the CorA or its homologs (4–7). The only structural features shared by all CorAs and their eukaryotic homologs are the presence of two transmembrane helices at the C termini, which are connected by a short extraplasmic loop. The loop contains an essentially conserved motif consisted of Gly-Met-Asn (GMN), which is believed to be important for the Mg2+ uptake (1, 2). The only reported structure of a CorA is that from Thermotoga maritima (TmCorA) (8–10). However, all these reports failed to reveal the structure of the loop, including the GMN motif. Furthermore, no functional model could be revealed by the structures of TmCorA and thus far its function has only been speculated. The speculations include the gating mechanism, where a hydrophobic gating, which opens and closes in an iris-like action, has been suggested (11). In addition, TmCorA was recently shown to be a Co2+ selective transporter with no ability to regulate the Mg2+ transport (12). Therefore, as the speculations of the functional mechanisms of this CorA homolog have been based on the functional and computational characterizations using Mg2+ as the substrate, a careful revision and reconsideration and certainly new structure of a Mg2+ transporter CorA are clearly needed. Here, we report the crystal structure of the Mg2+ transporter CorA from the Archaea Methanocaldococcus jannaschii (MjCorA) at 3.2 Å resolution, including its conserved extraplasmic loop. Thus, we provide a unique complete structure of a CorA protein, which is also Mg2+-selective. This structure presents previously undescribed details about the mode of Mg2+ uptake and transport, which is most likely applicable to the entire CorA family, including the distant eukaryotic homologs. Based on this structure, we have been able to suggest a unique gating mechanism.
Results
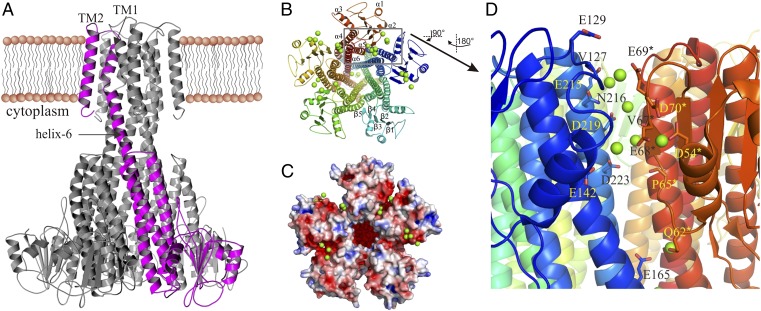
The MjCorA crystal structure, similar to the TmCorA crystal structures (8–10), shows a homopentameric arrangement with a funnel-shaped intracellular hydrophilic domain and two transmembrane helices (TMs), the first of which (TM1) forms the pore of the channel (Fig. 1A). Each monomer is composed of seven α-helices and five β-strands; helix-6 is a very long helix, which also makes the TM1, and helix-7 is the TM2 (Fig. 1B). MjCorA shares only 24.3% sequence identity with TmCorA and is also 39-amino acids shorter (Fig. S1A). The latter is reflected by the less dense N-terminal domain, where MjCorA is composed of five β-strands and five α-helices, in contrast to seven β-strands and seven α-helices in TmCorA (Fig. S1 B–D). Thus, MjCorA has a largely open monomer-monomer interface at the N-terminal side (Fig. 1 C and D and Fig. S1C).
Fig. 1.
The overall structure of MjCorA. (A) The side-view of the pentameric MjCorA with one monomer highlighted in purple. The helix-6 suggested to rotate during gating is indicated with an arrow. (B) The cytoplasmic view of MjCorA. The Mg2+ ions are shown as green spheres. α-Helices and β-strands are numbered. (C) The same view showing the electrostatic surface (± 5 kT/e) and the Mg2+-binding grooves. (D) The close-up of the metal binding groove between monomers A and E (the boxed area in B) showing the accumulation of Mg2+ (green spheres) in this region. The interacting residues are shown as sticks. All distances are in the range of 4–5 Å. *Denotes the residues from adjacent monomer.
Selectivity Filter.
