Abstract
Eukaryotic elongation factor 1α (eEF-1A) is a multifunctional protein. There are three known posttranslational modifications of eEF-1A that could potentially affect its function. Except for phosphorylation, the other posttranslational modifications have not been demonstrated in plants. Using matrix-assisted laser desorption/ionization-mass spectrometry and peptide mass mapping, we show that carrot (Daucus carota L.) eEF-1A contains a phosphoglycerylethanolamine (PGE) posttranslational modification. eEF-1A was the only protein labeled with [14C]ethanolamine in carrot cells and was the predominant ethanolamine-labeled protein in Arabidopsis seedlings and tobacco (Nicotiana tabacum L.) cell cultures. In vivo-labeling studies using [3H]glycerol, [32P]Pi, [14C]myristic acid, and [14C]linoleic acid indicated that the entire phospholipid phosphatidylethanolamine is covalently attached to the protein. The PGE lipid modification did not affect the partitioning of eEF-1A in Triton X-114 or its actin-binding activity in in vitro assays. Our in vitro data indicate that this newly characterized posttranslational modification alone does not affect the function of eEF-1A. Therefore, the PGE lipid modification may work in combination with other posttranslational modifications to affect the distribution and the function of eEF-1A within the cell.
eEF-1A is an abundant protein that constitutes 1 to 3% of the total soluble protein in the cell (Merrick and Hershey, 1996). Several different activities have been reported for this protein, ranging from its well-established role in protein synthesis (Merrick and Hershey, 1996) and its ability to bind and bundle actin (Demma et al., 1990; Edmonds et al., 1995), activate phosphatidylinositol 4-kinase (Yang et al., 1993), and bind (Durso and Cyr, 1994; Durso et al., 1996) and sever microtubules (Shiina et al., 1994), to its involvement in ubiquitin-dependent degradation of N-acetylated protein (Gonen et al., 1994).
Consistent with a predicted role of eEF-1A in regulating cytoskeletal structure, in situ studies have shown that eEF-1A is associated with both F-actin (Dharmawardhane et al., 1991; Collings et al., 1994; Clore et al., 1996; Sun et al., 1997) and tubulin (Durso and Cyr, 1994; Durso et al., 1996). Importantly, the distribution of eEF-1A changes when Dictyostelium discoideum. is stimulated with cAMP (Dharmawardhane et al., 1991). After adding cAMP, there is a rapid (within 25 s) decrease in eEF-1A associated with F-actin. As new filopodia form (by 90 s), eEF-1A relocalizes to the filopodia F-actin. Recent work (Edmonds et al., 1995; Liu et al., 1996; Murray et al., 1996) has shown that slight changes in pH will affect the actin binding, bundling, and translational activity of eEF-1A. This work suggests that eEF-1A is a very sensitive monitor of the status of protein synthesis and the cytoskeletal network. If eEF-1A serves as an internal cell sensor, changing its location within the cell, then there must be a mechanism regulating eEF-1A. One mechanism by which both the function and distribution of a protein can be altered is posttranslational modification.
At least three types of posttranslational modifications have been reported so far for eEF-1A: Lys residues are methylated (Hiatt et al., 1982; Van Hemert et al., 1984; Fonzi et al., 1985; Amons et al., 1993), Ser residues are phosphorylated (Yang et al., 1993), and Glu residues are modified by the attachment of PGE (Tisdale and Tartakoff, 1988; Dever et al., 1989; Rosenberry et al., 1989; Whiteheart et al., 1989). Even though the amino acid sequence of all eEF-1As remains highly conserved, the presence and/or location of the different posttranslational modifications are not (Cavallius et al., 1993; Browning, 1996). The number and type of methylation (mono, dimethyl, or trimethyl) vary depending on species (Dever et al., 1989). The PGE modification reported in rabbit, mouse, and human was not found in yeast (Cavallius et al., 1993). Although phosphorylation of the elongation factor complex is associated with increased translational activity (Venema et al., 1991; Chang and Traugh, 1997), to our knowledge, only one function of eEF-1A has been correlated with the requirement for a posttranslational modification. eEF-1A must be phosphorylated to be a functional activator of PI-4 kinase (Yang et al., 1993; Yang and Boss, 1994).
We have investigated the PGE posttranslational modification of eEF-1A. The PGE modification has the potential for facilitating association of eEF-1A with lipids or membranes. To study the PGE modification we labeled carrot (Daucus carota L.) suspension-cultured cells with [14C]ethanolamine. The incorporation of ethanolamine into proteins is a rare event and has only been observed in two types of proteins: those with GPI anchors (Aitken, 1992) and eEF-1A with a PGE attachment (Tisdale and Tartakoff, 1988; Dever et al., 1989; Rosenberry et al., 1989; Whiteheart et al., 1989). We will show the incorporation of [14C]ethanolamine into eEF-1A as part of a PGE modification and provide evidence for a phospholipid posttranslational modification, which could provide a mechanism for regulating the distribution of this multifunctional protein within the cell.
MATERIALS AND METHODS
Wild carrot (Daucus carota L.) cells were grown in suspension culture and transferred weekly as previously described (Chen and Boss, 1990). Chicken muscle actin, TCA, and Triton X-114 were purchased from Sigma. QMA Accell Plus and C18 Accell Sep-Pak cartridges were purchased from Waters. [14C]Ethanolamine (55 mCi/mmol), [14C]myristic acid (55 mCi/mmol), [3H]glycerol (20 Ci/mmol), and [32P]Pi (40 mCi mL−1) were purchased from American Radiolabeled Chemicals, Inc. (St. Louis, MO). [14C]Linoleic acid (55.6 mCi/mmol) and En3hance were purchased from DuPont NEN. [3H]Palmitoleic acid (60 Ci/mmol) was a gift from Dr. Leo Parks (North Carolina State University, Raleigh). Arabidopsis (ecotype Landsberg) seeds were grown in Murashige and Skoog liquid medium for 12 d, rotating at 1 rpm, at 23 ± 2°C under constant light from cool-white fluorescent 40-W bulbs. l-(Tosylamido-2-phenyl)ethyl chloromethyl ketone-treated trypsin was purchased from Worthington Biochemical Corporation (Freehold, NJ). Butylated hydroxytoluene was purchased from Calbiochem. TFA sequanal grade and Micro BCA protein assay reagent were purchased from Pierce. Bradford protein assay reagent was purchased from Bio-Rad. A phosphor imager (445SI, Molecular Dynamics) and a phosphor imager tritium screen were used for some of the radioisotopic analysis.
Protein Purification
A rapid method was developed for purifying eEF-1A protein from a small amount of cells using a modification of the method used by Yang et al. (1993). The cells were harvested by filtration (gravity) on Whatman No. 1 paper, weighed, and homogenized using ice-cold buffer at 1 mL/g fresh weight (buffer A: 30 mm Tris, 2 mm EGTA, 1 mm EDTA, 1 mm sodium molybdate, 25% glycerol [v/v], 1 mm PMSF, and 10 mm β-mercaptoethanol). All subsequent procedures were performed at 4°C unless noted otherwise. The homogenate was centrifuged at 700g for 5 min. The 700g supernatant was centrifuged at 40,000g, and the resulting supernatant was used as the soluble-protein fraction. eEF-1A was purified from the 40,000g supernatant using a Sep-Pak Accell QMA Classic cartridge (short body) attached to a 3-cc syringe. The cartridge was equilibrated with 3 mL of buffer B (30 mm Tris-HCl, pH 7.2) by adding buffer to the syringe and forcing it through the column by hand. The 40,000g (7.3 mg/mL, 500 μL) supernatant was placed in the syringe and forced through the cartridge. The protein in the 40,000g supernatant was determined using the modified Bradford assay (Bio-Rad) with BSA as a standard. After applying the protein, two additional 500-μL aliquots of buffer B were forced through the syringe and collected in 1.5-mL Eppendorf tubes to elute eEF-1A (fractions 2 and 3). Remaining protein was eluted with 500 μL of 100 mm KCl (buffer C). To remove noncovalently bound lipids, protein samples were extracted in acidic chloroform:methanol as previously described by Chen and Boss (1990). For trypsin digestion and MALDI-MS analysis the purification was scaled up to use 12-cc Sep-Pak Vac Accell QMA cartridges by increasing 2-fold the amount of 40,000g supernatant applied to the cartridge and by using a vacuum to facilitate elution. The 12-cc Sep-Pak Vac Accell QMA cartridge was equilibrated with buffer B, the 40,000g supernatant was applied, and the column was eluted with 6 mL of buffers B and C.
Isolation of cytoskeleton-associated eEF-1A was performed as described above, except for the use of a different homogenization buffer modified from Abe et al. (1992). The buffer contained 5 mm Hepes, pH 7.0, 10 mm MgCl2, 2 mm EGTA, 1 mm PMSF, and 1 mg/100 mL leupeptin (buffer D).
