Abstract
Eukaryotic translation termination results from the complex functional interplay between two eukaryotic release factors, eRF1 and eRF3, and the ribosome, in which GTP hydrolysis by eRF3 couples codon recognition with peptidyl-tRNA hydrolysis by eRF1. Here, using cryo-electron microscopy (cryo-EM) and flexible fitting, we determined the structure of eRF1–eRF3–guanosine 5′-[β,γ-imido]triphosphate (GMPPNP)-bound ribosomal pretermination complex (pre-TC), which corresponds to the initial, pre-GTP hydrolysis stage of factor attachment. Our results show that eukaryotic translation termination involves a network of interactions between the two release factors and the ribosome. Our structure provides mechanistic insight into the coordination between GTP hydrolysis by eRF3 and subsequent peptide release by eRF1.
Termination of translation occurs when a ribosome reaches the end of the coding region and a stop codon (UAA, UAG, or UGA) enters the aminoacyl tRNA binding site (A site), leaving peptidyl-tRNA in the peptidyl tRNA binding site (P site). It entails stop codon recognition by specialized release factors followed by hydrolysis of peptidyl-tRNA. In eukaryotes, termination is mediated by the concerted action of two directly interacting release factors, eRF1 and eRF3. eRF1 is responsible for stop codon recognition and inducing hydrolysis of peptidyl-tRNA, whereas eRF3, a ribosome-dependent GTPase, strongly stimulates peptide release by eRF1 in a GTP-dependent manner (for review, see ref. 1).
eRF1 also participates in ribosome recycling: after peptide release it remains associated with the ribosome and together with ATP-binding cassette sub-family E member 1 (ABCE1) promotes splitting the ribosome into free 60S and tRNA/mRNA-associated 40S subunits (2). eRF1 and eRF3 have paralogs, Dom34 (yeast)/Pelota (mammals) and Hbs1, respectively, which do not participate in termination but instead play a key role in No-go and nonstop decay surveillance mechanisms (e.g., see refs. 3, 4). Dom34/Pelota and Hbs1 do not induce peptide release in a mechanism similar to eRFs. Instead, they cooperate with ABCE1 to promote dissociation of stalled elongation complexes, which is accompanied by peptidyl-tRNA drop off (5–7).
eRF1 comprises N-terminal (N), middle (M), and C-terminal (C) domains (8). The N domain is responsible for stop codon recognition, which is achieved through a 3D network of conserved residues that include apical TASNIKS (amino acid sequence: threonine-alanine-serine-asparagine-isoleucine-lysine-serine) and YxCxxxF motifs (e.g., refs. 9–12). Domain M contains the universally conserved GGQ motif, which is critical for triggering peptide release: as shown for prokaryotes, its placement into the peptidyl transferase center (PTC) causes rRNA rearrangement, allowing a water molecule to enter (for review, see ref. 13). The rigid core of domain C forms an α–β sandwich (8) that deviates from the standard form by the presence of a small insertion, which forms a minidomain (14). eRF3 consists of an N-terminal sequence (residues 1–138) that is nonessential for termination (15), and the following essential region comprising a G domain and the β-barrel domains 2 and 3. These three domains share strong structural homology with elongation factors (EFs) eEF1α and EF-Tu (16). Domain 3 interacts directly with domain C of eRF1, predominantly through hydrophobic contacts (17).
Association with eRF1 specifically stabilizes binding of GTP to eRF3 by two orders of magnitude (18–20) so that eRF1 and eRF3 form a stable eRF1–eRF3–GTP ternary complex. eRF1 is also strictly required for stimulation of eRF3’s ribosome-dependent GTPase activity (21). Although the primary determinant of eRF1–eRF3 binding is the interaction between their C-terminal domains, eRF1’s domain M contributes additional affinity to the eRF1–eRF3 interaction and is strictly required for stimulation of eRF3’s GTP-binding and hydrolysis activities (22). Consistently, the model of eRF1–eRF3–GTP based on the crystal structure of the complex of eRF1 with domains 2 and 3 of eRF3 (eRF32–3) and the low-resolution small angle x-ray scattering (SAXS) structure of eRF1–eRF3–GTP (17), predicts that upon binding of eRF1 to eRF3, domain M shifts and rotates, resulting in adoption by eRF1 of a tRNA-like shape. In such a conformation, domain M would come in direct contact with the switch regions of the eRF3 G domain. Interestingly, binding of eRF1–eRF3–GTP to ribosomal pretermination complexes (pre-TCs) also induces a +2-nt forward shift in their mRNA toeprints (23), which could be due to additional conformational changes in the 80S ribosome.
Peptide release by eRF1 alone is very inefficient. eRF3 strongly stimulates the process in the presence of GTP, but this stimulation is completely abrogated in the presence of the nonhydrolyzable GTP analog guanosine 5′-[β,γ-imido]triphosphate (GMPPNP) (23). This indicates that eRF3’s GTPase activity couples stop codon recognition with hydrolysis of peptidyl-tRNA, as first suggested on the basis of genetic data (24). Eukaryotic termination can therefore be described by the following model. First, eRF1 and eRF3 bind to the A site of pre-TCs as an eRF1–eRF3–GTP ternary complex. Binding to the ribosome triggers GTP hydrolysis, which results in conformational changes, likely in eRF1, that enable the GGQ loop in its M domain to enter the PTC and hydrolyze peptidyl-tRNA.
Although this model is generally accepted, it has yet to be corroborated by structural data. To date, the eukaryotic ribosomal complexes corresponding to different stages in termination have not been visualized and ribosomal binding positions of eRFs, as well as their ribosome-bound conformations, remain unknown. Here, using cryo-EM, we investigated the structure of mammalian 80S–eRF1–eRF3–GMPPNP pre-TCs, which correspond to the pre-GTP hydrolysis stage in termination. Our model describes the complex interactions between eRF1 and eRF3 on the ribosome and provides insight into the way eRF3-catalyzed GTP hydrolysis accelerates peptide release by eRF1.
Results
Cryo-EM Reconstruction of the Individual Pretermination Complex.