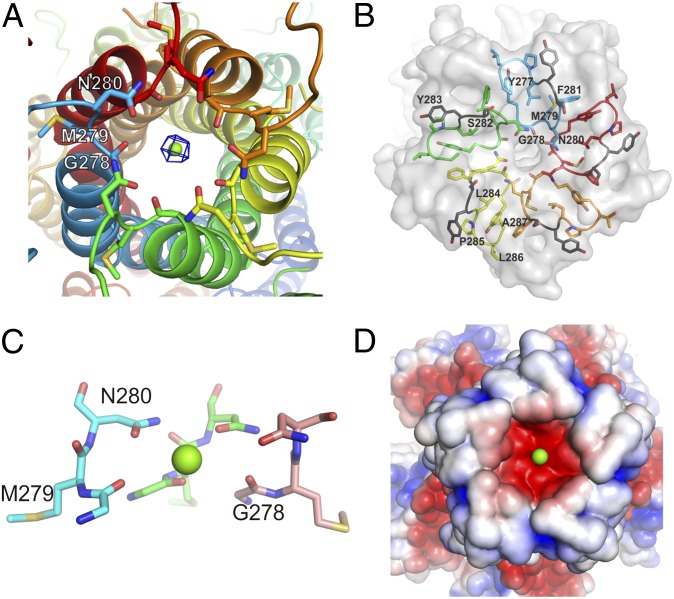
The most striking feature of the MjCorA structure is the arrangement of the conserved extracellular loop, which forms a concavity (Fig. 2 and Fig. S2A). The GMN motif (G278, M279, N280) is positioned at the bottom of this concavity. The side-chains of N280 from each monomer form a polar ring that creates the substrate entrance (Fig. 2 A and C). The ability of this ring to attract Mg2+ seems to be enhanced by an underlying carbonyl ring formed by G278 (Fig. 2C). We observed a spherical electron density within the N280-ring (Fig. 2A), which was interpreted to be Mg2+ because the crystals were grown in the presence of 60 mM MgCl2. The distance between Mg2+ and the hydroxyl group of each N280 is ∼4 Å. A fully hydrated Mg2+ has two hydration shells. The first shell has been shown, both theoretically and experimentally, to contain six water molecules organized in an octahedral geometry (13, 14). These molecules are tightly bound to the Mg2+ and make the ion partially hydrated. The second hydration shell is believed to contain 12 water molecules, which are loosely bound to the Mg2+ and substitute a fully hydrated ion (13, 14). Fully and partially hydrated Mg2+ have radii of ≤5 Å and ∼2.09 Å, respectively (13, 14). Therefore, the Mg2+ in the N280-ring is apparently partially hydrated and coordinates with the N280 hydroxyl groups via the water molecules in the first hydration shell. Although the role of the GMN motif had previously been addressed to be involved in ion transport (1, 14, 15), this is unique structural evidence for the direct role of the Asn of the GMN motif in Mg2+ uptake. The Asn-ring, because of its geometry, apparently allows the entry of only partially hydrated Mg2+. Additionally, the Asn-ring might be the binding area for cobalt(III)hexamine, the known inhibitor of CorA proteins (16), which could tightly seal the selectivity filter (Fig. S3). It was also shown that analogous compound nickel(II)hexamine [somewhat larger than cobalt(III)hexamine] failed to inhibit transport function (16), confirming the geometrical “narrowness” of selection. G278 creates a hinge that allows the hydrophobic M279 to be kept fixed in a highly hydrophobic environment. A mutation in any of these three residues results in the loss of CorA transport activity (15). Thus, this arrangement of the GMN motif appears to create a perfect entry for partially hydrated Mg2+, explaining why this motif has been maintained throughout the evolution of the CorA family and its eukaryotic homologs (1, 3).
Fig. 2.
The structure of the ion selectivity filter. (A) The selectivity filter made of the conserved GMN motif, where the N280-ring and the carbonyl groups of G278 are involved in ion selection and attraction, respectively, and M279 is fastening the filter in the hydrophobic interior. The 2fo-fc electron density for Mg2+ is contoured at 1.7 σ. (B) The architecture of the side-chains of the YGMNF as well as the LPLA motifs, which create the rigidity of the loop, and ultimately the selectivity filter. S282 and Y283 separating the two motifs are highlighted in gray. (C) The side view on GMN motif (two monomers are omitted for clarity). Note the widening of a second ring formed by carbonyls of G278. (D) Electrostatic potential surface (same as in Fig. 1C), showing the selectivity filter attractiveness for magnesium ions. LPLA motif and following residues form the shielding walls of concavity.
In prokaryotes, the conservation of the GMN motif is extended to the YGMNF sequence (Fig. 2B and Fig. S1A) (2). The aromatic groups in this sequence contribute to the hydrophobicity of the interior, where M279 is fixed (Fig. 2B). The residues in the LPLA motif (Fig. 2B) further strengthen this hydrophobic interior, which is located after the YGMNF sequence. These motifs ultimately form a very hydrophobic and thus rigid scaffold for the Asn-ring. Between the YGMNF and LPLA motifs, there is a highly conserved aromatic residue (Y283 of MjCorA) in the CorA family, which is involved in the stabilization of the hydrophobic interior. As seen in the MjCorA structure, the hydroxyl groups of S282 and the carbonyl groups of F281 are oriented toward the interior of the concavity. Apparently, this arrangement and the shape of the concavity together provide a suitable environment to attract and concentrate hydrated ions and move them toward the selectivity filter (Fig. 2 B and D). Other CorA homologs often have Asp or Glu, with carboxyl groups, instead of S282 (Fig. S1A) (2, 17); these residues most likely fulfill the same role but are more efficient in ion attraction (18).
Internal Cavity.