Isolation of Plasma Membranes
Cells were homogenized in buffer A as described above and centrifuged at 700g for 5 min. The 700g supernatant was centrifuged at 40,000g for 1 h and the resulting pellet was used as the microsomal fraction. Plasma membranes were further purified from the 40,000g pellet by aqueous two-phase partitioning (Wheeler and Boss, 1987). Protein concentration was determined using the microbicinchoninic acid assay.
In Vivo Labeling
Two days after transfer, carrot cells were incubated with [14C]ethanolamine (5–13 μCi/1 g fresh weight), [14C]myristic acid (3.6 μCi/g fresh weight), [3H]glycerol (3.5 μCi/g fresh weight), [14C]linoleic acid (8.8 mCi/g fresh weight), or [3H]palmitoleic acid (35 μCi/g fresh weight) for 48 h. The carrot cells were harvested and homogenized using buffer A, and soluble protein, purified eEF-1A, microsomes, and plasma membranes were obtained as described above.
Arabidopsis (ecotype Landsberg) seedlings were grown in Murashige and Skoog liquid culture for 12 d. Plants were incubated with [14C]ethanolamine (3.5 μCi/3–4 plants) in the liquid medium for 2 d. Plants were rinsed with fresh medium and harvested. The roots were excised from the shoots and leaves. The material was homogenized using buffer A, centrifuged at 700g for 6 min, and the supernatant was centrifuged at 40,000g for 1 h. Aliquots of soluble and microsomal proteins were analyzed by SDS-PAGE, or precipitated with TCA and delipidated as described below. For radioisotopic analysis of SDS-PAGE the polyacrylamide gel was treated with En3hance, dried, and exposed to film.
Tobacco (Nicotiana tabacum cv Wisconsin 38) suspension-cultured cells were grown as described in Roberts et al. (1992). Two days after transfer, cells were incubated with [14C]ethanolamine (4 μCi/g fresh weight) for two d. The cells were harvested and homogenized using buffer A, as described for carrot cells. A 40,000g supernatant and microsomal fraction was obtained for western-blot analysis using an antibody to D. discoideum eEF-1A (1:500 dilution of the antiserum; Demma et al., 1990). The western blot was exposed to film for 1 month.
Analysis of eEF-1A Peptides
[14C]et-eEF-1A ([14C]et-eEF-1A) and nonradioactive eEF-1A from carrot cells were excised from 10% SDS-polyacrylamide gels and digested with l-(tosylamido-2-phenyl)ethyl chloromethyl ketone-treated Trypsin (1:25, w/w) at 30°C for 24 h as described by Stone and Williams (1993). The resulting eEF-1A peptides were concentrated in a SpeedVac to 200 μL. The concentrated peptides were loaded onto a C18 Sep-Pak Classic cartridge of the short-body type (500 μg/200 μL) that had been pre-equilibrated with methanol (7 mL) and then with deionized water (7 mL). The peptides were eluted with 80% acetonitrile in 0.1% TFA. The eluted peptides were concentrated to 200 μL in a SpeedVac and separated on an Ultramex 5 C18 HPLC column (250 × 10 mm, Phenomenex, Torrance, CA) using a nonlinear gradient of 0 to 80% acetonitrile in 0.1% TFA at a flow rate of 0.8 mL/min for 200 min. From time 0 to 5 min a 0% gradient was used, and from 5 to 200 min a 0 to 80% gradient was used. Elution of the [14C]et-eEF-1A peptides was determined by counting an aliquot (200 μL) of each 1-mL fraction collected. Each aliquot was placed in a scintillation vial with Scintiverse II cocktail and counted in a scintillation counter (model LS 7000, Beckman). The fractions containing [14C]ethanolamine were analyzed by MALDI-MS.
MS
MALDI-MS analyses were performed on a Voyager Elite time-of-flight instrument (PerSeptive Biosystems, Framingham, MA) equipped with a N2 laser (337 nm, 2-ns pulse width). Data were acquired in the linear continuous-extraction mode with an acceleration potential of 25 kV. Samples were prepared for analysis by mixing 1 μL of the concentrated HPLC fraction with 1 μL of a saturated solution of 4-hydroxy-α-cyanocinnamic acid matrix in 1:1 acetonitrile: 0.1% aqueous TFA in a well on the sample stage. The solution was allowed to air dry before the sample stage was introduced through a vacuum lock into the instrument. Spectra were produced by averaging transients from 64 laser shots.
TCA Precipitation of [14C]et-eEF-1A
A method was developed for rapid analysis of covalently bound [14C]et-eEF-1A. We precipitated the protein with TCA and then removed noncovalently bound lipid using acetone. Samples (5–20 μg of protein) were spotted onto 1-cm square pieces of Micro Filtration Systems 1514A paper (Pleasanton, CA), placed in a beaker containing ice-cold TCA (5%, w/v; 200 mL/20 filters), and incubated for 5 min on a flat rotary shaker at 100 rpm and 25°C. The TCA was discarded and the filters were washed two more times with an equal volume of ice-cold TCA. After removing the final TCA wash, ice-cold acetone (200 mL/20 filter papers) was added to the beaker to remove noncovalently bound lipid, and filters were washed for 5 min at 200 rpm and 25°C. The acetone was discarded and the filters were washed again with an equal volume of ice-cold acetone. Each filter was dipped sequentially into a second beaker containing ice-cold acetone and then ether. The washed filters were air dried, placed in a scintillation vial with Scintiverse II cocktail (Fisher Scientific), and counted in a Beckman LS 7000 scintillation counter. Data are reported as dpm recovered minus a control of the same size filter paper that was incubated in the same beaker throughout the entire procedure to determine nonspecific binding to the filter paper. Additional controls were done to determine whether each of the isotopes incubated with protein would nonspecifically precipitate and be recovered in the delipidated product.
To test for removal of noncovalently bound lipid, we added [14C]PC to nonradiolabeled protein prior to TCA precipitation. By the third acetone wash, no [14C]PC was detected in the wash and only 23 dpm of the original 13,100 dpm added were recovered with the final washed precipitate (Table I). The entire acetone wash was dried, Scintiverse II cocktail was added, and the wash was counted in a Beckman LS 7000 scintillation counter. Other controls were [14C]ethanolamine plus nonradiolabeled eEF-1A, [14C]myristic acid plus nonradiolabeled eEF-1A, [3H]glycerol plus nonradiolabeled eEF-1A, and [3H]inositol plus nonradiolabeled eEF-1A (Table I). These controls were spotted onto filter paper and washed as described above. The same result observed for [14C]PC was obtained with the other controls except for [3H]inositol. When [3H]inositol was added to nonradiolabeled eEF-1A, the residual dpms indicated that noncovalently bound inositol was recovered. Therefore, this technique was not used for evaluating [3H]inositol incorporation into eEF-1A.
Table I.
A rapid method for analyzing isotope incorporation into eEF-1A
| Sample | Initial Aliquot | Delipidated Protein |
|---|---|---|
| dpm | ||
| [14C]PC + catalase | 1.3 × 104 | 23 ± 0.4 |
| [14C]Ethanolamine + eEF-1A | 3.2 × 105 | 0.19 ± 1.6 |
| [14C]Myristic acid + eEF-1A | 2.1 × 105 | 0 ± 2.7 |
| [3H]Glycerol + eEF-1A | 1.9 × 106 | 0.56 ± 4.0 |
| [3H]Inositol + eEF-1A | 2.8 × 106 | 8.0 ± 4.6 |
| Cold eEF-1A | 0 | 0 |
| [14C]et-EF-1A | 1.7 × 104 | 119.0 ± 31.0 |
[14C]PC, [14C]ethanolamine, [14C]myristate, and [3H]glycerol do not bind nonspecifically to eEF-1A or to catalase. [14C]PC was mixed with catalase (50 μg of protein), precipitated with TCA, and delipidated as described in Methods. Nonradioactive eEF-1A (5 μg of protein) was mixed with [14C]ethanolamine, [14C]myristate, or [3H]glycerol, TCA precipitated, and delipidated as described in Methods. [14C]et-eEF-1A (2 μg of protein) was TCA precipitated and delipidated as described in Methods. The numbers are the average of four samples for [14C]PC, two samples for [14C]ethanolamine, and three samples for [14C]myristic acid, [3H]glycerol, [3H]inositol, and [14C]et-eEF-1A.
Actin-Binding Assay
The actin-binding assay was performed according to the procedure of Demma et al. (1990) with minor modifications. A mixture of chicken muscle actin (0.2 or 0.4 mg/mL) and [14C]et-eEF-1A (0.1 mg/mL) or BSA (0.2 mg/mL), actin alone, or [14C]et-eEF-1A alone was incubated at room temperature for 30 min in 100 μL of actin sedimentation buffer (2 mm MgCl2, 2 mm EGTA, 50 mm KCl, 30 mm Tris-HCl, pH 7.2, 1 μm CaCl2, and 1 mm ATP). The incubated samples were centrifuged in a tabletop microfuge at 15,000 rpm for 30 min. The supernatant was removed and the actin pellet was resuspended in 20 μL of sedimentation buffer. The supernatant and pellet were analyzed by SDS-PAGE and exposed to a phosphor imager tritium screen for 4 weeks.
