Abstract
Tripartite motif protein isoform 5 alpha (TRIM5α) is a potent antiviral protein that restricts infection by HIV-1 and other retroviruses. TRIM5α recognizes the lattice of the retrovirus capsid through its B30.2 (PRY/SPRY) domain in a species-specific manner. Upon binding, TRIM5α induces premature disassembly of the viral capsid and activates the downstream innate immune response. We have determined the crystal structure of the rhesus TRIM5α PRY/SPRY domain that reveals essential features for capsid binding. Combined cryo-electron microscopy and biochemical data show that the monomeric rhesus TRIM5α PRY/SPRY, but not the human TRIM5α PRY/SPRY, can bind to HIV-1 capsid protein assemblies without causing disruption of the capsid. This suggests that the PRY/SPRY domain alone constitutes an important pattern-sensing component of TRIM5α that is capable of interacting with viral capsids of different curvatures. Our results provide molecular insights into the mechanisms of TRIM5α-mediated retroviral restriction.
TRIM5α potently inhibits infection by HIV-1 and other retroviruses at an early postentry stage in a species-specific manner (1). Rhesus TRIM5α (rhTRIM5α) potently blocks HIV-1 infection (2). In contrast, human TRIM5α (huTRIM5α) only weakly inhibits HIV-1, but potently restricts N-tropic murine leukemia viruses (N-MLV) (3, 4). TRIM5α is able to induce premature capsid disassembly (5) and activate downstream innate immune responses upon recognizing the retroviral capsid lattice (6).
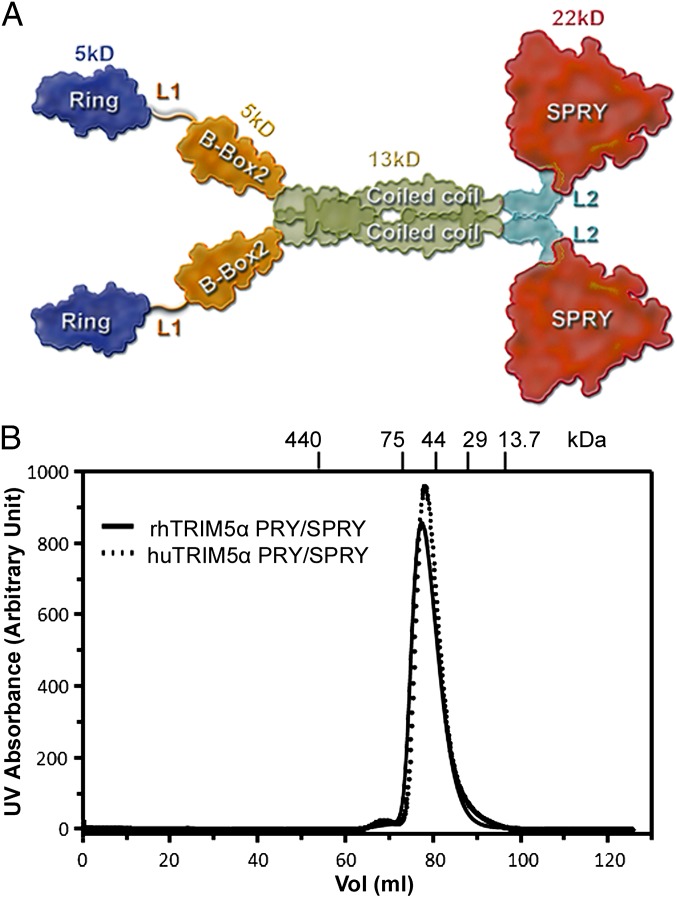
TRIM5α is composed of RING, B-box 2, coiled-coil (CC), and B30.2 (PRY/SPRY) domains (Fig. 1A), similar to many tripartite/RBCC motif (TRIM) family members (7). The RING domain functions as an E3 ubiquitin ligase (8); the B-box 2 domain mediates formation of higher-order structure and self-association (9, 10); and the coiled-coil domain mediates dimerization (11) and facilitates the formation of the hexagonal lattice (12). The PRY/SPRY domain is essential for recognition of retroviral capsids and determines the specificity of restriction (13, 14). Two linker regions, L1 and L2, separate RING/B-box 2 and coiled-coil/(PRY/SPRY) domains, respectively (Fig. 1A). The antiviral potency of TRIM5α has been shown to correlate with its affinity for the viral capsid lattice (5). Interestingly, a single amino acid change from arginine to proline at residue 332 (R332P) in the PRY/SPRY domain of huTRIM5α conferred the ability to restrict HIV-1 (14–16).
Fig. 1.
(A) Schematic depiction of dimeric TRIM5α. The four domains are colored differently and their respective molecular masses are indicated. The two linker regions are labeled L1 and L2. (B) Size-exclusion chromatograms of rhesus (solid) and human (dashed) MBP-TRIM5α PRY/SPRY. The molecular masses of protein standards are indicated at the top.