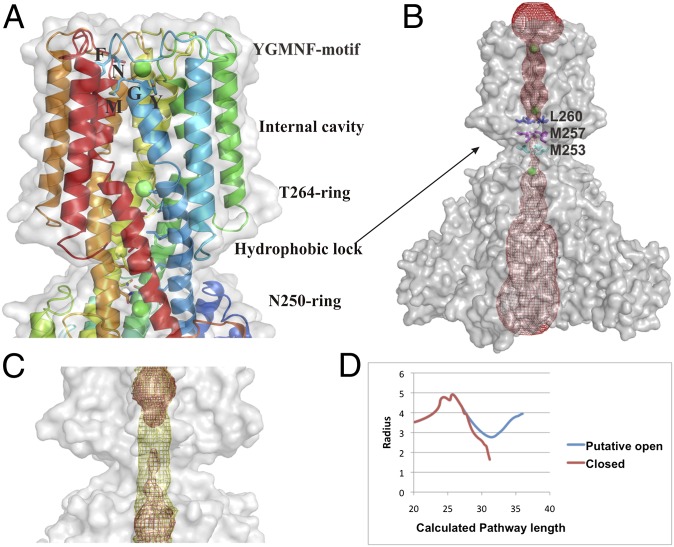
The Mg2+ bound to the N280-ring revealed that the uptake of the ion substrate by the selectivity filter is in the partially hydrated form. A relatively wide cavity (∼4–5 Å in radii) is located on the intracellular side of the N280-ring, which is wide enough to allow water movement (Fig. 3). The shape of the cavity is dictated by P270, which is positioned in the middle of the TM1 and creates a kink in the helix. The kink is further stabilized by the hydrophobic interactions between F267, P270, and W272* [here and elsewhere an asterisk (*) denotes a residue from an adjacent monomer] (Fig. S4). Furthermore, this cavity is ∼23 Å long, spans almost the entire membrane, and ends at a second hydroxyl ring composed of T264 (Fig. 3A). Similar to the selectivity filter, this ring is enhanced with an additional ring of the main-chain carbonyls of highly conserved F267 (2). The observed electron density within these polar rings was assigned to Mg2+, indicating that the partially hydrated Mg2+, once through the selectivity filter, passes through the pore as partially hydrated. A hydrophobic lock composed of the L260-ring, the M257-ring, and the M253-ring seems to block the movement of the partially hydrated Mg2+ toward the intracellular side of CorA immediately after the T264-ring (Fig. 3 A and B). The calculations of the pore profile revealed that the transmembrane passage is inevitably blocked with this hydrophobic lock and even water molecules cannot pass through (Fig. 3 B and D). Only after manual remodeling of the constriction site to become polar the passage widens to ∼3 Å in radius (Fig. 3 C and D), which is enough to allow the movement of water or ions, as demonstrated by Sansom and colleagues (19). In the same manner, calculations of the ion solvation energy along the permeation pathway of a channel revealed a considerably lowered energetic barrier in case of a putative polar passage (Fig. S5). These calculations show that the constriction site is indeed closed to water and Mg2+ because of its hydrophobic nature; however, it could allow Mg2+ movement if converted to become polar. The hydrophobic lock is positioned at the membrane/cytoplasm interface. Thus, the pore-forming transmembrane domain of MjCorA is arranged in a way that allows the free passage of partially hydrated Mg2+ through almost the entire membrane and this passage is stopped by the L260-ring in the closed form of MjCorA.
Fig. 3.
The internal cavity and the ion transport path. (A) The conserved YGMNF motif located at the bottom of the concavity at the extracellular entrance, where the side-chains of Y277, M279, and F281 make the hydrophobic interior, and the main-chain of G278 and side-chain of N280 trap a Mg2+ (green sphere). The T264-ring is holding a Mg2+ immediately before the hydrophobic lock. A Mg2+ ion held by the N250-ring at the cytoplasmic side, immediately after the hydrophobic lock, is also shown. (B) The calculated contour of the entire channel (red mesh). Mg2+ shown as green spheres. Note the gap at the area of the hydrophobic lock, where ion movement is blocked by M253, M257, and L260. (C). The computed channel profile for the putative open conformation shown as yellow mesh overlaid on B. (D) The change in the radius of the channel pore. The closed form is shown in red. The line break means program failed to calculate any possible route even with the smallest probe size (indicating the hydrophobic lock is not permeable even for water molecules). Values are in angstroms.
Mg2+ Binding Grooves.
At the N-terminal monomer-monomer interfaces spherical electron densities were identified and annotated as Mg2+ (Fig. 1D). In contrast to TmCorA, where the Mg2+ binding sites at the monomer-monomer interfaces are well defined, in MjCorA they are rather spread. Thus, we postulate them as the Mg2+-binding grooves, where several ions might bind and cause progressive structural impact ultimately leading to the pore closure (vide infra). The crystal structure shows heterogeneous accumulation of Mg2+ within the grooves, which might be caused by the crystal contacts. Each binding groove might provide a transient binding area for up to eight Mg2+ ions as found within the A-E groove (Fig. 1D). This groove primarily comprises the side-chains of the negatively charged D54*, E68*, E69*, D70*, E142, D219, E215, and D223 residues, the hydroxyl containing Q62*, Y137, N141, and N216 residues, and the main-chain carbonyls of P65*, V67*, and V127. The P65*, V67*, E68*, and D70* are bound to the rigid and bulky α-β-bundle, whereas the V127, Y137, N141, E142, N216, D219, and D223 bind mainly to the α-helices 5 and 6 of the adjacent monomer. The binding distances are ∼4 Å and thus indicate a partially hydrated Mg2+ to be bound, which in turn could indicate a weak and transient binding. To examine whether the presence of Mg2+ induces stability to MjCorA, we performed in vitro thermostability tests on the purified MjCorA. Because M. jannaschii is a hyperthermophile and can live at temperatures up to 95 °C (20), we monitored the stability of MjCorA from 25 °C to 95 °C. The tests revealed that the thermostability of MjCorA is unaffected by the presence of Mg2+ (Fig. S6). This finding further strengthens the argument that the Mg2+ bound to the binding grooves does not stabilize the protein and suggests that the binding of Mg2+ to these grooves is of regulatory purpose. Similar sites in the crystal structures of TmCorA (8–10) as well as the Mg2+ transporter MgtE (21) have been found, which have been postulated as the regulatory binding sites.
Gating.