Mammalian pre-TCs were obtained by in vitro reconstitution from purified rabbit 40S and 60S ribosomal subunits, initiation (2, 3, 1, 1A, 4A, 4B, 4G, 5, and 5B), and elongation factors (1A and 2), and aminoacylated tRNAs on MVHL-STOP (Methionine-valine-histidine-leucine-stop codon) mRNA comprising 12 unstructured 5′-terminal nucleotides (four CAA triplets) followed by the β-globin 5′-UTR, a coding sequence for the MVHL tetrapeptide, a UAA stop codon, and an ∼150-nt 3′-UTR consisting of the natural β-globin coding sequence (23). Assembled pre-TCs were purified by sucrose density gradient (SDG) centrifugation.
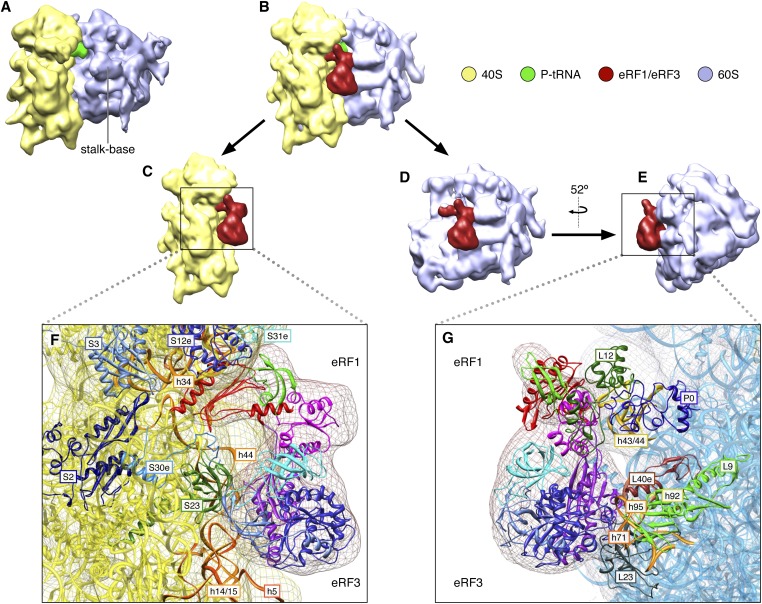
A cryo-EM reconstruction of the pre-TC was obtained using a density map of the human 80S ribosome (25) as a reference for projection matching procedures (26). After classification and angular refinement (Materials and Methods), a 17-Å cryo-EM map was generated for the programmed pre-TC. The complex contains P-site tRNA, but vacant A and E sites, and the ribosome is in the nonrotated conformation (Fig. 1A).
Fig. 1.
Structures of 80S–P-tRNA and 80S–P-tRNA–eRF1–eRF3–GMPPNP complexes. (A) Cryo-EM map of 80S–P-tRNA. (B) Cryo-EM map of the 80S–P-tRNA–eRF1–eRF3–GMPPNP complex. (C) The 60S portion removed from B to show 40S subunit’s interactions with the eRF1–eRF3–GMPPNP ternary complex. (D) The 40S portion removed from B to show 60S subunit’s interaction with the eRF1–eRF3–GMPPNP ternary complex. (E) Map shown in D rotated clockwise by 52°. (F) Detailed interactions between 40S rps and rRNA with individual domains of eRF1 and eRF3 (see Fig. 2A for the domain color scheme), inferred by fitting atomic structures into the density. Structure of 40S subunit (boxed portion in C) is shown as yellow ribbon. Proteins surrounding eRF1–eRF3 are shown as individually colored ribbon. (G) Detailed interactions between 60S rps and rRNA with individual domains of eRF1 and eRF3. Structure of 60S in boxed region as shown in E is displayed as blue ribbon. Large subunit proteins surrounding eRF1–eRF3 are shown as individually colored ribbons. In both F and G, individual domains of eRF1 and eRF3 are colored separately and the cryo-EM density is displayed in mesh.
Cryo-EM Reconstruction of the eRF1–eRF3–GMPPNP-Bound Pretermination Complex.
eRF1–eRF3-associated ribosomal complexes, corresponding to the pre-GTP hydrolysis stage of initial attachment of release factors, were obtained by incubation of SDG-purified pre-TCs formed on MVHL-STOP mRNA with the full-length eRF1 and truncated eRF3 lacking the nonessential 138 N-terminal amino acids (21, 23) in the presence of GMPPNP. The efficiency of pretermination complex formation, estimated by incorporation of [35S]Met, was ∼70%. The remaining ∼30% of ribosomes likely represented nonprogrammed particles, cosedimenting with the programmed population. The efficiency of eRFs binding to pre-TCs, which was determined in toeprinting experiments by the appearance of the characteristic +2-nt forward toeprint shift (23), was ∼85%.
Computational sorting was used to select images attributed to the programmed 80S ribosomal complexes (Figs. S1 and S2). The percentage (∼75%) of programmed versus vacant 80S ribosomes, as determined using single-particle reconstruction techniques, was in agreement with the biochemically estimated efficiency of pre-TC formation. Three-dimensional reconstruction and angular refinement revealed extra density, indicative of eRF1–eRF3 bound to the P-site tRNA-containing 80S ribosome (Fig. 1 B–G). The resolution of the reconstruction from 43,691 particles was 18 Å. Like the individual pre-TC described above, the 80S–eRF1–eRF3–GMPPNP pre-TC maintains the nonrotated conformation and a vacant E site. Density attributed to eRF1–eRF3 fills the ribosomal A site. Molecular interpretation of factor–factor and factor–ribosome interactions was facilitated by docking existing atomic models of the 80S ribosome (27), eRF1 (8, 14), eRF3 (16), and the eRF1–eRF32–3 complex (17) into the cryo-EM density. Fitting of the structures in a comprehensive model was subsequently optimized using molecular dynamics flexible fitting (MDFF) (Materials and Methods).
Conformation, Orientation, and Interactions of Ribosome-Bound eRF1–eRF3–GMPPNP.