HIV-1 capsid (CA) proteins can assemble into closed fullerene cones or helical tubes; other structurally homologous retrovirus CA proteins form cylindrical or spherical capsids (17–19). Despite the diverse array of retroviral capsids, different shapes are recognized by the same TRIM5α protein or highly homologous orthologs (20). The binding interaction requires an assembled capsid lattice as individual CA molecules do not have an appreciable affinity to TRIM5α (21). This broad, yet specific lattice pattern-sensing ability resides in the capsid-recognition PRY/SPRY domain of TRIM5α (5, 13, 14, 16, 22). A TRIM5α truncation construct containing the coiled-coil and the PRY/SPRY domains (CC-SPRY) is sufficient to bind and disrupt HIV-1 CA assembly (23). However, the lack of detailed structural data on TRIM5α PRY/SPRY poses an obstacle to understanding the pattern-sensing mechanism by which TRIM5α interacts with the retrovirus capsid. Here we report the crystal structure of the rhTRIM5α PRY/SPRY domain that reveals important features for viral capsid recognition. Both our EM and biochemical data demonstrate that rhTRIM5α PRY/SPRY (PRY/SPRYrh) alone is able to recognize HIV-1 capsid tubes, whereas huTRIM5α PRY/SPRY (PRY/SPRYhu) cannot. These findings provide a structural framework that enables us to begin understanding the capsid pattern-recognition mechanisms of TRIM5α.
Results
TRIM5α PRY/SPRY Is a Monomer in Solution.
It has been shown that a full-length TRIM5α chimera (TRIM5Rh-21R) exists as a mixture of monomers and dimers whereas a truncated TRIM5α (CC-SPRY) is a dimer (11, 23, 24). We investigated the oligomerization state of PRY/SPRYrh and PRY/SPRYhu. To overcome the poor yield and solubility problem of TRIM5α PRY/SPRY, either PRY/SPRYrh or PRY/SPRYhu was fused to the end of the C-terminal helix of the maltose binding protein (MBP) (25), resulting in MBP-PRY/SPRYrh275 (residues 275–493) and MBP-PRY/SPRYhu273 (residues 273–489). Typical yields for these fusion proteins were over 10 mg/L of Escherichia coli cells. Both fusion proteins are very soluble (>10 mg/mL) and elute as monodispersed peaks from a size exclusion column (Fig. 1B), with the elution volumes precisely matching those expected for monomers. These data confirm that PRY/SPRYrh and PRY/SPRYhu are monomeric in solution and are consistent with the notion that the coiled-coil domain is critical for TRIM5α dimerization.
Crystal Structure of MBP-PRY/SPRYrh.
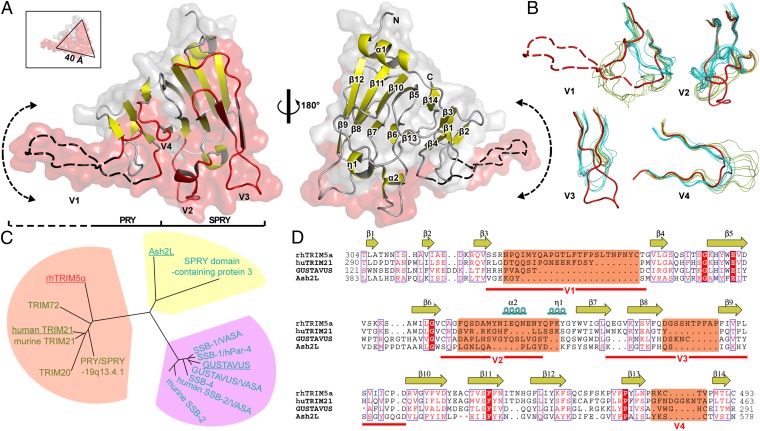
We determined the crystal structure of MBP-PRY/SPRYrh275 at a resolution of 3.3 Å (Table S1 and Fig. S1). Two independent fusion protein molecules with an identical conformation (rmsd 0.1 Å) were observed in the asymmetric unit of the crystal. Similar to the known structures of the PRY/SPRY domain of other proteins, PRY/SPRYrh adopts a highly distorted β-sandwich fold, comprising two, seven-stranded antiparallel β-sheets with an α-helix at the N terminus, beginning with residue 287 (Fig. 2A and Fig. S2). TRIM5α residues before 287 were not observed in the crystal structure and are most likely disordered, except residues 275–279 that connect PRY/SPRYrh to the C-terminal helix of MBP. This segment is part of the L2 region (residues 234–296) of TRIM5α (28) and is presumably unstructured because the majority of the L2 residues are not included in the construct.
Fig. 2.
PRY/SPRYrh is a conserved module with unique structural features. (A) Two views of the crystal structure of PRY/SPRYrh in ribbon and surface representations. Density for V1 is not observed. An example conformation of the V1 loop obtained by homology modeling is shown by the black dashed line in the molecular surface (red). The arrowed line around this part of the structure indicates the conformational space available to V1. The variable regions (V1–V4) are colored in red. (Inset) Dimensions of the triangular-shaped core. (B) Four variable regions are shown separately by superimposing 15 SPRY-containing protein structures. TRIM5α is colored red and the others are grouped into two clusters, colored cyan and green, respectively. (C) Structure-based phylogenetic tree of the 15 SPRY-containing proteins [using the program SHP (26) and PHYLIP (27)]. The following structures with PDB ID in parentheses are included: PRY/SPRY-19q13.4.1 (2FBE), human TRIM21 (2IWG), murine TRIM21 (2VOK), TRIM20 (2WL1), TRIM72 (2KB5), Ash2L (3TOJ), SPRY-containing protein3 (2YYO), GUSTAVUS (2FNJ), GUSTAVUS/VASA (2IHS), SSB-1/VASA (3F2O), SSB-1/hPar-4 (2JK9), SSB-4 (2V24), human SSB-2/VASA (3EMW), and murine SSB-2 (3EK9). Protein names are color-coded according to B. Proteins analyzed in D are underlined. (D) Structure-based amino acid sequence alignment. The secondary structure elements of rhTRIM5α are indicated. Fully conserved residues are marked in the solid red box; similar residues are shown in red type. Residues that do not fit into the structure-based alignment are shaded in semitransparent red. Large protein-specific insertions were omitted in regions where orange dots are shown.