Triton X-114 Partitioning
Triton X-114 partitioning was performed as described by Bordier (1981). The Triton X-114 was precondensed before use with Tris buffer and butylated hydroxytoluene (Bordier, 1981). The protein samples (15 μg) were prepared in 200 μL of TBS buffer (10 mm Tris-HCl, pH 7.5, 150 mm NaCl) and 0.5% Triton X-114 (v/v) at 0°C. For separation of the proteins, the protein in TBS buffer plus Triton X-114 was layered onto a 6% Suc cushion consisting of 6% Suc (w/v), 10 mm Tris-HCl, pH 7.4, 150 mm NaCl, and 0.6% Triton X-114 in a 1.5-mL Eppendorf microfuge tube. The tube was incubated at 30°C for 5 min then centrifuged for 5 min at 300g. The detergent-rich phase collects as an oily drop at the bottom of the tube. The aqueous phase was removed to a clean Eppendorf tube and mixed with fresh 0.5% Triton X-114. The aqueous phase was incubated at 0°C for 5 min and then relayered onto the Suc cushion, incubated at 30°C for 5 min, and centrifuged for 5 min at 300g. The aqueous phase was removed to a clean Eppendorf tube and mixed with fresh 2% Triton X-114, incubated at 30°C for 5 min, and centrifuged for 5 min at 300g. The aqueous phase was removed to a clean Eppendorf tube, and the protein was precipitated with ice-cold acetone overnight at −20°C and centrifuged at 15,000 rpm for 15 min. The Suc cushion was removed and discarded. The detergent-rich phase and the acetone-precipitated aqueous phase were analyzed by SDS-PAGE and exposed to a phosphor imager tritium screen for 11 d.
RESULTS
[14C]Ethanolamine Is Incorporated into a 49-kD Peptide in Plants
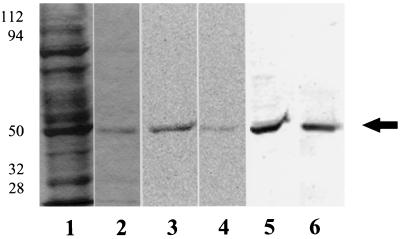
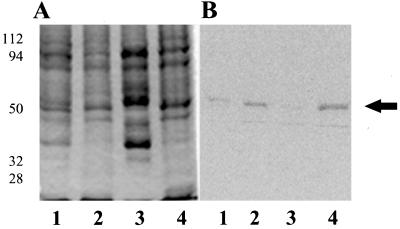
To determine whether [14C]ethanolamine is incorporated into plant proteins, Arabidopsis plants and carrot suspension-cultured cells were incubated with [14C]ethanolamine for 2 d. When proteins from carrot cells were analyzed by SDS-PAGE and autoradiography, only one polypeptide band had incorporated the [14C]ethanolamine (Fig. 1). The apparent molecular mass of the [14C]-labeled band was 49 kD based on molecular mass markers (Fig. 1). The [14C]ethanolamine-labeled protein copurified with carrot eEF-1A, and was recognized by antibodies to D. discoideum eEF-1A (Fig. 1).
Figure 1.
Carrot cells were incubated with [14C]ethanolamine (7 μCi/g fresh weight) and harvested as described in Methods. The 40,000g carrot supernatant (lanes 1, 3, and 5) and purified eEF-1A (lanes 2, 4, and 6) were separated by 10% SDS-PAGE and visualized using Coomassie brilliant blue staining (lanes 1 and 2), a phosphor imager tritium screen exposed for 5 d (lanes 3 and 4), and a western blot with eEF-1A antibody (lanes 5 and 6). Equal protein (1 μg total) was used per lane for the 40,000g supernatant and purified eEF-1A. Molecular mass standards are shown at left (in kD). The experiment was repeated multiple times. The arrow indicates the migration of the 50-kD standard.
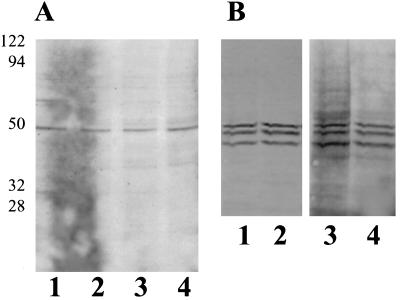
[14C]Ethanolamine-labeled peptides were compared in Arabidopsis and several cell culture lines of carrot, tobacco, and maize endosperm cells. When [14C]ethanolamine-labeled proteins from Arabidopsis seedlings were analyzed, several radiolabeled bands could be detected (Fig. 2A). The predominant [14C]ethanolamine band on a gel was 49 kD and cross-reacted with antibodies to eEF-1A (data not shown). [14C]Ethanolamine was incorporated equally into soluble and microsomal peptides in the root as well as the shoot and leaf fractions. Little or no labeled protein was recovered from rapidly growing carrot cells (5 g harvested/4 d in culture) or from maize endosperm cultures (data not shown). With tobacco cells grown in suspension culture a [14C]ethanolamine-labeled 49-kD peptide was observed on an autoradiograph of a western blot (Fig. 2B).
Figure 2.
Arabidopsis seedlings and tobacco cells grown in suspension culture were incubated with [14C]ethanolamine for 2 d and harvested as described in Methods. A, Arabidopsis protein. Autoradiograph of 10% SDS-PAGE of 40,000g pellet (microsomes) (lanes 1 and 2) and supernatant (lanes 3 and 4) fractions (26 μg of total protein per lane). Lanes 1 and 3, Roots; lanes 2 and 4, leaves and shoots. B, Tobacco protein. Western blot using eEF-1A antibodies (lanes 1 and 2) and autoradiograph of the western blot exposed to film for 1 month (lanes 3 and 4). Lanes 1 and 3, 40,000g supernatant; lanes 2 and 4, 40,000g microsomes. The cross-reacting polypeptides below 49 kD are typical of proteolytic breakdown products. Molecular mass standards are shown at left (in kD).
The most efficient labeling of eEF-1A was found in two slowly growing carrot cell lines. A newly isolated embryogenic cell line, which contained large (1 mm) clusters of cells (10–20 μm in diameter) that were lipid rich based on the fact that a layer of fat was collected during membrane isolation, consistently yielded a high specific activity of purified [14C]et-eEF-1A (50,000 ± 400 dpm/mg). The other culture that had a high specific activity of purified [14C]et-eEF-1A (70,000 ± 8000 dpm/mg) was a slowly growing cell line derived from embryogenic cultures, which had been in culture for over 10 years (Chen and Boss, 1990). The cells appeared as a fine suspension and yielded 0.4 g fresh weight after 4 d in culture. Because eEF-1A was the only protein that incorporated ethanolamine in the carrot cell culture system (Fig. 1), and because the specific activity was highest in the slower-growing cell lines, we used the carrot cells to characterize this posttranslational modification.
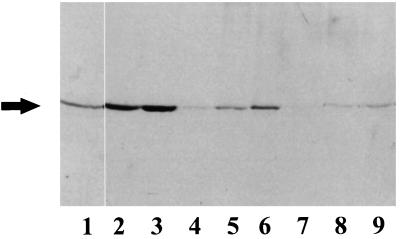
To determine the best time during the cell growth cycle for incorporation of [14C]ethanolamine, carrot cells were labeled for 2-d periods at various times during the cell's 7-d growth cycle and harvested to obtain a 40,000g supernatant and membrane fractions, as described in Methods. The amount of [14C]ethanolamine incorporated into the 49-kD peptide increased over the growth cycle of the culture (Fig. 3). [14C]et-eEF-1A was found in the soluble, endomembrane, and plasma membrane fractions. The specific activity of [14C]et-eEF-1A increased with time of incubation and/or increased concentration of isotope per gram fresh weight of cells. We routinely obtained good incorporation if we incubated log-phase cells for 2 d with 10 μCi [14C]ethanolamine/g fresh weight.
Figure 3.
Carrot cells were incubated with [14C]ethanolamine for 2 d during different times of the growth cycle. Cells were weighed, homogenized, and centrifuged to obtain a 40,000g supernatant, endomembrane, and plasma membrane fraction. The final specific activity recovered was 30 μCi [14C]ethanolamine/g fresh weight for cells labeled from d 1 to 3, 25 μCi/g fresh weight for cells labeled from d 2 to 5, and 22 μCi/g fresh weight for cells labeled from d 5 to 7. Shown is an autoradiograph of 10% SDS-PAGE with 40 μg of total protein/lane for supernatant and endomembrane fractions, 10 μg of total protein for the plasma membrane fraction. Lanes 1 to 3, 40,000g supernatant from cells labeled from d 1 to 3, 2 to 5, and 5 to 7, respectively; lanes 4 to 6, endomembranes; and lanes 7 to 9, plasma membrane of cells from the same respective culture times. The experiment was repeated twice with duplicate samples. A representative experiment is shown. The arrow indicates the migration of the 50-kD standard.