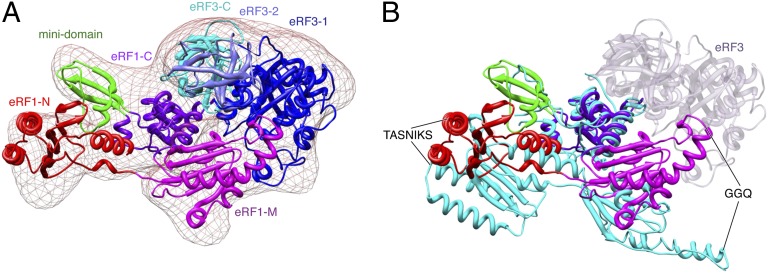
Compared with the crystal structure of individual eRF1 (8), eRF1’s domains N and M are shifted and rotated substantially in the 80S–eRF1–eRF3–GMPPNP complex (Fig. 2 and Fig. S3). Consistent with biochemical studies (22) and the eRF1–eRF3–GTP model (17), domain M rotates by 37° with respect to eRF1’s C-terminal domain, which positions it near eRF3. Domain N is rotated by ∼100° as it reaches into the decoding center. As a result of these changes, the ribosome-bound eRF1–eRF3–GMPPNP complex adopts a conformation that is similar to those observed for bacterial 70S-bound tRNA–EF-Tu and yeast 80S-bound Dom34–Hbs1 complexes (28, 29) (Fig. S3).
Fig. 2.
Structural dynamics of the eRF1–eRF3 complex. (A) Conformation of the eRF1–eRF3–GMPPNP ternary complex when bound to the pretermination 80S ribosome. Individual domains for each factor are labeled and colored separately. Models of eRF1 and eRF3 determined by X-ray crystallography were docked into the cryo-EM map. (B) Superimposition of human apo-eRF1 (cyan) with that of the observed eRF1 structure in the 80S–P-tRNA–eRF1–eRF3–GMPPNP complex. The latter is labeled and color coded by domain. Alignment of the two structures was facilitated by superimposing the C-terminal domain of eRF1. The position of eRF3 in the ternary complex is shown in transparent blue.
The two factors contact both ribosomal subunits at multiple sites, through a complex interaction network (Table S1 and Fig. 1 F and G). eRF3 binds near the universal GTPase-associated center (GAC) of the ribosome. eRF3’s G domain contacts the sarcin–ricin loop (SRL) (H95) of 28S rRNA, and is near the P stalk. rpL40e and rpL9 (rpL6p) are in the immediate vicinity, as well. On the 40S subunit interface, the G domain interacts with h14 of 18S rRNA. eRF3’s domain 2 projects toward the 40S subunit, where it contacts h5 and h15 of 18S rRNA. Both the G domain and domain 2 of eRF3 interact with domain M of eRF1, which is nestled between the two domains of eRF3 (see below). Domain 3 of eRF3 appears to interact exclusively with the C-terminal domain of eRF1.
eRF1 forms four well-defined lobes corresponding to its N-, M-, C-, and mini-domains, with an overall shape similar to that of a tRNA molecule (Fig. 2 and Fig. S3C). eRF1’s domain N reaches deep into the decoding center of the 40S subunit (Fig. 1F). There are multiple points of contact between domain N and regions of the 40S subunit, including helices 18, 30, 31, 34, and 44 of 18S rRNA and eukarya/archaea-specific ribosomal proteins rpS30e and rpS31e.
Domain M of eRF1 is folded back so that the tip of its extended α5 helix, containing the universal GGQ motif, is tucked between eRF3 and the 60S subunit within the intersubunit space of the 80S ribosome, and positioned precisely between the G domain and domain 2 of eRF3 (Figs. 1 and 2). In this orientation, the GGQ motif is situated more than 80 Å away from the PTC of the 60S subunit, which is consistent with the biochemical data, indicating that its accommodation into the PTC occurs only after GTP hydrolysis (23). A similar position has been described for the central domain of Dom34 in the yeast Hbs1–Dom34-bound No-go decay complex, also stalled in a pre-GTP hydrolysis state (29). In the docked structure of eRF1–eRF3, rpS23 (rpS12p) is nestled between the base of eRF1’s domain M and domain 2 of eRF3. Domain M is also proximal to the regions of the 60S subunit that encompass H71 and H92 and rpL23 (rpL14p). The tip of the domain approaches the area where the G domain of eRF3 interacts with the SRL. This conformation also positions the M domain of eRF1 in the immediate vicinity of the switch 1 region of eRF3.
Domain C of eRF1 protrudes away from the intersubunit space of the ribosome, ultimately forming a contact with the P-stalk base of the 60S subunit. The interaction is coordinated via rpL12 (L11p), H43 and H44. Consistent with previous reports (17, 30), domain C of eRF1 interacts intimately with domain 3 of eRF3.
Although the ribosomal interactions of the eRF1–eRF3 complex described thus far are similar to those that were previously reported for the Hbs1–Dom34 and EF-Tu–tRNA complexes (28, 29), there is an additional specific point of contact that involves the 40S subunit and eRF1’s minidomain (Fig. 1 C and F). This minidomain is formed by a specific insert in the C-terminal domain of eRF1, which does not exist in the C-terminal domain of Pelota/Dom34, even though their C-terminal domains are homologous (31). Whereas this region is disordered in all available crystal structures of eRF1, the NMR structure of eRF1’s C-terminal domain showed that in solution it forms an α-helix and three β-strands (14). This structure closely matches our cryo-EM map. The minidomain extends away from the core of eRF1 density and protrudes toward the beak of the 40S subunit, where it interacts with rpS31e and 18S rRNA (Fig. 1F). The long tail of rpS31e contacts both the minidomain and domain N of eRF1.
80S Structural Rearrangements Induced by Binding of eRF1–eRF3–GMPPNP.