A distinct feature revealed by the structure is that the variable regions (V1–V3) that harbor the viral capsid-interacting residues (29–31) form a surface on one side of the molecule (Fig. 2A; Figs. S2 and S3). This surface likely constitutes the capsid-binding interface. This surface includes a triangular shaped, ∼40 Å large core region that is largely flat. The bulk of the variable regions is composed of loops that extend out from the core fold. Most of the V1 loop (residues 328–347) is disordered and is not observed in the crystal, highlighting most likely its intrinsic flexibility. The electron density for residues 384–388 in V2 is observed, but poorly defined. The V3 loop is well ordered by packing interactions with a neighboring molecule in the crystal. The V4 loop that has not been implicated in capsid binding is also well structured. The flexible V1–V3 loops have the ability to adopt a variety of different conformations, thereby enlarging the putative capsid-binding interface to up to ∼70 Å in dimension. This ability may have profound implications in recognizing viral capsids of different curvatures (discussed below).
We carried out homology modeling to gain more insight into the V1 loop. The I-TASSER program (32) was used to model the PRY/SPRYrh structure based on the crystal structures of multiple homologous proteins. As expected, the core fold of PRY/SPRYrh is predicted consistently, whereas the predicted conformation of the V1 loop varies significantly among the top homology models produced. This is consistent with the flexible nature of the V1 loop. An example of the conformation is shown in Fig. 2. The V1 loop contains many important residues in retroviral capsid interaction, and its flexibility may allow these residues to recognize various viral capsids of different shapes (discussed below). Regardless of the conformation of the V1 loop, some critical residues (residues 332, 335, and 336 of huTRIM5α) that confer restriction specificity (15, 16, 33, 34) are located in the middle of V1. This location could place the residues at the tip of the V1 loop when it is extended (Fig. 2D and Fig. S3). It has been demonstrated that a swap of the V1 region of rhTRIM5α and huTRIM5α can change the restriction specificity toward MLV CA mutants harboring mutations in the β1/β2 region (30), supporting a potential interaction between TRIM5α V1 and CA β1/β2.
We compared the conformations of the variable regions of PRY/SPRYrh with those in homologous proteins to gain further structural insight. The crystal structures of 14 SPRY-containing proteins of different functions are available in the Protein Data Bank (PDB). Structural superposition of all 14 structures reveals that the disposition of the variable regions falls roughly into two groups (Fig. 2B), indicating their well-defined structures and low degree of variability within each group. This analysis also suggests that the positions of the loops from the TRIM family cluster, except TRIM5α, may be inherited from a common ancestor, whereas those from the other two clusters may derive from another origin (Fig. 2 B and C). In contrast, V1–V3 of PRY/SPRYrh do not fall into either group, and V1 is significantly longer. These findings reinforce the notion that the variable regions of TRIM5α PRY/SPRY are mostly flexible and have evolved for antiviral activity (29–31).
TRIM5α PRY/SPRY Is an Evolutionarily Conserved Module That Acquired Unique Features for Retrovirus Capsid Recognition.
Our crystal structure allowed us to carry out a structure-based sequence alignment to further examine the details of the antiviral elements of TRIM5α PRY/SPRY. The SPRY domain is a commonly used protein–protein interaction module in many proteins from a variety of species. Distinct from its evolutionary ancestor SPRY, the B30.2 (PRY/SPRY) domain is found only in vertebrates and contains an additional PRY, N-terminal to SPRY (35). Over 150 human proteins contain B30.2 and SPRY domains, functioning in diverse cellular processes (7). We analyzed the evolutionary relationship between TRIM5α PRY/SPRY and other B30.2 and SPRY domains by structural alignment. Despite the extensive amino acid sequence variation (identity ranges from below 10% to 40%), the fold of the SPRY domain is highly conserved (backbone rmsd of 0.42–1.93 Å). This suggests that the SPRY fold is a robust scaffold that can be modulated by sequence variation for distinct cellular functions.
We generated a structure-based phylogenetic tree for the PRY/SPRY and SPRY lineage among all available structures (Fig. 2C), using the Structure Homology Program (26). This tree shows that all of the TRIM family members that contain PRY, including rhTRIM5α, are clustered together. Those lacking PRY (SSB-2) or exhibiting an ambiguous PRY (GUSTAVUS) fall into a separate group. The structure of PRY/SPRYrh reveals that some regions implicated in viral capsid recognition, i.e., V2–V3, are located in the SPRY subdomain whereas the most critical V1 resides in the PRY subdomain (Fig. 2 A and D). This suggests that the acquisition of the PRY subdomain into the more ancient SPRY domain may correlate with the emergence of a viral capsid-sensing capacity in vertebrates.
Monomeric PRY/SPRYrh Interacts with Individual CA Hexamers Very Weakly.