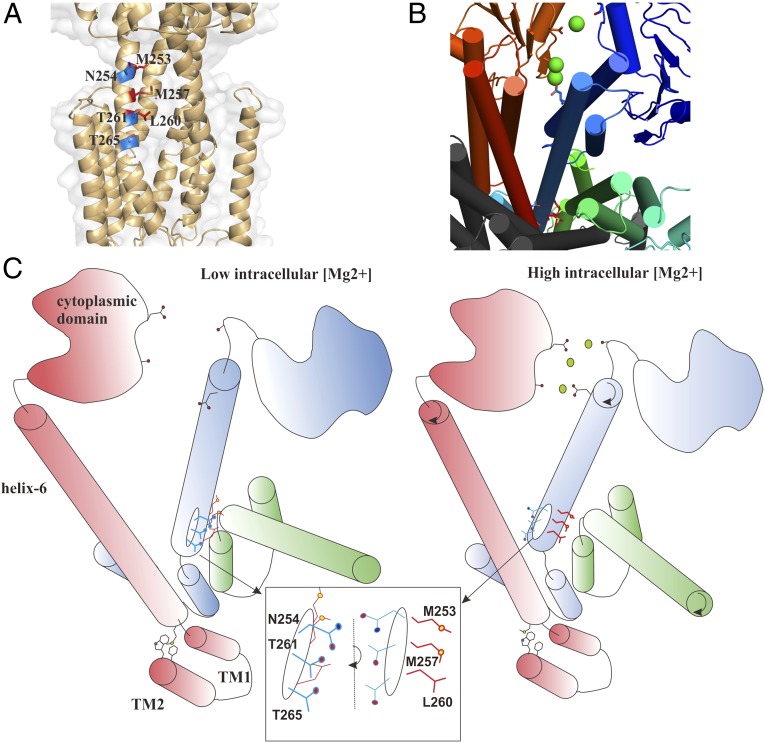
In both TmCorA and MgtE, the involvement of the metal ion binding sites in the regulation of gating has been further confirmed by biochemical studies (15, 22) and molecular dynamics simulations (11, 23). Based on these studies, the suggested gating mechanism for TmCorA proposes that a hydrophobic gate, sufficiently wide to allow the passage of hydrated Mg2+, closes as the result of a decrease in the pore size upon Mg2+ binding to the aforementioned binding sites (11). This gating supposedly occurs as the result of the sideway, iris-like, movement of the body of the pore-forming helices. However, it is unclear how the ion binding will induce such a large movement. As observed in the MjCorA crystal structure, polar hydroxyl groups coordinate the uptake and movement of partially hydrated Mg2+ through the pore. MjCorA contains three other polar residues (N254, T261, and T265) on the TM1, which all point away from the pore (Fig. 4A). N254 is highly conserved in the CorA family, and T261 is conserved as Thr and Ser in subgroups A and B, respectively (2). In TmCorA, the corresponding residues are N288, T295, and T299, and superposition of the crystal structures of MjCorA and TmCorA clearly shows similar spatial arrangement of these residues (Fig. S7). However, if these residues were pointing toward the interior of the pore in the open state, either completely or partially, they would create a more suitable environment for the passage of Mg2+. Because all of these residues are vertically aligned, turning TM1 in the closed state anticlockwise would not only move these residues toward the pore but would also move the hydrophobic residues away from the pore. This process would create a notably more polar environment that is open to Mg2+, as shown by the calculations (Fig. 3D and Fig. S5). To further examine this finding, we explored the possibility of mutating the conserved Asn to a hydrophobic residue. Such mutation would thus force CorA into a closed state. We chose TmCorA as a model system for this purpose by examining its Co2+ transport activity, which is remarkable for this transporter (12). Therefore, we created the N288L mutant of TmCorA and followed its Co2+ transport activity by evaluating the Co2+ resistance of a corA-deficient Escherichia coli strain. The complete resistance of this E. coli strain was a clear indication of a completely abolished Co2+ transport (Fig. S8). This observation could further strengthen our hypothesis. Based on both MjCorA and TmCorA structures, such an anticlockwise turn to open the channel would seem possible if there were no Mg2+ bound to the Mg2+-binding groove and metal binding sites, respectively. Therefore, as illustrated in Fig. 4, we believe that when CorA is open, the pore is polar and allows the passage of a partially hydrated Mg2+ (19). An increase in the intracellular concentration of Mg2+ leads to the accumulation of Mg2+ at the interfacial cavities between monomers because of the presence of negative charges. The partially hydrated Mg2+ will eventually bind to D54*, P65*, V67*, E68*, E69*, and D70*, and thus pull the E215, N216, D219, and D223 by attractive forces through the water molecules in its hydration shell, which will cause a clockwise turn of helix 6 along its axis. This turn will move the polar residues away from the pore and replace them with the hydrophobic residues, thus blocking the pore. D54*, P65*, V67*, E68*, E69*, and D70* reside on the α-β-bundle and are therefore more rigid in movement than the E215, N216, D219, and D223 on the long helix-6; the latter is more flexible and thus prone to turn.
Fig. 4.
The model of the gating mechanism. (A) Alignment of M253, M257, and L260 (in red) that point inside the channel and create the hydrophobic lock, and the polar side-chains of N254, T261, and T265 (in blue) that face away from the channel. One monomer is removed for clarity. (B) The cytoplasmic view of the arrangement of the metal binding groove, as well as the hydrophobic lock, used as the basis for (C) an illustrative model for the gating mechanism of CorA. (Left) Illustration of the predicted model of CorA during the open state at low intracellular Mg2+ concentration. The helix 6 is positioned to point N254, T261, and T265 of the TM1 toward the channel to create a hydrophilic pore. Simultaneously, M253, M257, and L260 are facing away from the pore. Upon elevation in the intracellular Mg2+ concentration (Right), magnesium ions (green circles) accumulate in the metal binding groove and bind to the carboxyl and carbonyl groups on the helix 6 by pulling them with ionic forces. Consequently, the helix 6 will turn clockwise (the black arrows), which will result in moving the hydroxyl groups of the TM1 away from the channel and their replacement by the hydrophobic residues. Now, the gate is too hydrophobic to allow the ion passage.