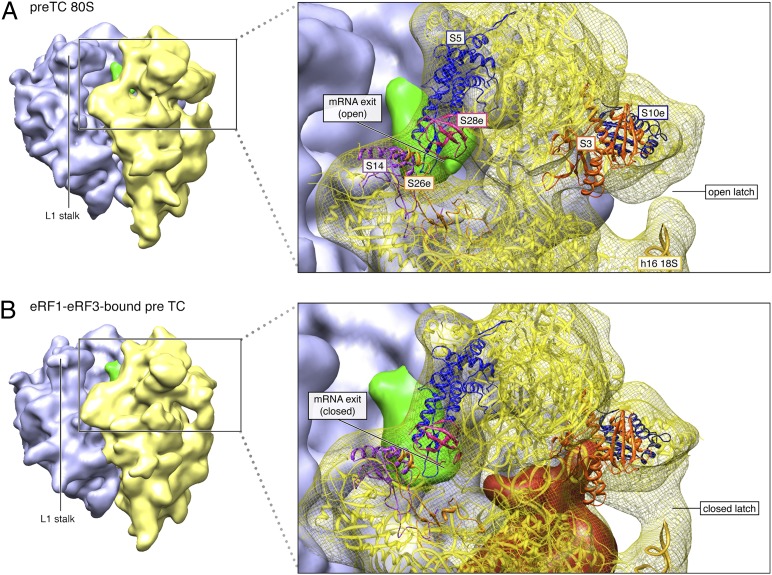
Comparison of the cryo-EM reconstructions of the pre-TC and the 80S–eRF1–eRF3–GMPPNP complex also reveals that binding of release factors induces conformational changes in both 40S and 60S subunits. In the 40S subunit, the first notable rearrangement occurs at the entrance of the mRNA-binding channel (Fig. 3). Thus, upon binding of eRF1–eRF3, h16 of 18S rRNA in the shoulder of the 40S subunit, which points into the solvent in the vacant pre-TC, moves toward the head. This movement of h16 occurs in concert with a shift of the head of the 40S subunit, where rpS3 (rpS3p) resides, toward the body. The overall outcome is the establishment of a new head–body connection, most likely between h16 and rpS3, and considerable constriction of the mRNA entrance (Fig. 3). In addition, binding of release factors also induces constriction of the mRNA exit tunnel, which on one side is lined by multiple ribosomal proteins, including rpS5 (rpS7p), rpS14 (rpS11p), rpS26e, and rpS28e, and is occupied by 18S rRNA on the other (Fig. 3).
Fig. 3.
Structural rearrangements within the 80S pre-TC incurred upon eRF1–eRF3–GMPPNP binding. Boxed portion of the 40S subunit is shown with yellow mesh for the density and yellow ribbon for the fitted structure. Green map is for P-tRNA and red for eRF1–eRF3. (A) Cryo-EM map and the MDFF structure of the pre-TC. (B) Cryo-EM map and the MDFF structure of 80S–eRF1–eRF3–GMPPNP complex. Comparison between the two structures reveals significant structural rearrangements in the 80S pre-TC, upon binding of eRF1–eRF3–GMPPNP. Most notable differences occur in the 40S subunit and include the closing of an h16–rpS3 latch at the mRNA entrance and a constriction at the mRNA exit channel, each upon eRF1–eRF3–GMPPNP binding. In the 60S subunit, the L1 stalk adopts a slightly different configuration upon eRF1–eRF3–GMPPNP binding.
Structural rearrangements of the 60S subunit induced upon eRF1–eRF3–GMPPNP binding appear primarily in its two stalks. The P stalk (L7/L12 stalk in prokaryotes) is a highly dynamic structure, which is essential for binding of multiple ribosomal GTPases (e.g., refs. 32, 33). It comprises H43 and H44 and rpL12 (L11p), rpP0 (L10p), and rpP1 and rpP2 which are equivalent to prokaryotic rpL7/L12p. In our fitted structure, rpL12 (L11p) and H43 and H44, which form the stalk base, shift away from the 40S subunit, to accommodate binding of eRF1–eRF3, particularly the C-domain of eRF1 (compare panels in Fig. 1 A and B).
The L1 stalk is a highly mobile domain, near the E site, which moves between at least three distinct conformational states (34, 35). In the inward-facing closed and half-closed conformations, the stalk interacts with the elbow of deacylated tRNA in hybrid (P/E) and classical (E/E) orientations, respectively. The open conformation of the L1 stalk facilitates release of deacylated tRNA from the E site. The position of the L1 stalk is generally correlated with the intersubunit (“ratchet-like”) rotation of the ribosome (35). When the ribosome is in the unrotated conformation, the L1 stalk occupies the open position. Conversely, the rotated state of the ribosome is concurrent with the L1 stalk in the closed position. Consistent with these observations and the nonrotated state of the 80S ribosome, the L1 stalk is in the same open position for both the pre-TC and the 80S–eRF1–eRF3 pre-TC complexes, although its overall shape suggests subtle differences between the two complexes (Fig. 3).
Discussion
Our cryo-EM density maps, combined with flexible fitting of individual components determined by X-ray crystallography, revealed that eRF1–eRF3–GMPPNP bound to the pre-TC adopts a conformation that is similar to the predicted model for the eRF1–eRF3–GTP complex in solution (17), and resembles those of ribosome-bound Dom34–Hbs1–GMPPNP and tRNA–EF-Tu (28, 29). In this conformation, domains M and N of eRF1 shift and rotate compared with the structure of individual eRF1 (8). In addition to interactions between the C-terminal domains of eRF1 and eRF3, eRF1’s domain M also establishes contacts with eRF3’s G-domain, in the immediate vicinity of the switch 1 region. The higher-resolution (∼9.5 Å) cryo-EM structure of the yeast 80S–Dom34–Hbs1–GMPPNP complex revealed a homologous interaction between the middle domain of Dom34 (which is structurally homologous to domain M of eRF1 except that it lacks the GGQ motif) and the G-domain of Hbs1, with the positively charged loop β10–α3 loop of Dom34 directly contacting the switch-1 region of Hbs1 (29). The similarities in overall architecture of the eRF1–eRF3- and Dom34–Hbs1-bound ribosomal complexes in a pre-GTP hydrolysis state suggest a conserved mechanism of ribosome binding and GTP hydrolysis and are consistent with the essential roles of eRF1 and Dom34 in promoting GTP binding and hydrolysis by eRF3 and Hbs1, respectively (5, 18–22, 36).