The [14C]Ethanolamine Is Part of a PGE Posttranslational Modification
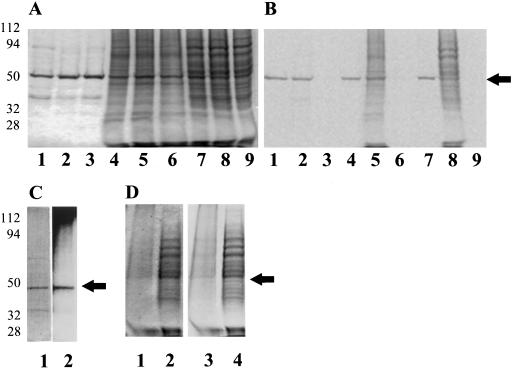
To determine whether [14C]ethanolamine incorporated into eEF-1A is part of the PGE modification, as previously reported in animal cells (Dever et al., 1989; Rosenberry et al., 1989; Whiteheart et al., 1989), or whether it is part of a GPI anchor, cells were incubated with [3H]glycerol, [32P]Pi, [14C]myristic acid, [3H]palmitoleic acid, [14C]linoleic acid, or [3H]inositol, and the incorporation into eEF-1A was compared with that of [14C]ethanolamine. For these experiments, eEF-1A was purified from the 40,000g soluble-protein fraction, and the purified protein was precipitated with TCA, washed with acetone to remove all noncovalently bound lipids, and counted in a scintillation counter or analyzed by SDS-PAGE and autoradiography. Analysis of the protein by SDS-PAGE and autoradiography indicated comigration of [14C]ethanolamine, [14C]myristic acid, [14C]linoleic acid, and [32P]Pi with eEF-1A in both the purified and the 40,000g soluble fractions (Fig. 4). When the amount of [14C]ethanolamine and [3H]glycerol was quantitated per milligram of delipidated protein, there was an increase in incorporation in the column-purified protein compared with the soluble fraction (Table II). This means that eEF-1A was the only or primary protein incorporating these isotopes. The potential for nonspecific lipid binding to the TCA precipitate was tested by incubating [14C]PC with nonradiolabeled protein prior to TCA precipitation. Less than 1% of the total PC added was recovered after TCA precipitation, indicating that the extraction was effective and that noncovalently bound lipids were removed (Table I).
Figure 4.
Carrot cells (d 2) were incubated with [14C]ethanolamine (7.5 μCi/g fresh weight), [14C]myristic acid (25 μCi/g fresh weight), [3H]inositol (50 μCi/g fresh weight), or with [14C]linoleic acid 0.1 mCi/g fresh weight) for 2 d. Cells were placed into phosphate-free medium 30 min before the addition of [32P]Pi (0.4 mCi/g fresh weight) and incubated with isotope for 16 h. Cells were harvested as described in Methods. A, Proteins were separated by 10% SDS-PAGE and visualized by Coomassie brilliant blue staining. Lanes 1 to 3, Partially purified eEF-1A fraction (2 μg of total protein); lanes 4 to 6, microsomal fraction (20 μg of total protein); and lanes 7 to 9, supernatant fraction (20 μg of total protein). In lanes 1, 4, and 7, cells were labeled with [14C]ethanolamine; in lanes 2, 5, and 8 with [14C]myristic acid; and in lanes 3, 6, and 9 with [3H]inositol. B, Image of gel shown in A using a phosphor imager tritium screen exposed for 13 d. C, Purified eEF-1A labeled with [32P]Pi. Lane 1, 10% SDS-PAGE visualized by Coomassie brilliant blue staining (2 μg); lane 2, autoradiograph of [32P]Pi eEF-1A exposed to film for 5 d. D, [14C]Linoleic acid-labeled cells: 40,000g supernatant protein and microsomes. Lanes 1 and 2, 10% SDS-PAGE visualized by Coomassie brilliant blue staining. Lane 1, Microsomes (20 μg of total protein); lane 2, soluble protein (20 μg of total protein); and lanes 3 and 4, autoradiograph of SDS-PAGE. Arrows indicate migration of 50-kD standard. Molecular mass standards are shown at left (in kD). In vivo-labeling experiments were repeated at least twice with duplicates.
Table II.
Components of the carrot eEF-1A PGE posttranslational modification
| Fraction | [14C]Ethanolamine
|
[3H]Glycerol
|
[14C]Myristate
|
|||
|---|---|---|---|---|---|---|
| Initial aliquot | Delipidated product | Initial aliquot | Delipidated product | Initial aliquot | Delipidated product | |
| dpm | dpm/mg protein | dpm | dpm/mg protein | dpm | dpm/mg protein | |
| Supernatant | 3.8 × 105 | 2.8 × 104 ± 2.8 × 103 | 2.8 × 105 | 1.7 × 104 ± 4.3 × 102 | 5.2 × 105 | 1.2 × 105 ± 1.7 × 104 |
| Purified eEF-1A | 1.2 × 107 | 6.9 × 104 ± 8.1 × 103 | 4.6 × 105 | 5.6 × 104 ± 1.3 × 104 | 4.4 × 105 | 1.8 × 104 ± 4.8 × 103 |
| Microsomes | 3.1 × 105 | 6.4 × 104 ± 6.3 × 103 | 3.1 × 105 | 3.7 × 104 ± 3.1 × 103 | 2.5 × 106 | 2.5 × 105 ± 2.1 × 104 |
Carrot cells were labeled with [14C]ethanolamine, [14C]myristic acid, and [3H]glycerol as described in Methods. Supernatant (40,000g), microsomal pellet, and purified eEF-1A fractions were TCA precipitated and delipidated. The numbers for the delipidated protein represent the average of three samples.
When [14C]myristic acid was added to the cells, the specific activity of eEF-1A did not increase with purification. In fact, purified eEF-1A had a lower specific activity than the mixture of myristoylated proteins in the soluble fraction (Table II). This would be anticipated if myristate entered the fatty acid biosynthesis pathway before attachment to eEF-1A. For example, myristate could form a CoA intermediate, be converted to palmitic or linoleic acid, and then be incorporated as one of two fatty acids on the PGE glycerol backbone.
Like myristate, the incorporation of [14C]linoleic acid is consistent with the presence of fatty acids bound via a phospholipid. To test for nonspecific binding of fatty acids, carrot cells were incubated with [3H]palmitoleic acid, a fatty acid only found in a rare lipid modification of proteins (Casey et al., 1994), and the incorporation into purified, delipidated eEF-1A was monitored. Very little incorporation was found (3 dpm above background) even when the cells were incubated with 35 μCi/g fresh weight.
Because inositol phosphates and other inositol metabolites that may coelute with eEF-1A on the mini-columns would precipitate with TCA (Cho et al., 1995), we could not use this method to assess the presence of inositol or phosphate in purified eEF-1A. We could never detect [3H]inositol comigrating with eEF-1A by SDS-PAGE and autoradiography. On one occasion, after a 6-month exposure, a very faint band was detected on an autoradiograph of a western blot with [3H] inositol-labeled partially purified eEF-1A (data not shown). This may have resulted from noncovalently bound phosphatidylinositol, which migrates in the same region on SDS-PAGE (I. Brglez and W.F. Boss, unpublished results).
The incorporation of ethanolamine, glycerol, and phosphate is consistent with a PGE modification. Incorporation of [14C]myristic and [14C]linoleic acid (Fig. 4, B and D) suggests the attachment of one or two fatty acids to the glycerol backbone of the PGE modification. Evidence for the presence of a complete phospholipid moiety, PE, covalently attached to eEF-1A has only been reported in Chinese hamster fibroblast cells (Hayashi et al., 1989). In rabbit myeloma and human T-lymphocyte cells, incorporation of fatty acid into eEF-1A was not reported. With the carrot eEF-1A, the incorporation of [14C]myristic acid into the purified, delipidated protein and comigration of [14C]myristic acid and [14C]linoleic acid with eEF-1A on SDS-PAGE suggests that the entire phospholipid is present.
Because the incorporation of [32P] into eEF-1A could also result from phosphorylation of amino acids (Venema et al., 1991; Yang et al., 1993), and because even a low percentage of radiolabeled lipid adhering to eEF-1A could potentially affect the interpretation of the data, we attempted to hydrolyze the phospholipid moiety and analyze it by TLC. However, after delipidation and hydrolysis of the protein with 6 n HCl for 16 h at 110°C we could not recover a significant amount of the released PGE product for further analysis. Enzymatic hydrolysis of the proteolipid also did not yield sufficient lipid for analysis. For these reasons, we used MS analysis to confirm the presence of a covalently bound PGE modification.