TM2 is highly hydrophobic, binds strongly to lipid bilayers, and is the only support providing stability to TM1 (and the entire protein) in the membrane. Moreover, the lateral movement of TM2 is probably restricted by the rigid scaffold of the selectivity filter, as well as tight hydrophobic interactions with TM1 (Fig. S9) and the lipid bilayer. Thus, the sideways movement, as suggested in the iris-like mechanism (11), seems rather unlikely. Upon such consideration, a turn along the helical axis of TM1 might be more feasible than sideways helical movements. As seen from the channel profile and the channel-permeation energy calculations, a simple rearrangement of side-chains of amino acids forming the hydrophobic lock can be enough to permit the ion transport. Thus, the postulated turn should not be very considerable; a slight rotation would be sufficient. At the C terminus of TM2 there are two highly conserved residues: F311 and W316. As shown by the crystal structure, these residues form hydrophobic interactions with the residues from TM1 at the membrane/cytoplasm surface (Fig. S10). These interactions may hold TM1 in place to ensure that its hydrophobic residues at the constriction site in the closed state are far enough from each other to prevent irreversible closure. This assembly is further stabilized by the conserved positively charged residues at the end of the C terminus of CorA; these residues most likely form salt bridges with the phospholipids from the membrane (Fig. S10). This mechanism, in which the encapsulating helices interact with the interior and pore-forming helices and stabilize their helical turn, has been observed in the acetylcholine receptor (24).
Discussion
In this study we have presented a unique complete structure of a CorA Mg2+ transporter, including the conserved extraplasmic loop. Until now, it has been a mystery why the YGMNF motif, and in particular the GMN motif, have been so strictly conserved throughout the life kingdoms. Asn is the only polar amino acid in the YGMNF motif and it appears to be the key residue for coordinating the Mg2+ uptake by replacing the water molecules in the second hydration shell. It seems that neutral Asn fulfills this role better than, for example, an acidic Asp, because the latter might interfere strongly with the first hydration shell of Mg2+ causing the blockage of the entry. Additionally, weaker interaction provided by Asn is perhaps to maintain a highly transient binding to facilitate the transport. The only carbonyl that is involved in the Mg2+ uptake is from the G278 main chain, which is distant enough from the ion pathway and this is likely to avoid a strong binding to the Mg2+.
Whether the N280-ring, or the Asn-ring in general, acts also as a fine selectivity filter to distinguish among similar divalent ions (Mg2+, Co2+, Ni2+) is unclear. Perhaps additional residues are involved in the fine-tuning of the selection. However, the filter does not allow other divalent cations, such as Ca2+ and Mn2+, to enter. This finding is probably because of the differences in the radii of the first hydration shells, which is fairly similar for Mg2+, Co2+, and Ni2+ (2.09 Å, 2.1 Å, and 2.05 Å, respectively) but relatively larger for Ca2+ and Mn2+ (2.46 Å and 2.2 Å, respectively) (25). Because Mg2+ and Co2+ are the most suitable substrates for CorA, we can assume that the filter selects cations with a very narrow range in first hydration shell radii.
The hydrophobic residues in the YGMNF motif are clearly involved in creating a stable and rigid scaffold to ensure the position of the Asn-ring is kept fixed. In MjCorA, this rigidity formed by Y277, M279, and F281 is further strengthened by the LPLA motif. MjCorA is one of the rare CorA homologs that contain the LPLA motif. In most microorganisms this motif is conserved as MPEL (Fig. S1) (2, 17). LPLA and MPEL are similar with respect to their hydrophobic features and differ with respect to the negatively charged Glu in MPEL. Therefore, it is reasonable to assume that the Met, Pro, and Leu residues of the MPEL motif in the rest of the CorA family are positioned similarly to the corresponding Leu, Pro, and Ala residues of the LPLA motif. However, such positioning of the given residues would orient the charged Glu of the MPEL motif toward the unfavorable hydrophobic interior. Nonetheless, assuming spatial similarity between the MjCorA and other CorA loops, sequence alignment indicates that the negatively charged Glu of MPEL might be compensated by a neighboring positively charged Arg/Lys/His residue via a salt bridge. Certainly, evolution has chosen the MPEL motif in most cases. Whether this choice is a result of either structural or functional factors remains unknown. In addition to the highly rigid loop formed by the hydrophobic interactions, the loop is kept unaffected from any structural rearrangement as directed from the cytoplasmic side, by creating a kink in the middle of the TM1. A similar kink was observed in the TmCorA structure (8), which was created by the hydrophobic interactions between a Pro residue from one monomer and the Phe residue from the adjacent monomer.
Until now, it has been unknown in what shape Mg2+ is taken up and transported by CorA. Recently, it was reported that Mg2+ is selected as fully hydrated by the Salmonella typhimurium CorA (StCorA) (17). Indeed, more structural evidences from StCorA (or any of its close homologs) are needed to further justify this. Certainly, the differences between the LPLA motif of MjCorA and the MPEL motif of StCorA, as well as the extra Glu present in StCorA (S282 in MjCorA) positioned between the YGMNF and MPEL motifs, may lead to the ability of the StCorA to select Mg2+ as fully hydrated before the actual uptake. However, these parts are not involved in the final uptake and it is most likely that all CorAs take up their substrates as partially hydrated similar to MjCorA; this is because of the entry is made up of the extremely conserved YGMNF motif.