Interaction of eRF1’s domain M with eRF3 in termination complexes stalled in a pre-GTP hydrolysis state results in positioning of the GGQ motif far away from the PTC, consistent with biochemical data indicating that accommodation of the GGQ motif into the PTC occurs only after GTP hydrolysis (23). To accommodate the GGQ motif in the PTC, eRF1 must undergo substantial conformational changes. GTP hydrolysis by translational GTPases generally involves a reorganization of the GAC of the ribosome (32). Also, universally consistent with GTPase mechanisms, GTP hydrolysis by eRF3 would result in altered conformations of the switch-1 and -2 regions of its G domain (37). The combination of these events would release domain M of eRF1 from its interaction with eRF3, allowing it to swing into the PTC and to induce peptide release. In common with other translational GTPases, eRF3 in its GDP-bound form likely has a lower affinity to ribosomal complexes and dissociates after GTP hydrolysis, so that accommodation of domain M could be preceded or accompanied by dissociation of eRF3. However, the observation that eRF1 associates more firmly with posttermination complexes in the presence of eRF3 (2) suggests that after GTP hydrolysis, eRF3 might not dissociate entirely from ribosomal complexes. If this is the case, efficient release of eRF3 could be promoted by ABCE1, whose ribosome-binding site overlaps with that of eRF3 (38). Dissociation of eRF3 might in turn accelerate accommodation of eRF1’s domain M, which would at least in part account for stimulation of peptide release by ABCE1 (6, 39).
Although in contrast to the Dom34–Hbs1–GTP mRNA surveillance complex, which targets stalled elongation complexes in an A-site codon-independent manner (5, 7), the eRF1–eRF3–GTP complex recognizes stop codons exclusively, and the N domains of eRF1 and Dom34 are not structurally related (8, 40), in both cases the N domains of eRF1 and Dom34 penetrate deeply into the decoding center and establish multiple interactions with the 40S subunit (present study and ref. 29). However, our data revealed that in addition to the connection with the decoding center via the N domain, eRF1 also establishes a specific interaction with 18S rRNA in the beak of the 40S subunit via its unique minidomain. The NMR structure of eRF1’s C-terminal domain showed that the minidomain exists in two distinct conformational states (14). The cryo-EM density of the minidomain is well ordered where it makes interactions with the beak of the 40S subunit, suggesting that although it is dynamic in solution, the minidomain is stabilized in a specific conformation upon binding of eRF1 to the pre-TC, possibly due to its contact with the 40S subunit. Thus, by establishing an additional contact with the 40S subunit, the minidomain could either assist in probing for a stop codon or stabilize binding of eRF1–eRF3 to the pre-TC once a stop codon has been recognized. Consistent with this hypothesis are data showing that mutations to individual residues in the minidomain affect stop codon specificity and enhance termination on UAG (14). However, another function of the interaction of the minidomain with the beak of the 40S subunit may be to limit intersubunit rotation and movement of the head of the 40S subunit for efficient peptide release and/or subsequent ribosome recycling mediated by eRF1 and ABCE1.
Our structure also revealed that binding of eRF1–eRF3–GMPPNP induces conformational changes within both 40S and 60S ribosomal subunits. Conformational changes in the large subunit are primarily confined to the P-stalk base. A rearrangement at the stalk base is likely important for catalyzing GTP hydrolysis and has been described for many of both eukaryotic and bacterial ribosomal GTPases (e.g., ref. 32). Conformational changes in the 40S subunit involve a movement of h16 of 18S rRNA and the N-terminal domain (NTD) of rpS3 toward each other, which results in the establishment of a new head–body connection on the solvent side of the 40S subunit and a constriction of the mRNA entrance. These conformational changes are likely the cause of the +2-nt shift in the toeprint of the pre-TC caused by binding of eRF1–eRF3 (23), as constriction at the mRNA entrance would feasibly prevent reverse transcriptase from penetrating further.
Binding of Dom34–Hbs1 to stalled yeast ribosomes also led to the appearance of density bridging h16 and rpS3 (29). Whereas this density was assigned to the Hbs1 NTD, it is possible, given the lack of a complete structural model for this domain, that the additional density may include conformational changes in the 40S subunit comparable to those induced by eRF1–eRF3. Moreover, the purpose of binding of the Hbs1 NTD to this region may indeed be to induce these changes. Interestingly, similar conformational changes involving h16 and rpS3 are also induced by binding to the 40S subunit of the initiation factors eIF1 and eIF1A (41), and the structurally distinct internal ribosomal entry sites (IRESs) of hepatitis C virus and cricket paralysis virus (25, 42). In the case of these 40S initiation complexes, conformational changes involving h16 and rpS3 are concomitant with the opening of the mRNA entry channel “latch” on the intersubunit side, which is formed by h18 in the body and h34 and rpS3 in the neck of the 40S subunit. As noted previously (25), the orientation of h16 in eukaryotes differs radically from that in prokaryotes, and the rpS3–h16 region may constitute an adaptation that facilitates a variety of eukaryote-specific aspects of the translation process, by enabling opening and closing of the mRNA entrance and positioning of mRNA in the channel to be regulated.
In the case of termination, the constriction at the mRNA entrance, particularly combined with a similar constriction at the mRNA exit tunnel, may serve the purpose of clamping onto the mRNA to stabilize the complex as it prepares for peptide release. In addition to clamping the ribosome more firmly on the mRNA, conformational changes in the ribosome induced by binding of eRF1–eRF3 may also promote subsequent steps in their own activation. Conversely, antagonism of these induced conformational changes in the 40S subunit would be predicted to impair the termination process, and this could underlie the stop codon “read-through” mechanism that is used by numerous viral mRNAs and a number of cellular mRNAs for the synthesis of C-terminally extended polypeptides. Read-through on these mRNAs is strongly stimulated by stable structural elements that are commonly located ∼8 nt downstream of the recoded termination codon, such as a pseudoknot in murine leukemia virus (43) and hairpin or other stable stem–loop structures in e.g., alphavirus and Drosophila Headcase mRNAs (44, 45). The location of these elements in ribosomal complexes stalled at the termination codon suggests that they could antagonize factor-induced constriction of the mRNA entrance channel, leading to abortive interaction with the eRF1–eRF3 complex and instead permitting misreading of the stop codon by an aminoacylated suppressor tRNA.