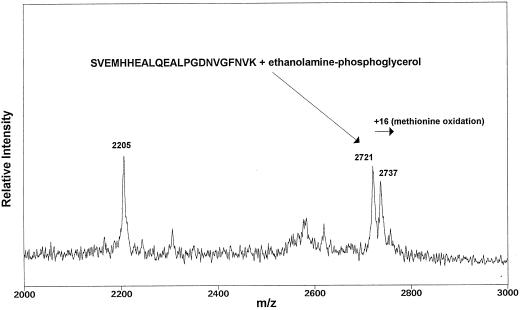
For the MS studies [14C]et-eEF-1A and nonradiolabeled carrot eEF-1A were purified, separated by SDS-PAGE, and digested with trypsin as described in Methods. The tryptic peptides were separated by reverse-phase HPLC, as described in Methods. HPLC fractions containing the [14C]ethanolamine peptides were identified by counting an aliquot of each fraction in Scintiverse II cocktail in a scintillation counter and resulted in a profile identical to that obtained by Whiteheart et al. (1989; data not shown). The major radioactive peaks were combined into three fractions and analyzed by MALDI-MS. Similar spectra were obtained for fractions 2 and 3. Two signals at m/z 2721 and 2737, representing intact molecule plus one proton, were identified as corresponding to the tryptic peptide containing amino acid residues 279–301 with one PGE molecule attached (Fig. 5). The PGE moiety adds 197 mass units to the peptide, which has an intact molecule plus one proton at m/z 2524. The amino acid sequence for the tryptic peptide spanning residues 279 to 301 is SVEMHHEALQEALPGDNVGFNVK. The predicted PGE attachment site is glutamate no. 285 (shown in italic; Dever et al., 1989; Rosenberry et al., 1989; Whiteheart et al., 1989). The second signal at m/z 2737 corresponds to the same peptide where Met oxidation took place, adding an additional 16 mass units to the peptide.
Figure 5.
MS analysis of [14C]et-eEF-1A tryptic peptides. MALDI-MS of concentrated HPLC fractions from two peaks of the [14C]ethanolamine HPLC profile. A signal at m/z 2721 was detected, which corresponds to the predicted mass of tryptic peptide amino acids 279 to 301 covalently bound to one PGE molecule. The signal for peptide 279 to 301 plus PGE moiety was detected from two separate sample preparations.
Conservation of the PGE Attachment Site
A comparison of the amino acid sequences of eEF-1A from other organisms is shown in Table III. Two attachment sites for the PGE posttranslational modification have been identified in rabbit, mouse, and human (analogous to Glu residues 285 and 362), as determined by amino acid sequencing and fast-atom-bombardment MS analysis of eEF-1A tryptic peptides (Dever et al., 1989; Rosenberry et al., 1989; Whiteheart et al., 1989). These two sites are highly conserved in plants with sequence motifs of SVEMHHEA/SLL/QEALPGDNVGFNVK (amino acids 279–301) and KFAEXXK (amino acids 282–288). It is interesting that no ethanolamine labeling has been reported in yeast, and yeast do not have a glutamate residue at position 299 (285 in plants) or the KFAEXXK motif of the second attachment site (amino acids 361–369) (Table III; Cavallius et al., 1993).
Table III.
Sequence comparison of different eEF-1As showing the two putative PGE attachment sites
| Modified Trypsin Peptide | Ethanolamine Incorporation in Vivo | Amino Acids 279 to 301 | Amino Acids 361 to 369 | Reference |
|---|---|---|---|---|
| Carrot1 | Yes | SVEMHHEALQEALPGDNVGFNVK | KFAEIQTK | Kawahara et al., 1992 |
| Carrot2 | ? | –––––––S–L––––––––––––– | ––––LV–– | Apuya and Zimmerman, 1991 |
| PP50 | ? | –––––––S––––––––––––––– | NA | Durso and Cyr, 1994 |
| Tomato | ? | ––––––––––––––––––––––– | –––––L–– | Pokalsky et al., 1989 |
| Wheat | ? | –––––––S–L––––––––––––– | ––––LV–– | Metz et al., 1992 |
| Arabidopsis | Yes | –––––––S–L––––––––––––– | ––S––L–– | Axelos et al., 1989 |
| Maize | ? | ––––––––––––––––––––––– | ––––LV–– | Berberich et al., 1995 |
| Barley1 | ? | –––V–––S–L––––––––––––– | –––––––– | Sutton and Kenefick, 1994 |
| Barley2 | ? | –––––––S–L––––––––––––– | ––––LV–– | Nielsen et al., 1995 |
| Barley3 | ? | –––––––S–L––––––––––––– | –––––––– | Dunn et al., 1993 |
| Soybean | ? | –––––––S–T––H–––––––––– | ––––LM–– | Aguilar et al., 1991 |
| Tobacco | Yes | ––––––––––––––––––––––– | –––––L–– | Kumagai et al., 1995 |
| Bean | ? | –––––––––I––––––––––––– | ––––LL–– | Axelos et al., 1989 |
| D. discoideum | ? | –––––––Q–P––––––––––––– | ––T––VD– | Yang et al., 1990 |
| Schizosaccharomyces pombe | No | –––––––S–DAG––––––––––– | ––––LIE– | Wiebe and Rasmassen, 1995 |
| Saccharomyces cerevisiae | No | –––––––Q–EQGV–––––––––– | R–D–LLE– | Cavallius et al., 1993 |
| Artemia salina | ? | –––––––S–EQ–S–––––––––– | –––Q–KE– | VanHemert et al., 1984 |
| ? | –––––––S–EQ–S–––––––––– | –––––KE– | Lenstra et al., 1986 | |
| Rabbit, mouse, humana | Yes | –––––––––S––––––––––––– | ––––LKE– | Roth et al., 1987; Brands et al., 1986; Uetuski et al., 1989 |
What Is the Distribution of PGE eEF-1A?
To gain some insight into the possible subcellular distribution of the PGE posttranslational modification of eEF-1A, we wondered if [14C]et-eEF-1A was preferentially associated with membranes or the cytoskeleton. The in vivo-labeling studies of carrot cells using [14C]ethanolamine indicated the presence of [14C]et-eEF-1A in the soluble, endomembrane, and plasma membrane fractions of the cell (Fig. 3). The fatty acids on the PGE modification could form a lipid anchor holding eEF-1A to the membranes and cytoskeleton or lipid-associated proteins. Under the homogenizing conditions optimized for purifying soluble protein, most of the eEF-1A was soluble. However, if a homogenizing buffer was used that stabilizes actin filaments (Abe et al., 1992), most (>90%) of the [14C]et-eEF-1A was recovered in the microsomes (Fig. 6). These data suggest that [14C]et-eEF-1A is capable of binding actin.
Figure 6.
The recovery of [14C]et-eEF-1A in the 40,000g microsomal pellet and supernatant was compared using cytoskeleton isolation buffer or regular buffer as described in Methods. A, 40,000g supernatant and microsomal pellet fractions from two different homogenizing buffers separated on 10% SDS-PAGE and visualized with Coomassie brilliant blue staining. Lanes 1and 2, Homogenizing with regular buffer: lane 1, supernatant; lane 2, microsomes. Lanes 3 and 4, Homogenizing with cytoskeleton isolation buffer: lane 3, supernatant; lane 4, microsomes. B, Image of the gel in A using a phosphor imager tritium screen exposed for 4 d. The experiment was repeated two times with duplicates. The arrow indicates the migration of the 50-kD standard.
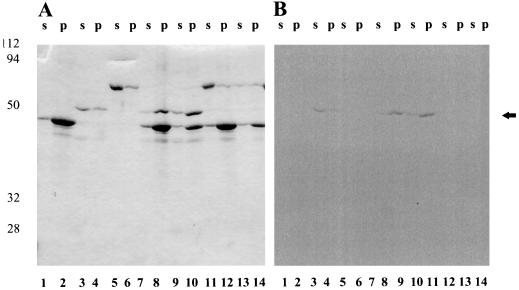
To test F-actin binding, [14C]et-eEF-1A was incubated with different concentrations of actin under actin sedimentation conditions. The supernatant (G-actin) and pellet (F-actin) fractions were analyzed by SDS-PAGE and visualized by staining with Coomassie brilliant blue (Fig. 7) and with a phosphor imager tritium screen. The relative distribution of Coomassie blue-stained protein and [14C]ethanolamine was similar. These data indicate that [14C]et-eEF-1A, like eEF-1A, will bind F-actin, but they do not show preferential actin binding by the [14C]et-eEF-1A.
Figure 7.