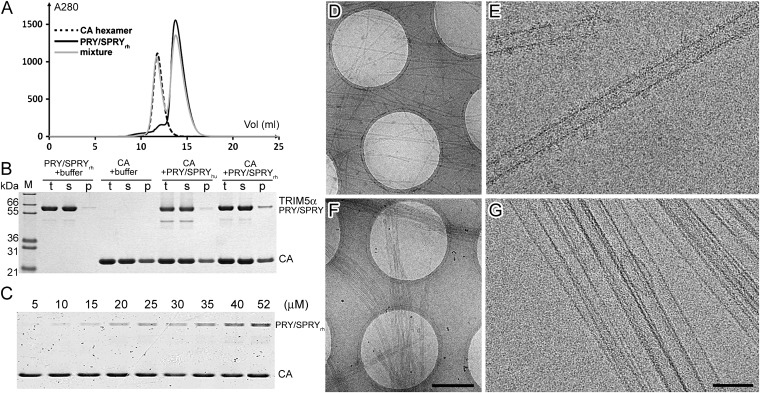
To investigate the binding strength between monomeric PRY/SPRYrh and hexameric CA, we examined the binding affinity between monomeric PRY/SPRYrh and individual CA hexamers (A14C/E45C/W184A/M185A) (36) in solution by size-exclusion chromatography. No complex formation was detected even at very high concentrations of both proteins (400 μM CA and 200 μM PRY/SPRYrh). The elution profiles of the mixture of the CA hexamers and PRY/SPRYrh overlay precisely with their individual profiles (Fig. 3A), without any shift of peak positions expected for complex formation. The results demonstrate that monomeric PRY/SPRYrh has a very weak binding capacity to individual CA hexamers, implying that higher-order assemblies of CA and/or TRIM5α are required for efficient capsid interaction.
Fig. 3.
Interaction of TRIM5α PRY/SPRY domains with CA hexamers and wild-type CA tubular assemblies. (A) Size-exclusion chromatographic profiles of individual CA hexamers (CA concentration 400 μM) (black dashed line), PRY/SPRYrh (200 μM) (black solid line), and their mixture (gray solid line). (B and C) Binding of MBP-TRIM5α PRY/SPYR to preassembled wild-type CA tubes. Binding reactions were analyzed by SDS/PAGE using CA tubular assemblies (69 μM), incubated with MBP-PRY/SPRYrh (24 μM), MBP-PRY/SPRYhu (21 μM), and binding buffer (B) or MBP-PRY/SPRYrh at various concentrations of 5–52 μM (C). Samples of the reaction mixture before centrifugation (t), of supernatant (s), and of pellet (p) are shown. (D–G) Cryo-EM images of the reaction mixture. Low-dose projection image of wild-type CA tubes (72 µM) incubated with MBP-TRIM5α PRY/SPRY at nominal magnifications of 4,700× (D and F) and 59,000× (E and G). CA tubes are well separated (D) and decorated with protein density (E) upon binding of MBP-PRY/SPRYrh (24 µM). Incubation with MBP-PRY/SPRYhu (21 µM) yields bundled CA tubes (F) similar to CA assembly alone, and no additional protein density is observed on the CA tube surface (G). (Scale bars: 1 µm in D and F; 100 nm in E and G.)
Monomeric PRY/SPRYrh Is Sufficient to Recognize HIV-1 Capsid but Does Not Disrupt the CA Tubular Assembly.
We performed further dissection of the TRIM5α-CA interaction to address the mechanistic question of whether the PRY/SPRYrh domain alone is sufficient to recognize the HIV-1 capsid. It had been demonstrated previously that rhTRIM5α (11, 37) and its truncation containing the coiled-coil and PRY/SPRY domains (23) can bind and disrupt HIV-1 CA/CA-NC assembled into tubes or purified viral cores. We used monomeric MBP-PRY/SPRYrh (24 μM) or MBP-PRY/SPRYhu (21 μM) in a precipitation assay (23) with preassembled CA tubes (69 μM). MBP-PRY/SPRYrh copelleted with assembled CA, demonstrating binding, whereas essentially no binding was observed for MBP-PRY/SPRYhu under the same assay conditions (Fig. 3B). Similar to rhTRIM5α CC-SPRY, the binding of MBP-PRY/SPRYrh to CA tubes exhibited a clear dose dependence (Fig. 3C). Compared with the dimeric rhTRIM5α CC-SPRY (23), monomeric MBP-PRY/SPRYrh displayed a lower binding ratio, consistent with previous findings that showed an increase in binding affinity upon dimerization (11).
Cryo-electron microscopy (cryo-EM) was used to confirm binding of PRY/SPRYrh to HIV-1 CA tubes and to probe whether monomeric PRY/SPRYrh can disrupt these tubes (Fig. 3 D–G). We obtained cryo-EM images that clearly show decoration of the tubular CA assemblies by MBP-PRY/SPRYrh on the surface of the tubes (Fig. 3E). In addition, these CA tubes are well separated (Fig. 3D), which differs from the bundled appearance of the CA tubes without TRIM5α (38). In contrast, using MBP-PRY/SPRYhu in the same assay, the CA tubes remain bundled (Fig. 3F), and no additional density can be observed on their surfaces (Fig. 3G). Unlike full-length rhTRIM5α and rhTRIM5α CC-SPRY, the binding of MBP-PRY/SPRYrh does not cause shortening or destruction of the CA helical assemblies (Fig. 3 D and E). Thus, the above findings demonstrate that the monomeric PRY/SPRY likely constitutes an important part of the pattern-sensing module of TRIM5α that is capable of recognizing assembled CA, but cannot disrupt the HIV-1 capsid.
Discussion
TRIM5α has evolved to be a potent antiviral restriction factor with very unique properties. It has strong species specificity that determines the tropism of retroviruses such as HIV/SIV and MLV (3, 4, 39, 40). At the same time, it possesses a pattern-sensing ability that recognizes retroviral capsids of a broad range of shapes. The viral capsid proteins are highly similar in structure, with the predicted TRIM5α-binding regions located at the same surface areas (Fig. S4), even though the L4/5 region of HIV CA is much longer than the corresponding region in MLV CA. An emerging consensus is that a conserved TRIM5α-binding mode exists for various viral capsids and that the surface details of the CA lattice dictate the species-specific interaction with TRIM5α (5, 14, 22, 41–43). On the other hand, TRIM5α possesses an intriguing curvature-independent pattern-recognition property. Given that TRIM5α proteins bind to the contiguous surface of the HIV capsid that comprises varying interhexamer interfaces, the flat CA lattice formed in vitro, as well as the spherical surface of the N-MLV capsid (4, 12), it must be able to recognize CA assemblies with different curvatures.