Once the Mg2+ is inside the channel, it can either remain as partially hydrated or return to a fully hydrated state. The latter is because of the wide cavity, which could accommodate water molecules and a fully hydrated Mg2+. It is also a more plausible scenario, as the N280-ring will also be able to allow water to enter the cavity. This cavity of ∼23 Å long, together with the concave entry, reduces the distance inside the membrane that the Mg2+ needs to pass as partially hydrated. Thus, it is only at the extraplasmic entry, and most likely at the exit to the cytoplasm, where partial dehydration of Mg2+ becomes necessary. Such reduction in the traveling distance through the membrane is an important, but also a common feature of ion channels (26, 27).
In the crystal structure, the Mg2+ at the cytoplasmic-membrane interface is coordinated by the T264-ring. Based on our gating model, a helical turn will then move the T264 residues away from the pore upon opening. However, the turn will also move the T265 residues inside the channel, and thus one Thr is replaced by another Thr. Hence, to pass through the narrow gate on the cytoplasmic side, Mg2+ will remain in a partially hydrated state. At the exit, Mg2+ will pass through another highly conserved Asn-ring, the N254-ring. It is feasible to assume that the similar ring to the N280-ring at the entrance will form upon the pore opening. Altogether, this means that once Mg2+ has passed the cavity it will pass by the T265-, T261-, and N254 rings in a partially hydrated form. Therefore, Mg2+ that passes through the MjCorA channel is never completely dehydrated. Mg2+ holds to its water molecules of the first hydration shell very strongly and, hence, a complete dehydration is both energetically unfavorable and time-consuming. Partial dehydration of Mg2+, on the other hand, requires much less energy and time. The structural arrangement of the MjCorA has thus created a polar environment that is cost-efficient energetically and allows partially hydrated Mg2+ to freely pass through the membrane without the need of complete dehydration. It also defines how the Mg2+ binding to the cytoplasmic side closes the channel gate and stops the uptake of Mg2+. Concluding from the binding distances (∼4 Å), the binding of Mg2+ in case of MjCorA will probably be weaker compared with TmCorA, in which clearly defined binding sites (with ∼2 Å binding distances) are located (8, 10). Thus, the Mg2+ ions in the MjCorA sites are partially hydrated and bind weakly. Such difference could be because of physiological dissimilarities between these homologs; TmCorA is a bacterial Co2+ transporter, whereas the archaeal MjCorA is a Mg2+ transporter. However, more specialized experiments are needed to in detail examine the degree of a possible main-chain rotation and explore other possibilities: for example, whether the mechanism of gating is a combination of slight main chain movements with rotation.
The presented MjCorA crystal structure clarifies the general role of the essential and conserved GMN motif as the entry for partially hydrated ion substrate and provides the basis for a plausible novel gating mechanism involving helical turn in a substrate feedback mechanism, which converts an open hydrophilic channel into a closed hydrophobic pore. Impairing this mechanism could be used to develop novel and specific antivirulent agents.
Methods
Production of MjCorA.
MjCorA was cloned into pNIC28-Bsa4 vector encoding an N-terminal 6xHis-tag and a tobacco etch virus protease cleavage site. The protein was overexpressed in E. coli BL21(DE3)Rosetta grown in terrific broth at 37 °C by 0.1 mM isopropyl-b-d-thiogalactoside for 3 h. The cells were resuspended in 20 mM Tris•HCl pH 8.5, 300 mM NaCl, and 0.5 mM Tris(2-carboxyethyl)phosphine (TCEP) and lysed using a high-pressure homogenizer. The lysed cells were centrifuged at 10,000 × g for 30 min and the supernatants were centrifuged at 135,000 × g for 1 h. The pellet was solubilized with 1% (wt/vol) n-Undecyl-β-d-Maltopyranoside (UDM) (Anatrace) in 20 mM Tris•HCl pH 8.5, 300 mM NaCl, 0.5 mM TCEP. The proteins were purified by immobilized metal ion affinity chromatography, where 0.1% UDM was added to all buffers. The proteins were further purified by Superdex 200 16/60 (GE Healthcare). The purified proteins were concentrated by ultrafiltration using Vivaspin columns (Sartorius) and kept at 4 °C until further use. SeMet construct of MjCorA was expressed using M9 SeMet growth kit (Medicilon) according to the manufacturer’s protocol, followed by purification as mentioned above.
Crystallization.
Native crystals of soluble MjCorA were obtained by the vapor-diffusion method, in the sitting-drop plate in 28% (wt/vol) PEG8000, Hepes-Mops, pH 7.5 with the addition of alcohols–morpheus additives (Molecular Dimensions), whereas SeMet-derivative crystals were grown in 31% (vol/vol) PEG550MME, Imidazole-Mes, pH 6.5 with monosaccharides (Molecular Dimensions). Full-length MjCorA was crystallized in 30% (wt/vol) PEG8000, Hepes•NAOH, 60 mM MgCl2.
Structure Determination and Refinement.
Datasets from native, SeMet, and full-length native crystals were collected at BL13C1 beamline (National Synchrotron Radiation Research Center, Taiwan), MX2 beamline (Australian Synchrotron, Australia), and I04 beamline (Diamond, United Kingdom), respectively. All datasets were processed and scaled with XDS (28). Experimental phases were determined by the SAD method, using SeMet-derivative crystals. Initial model was obtained using AutoRickshaw software (29). The structure of the full-length protein was solved by molecular replacement using the model obtained at the previous step. Model building was performed with Coot (30). Refinement was carried out with Phenix (31). All figures were generated with Pymol (32). The data collection and refinement statistics are shown in Table S1.
Structure Analysis and Calculations.