Different concentrations of actin and [14C]et-eEF-1A were mixed together in sedimentation buffer for an actin-binding assay as described in Methods. The mixture was separated at 15,000 rpm into supernatant and pellet fractions by centrifugation. A, The supernatant (s) and pellet (p) fractions were analyzed by 12% SDS-PAGE and visualized with Coomassie brilliant blue staining. Lanes 1 and 2, Actin alone (40 μg); lanes 3 and 4, eEF-1A alone (10 μg); lanes 5 and 6, BSA alone (10 μg); lanes 7 and 8, actin (40 μg) plus eEF-1A; lanes 9 and 10, actin (20 μg) plus eEF-1A; lanes 11 and 12 actin (40 μg) plus BSA; and lanes 13 and 14, actin alone (20 μg). B, Image of the gel in A using a phosphor imager tritium screen exposed for 4 weeks. The experiment was repeated three times with duplicates. A representative experiment is shown. The arrow indicates the migration of the 50-kD standard.
Triton X-114 Partitioning of eEF-1A
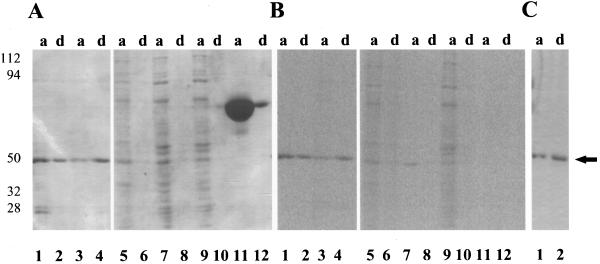
Triton X-114 is a detergent used to separate hydrophilic proteins from amphipathic integral membrane proteins in a temperature-dependent phase separation (Bordier, 1981). Hydrophilic proteins (>80%) from lysed cell extracts and isolated membranes or purified proteins without a lipid anchor remain in the aqueous phase, whereas amphipathic integral membrane and lipid-anchored proteins (>80%) enter the detergent-rich phase after separation (Hooper, 1992). Degradation or cleavage of the protein's lipid anchor will shift partitioning of the protein from the detergent-rich to the detergent-poor phase. Previous work showed that [3H]ethanolamine-labeled eEF-1A from mouse cell lysates partitioned into the aqueous phase (Tisdale and Tartakoff, 1988). To determine the effect of the PGE attachment on carrot eEF-1A Triton X-114 partitioning, eEF-1A labeled with [14C]ethanolamine and [14C]myristic acid was mixed with Triton X-114, and the aqueous and detergent-rich phases were analyzed by SDS-PAGE and exposed to a phosphor imager tritium screen for 11 d. Purified [14C]et-eEF-1A partitioned equally between the aqueous and the detergent-rich phases. The distribution of [14C]myristic acid-labeled purified eEF-1A analyzed by the phosphor imager was consistent with distribution of the Coomassie blue-stained protein. The identical patterns indicate that there was no preference for the [14C]myristoylated isoform to partition into the detergent phase (Fig. 8). Crude eEF-1A from a 40,000g soluble fraction of [14C]ethanolamine-labeled and [14C]myristic acid-labeled cells also partitioned mostly into the aqueous phase. Even when the eEF-1A in the 40,000g microsomal pellet was used, both [14C]et-eEF-1A and [14C]myristic acid-labeled eEF-1A were observed in the aqueous phase. In a separate experiment, reEF-1A from tomato expressed in Escherichia coli with a 6× His tag partitioned equally between the aqueous and detergent-rich phases (Fig. 8C). The increase in the percentage of protein partitioning into the detergent-rich phase with the E. coli-expressed protein, which would not have a PGE modification, probably resulted from an increase in the percentage of denatured protein. Importantly, the reEF-1A was active in an actin-binding assay and, when phosphorylated, activated PI 4-kinase, indicating that the PGE modification was not essential for either actin binding or for PI 4-kinase activation (data not shown).
Figure 8.
Purified [14C]et-eEF-1A, purified [14C]myristate eEF-1A, 40,000g microsomal proteins, BSA, and tomato reEF-1A (15 μg) were partitioned into the aqueous (a) and detergent-rich (d) phase as described in Methods. A, Aqueous and detergent-rich fractions were separated by 10% SDS-PAGE and visualized by Coomassie brilliant blue staining. Lanes 1 and 2, [14C]et-eEF-1A; lanes 3 and 4, [14C]myristic acid eEF-1A; lanes 5 and 6, [14C]myristic acid-labeled 40,000g microsomal pellet; lanes 7 and 8, [14C]ethanolamine-labeled 40,000g supernatant; lanes 9 and 10, [14C]myristic acid-labeled 40,000g supernatant; and lanes 11 and 12, BSA. B, Image of the gel in A using a tritium phosphor imager screen exposed for 4 weeks. C, Tomato reEF-1A visualized by staining with Coomassie brilliant blue staining. Lane 1, Aqueous phase; and lane 2, detergent-rich phase. The experiment was repeated two times with duplicates. A representative experiment is shown. The arrow indicates the migration of the 50-kD standard.
BSA was used as a positive control for aqueous partitioning. Greater than 90% of the BSA was observed in the aqueous phase as visualized by SDS-PAGE stained with Coomassie blue (Fig. 8A). If the PGE had a significant effect on hydrophobicity, [14C]et-eEF-1A and [14C]myristic acid-labeled eEF-1A would have gone preferentially into the detergent-rich phase relative to the amount of protein. There was no evidence of preferential partitioning based on Coomassie blue staining that visualized the total protein and analysis of [14C]et-EF-1A and [14C]myristoylated-eEF-1A by the phosphor imager. Clearly, the PGE modification is not the limiting factor in how eEF-1A partitions.
DISCUSSION
We have shown that [14C]ethanolamine is incorporated into a 49-kD polypeptide in both Arabidopsis and carrot cells grown in suspension culture. In the carrot cells the 49-kD peptide is the only ethanolamine-labeled protein recovered. The [14C]ethanolamine-labeled protein copurified with eEF-1A and cross-reacted with eEF-1A antibodies. Because of previous reports of ethanolamine-labeled and GPI-anchored proteins in plants (Morita et al., 1996; Takos et al., 1997), it was essential to use MS to analyze the composition of the modification. MALDI-MS analysis of tryptic peptides revealed that the purified carrot eEF-1A has a PGE posttranslational modification similar to that reported for rabbit and human eEF-1A (Dever et al., 1989; Rosenberry et al., 1989; Whiteheart et al., 1989). The MALDI-MS data are consistent with the in vivo-labeling data. In addition, the in vivo-labeling studies but not the MALDI-MS data provide evidence for the presence of long-chain fatty acids on the glycerol backbone of the molecule. Both [14C]myristate and [14C]linoleic acid-labeled polypeptides comigrated with eEF-1A in SDS-PAGE and copurified with the delipidated protein. These data are consistent with myristate first being elongated, forming a CoA intermediate, and then being incorporated into the glycerol backbone of the phospholipid. [14C]Linoleic acid was incorporated into several proteins, including eEF-1A. The incorporation of [14C]linoleic acid is less than that of [14C]myristate. This is typical for the incorporation of exogenously added fatty acid into phospholipids and reflects the decreased synthesis of the fatty acyl CoA intermediate by the longer-chain fatty acid. eEF-1A does not contain the typical MGXXXSXX motif considered necessary for N-myristoylation (Johnson et al., 1994). These data, in addition to the observed decreased specific activity of [14C]myristic acid eEF-1A, indicate that myristic acid was not directly bound to the protein but rather entered fatty acid or phospholipid metabolic pathways prior to binding eEF-1A. Previous MS data and labeling studies have demonstrated that rat, rabbit, and human eEF-1A have a PGE group covalently linked via an amide bond to Glu through the ethanolamine nitrogen (Dever et al., 1989; Rosenberry et al., 1989; Whiteheart et al., 1989). Our data indicate that a similar PGE modification to a peptide containing this conserved Glu consensus sequence is present in carrot eEF-1A. The fatty acid ester bond might not have been stable during sample preparation. Alternatively, PE modified peptides (incorporating fatty acyl groups) are not detected because they may have a significantly lower response in MALDI-MS analysis relative to the PGE-modified and unmodified peptides present in the HPLC fractions. However, in vivo-labeling studies confirm the presence of covalently bound fatty acids forming a phospholipid attachment. This type of covalently bound phospholipid could facilitate association of the protein with membrane lipids and hydrophobic proteins in vivo.
Triton X-114 partitioning of both [14C]ethanolamine and [14C]myristic acid eEF-1A did not indicate a clear preference of the lipid-modified proteins for the detergent-rich phase. GPI-anchored proteins, which contain several sugars in addition to PI, preferentially partition to the detergent-rich phase (Hooper, 1992), whereas myristoylated and PGE-modified proteins partition to the aqueous phase (Tisdale and Tartakoff, 1988). Our data indicate that the additional fatty acids on the PGE moiety were not sufficient to enhance partitioning to the detergent-rich phase. The fact that a greater percentage of the E. coli-expressed recombinant protein partitioned into the detergent-rich phase indicated that partitioning correlated with the increase in the percentage of denatured protein and not the PGE modification.