Materials and Methods
Purification of Ribosomal Subunits, Initiation, Elongation, and Termination Factors, and Aminoacylation of initiator methyionyl-tRNA (tRNAiMet).
Native eIF2, eIF3, eIF5B, eEF2, aminoacyl-tRNA synthetases, and 40S and 60S ribosomal subunits were purified from rabbit reticulocyte lysate, and recombinant eIF1, eIF1A, eIF4A, eIF4B, eIF4G736–1115, eIF5, eRF1, and eRF3aC lacking the N-terminal 138 aa and recombinant Escherichia coli methionyl tRNA synthetase were expressed and purified from E. coli as described (refs. 23 and 46 and references therein). In vitro transcribed tRNAiMet was aminoacylated using recombinant E. coli methionyl tRNA synthetase, whereas total native tRNAs were aminoacylated with Met, Val, His, and Leu using native aminoacyl-tRNA synthetases (46).
Assembly and Purification of Pretermination Complexes.
Pre-TCs were assembled on MVHL-STOP mRNA essentially as described (23). First, 48S initiation complexes were formed by incubating triple aliquots of 100 pmol MVHL-STOP mRNA, 30 pmol 40S subunits, 100 pmol eIF2, 70 pmol eIF3, 130 pmol eIF4A, 50 pmol eIF4B, 150 pmol eIF4G736–1115, 300 pmol eIF1, 300 pmol eIF1A, and 70 pmol Met-tRNAiMet in a 400 μL buffer A (20 mM Tris, pH 7.5, 100 mM KAc, 2.5 mM MgCl2, 2 mM DTT, and 0.25 mM spermidine) supplemented with 200 units RNasin, 0.4 mM ATP, and 0.2 mM GTP for 15 min at 37 °C. The 48S complexes were then incubated for 15 min with 200 pmol eIF5, 15 pmol eIF5B, and 60 pmol 60S subunits to form 80S initiation complexes. Pre-TCs were obtained by incubating 80S complexes with 200 pmol yeast eEF1A, 30 pmol eEF2, and 150 μg total tRNA (aminoacylated with Met, Val, His, and Leu) for 15 min and then purified by centrifugation through 10–30% (wt/vol) linear sucrose density gradients prepared in buffer A in a Beckman SW55 rotor for 1 h 35 min at 4 °C and 50,000 rpm. Fractions that corresponded to pre-TCs were combined, concentrated, and transferred into buffer A using Microcon YM-10 filter units. Purified pre-TCs (3.2 pmol) were then incubated with 32 pmol eRF1 and 32 pmol eRF3 in 100 μL buffer A supplemented with 3 mM Mg-GMPPNP.
Electron Microscopy and Image Processing.
After assembly, purification, and biochemical verification of the 80S pretermination complex, specimens were vitrified and micrographs were recorded under cryo conditions (47). In total, 430,167 particles were selected using reference-based selection methods (48) and multivariate data analysis and classification procedures, followed by manual verification of the aligned and classified projections (49).
For the pre-TC, 22,816 total projections were used for a 3D reconstruction using single-particle–based methods. As with the PTC complex, image projections of the 80S–eRF1–eRF3–GMPPNP complex were first sorted based on cross-correlation coefficient (CCC) to PTC reference, which displayed a bimodal distribution. Selection of the mode with the higher CCC resulted in 322,646 particles, or ∼75% of the total projections. The particles were further sorted based on density in the A site, indicative of eRF1–eRF3 bound to the release complex, using supervised classification (50). 80S–eRF1–eRF3 populations were further sorted using a multireference classification algorithm (51).
Flexible Fitting.
The atomic models of both the 80S pre-TC ribosome, with and without the release complex, were obtained by performing MDFF (52), an approach based on adding external forces proportional to the gradient of the cryo-EM density map into a molecular dynamics (MD) simulation of the atomic structure. The starting model was composed of the X-ray structure of the 80S ribosome from the yeast Saccharomyces cerevisiae (Protein Data Base ID codes 3U5B, 3U5C, 3U5D, and 3U5E) (27) and the X-ray structure of human eRF1 bound with domains 2 and 3 of human eRF3 (17).
For the 80S pre-TC model, MDFF was performed to fit the 80S ribosome X-ray structure into the cryo-EM density map. For the 80S–eRF1–eRF3 structure, MDFF was first performed on the 80S ribosome X-ray structure of S. cerevisiae alone. In this first step, densities attributed to eRF1 and eRF3 were approximately masked. The coordinates of eRF1 and eRF3 were then fitted into their respective density within the cryo-EM volume. Finally the 80S ribosome, with both factors attached, was subjected to EM density-based MDFF fitting, allowing necessary structural adjustments at the molecular boundaries. Because of the relatively low resolution of the density map, all of the MDFF computations were performed with the molecule placed in vacuum.
Supplementary Material
Acknowledgments
We thank Melissa Thomas for assistance in generating illustrations and Terry Kinzy for yeast eEF1A. This work was supported by Howard Hughes Medical Institute and National Institutes of Health Grants R01 GM29169 and GM55440 (to J.F.) and GM80623 (to T.V.P.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3J2K). The cryo-EM map has been deposited in the EMDataBank (accession code EMD-5518).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1216730109/-/DCSupplemental.