The channel profiles were calculated with MOLE software (33). The putative open conformation was constructed in Coot by choosing those rotamers for amino acids forming hydrophobic lock. The visualization of charge distribution was done with Adaptive Poisson-Boltzmann Solver (APBS)-plugin (34) in Pymol. The contribution to the electrostatic potential of Mg2+ along the pore axis of the channel was calculated with the APBSmem software (35), based on the original APBS (36), but optimized for membrane protein calculations. The Mg2+ pathway and axis were defined using APBSmem. Charges and radii were assigned with PDB2PQR server (37). Energies were calculated using a NaCl bath with an ionic strength of 0.15 M at 298 K. The protein was embedded into a dielectric slab (ɛ = 2), mimicking the membrane environment, with membrane thickness set to 35 Å. The pore itself was excluded from the membrane and assigned a solvent dielectric (ɛ = 80).
Co2+ Transport Assay.
The assay was performed as previously described (12), with some modifications. The E. coli MG1655 with a knock-out corA (National Institute of Genetics) was transformed with either wild-type or mutated corA in pBAD or empty pBAD vector. Cultures were grown overnight at 37 °C in Luria Bertani medium (LB). The overnight culture was diluted 12 times into 10 mL fresh LB. Cells were grown at 37 °C for 1 h. The expression was then induced with 0.02% l-arabinose (Sigma-Aldrich) for 2 h. Subsequently, the cells were harvested at 3,000 × g for 10 min at 25 °C and washed twice by resuspension in 10 mL of N-buffer [7.5 mM (NH4)2SO4, 5 mM KCl, 1 mM KH2PO4, 0.5 mM K2SO4 and 0.1 M Tris•HCl, pH 7.4]. The cells were then normalized to an OD600 of 0.2 by dilution with N-buffer containing various concentrations of Co2+. The mixture was then incubated for 10 min at 37 °C. Three equivalents of LB containing the same concentrations of Co2+ were added into each mixture, and the solution was incubated at 37 °C for an additional 3 h. The final OD600 was recorded and analyzed in comparison with the starting OD600.
Thermostability Assay.
Purified MjCorA in solubilization buffer was diluted to a concentration of about 0.5 mg/mL. The solution was divided in 50-μL aliquots and 1 μL of stock solution of different metals was added. The samples were incubated at room temperature for about 30 min before heating in a thermocycler (Agilent Technologies). Samples were incubated for 10 min at each temperature. The samples were filtered on a 96-well filter-plate (0.65 μm) (Millipore). The filtrate was analyzed by SDS/PAGE followed by Coomassie staining.
Thermal Aggregation Shift Assay.
Purified MjCorA in solubilization buffer was diluted to a concentration of 0.2 mg/mL in various concentrations of Mg2+ or Co2+ and aliquoted to a final volume of 50 μL in a clear-bottom 384-well plate (Nunc). The mixture was then covered with 45 μL mineral oil to prevent evaporation and heated up gently from 25 °C to 80 °C using Stargazer-384 (Harbinger Biotech). Data collated was analyzed with Bioactive software (Harbinger Biotech).
Supplementary Material
Acknowledgments
The authors thank Lu Si Yan, Hojjat Eshaghi, and Newsha Sahaf for their technical support; and the beamline personnel at BL13C1 beamline (National Synchrotron Radiation Research Center, Taiwan), MX2 beamline (Australian Synchrotron, Australia), and I04 beamline (Diamond, United Kingdom). A.G. thanks Dr. S. Panjikar for help with AutoRickshaw software (Australian synchrotron). This work was supported by funding from Nanyang Technological University, National Research Foundation Grant NRF-CRP4-2008-02, and the Biomedical Research Council.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org [PDB ID codes 4EGW (2.5 Å) and 4EV6 (3.2 Å)]
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1210076109/-/DCSupplemental.
References
- 1.Knoop V, Groth-Malonek M, Gebert M, Eifler K, Weyand K. Transport of magnesium and other divalent cations: Evolution of the 2-TM-GxN proteins in the MIT superfamily. Mol Genet Genomics. 2005;274(3):205–216. doi: 10.1007/s00438-005-0011-x. [DOI] [PubMed] [Google Scholar]
- 2.Niegowski D, Eshaghi S. The CorA family: Structure and function revisited. Cell Mol Life Sci. 2007;64(19–20):2564–2574. doi: 10.1007/s00018-007-7174-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Papp-Wallace KM, Maguire ME. Bacterial homologs of eukaryotic membrane proteins: The 2-TM-GxN family of Mg(2+) transporters. Mol Membr Biol. 2007;24(5-6):351–356. doi: 10.1080/09687680701441883. [DOI] [PubMed] [Google Scholar]
- 4.Faruk MI, Eusebio-Cope A, Suzuki N. A host factor involved in hypovirus symptom expression in the chestnut blight fungus, Cryphonectria parasitica. J Virol. 2008;82(2):740–754. doi: 10.1128/JVI.02015-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Papp-Wallace KM, et al. The CorA Mg2+ channel is required for the virulence of Salmonella enterica serovar typhimurium. J Bacteriol. 2008;190(19):6517–6523. doi: 10.1128/JB.00772-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu Y, Davis A, Smith BJ, Curtis J, Handman E. Leishmania major CorA-like magnesium transporters play a critical role in parasite development and virulence. Int J Parasitol. 2009;39(6):713–723. doi: 10.1016/j.ijpara.2008.11.010. [DOI] [PubMed] [Google Scholar]