Because eEF-1A is found widely distributed throughout the cell, it is hypothesized that its function and or location may be regulated by different posttranslational modifications. However, except for a requirement for phosphorylation for activation of PI 4-kinase (Yang and Boss, 1994), the effects of posttranslational modifications of eEF-1A have not been demonstrated either in vivo (Cavallius et al., 1997) or in vitro (Venema et al., 1991). We have shown that the PGE modification alone does not appear to be a limiting factor for either actin binding or Triton X-114 partitioning, and that the PGE modification is not essential for PI-4 kinase activation, because phosphorylated reEF-1A, which has no PGE, will activate PI-4 kinase. It may be that the posttranslational modifications, PGE, phosphorylation, and methylation, work in combination to influence the translocation and function of eEF-1A within the cell. We are currently using site-directed mutagenesis of the PGE attachment to study the roles of this posttranslational modification alone and in combination with phosphorylation sites.
ACKNOWLEDGMENTS
We would like to thank Drs. Steve Huber and Marcus Bachman (North Carolina State University, Raleigh) for use of the HPLC and instruction on purification of peptides; Dr. Leo Parks (North Carolina State University, Raleigh) for the gift of the [3H]palmitoleic acid; Drs. Brian Edmonds and John S. Condeelis (Albert Einstein College of Medicine, Bronx, NY) for the antibody to D. discoideum eEF-1A (ABP-50); and Dr. Christine K. Shewmaker (Calgene, Inc., Davis, CA) for the tomato eEF-1A clone.
Abbreviations:
- [14C]et-eEF-1A
[14C]ethanolamine-labeled eEF-1A
- eEF-1A
eukaryotic elongation factor 1α
- GPI
glycosylphosphatidylinositol
- MALDI-MS
matrix-assisted laser desorption/ionization-MS
- PC
phosphatidylcholine
- PE
phosphatidylethanolamine
- PGE
phosphoglycerylethanolamine
- PI-4 kinase
phosphatidylinositol-4 kinase
- reEF-1A
recombinant eEF-1A
- TFA
trifluoroacetic acid
Footnotes
This research was supported by the National Science Foundation (grant no. MCB-9604285 to W.F.B.) and by a Patricia Roberts Harris fellowship to W.D.R. Acquisition of mass spectral data at Michigan State University-National Institutes of Health (NIH) Mass Spectrometry Facility was supported in part by the NIH (grant no. RR00480).
LITERATURE CITED
- Abe S, Ito Y, Davies E. Co-sedimentation of actin, tubulin and membranes in the cytoskeleton fractions from peas and mouse 3T3 cells. J Exp Botany. 1992;43:941–949. [Google Scholar]
- Aguilar F, Montandon P-E, Stutz E. Two genes encoding the soybean translation elongation factor eEF-1α are transcribed in seedling leaves. Plant Mol Biol. 1991;17:351–360. doi: 10.1007/BF00040630. [DOI] [PubMed] [Google Scholar]
- Aitken A (1992) Structural determination of acylated proteins. In NW Hooper, AJ Turner, eds, Lipid Modification of Proteins. IRL Press, New York, pp 63–88
- Amons R, Plujims W, Roobol K, Moller W. Sequence homology between EF-1 alpha, the alpha chain of elongation factor 1 from Artemia salina and elongation factor EF-Tu from Escherichia coli. FEBS Lett. 1993;153:37–42. doi: 10.1016/0014-5793(83)80115-x. [DOI] [PubMed] [Google Scholar]
- Axelos M, Bardet C, Liboz T, Le Van Thai A, Curie C, Lescure B. The gene family encoding the Arabidopsis thaliana translation elongation factor EF-1α: molecular cloning, characterization and expression. Mol Gen Genet. 1989;219:106–112. doi: 10.1007/BF00261164. [DOI] [PubMed] [Google Scholar]
- Berberich T, Sugawara K, Harada M, Kusano T. Molecular cloning, characterization and expression of an elongation factor 1α gene in maize. Plant Mol Biol. 1995;29:611–615. doi: 10.1007/BF00020988. [DOI] [PubMed] [Google Scholar]
- Bordier C. Phase separation of integral proteins in Triton X-114 solution. J Biol Chem. 1981;256:1604–1607. [PubMed] [Google Scholar]
- Brands JHGM, Maassen JA, Van Hemert FJ, Amons R, Moller W. The primary structure of the alpha subunit of human elongation factor 1: structural aspects of guanine-nucleotide-binding sites. Eur J Biochem. 1986;155:167–171. doi: 10.1111/j.1432-1033.1986.tb09472.x. [DOI] [PubMed] [Google Scholar]
- Browning K. The plant translational apparatus. Plant Mol Biol. 1996;32:107–144. doi: 10.1007/BF00039380. [DOI] [PubMed] [Google Scholar]
- Casey WM, Gibson KJ, Parks LW. Covalent attachment of palmitoleic acid (C16:1 Δ9) to proteins in Saccharomyces cerevisiae. J Biol Chem. 1994;269:2082–2085. [PubMed] [Google Scholar]
- Cavallius J, Popkie AP, Merrick WC. Site-directed mutants of post-translationally modified sites of yeast eEF-1A using a shuttle vector containing a chromogenic switch. Biochim Biophys Acta. 1997;1350:345–358. doi: 10.1016/s0167-4781(96)00181-9. [DOI] [PubMed] [Google Scholar]
- Cavallius J, Zoll W, Chakraburtty K, Merrick WC. Characterization of yeast EF-1α: non-conservation of post-translational modifications. Biochim Biophys Acta. 1993;1163:75–80. doi: 10.1016/0167-4838(93)90281-u. [DOI] [PubMed] [Google Scholar]
- Chang YE, Traugh JA. Phosphorylation of elongation factor 1 and ribosomal protein S6 by multipotential S6 kinase and insulin stimulation of translational elongation. J Biol Chem. 1997;272:28252–28257. doi: 10.1074/jbc.272.45.28252. [DOI] [PubMed] [Google Scholar]
- Chen Q, Boss WF. Short-term treatment with cell wall degradation enzymes increases the activity of the inositol phospholipid kinases and the vanadate-sensitive ATPase of carrot cells. Plant Physiol. 1990;94:1820–1829. doi: 10.1104/pp.94.4.1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho MH, Zheng T, Erneux C, Shears SB, Boss WF. The effects of mastoparan on the carrot cell plasma membrane polyphosphoinositide phospholipase C. Plant Physiol. 1995;107:845–856. doi: 10.1104/pp.107.3.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clore AM, Dannenhoffer JM, Larkins BA. EF-1α is associated with a cytoskeletal network surrounding protein bodies in maize endosperm cells. Plant Cell. 1996;8:2003–2014. doi: 10.1105/tpc.8.11.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collings DA, Wasteneys GO, Miyazaki M, Williamson RE. Elongation factor 1α is a component of the subcortical actin bundles of Characean algae. Cell Biol Int. 1994;18:1019–1024. doi: 10.1006/cbir.1994.1025. [DOI] [PubMed] [Google Scholar]
- Demma M, Warren V, Hock R, Dharmawardhane S, Condeelis J. Isolation of an abundant 50,000-dalton actin filament bundling protein from Dictyostelium amoebae. J Biol Chem. 1990;265:2286–2291. [PubMed] [Google Scholar]
- Dever TE, Costello CE, Owens CL, Rosenberry TL, Merrick WC. Location of seven post-translational modifications in rabbit elongation factor 1α including dimethyllysine, trimethyllysine, and glycerylphosphorylethanolamine. J Biol Chem. 1989;264:20518–20525. [PubMed] [Google Scholar]
- Dharmawardhane M, Demma M, Yang F, Condeelis J. Compartmentalization and actin binding properties of ABP-50: the elongation factor-1 alpha of Dictyostelium. Cell Motil Cytoskeleton. 1991;20:279–288. doi: 10.1002/cm.970200404. [DOI] [PubMed] [Google Scholar]
- Dunn MA, Morris A, Jack PL, Hughes MA. A low-temperature-responsive translation elongation factor 1α from barley (Hordeum vulgare L.) Plant Mol Biol. 1993;23:221–225. doi: 10.1007/BF00021434. [DOI] [PubMed] [Google Scholar]
- Durso NA, Cyr RJ. A calmodulin-sensitive interaction between microtubules and a higher plant homolog of elongation factor-1α. Plant Cell. 1994;6:893–905. doi: 10.1105/tpc.6.6.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durso NA, Leslie JD, Cyr RJ. In situ immunocytochemical evidence that a homolog of protein translation elongation factor EF-1α is associated with microtubules in carrot cells. Protoplasma. 1996;190:141–150. [Google Scholar]