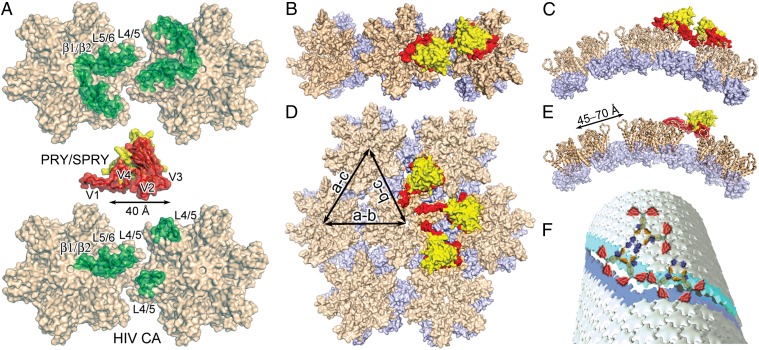
Our results and the large body of available data on TRIM5α-CA-capsid recognition allow for a mechanistic interpretation of this interaction at a molecular level. First, our crystal structure reveals the most likely CA-binding interface on PRY/SPRYrh that contains all elements of positive selection (41) and mutational hot spots identified previously (15, 16, 29–31, 40–42, 44) (Figs. 2A and 4A; Fig. S3). Second, the TRIM5α-binding site has been mapped to encompass the entire outer surface of CA (Fig. 4A and Fig. S4), including the β1/β2 hairpin, L5/6, and L4/5 regions in HIV CA and the corresponding sites on MLV CA (30, 33, 45–50). Specifically, TRIM5α V1 may interact with CA β1/β2 at the center of the CA hexamer, as swapping the V1 region of rhTRIM5α and huTRIM5α resulted in a change of the restriction specificity toward MLV CA with mutations in the β1/β2 region (30). Third, our cryo-EM and cosedimentation experiments reveal that the PRY/SPRYrh domain retains the ability to bind HIV-1 CA helical tubes without breaking the assembly. Importantly, our crystal structure of PRY/SPRYrh, the crystal structure of the HIV-1 CA hexamer lattice (36, 51), and the cryo-EM maps of HIV-1 capsid and helical tubes (18, 38) together establish a structural framework for further detailed investigation of this lattice-specific interaction.
Fig. 4.
Analysis of the interaction of the PRY/SPRYrh domain and assembled HIV CA tubes. (A) The potential binding surfaces on the PRY/SPRY domain and two neighboring HIV CA hexamers are colored in red and green, respectively, in intrahexamer (Upper) and interhexamer (Lower) binding scenarios. The molecules are drawn to the same scale and are shown in surface representations with the interaction elements marked. The HIV-1 CA model is from the hexamer crystal structure (PDB ID: 3H4E). (B) A conceptual model of two PRY/SPRY molecules (red and yellow surface) with each binding to a neighboring CA hexamer (tan: N-terminal domain; light blue: C-terminal domain) separately. (C) Side view of B. (D) A conceptual model for binding between the PRY/SPRY domain and interhexamer CA interfaces in different directions on an HIV-1 CA tubular assembly. Note that the binding surface in the PRY/SPRY core (red) fits well at the CA hexamer interface (buried in the top view of the CA hexamer assembly) and that the flexible V1 loop adopts various conformations to fit the varying curvature along different directions. The CA tube model was created by docking the crystal structure of HIV-1 CA hexamer (PDB ID: 3H4E) to the EM map of the HIV-CA helical tube (Electron Microscopy Data Bank accession code: EMD-5136). (E) A side view of PRY/SPRY binding to the interhexamer interface in the a-b direction in D. The CA-NTD is shown in ribbon representation. The arrow marks the range of distances that PRY/SPRY needs to span across neighboring CA hexamers in the HIV capsid. (F) A schematic depiction of how the dimeric TRIM5α causes the disruption of the HIV CA tubular assembly.
A potential binding mode is that the PRY/SPRY domain primarily interacts with a hexamer CA unit and that the dimerization or higher-order oligomerization of TRIM5α allows for the recognition of the capsid lattice (Fig. 4 A–C). It is interesting to note that the putative interaction interface on PRY/SPRY appears to be larger than that on a single CA protein (Fig. 4A). TRIM5α PRY/SPRY therefore may contact more than one CA monomer in the hexamer unit, or even in a second CA hexamer nearby. A TRIM5α dimer can then bridge two CA hexamers, each of which is primarily recognized by one PRY/SPRY domain (Fig. 4 B and C). The L2 linker region may have a certain degree of flexibility that allows the PRY/SPRY dimer to adapt to the capsid lattice with varying curvatures (Fig. S5A). The higher oligomerization or the lattice formation of TRIM5α (10–12) may further enhance the capsid pattern-sensing ability of the molecule.