References
- 1.Jackson RJ, Hellen CU, Pestova TV. Termination and post-termination events in eukaryotic translation. Adv Protein Chem Struct Biol. 2012;86:45–93. doi: 10.1016/B978-0-12-386497-0.00002-5. [DOI] [PubMed] [Google Scholar]
- 2.Pisarev AV, et al. The role of ABCE1 in eukaryotic posttermination ribosomal recycling. Mol Cell. 2010;37(2):196–210. doi: 10.1016/j.molcel.2009.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Doma MK, Parker R. Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature. 2006;440(7083):561–564. doi: 10.1038/nature04530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kobayashi K, et al. Structural basis for mRNA surveillance by archaeal Pelota and GTP-bound EF1α complex. Proc Natl Acad Sci USA. 2010;107(41):17575–17579. doi: 10.1073/pnas.1009598107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shoemaker CJ, Eyler DE, Green R. Dom34:Hbs1 promotes subunit dissociation and peptidyl-tRNA drop-off to initiate no-go decay. Science. 2010;330(6002):369–372. doi: 10.1126/science.1192430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shoemaker CJ, Green R. Kinetic analysis reveals the ordered coupling of translation termination and ribosome recycling in yeast. Proc Natl Acad Sci USA. 2011;108(51):E1392–E1398. doi: 10.1073/pnas.1113956108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pisareva VP, Skabkin MA, Hellen CU, Pestova TV, Pisarev AV. Dissociation by Pelota, Hbs1 and ABCE1 of mammalian vacant 80S ribosomes and stalled elongation complexes. EMBO J. 2011;30(9):1804–1817. doi: 10.1038/emboj.2011.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Song H, et al. The crystal structure of human eukaryotic release factor eRF1—mechanism of stop codon recognition and peptidyl-tRNA hydrolysis. Cell. 2000;100(3):311–321. doi: 10.1016/s0092-8674(00)80667-4. [DOI] [PubMed] [Google Scholar]
- 9.Bertram G, Bell HA, Ritchie DW, Fullerton G, Stansfield I. Terminating eukaryote translation: Domain 1 of release factor eRF1 functions in stop codon recognition. RNA. 2000;6(9):1236–1247. doi: 10.1017/s1355838200000777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seit-Nebi A, Frolova L, Kisselev L. Conversion of omnipotent translation termination factor eRF1 into ciliate-like UGA-only unipotent eRF1. EMBO Rep. 2002;3(9):881–886. doi: 10.1093/embo-reports/kvf178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fan-Minogue H, et al. Distinct eRF3 requirements suggest alternate eRF1 conformations mediate peptide release during eukaryotic translation termination. Mol Cell. 2008;30(5):599–609. doi: 10.1016/j.molcel.2008.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Conard SE, et al. Identification of eRF1 residues that play critical and complementary roles in stop codon recognition. RNA. 2012;18(6):1210–1221. doi: 10.1261/rna.031997.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Korostelev AA. Structural aspects of translation termination on the ribosome. RNA. 2011;17(8):1409–1421. doi: 10.1261/rna.2733411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mantsyzov AB, et al. NMR solution structure and function of the C-terminal domain of eukaryotic class 1 polypeptide chain release factor. FEBS J. 2010;277(12):2611–2627. doi: 10.1111/j.1742-4658.2010.07672.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kushnirov VV, et al. Nucleotide sequence of the SUP2 (SUP35) gene of Saccharomyces cerevisiae. Gene. 1988;66(1):45–54. doi: 10.1016/0378-1119(88)90223-5. [DOI] [PubMed] [Google Scholar]
- 16.Kong C, et al. Crystal structure and functional analysis of the eukaryotic class II release factor eRF3 from S. pombe. Mol Cell. 2004;14(2):233–245. doi: 10.1016/s1097-2765(04)00206-0. [DOI] [PubMed] [Google Scholar]
- 17.Cheng Z, et al. Structural insights into eRF3 and stop codon recognition by eRF1. Genes Dev. 2009;23(9):1106–1118. doi: 10.1101/gad.1770109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hauryliuk V, Zavialov A, Kisselev L, Ehrenberg M. Class-1 release factor eRF1 promotes GTP binding by class-2 release factor eRF3. Biochimie. 2006;88(7):747–757. doi: 10.1016/j.biochi.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 19.Mitkevich VA, et al. Termination of translation in eukaryotes is mediated by the quaternary eRF1*eRF3*GTP*Mg2+ complex. The biological roles of eRF3 and prokaryotic RF3 are profoundly distinct. Nucleic Acids Res. 2006;34(14):3947–3954. doi: 10.1093/nar/gkl549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pisareva VP, Pisarev AV, Hellen CU, Rodnina MV, Pestova TV. Kinetic analysis of interaction of eukaryotic release factor 3 with guanine nucleotides. J Biol Chem. 2006;281(52):40224–40235. doi: 10.1074/jbc.M607461200. [DOI] [PubMed] [Google Scholar]
- 21.Frolova L, et al. Eukaryotic polypeptide chain release factor eRF3 is an eRF1- and ribosome-dependent guanosine triphosphatase. RNA. 1996;2(4):334–341. [PMC free article] [PubMed] [Google Scholar]
- 22.Kononenko AV, et al. Role of the individual domains of translation termination factor eRF1 in GTP binding to eRF3. Proteins. 2008;70(2):388–393. doi: 10.1002/prot.21544. [DOI] [PubMed] [Google Scholar]
- 23.Alkalaeva EZ, Pisarev AV, Frolova LY, Kisselev LL, Pestova TV. In vitro reconstitution of eukaryotic translation reveals cooperativity between release factors eRF1 and eRF3. Cell. 2006;125(6):1125–1136. doi: 10.1016/j.cell.2006.04.035. [DOI] [PubMed] [Google Scholar]