- Edmonds BT, Murray J, Condeelis J. pH regulation of the F-actin binding properties of Dictyostelium elongation factor 1α. J Biol Chem. 1995;271:15222–15230. doi: 10.1074/jbc.270.25.15222. [DOI] [PubMed] [Google Scholar]
- Fonzi WA, Katayama C, Leathers T, Sypherd PS. Regulation of protein synthesis factor EF-1α in Mucor racemosus. Mol Cell Biol. 1985;5:1100–1103. doi: 10.1128/mcb.5.5.1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonen H, Smith CE, Siegel NR, Kahana C, Merrick WC, Chakraburtty K, Schwartz AL, Ciechanover A. Protein synthesis elongation factor EF-1α is essential for ubiquitin-dependent degradation of certain Nα-acetylated proteins and may be substituted for by the bacterial elongation factor EF-Tu. Proc Natl Acad Sci USA. 1994;91:7648–7652. doi: 10.1073/pnas.91.16.7648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi Y, Urade R, Utsumi S, Kito M. Anchoring of peptide elongation factor EF-1α by phosphatidylinositol at the endoplasmic reticulum membrane. J Biochem. 1989;106:560–563. doi: 10.1093/oxfordjournals.jbchem.a122895. [DOI] [PubMed] [Google Scholar]
- Hiatt WR, Roberto G, Merrick WC, Sypherd PS. Methylation of elongation factor 1α from the fungus Mucor. Proc Natl Acad Sci USA. 1982;79:3433–3437. doi: 10.1073/pnas.79.11.3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper NM (1992) Identification of a glycosyl-phosphatidylinositol anchor on membrane proteins. In NW Hooper, AJ Turner, eds, Lipid Modification of Proteins. IRL Press, New York, pp 89–115
- Johnson DR, Bhatnagar RS, Knoll LJ, Gordon JI. Genetic and biochemical studies of protein N-myristoylation. Annu Rev Biochem. 1994;63:869–914. doi: 10.1146/annurev.bi.63.070194.004253. [DOI] [PubMed] [Google Scholar]
- Kawahara R, Sunabori S, Fukuda H, Komamine A. A gene expressed preferentially in the globular stage of somatic embryogenesis encodes elongation-factor 1α in carrot. Eur J Biochem. 1992;209:157–162. doi: 10.1111/j.1432-1033.1992.tb17272.x. [DOI] [PubMed] [Google Scholar]
- Kumagai F, Hasezawa S, Takahashi Y, Nagata T. The involvement of protein synthesis elongation factor 1α in the organization of microtubules on the perinuclear region during the cell cycle transition from M phase to G1 phase in tobacco BY-2 cells. Bot Acta. 1995;108:467–473. [Google Scholar]
- Lenstra JA, van Vliet A, Arnberg AC, van Hemert FJ, Moller W. Genes coding for the elongation factor EF-1α in Artemia. Eur J Biochem. 1986;155:475–483. doi: 10.1111/j.1432-1033.1986.tb09514.x. [DOI] [PubMed] [Google Scholar]
- Liu G, Tang J, Edmonds BT, Murray J, Levin S, Condeelis J. F-actin sequesters elongation factor 1α from interaction with aminoacyl-tRNA in a pH-dependent reaction. J Cell Biol. 1996;135:953–963. doi: 10.1083/jcb.135.4.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrick WC, Hershey JWB. The pathway and mechanism of eukaryotic protein synthesis. In: Hershey JWB, Mathews MB, Sonenberg N, editors. Translational Control. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 1996. pp. 31–69. [Google Scholar]
- Metz AM, Timmer RT, Allen ML, Browning KS. Sequence of a cDNA encoding the α-subunit of wheat translation elongation factor 1. Gene. 1992;120:315–316. doi: 10.1016/0378-1119(92)90113-4. [DOI] [PubMed] [Google Scholar]
- Morita N, Nakazato h, Okuyama H, Kim Y, Thompson, GA (1996) Evidence for a glycosylinositolphospholipid-anchored alkaline phosphatase in the aquatic plant Spirodela oligorrhiza. Biochim Biophys Acta 1290: 53–62 [DOI] [PubMed]
- Murray JW, Edmonds BT, Liu G, Condeelis J. Bundling of actin filaments by elongation factor 1 α inhibits polymerization at filament ends. J Cell Biol. 1996;135:1309–1321. doi: 10.1083/jcb.135.5.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokalsky AR, Hiatt WR, Ridge N, Rasmussen R, Houck CM, Shewmaker CK. Structure and expression of elongation factor 1α in tomato. Nucleic Acids Res. 1989;17:4661–4673. doi: 10.1093/nar/17.12.4661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts DM, Besl L, Oh S, Masterson RV, Schell J, Stacey G. Expression of a calmodulin methylation mutant affects the growth and development of transgenic tobacco plants. Proc Natl Acad Sci USA. 1992;89:8394–8398. doi: 10.1073/pnas.89.17.8394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberry TL, Krall JA, Dever TE, Haas R, Louvard D, Merrick WC. Biosynthetic incorporation of [3H]ethanolamine into protein synthesis elongation factor 1α reveals a new post-translational protein modification. J Biol Chem. 1989;264:7096–7099. [PubMed] [Google Scholar]
- Roth WW, Bragg PW, Corrias MV, Reddy NS, Dholakia JN, Wahba AJ. Expression of a gene for mouse eukaryotic elongation factor Tu during murine erythroleukemic cell differentiation. Mol Cell Biol. 1987;7:3929–3936. doi: 10.1128/mcb.7.11.3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiina N, Gotoh Y, Kubomura N, Iwamatsu A, Nishida E. Microtubule severing by elongation factor 1α. Science. 1994;266:282–285. doi: 10.1126/science.7939665. [DOI] [PubMed] [Google Scholar]
- Stone KL, Williams KR. Enzymatic digestion of proteins and HPLC peptide isolation. In: Matsudaira P, editor. A Practical to Protein and Peptide Purification for Microsequencing, Ed 2. San Diego, CA: Academic Press; 1993. pp. 43–69. [Google Scholar]
- Sun Y, Carneiro N, Clore AM, Gloverson ML, Habben JE, Larkins BA. Characterization of maize elongation factor 1A and its relationship to protein quality in the endosperm. Plant Physiol. 1997;115:1101–1107. doi: 10.1104/pp.115.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton F, Kenefick DG. Nucleotide sequence of a cDNA encoding an elongation factor (EF-1α) from barley primary leaf. Plant Physiol. 1994;104:807. doi: 10.1104/pp.104.2.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takos AM, Dry IB, Soole KL. Detection of glycosyl-phosphatidylinositol-anchored proteins on the surface of Nicotiana tabacum protoplasts. FEBS Lett. 1997;405:1–4. doi: 10.1016/s0014-5793(97)00064-1. [DOI] [PubMed] [Google Scholar]
- Tisdale EJ, Tartakoff AM. Extensive labeling with [3H]ethanolamine of a hydrophilic protein of animal cells. J Biol Chem. 1988;263:8244–8252. [PubMed] [Google Scholar]
- Uetuski T, Naito A, Nagata S, Kaziro Y. Isolation and characterization of the human chromosomal gene for polypeptide chain elongation factor-1α. J Biol Chem. 1989;264:5791–5798. [PubMed] [Google Scholar]
- Van Hemert FJ, Amons R, Pluijms WJM, van Ormondt H, Moller W. The primary structure of elongation factor EF-1α from the brine shrimp Artemia. EMBO J. 1984;3:1109–1113. doi: 10.1002/j.1460-2075.1984.tb01937.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venema RC, Holme IP, Traugh JA. Phosphorylation of elongation factor 1 (EF-1) and valyl-tRNA synthetase by protein kinase C and stimulation of EF-1 activity. J Biol Chem. 1991;266:12574–12580. [PubMed] [Google Scholar]
- Wheeler JJ, Boss WF. Polyphosphoinositides are present in plasma membranes isolated from fusogenic carrot cells. Plant Physiol. 1987;85:389–392. doi: 10.1104/pp.85.2.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteheart SW, Shenbagamurthi P, Chen L, Cotter RJ, Hart GW. Murine elongation factor 1α (EF-1α) is posttranslationally modified by novel amide-linked ethanolamine-phosphoglycerol moieties. J Biol Chem. 1989;264:14334–14341. [PubMed] [Google Scholar]
- Yang F, Demma M, Warren V, Dharmawardhane S, Condeelis J. Identification of an actin-binding protein from Dictyostelium as elongation factor 1a. Nature. 1990;347:494–496. doi: 10.1038/347494a0. [DOI] [PubMed] [Google Scholar]
- Yang W, Burkhart W, Cavallius J, Merrick WC, Boss WF. Purification and characterization of a phosphatidylinositol 4-kinase activator in carrot cells. J Biol Chem. 1993;268:392–398. [PubMed] [Google Scholar]
- Yang W, Boss WF. Regulation of phosphatidylinositol 4-kinase by the protein activator PIK-A49: activation requires phosphorylation of PIK-A49. J Biol Chem. 1994;269:3852–3857. [PubMed] [Google Scholar]