Alternatively, a single PRY/SPRY domain may constitute an important part of the pattern-sensing module of TRIM5α and may be capable of binding capsids of various shapes. It has been established that individual CA molecules do not have an appreciable affinity to TRIM5α (21), and our data show that the CA hexamer interacts with PRY/SPRYrh very weakly (Fig. 3A). In contrast, the preassembled CA lattice has a stronger interaction with PRY/SPRYrh, as we could detect substantial binding to CA tubular assemblies (Fig. 3 B and D). This corroborates the observation that monomeric TRIM5-21R retains its ability to interact with CA tubes, where the binding ratio of monomeric TRIM5-21R to CA has only a modest reduction (two- to threefold less at 0.15–0.6 μM TRIM5-21R) (11) compared with that of dimeric TRIM5-21R to CA. The increased binding strength to the CA lattice suggests that monomeric PRY/SPRY likely engages in interhexamer binding of CA, indicating an intrinsic capsid lattice sensing ability in the PRY/SPRY monomer.
To achieve binding across the interhexamer CA interface, a single rhTRIM5α PRY/SPRY domain (∼40 Å across the core fold) must simultaneously interact with an extended surface area on CA including β1/β2, L5/6, and L4/5, as well as at least one additional binding element (such as a neighboring L4/5) from a second CA hexamer (Fig. 4A). This binding interface spans a distance ranging from ∼45 Å in the flat lattice to ∼70 Å in the CA assembly with the largest curvature at the tip of the HIV capsid cone (Fig. S5) (17, 18). The most likely scenario is that an extended V1 latches onto the binding surface on one CA molecule, as it has been implicated in interaction with β1/β2 (30) and residue 110 of MLV CA (corresponding to the L5/6 region in HIV CA) (47), whereas V2 and V3 interact with CA molecules in a neighboring hexamer (Fig. 4 D and E). In this binding mode, the flexible V1–V3 regions of PRY/SPRYrh, especially V1, most likely undergo conformational changes to adapt to the varying curvatures in the retrovirus capsids (Fig. 4 D and E; Fig. S5). Interestingly, the maximum distance that the PRY/SPRY domain can cover when the V1 loop is fully extended is about 70 Å, which coincides with the largest separation of CA-binding elements in the capsid cone (Fig. S5). The flexibility of the variable regions in PRY/SPRY has the ability to make TRIM5α a robust pattern-recognition sensor for diverse retroviral capsids.
TRIM5α achieves both recognition and disruption of viral capsids. Regardless of whether the initial capsid sensing is achieved by monomer, dimer, or a combination involving higher-order oligomers of the PRY/SPRY domain, binding to the CA lattice is likely to be an early step in the process of TRIM5α restriction. Monomeric PRY/SPRY binds to CA assemblies, but cannot disrupt them. In contrast, full-length TRIM5α or a dimeric CC-SPRY is able to break CA tubular assemblies via simultaneous binding of the two monomeric units in the dimer (Fig. 4F). Oligomerization or lattice formation by TRIM5α may increase the binding affinity of each PRY/SPRY domain sufficiently to promote capsid disruption, with avidity compensating for the intrinsically low affinity of the monomeric units and contributing significantly to the antiviral function of this important host restriction factor.
Materials and Methods
MBP-tagged PRY/SPRYrh or PRY/SPRYhu was expressed in E. coli and purified using affinity and size-exclusion chromatography. The CA hexamer (A14C/E45C/W184A/M185A) was expressed and purified as previously described (36). MBP-PRY/SPRYrh crystals were grown by the microbatch-under-oil and the hanging-drop vapor diffusion methods. Diffraction data were collected at the Advanced Photon Source beamline 24-ID and the National Synchrotron Light Source beamline X29. The structure was solved by molecular replacement using the coordinates of MBP (1ANF) and murine TRIM21 PRY/SPRY domain (2VOL) as search models. Diffraction data and refinement statistics are summarized in Table S1. Cosedimentation and cryo-EM experiments were performed as described previously (23), using preassembled wild-type HIV-1 CA tubes and PRY/SPRYrh or PRY/SPRYhu protein at various concentrations.
Supplementary Material
Acknowledgments
We thank Walther Mothes, Geoff Sutton, William Eliason, Xiaofei Jia, Erin Weber, Yuanyuan Xu, Jenny Fribourgh, and Henry Nguyen for assistance and discussion and Jing Zhou for technical assistance. We also thank the staff at the Advanced Photon Source beamline 24-ID and the National Synchrotron Light Source beamlines X29A. This work was supported by National Institutes of Health Grants AI097064 (to Y.X.), GM085043 (to P.Z.), P50GM82251 (to A.M.G.), and AI089401 (to C.A.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Crystallography, atomic coordinates, and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 4B3N).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1210903109/-/DCSupplemental.