- 7.Kersey CM, Agyemang PA, Dumenyo CK. CorA, the magnesium/nickel/cobalt transporter, affects virulence and extracellular enzyme production in the soft rot pathogen Pectobacterium carotovorum. Mol Plant Pathol. 2012;13(1):58–71. doi: 10.1111/j.1364-3703.2011.00726.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eshaghi S, et al. Crystal structure of a divalent metal ion transporter CorA at 2.9 angstrom resolution. Science. 2006;313(5785):354–357. doi: 10.1126/science.1127121. [DOI] [PubMed] [Google Scholar]
- 9.Lunin VV, et al. Crystal structure of the CorA Mg2+ transporter. Nature. 2006;440(7085):833–837. doi: 10.1038/nature04642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Payandeh J, Pai EF. A structural basis for Mg2+ homeostasis and the CorA translocation cycle. EMBO J. 2006;25(16):3762–3773. doi: 10.1038/sj.emboj.7601269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chakrabarti N, Neale C, Payandeh J, Pai EF, Pomès R. An iris-like mechanism of pore dilation in the CorA magnesium transport system. Biophys J. 2010;98(5):784–792. doi: 10.1016/j.bpj.2009.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xia Y, et al. Co2+ selectivity of Thermotoga maritima CorA and its inability to regulate Mg2+ homeostasis present a new class of CorA proteins. J Biol Chem. 2011;286(18):16525–16532. doi: 10.1074/jbc.M111.222166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marcus Y. Ionic radii in aqueous solutions. Chem Rev. 1988;88(8):1475–1498. [Google Scholar]
- 14.Ohtaki H, Radnai T. Structure and dynamics of hydrated ions. Chem Rev. 1993;93(3):1157–1204. [Google Scholar]
- 15.Payandeh J, et al. Probing structure-function relationships and gating mechanisms in the CorA Mg2+ transport system. J Biol Chem. 2008;283(17):11721–11733. doi: 10.1074/jbc.M707889200. [DOI] [PubMed] [Google Scholar]
- 16.Kucharski LM, Lubbe WJ, Maguire ME. Cation hexaammines are selective and potent inhibitors of the CorA magnesium transport system. J Biol Chem. 2000;275(22):16767–16773. doi: 10.1074/jbc.M001507200. [DOI] [PubMed] [Google Scholar]
- 17.Moomaw AS, Maguire ME. Cation selectivity by the CorA Mg2+ channel requires a fully hydrated cation. Biochemistry. 2010;49(29):5998–6008. doi: 10.1021/bi1005656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dalmas O, et al. Structural dynamics of the magnesium-bound conformation of CorA in a lipid bilayer. Structure. 2010;18(7):868–878. doi: 10.1016/j.str.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beckstein O, et al. Ion channel gating: Insights via molecular simulations. FEBS Lett. 2003;555(1):85–90. doi: 10.1016/s0014-5793(03)01151-7. [DOI] [PubMed] [Google Scholar]
- 20.Tsoka S, Simon D, Ouzounis CA. Automated metabolic reconstruction for Methanococcus jannaschii. Archaea. 2004;1(4):223–229. doi: 10.1155/2004/324925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hattori M, Tanaka Y, Fukai S, Ishitani R, Nureki O. Crystal structure of the MgtE Mg2+ transporter. Nature. 2007;448(7157):1072–1075. doi: 10.1038/nature06093. [DOI] [PubMed] [Google Scholar]
- 22.Hattori M, et al. Mg(2+)-dependent gating of bacterial MgtE channel underlies Mg(2+) homeostasis. EMBO J. 2009;28(22):3602–3612. doi: 10.1038/emboj.2009.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ishitani R, et al. Mg2+-sensing mechanism of Mg2+ transporter MgtE probed by molecular dynamics study. Proc Natl Acad Sci USA. 2008;105(40):15393–15398. doi: 10.1073/pnas.0802991105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miyazawa A, Fujiyoshi Y, Unwin N. Structure and gating mechanism of the acetylcholine receptor pore. Nature. 2003;423(6943):949–955. doi: 10.1038/nature01748. [DOI] [PubMed] [Google Scholar]
- 25.Persson I. Hydrated metal ions in aqueous solution: How regular are their structures? Pure Appl Chem. 2010;82(10):1901–1917. [Google Scholar]
- 26.Doyle DA. Molecular insights into ion channel function (Review) Mol Membr Biol. 2004;21(4):221–225. doi: 10.1080/09687680410001716844. [DOI] [PubMed] [Google Scholar]
- 27.Doyle DA. Structural themes in ion channels. Eur Biophys J. 2004;33(3):175–179. doi: 10.1007/s00249-003-0382-z. [DOI] [PubMed] [Google Scholar]
- 28.Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Panjikar S, Parthasarathy V, Lamzin VS, Weiss MS, Tucker PA. On the combination of molecular replacement and single-wavelength anomalous diffraction phasing for automated structure determination. Acta Crystallogr D Biol Crystallogr. 2009;65(Pt 10):1089–1097. doi: 10.1107/S0907444909029643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Anonymous (2010) The PyMOL Molecular Graphics System (Schrödinger, Cambridge, MA), 1.5.0.3.
- 33.Berka K, et al. MOLEonline 2.0: Interactive web-based analysis of biomacromolecular channels. Nucleic Acids Res. 2012;40(Web Server issue):W222–W227. doi: 10.1093/nar/gks363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lerner MG, Carlson HA. APBS Plugin for PyMOL. Ann Arbor: Univ of Michigan; 2006. [Google Scholar]
- 35.Callenberg KM, et al. APBSmem: A graphical interface for electrostatic calculations at the membrane. PLoS ONE. 2010;5(9) doi: 10.1371/journal.pone.0012722. pii:e12722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc Natl Acad Sci USA. 2001;98(18):10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dolinsky TJ, et al. PDB2PQR: Expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res. 2007;35(Web Server issue):W522–W555. doi: 10.1093/nar/gkm276. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.