References
- 1.Sastri J, Campbell EM. Recent insights into the mechanism and consequences of TRIM5α retroviral restriction. AIDS Res Hum Retroviruses. 2011;27(3):231–238. doi: 10.1089/aid.2010.0367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stremlau M, et al. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature. 2004;427(6977):848–853. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- 3.Perron MJ, et al. TRIM5alpha mediates the postentry block to N-tropic murine leukemia viruses in human cells. Proc Natl Acad Sci USA. 2004;101(32):11827–11832. doi: 10.1073/pnas.0403364101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yap MW, Nisole S, Lynch C, Stoye JP. Trim5alpha protein restricts both HIV-1 and murine leukemia virus. Proc Natl Acad Sci USA. 2004;101(29):10786–10791. doi: 10.1073/pnas.0402876101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stremlau M, et al. Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5alpha restriction factor. Proc Natl Acad Sci USA. 2006;103(14):5514–5519. doi: 10.1073/pnas.0509996103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pertel T, et al. TRIM5 is an innate immune sensor for the retrovirus capsid lattice. Nature. 2011;472(7343):361–365. doi: 10.1038/nature09976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nisole S, Stoye JP, Saïb A. TRIM family proteins: Retroviral restriction and antiviral defence. Nat Rev Microbiol. 2005;3(10):799–808. doi: 10.1038/nrmicro1248. [DOI] [PubMed] [Google Scholar]
- 8.Diaz-Griffero F, et al. Rapid turnover and polyubiquitylation of the retroviral restriction factor TRIM5. Virology. 2006;349(2):300–315. doi: 10.1016/j.virol.2005.12.040. [DOI] [PubMed] [Google Scholar]
- 9.Li X, Sodroski J. The TRIM5alpha B-box 2 domain promotes cooperative binding to the retroviral capsid by mediating higher-order self-association. J Virol. 2008;82(23):11495–11502. doi: 10.1128/JVI.01548-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Diaz-Griffero F, et al. A B-box 2 surface patch important for TRIM5alpha self-association, capsid binding avidity, and retrovirus restriction. J Virol. 2009;83(20):10737–10751. doi: 10.1128/JVI.01307-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Langelier CR, et al. Biochemical characterization of a recombinant TRIM5alpha protein that restricts human immunodeficiency virus type 1 replication. J Virol. 2008;82(23):11682–11694. doi: 10.1128/JVI.01562-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ganser-Pornillos BK, et al. Hexagonal assembly of a restricting TRIM5alpha protein. Proc Natl Acad Sci USA. 2011;108(2):534–539. doi: 10.1073/pnas.1013426108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sebastian S, Luban J. TRIM5alpha selectively binds a restriction-sensitive retroviral capsid. Retrovirology. 2005;2:40. doi: 10.1186/1742-4690-2-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stremlau M, Perron M, Welikala S, Sodroski J. Species-specific variation in the B30.2(SPRY) domain of TRIM5alpha determines the potency of human immunodeficiency virus restriction. J Virol. 2005;79(5):3139–3145. doi: 10.1128/JVI.79.5.3139-3145.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Y, Li X, Stremlau M, Lee M, Sodroski J. Removal of arginine 332 allows human TRIM5alpha to bind human immunodeficiency virus capsids and to restrict infection. J Virol. 2006;80(14):6738–6744. doi: 10.1128/JVI.00270-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yap MW, Nisole S, Stoye JP. A single amino acid change in the SPRY domain of human Trim5alpha leads to HIV-1 restriction. Curr Biol. 2005;15(1):73–78. doi: 10.1016/j.cub.2004.12.042. [DOI] [PubMed] [Google Scholar]
- 17.Ganser BK, Li S, Klishko VY, Finch JT, Sundquist WI. Assembly and analysis of conical models for the HIV-1 core. Science. 1999;283(5398):80–83. doi: 10.1126/science.283.5398.80. [DOI] [PubMed] [Google Scholar]
- 18.Li S, Hill CP, Sundquist WI, Finch JT. Image reconstructions of helical assemblies of the HIV-1 CA protein. Nature. 2000;407(6802):409–413. doi: 10.1038/35030177. [DOI] [PubMed] [Google Scholar]
- 19.Ganser-Pornillos BK, Yeager M, Sundquist WI. The structural biology of HIV assembly. Curr Opin Struct Biol. 2008;18(2):203–217. doi: 10.1016/j.sbi.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakayama EE, Shioda T. Role of human TRIM5α in intrinsic immunity. Front Microbiol. 2012;3:97. doi: 10.3389/fmicb.2012.00097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luban J. Cyclophilin A, TRIM5, and resistance to human immunodeficiency virus type 1 infection. J Virol. 2007;81(3):1054–1061. doi: 10.1128/JVI.01519-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perez-Caballero D, Hatziioannou T, Yang A, Cowan S, Bieniasz PD. Human tripartite motif 5alpha domains responsible for retrovirus restriction activity and specificity. J Virol. 2005;79(14):8969–8978. doi: 10.1128/JVI.79.14.8969-8978.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao G, et al. Rhesus TRIM5α disrupts the HIV-1 capsid at the inter-hexamer interfaces. PLoS Pathog. 2011;7(3):e1002009. doi: 10.1371/journal.ppat.1002009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kar AK, Diaz-Griffero F, Li Y, Li X, Sodroski J. Biochemical and biophysical characterization of a chimeric TRIM21-TRIM5alpha protein. J Virol. 2008;82(23):11669–11681. doi: 10.1128/JVI.01559-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang H, et al. Structural insight into the mechanisms of enveloped virus tethering by tetherin. Proc Natl Acad Sci USA. 2010;107(43):18428–18432. doi: 10.1073/pnas.1011485107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stuart DI, Levine M, Muirhead H, Stammers DK. Crystal structure of cat muscle pyruvate kinase at a resolution of 2.6 A. J Mol Biol. 1979;134(1):109–142. doi: 10.1016/0022-2836(79)90416-9. [DOI] [PubMed] [Google Scholar]
- 27.Felsenstein J. An alternating least squares approach to inferring phylogenies from pairwise distances. Systematic biology. 1997;46(1):101–111. doi: 10.1093/sysbio/46.1.101. [DOI] [PubMed] [Google Scholar]
- 28.Li X, Yeung DF, Fiegen AM, Sodroski J. Determinants of the higher order association of the restriction factor TRIM5alpha and other tripartite motif (TRIM) proteins. J Biol Chem. 2011;286(32):27959–27970. doi: 10.1074/jbc.M111.260406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perron MJ, Stremlau M, Sodroski J. Two surface-exposed elements of the B30.2/SPRY domain as potency determinants of N-tropic murine leukemia virus restriction by human TRIM5alpha. J Virol. 2006;80(11):5631–5636. doi: 10.1128/JVI.00219-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ohkura S, et al. Novel escape mutants suggest an extensive TRIM5α binding site spanning the entire outer surface of the murine leukemia virus capsid protein. PLoS Pathog. 2011;7(3):e1002011. doi: 10.1371/journal.ppat.1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ohkura S, Yap MW, Sheldon T, Stoye JP. All three variable regions of the TRIM5alpha B30.2 domain can contribute to the specificity of retrovirus restriction. J Virol. 2006;80(17):8554–8565. doi: 10.1128/JVI.00688-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9(1):40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maillard PV, Reynard S, Serhan F, Turelli P, Trono D. Interfering residues narrow the spectrum of MLV restriction by human TRIM5alpha. PLoS Pathog. 2007;3(12):e200. doi: 10.1371/journal.ppat.0030200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pham QT, Bouchard A, Grütter MG, Berthoux L. Generation of human TRIM5alpha mutants with high HIV-1 restriction activity. Gene Ther. 2010;17(7):859–871. doi: 10.1038/gt.2010.40. [DOI] [PubMed] [Google Scholar]
- 35.Rhodes DA, de Bono B, Trowsdale J. Relationship between SPRY and B30.2 protein domains. Evolution of a component of immune defence? Immunology. 2005;116(4):411–417. doi: 10.1111/j.1365-2567.2005.02248.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pornillos O, et al. X-ray structures of the hexameric building block of the HIV capsid. Cell. 2009;137(7):1282–1292. doi: 10.1016/j.cell.2009.04.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Black LR, Aiken C. TRIM5alpha disrupts the structure of assembled HIV-1 capsid complexes in vitro. J Virol. 2010;84(13):6564–6569. doi: 10.1128/JVI.00210-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Byeon IJ, et al. Structural convergence between Cryo-EM and NMR reveals intersubunit interactions critical for HIV-1 capsid function. Cell. 2009;139(4):780–790. doi: 10.1016/j.cell.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Song B, et al. Retrovirus restriction by TRIM5alpha variants from Old World and New World primates. J Virol. 2005;79(7):3930–3937. doi: 10.1128/JVI.79.7.3930-3937.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakayama EE, Miyoshi H, Nagai Y, Shioda T. A specific region of 37 amino acid residues in the SPRY (B30.2) domain of African green monkey TRIM5alpha determines species-specific restriction of simian immunodeficiency virus SIVmac infection. J Virol. 2005;79(14):8870–8877. doi: 10.1128/JVI.79.14.8870-8877.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sawyer SL, Wu LI, Emerman M, Malik HS. Positive selection of primate TRIM5alpha identifies a critical species-specific retroviral restriction domain. Proc Natl Acad Sci USA. 2005;102(8):2832–2837. doi: 10.1073/pnas.0409853102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Song B, et al. The B30.2(SPRY) domain of the retroviral restriction factor TRIM5alpha exhibits lineage-specific length and sequence variation in primates. J Virol. 2005;79(10):6111–6121. doi: 10.1128/JVI.79.10.6111-6121.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wilson SJ, et al. Rhesus macaque TRIM5 alleles have divergent antiretroviral specificities. J Virol. 2008;82(14):7243–7247. doi: 10.1128/JVI.00307-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Diaz-Griffero F, et al. A human TRIM5alpha B30.2/SPRY domain mutant gains the ability to restrict and prematurely uncoat B-tropic murine leukemia virus. Virology. 2008;378(2):233–242. doi: 10.1016/j.virol.2008.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kuroishi A, Bozek K, Shioda T, Nakayama EE. A single amino acid substitution of the human immunodeficiency virus type 1 capsid protein affects viral sensitivity to TRIM5 alpha. Retrovirology. 2010;7:58. doi: 10.1186/1742-4690-7-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miyamoto T, et al. A single amino acid of human immunodeficiency virus type 2 capsid protein affects conformation of two external loops and viral sensitivity to TRIM5α. PLoS ONE. 2011;6(7):e22779. doi: 10.1371/journal.pone.0022779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pacheco B, Finzi A, Stremlau M, Sodroski J. Adaptation of HIV-1 to cells expressing rhesus monkey TRIM5α. Virology. 2010;408(2):204–212. doi: 10.1016/j.virol.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ulm JW, Perron M, Sodroski J, C Mulligan R. Complex determinants within the Moloney murine leukemia virus capsid modulate susceptibility of the virus to Fv1 and Ref1-mediated restriction. Virology. 2007;363(2):245–255. doi: 10.1016/j.virol.2006.09.048. [DOI] [PubMed] [Google Scholar]
- 49.Kootstra NA, Navis M, Beugeling C, van Dort KA, Schuitemaker H. The presence of the Trim5alpha escape mutation H87Q in the capsid of late stage HIV-1 variants is preceded by a prolonged asymptomatic infection phase. AIDS. 2007;21(15):2015–2023. doi: 10.1097/QAD.0b013e3282effa87. [DOI] [PubMed] [Google Scholar]
- 50.Song H, et al. A single amino acid of the human immunodeficiency virus type 2 capsid affects its replication in the presence of cynomolgus monkey and human TRIM5alphas. J Virol. 2007;81(13):7280–7285. doi: 10.1128/JVI.00406-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ganser-Pornillos BK, Cheng A, Yeager M. Structure of full-length HIV-1 CA: A model for the mature capsid lattice. Cell. 2007;131(1):70–79. doi: 10.1016/j.cell.2007.08.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.