- 24.Salas-Marco J, Bedwell DM. GTP hydrolysis by eRF3 facilitates stop codon decoding during eukaryotic translation termination. Mol Cell Biol. 2004;24(17):7769–7778. doi: 10.1128/MCB.24.17.7769-7778.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spahn CM, et al. Cryo-EM visualization of a viral internal ribosome entry site bound to human ribosomes: The IRES functions as an RNA-based translation factor. Cell. 2004;118(4):465–475. doi: 10.1016/j.cell.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 26.Penczek PA, Grassucci RA, Frank J. The ribosome at improved resolution: New techniques for merging and orientation refinement in 3D cryo-electron microscopy of biological particles. Ultramicroscopy. 1994;53(3):251–270. doi: 10.1016/0304-3991(94)90038-8. [DOI] [PubMed] [Google Scholar]
- 27.Ben-Shem A, et al. The structure of the eukaryotic ribosome at 3.0 Å resolution. Science. 2011;334(6062):1524–1529. doi: 10.1126/science.1212642. [DOI] [PubMed] [Google Scholar]
- 28.Schmeing TM, et al. The crystal structure of the ribosome bound to EF-Tu and aminoacyl-tRNA. Science. 2009;326(5953):688–694. doi: 10.1126/science.1179700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Becker T, et al. Structure of the no-go mRNA decay complex Dom34-Hbs1 bound to a stalled 80S ribosome. Nat Struct Mol Biol. 2011;18(6):715–720. doi: 10.1038/nsmb.2057. [DOI] [PubMed] [Google Scholar]
- 30.Ito K, Ebihara K, Nakamura Y. The stretch of C-terminal acidic amino acids of translational release factor eRF1 is a primary binding site for eRF3 of fission yeast. RNA. 1998;4(8):958–972. doi: 10.1017/s1355838298971874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Atkinson GC, Baldauf SL, Hauryliuk V. Evolution of nonstop, no-go and nonsense-mediated mRNA decay and their termination factor-derived components. BMC Evol Biol. 2008;8:290. doi: 10.1186/1471-2148-8-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Frank J, Gao H, Sengupta J, Gao N, Taylor DJ. The process of mRNA-tRNA translocation. Proc Natl Acad Sci USA. 2007;104(50):19671–19678. doi: 10.1073/pnas.0708517104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Uchiumi T, Hori K, Nomura T, Hachimori A. Replacement of L7/L12.L10 protein complex in Escherichia coli ribosomes with the eukaryotic counterpart changes the specificity of elongation factor binding. J Biol Chem. 1999;274(39):27578–27582. doi: 10.1074/jbc.274.39.27578. [DOI] [PubMed] [Google Scholar]
- 34.Korostelev A, et al. Crystal structure of a translation termination complex formed with release factor RF2. Proc Natl Acad Sci USA. 2008;105(50):19684–19689. doi: 10.1073/pnas.0810953105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Valle M, et al. Locking and unlocking of ribosomal motions. Cell. 2003;114(1):123–134. doi: 10.1016/s0092-8674(03)00476-8. [DOI] [PubMed] [Google Scholar]
- 36.Chen L, et al. Structure of the Dom34-Hbs1 complex and implications for no-go decay. Nat Struct Mol Biol. 2010;17(10):1233–1240. doi: 10.1038/nsmb.1922. [DOI] [PubMed] [Google Scholar]
- 37.Vetter IR, Wittinghofer A. The guanine nucleotide-binding switch in three dimensions. Science. 2001;294(5545):1299–1304. doi: 10.1126/science.1062023. [DOI] [PubMed] [Google Scholar]
- 38.Becker T, et al. Structural basis of highly conserved ribosome recycling in eukaryotes and archaea. Nature. 2012;482(7386):501–506. doi: 10.1038/nature10829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Khoshnevis S, et al. The iron-sulphur protein RNase L inhibitor functions in translation termination. EMBO Rep. 2010;11(3):214–219. doi: 10.1038/embor.2009.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee HH, et al. Structural and functional insights into Dom34, a key component of no-go mRNA decay. Mol Cell. 2007;27(6):938–950. doi: 10.1016/j.molcel.2007.07.019. [DOI] [PubMed] [Google Scholar]
- 41.Passmore LA, et al. The eukaryotic translation initiation factors eIF1 and eIF1A induce an open conformation of the 40S ribosome. Mol Cell. 2007;26(1):41–50. doi: 10.1016/j.molcel.2007.03.018. [DOI] [PubMed] [Google Scholar]
- 42.Spahn CM, et al. Hepatitis C virus IRES RNA-induced changes in the conformation of the 40s ribosomal subunit. Science. 2001;291(5510):1959–1962. doi: 10.1126/science.1058409. [DOI] [PubMed] [Google Scholar]
- 43.Wills NM, Gesteland RF, Atkins JF. Evidence that a downstream pseudoknot is required for translational read-through of the Moloney murine leukemia virus gag stop codon. Proc Natl Acad Sci USA. 1991;88(16):6991–6995. doi: 10.1073/pnas.88.16.6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Steneberg P, Samakovlis C. A novel stop codon readthrough mechanism produces functional Headcase protein in Drosophila trachea. EMBO Rep. 2001;2(7):593–597. doi: 10.1093/embo-reports/kve128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Firth AE, Wills NM, Gesteland RF, Atkins JF. Stimulation of stop codon readthrough: Frequent presence of an extended 3′ RNA structural element. Nucleic Acids Res. 2011;39(15):6679–6691. doi: 10.1093/nar/gkr224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pisarev AV, Hellen CU, Pestova TV. Recycling of eukaryotic posttermination ribosomal complexes. Cell. 2007;131(2):286–299. doi: 10.1016/j.cell.2007.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dubochet J, et al. Cryo-electron microscopy of vitrified specimens. Q Rev Biophys. 1988;21(2):129–228. doi: 10.1017/s0033583500004297. [DOI] [PubMed] [Google Scholar]
- 48.Rath BK, Frank J. Fast automatic particle picking from cryo-electron micrographs using a locally normalized cross-correlation function: A case study. J Struct Biol. 2004;145(1-2):84–90. doi: 10.1016/j.jsb.2003.11.015. [DOI] [PubMed] [Google Scholar]
- 49.Shaikh TR, Trujillo R, LeBarron JS, Baxter WT, Frank J. Particle-verification for single-particle, reference-based reconstruction using multivariate data analysis and classification. J Struct Biol. 2008;164(1):41–48. doi: 10.1016/j.jsb.2008.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gao H, Valle M, Ehrenberg M, Frank J. Dynamics of EF-G interaction with the ribosome explored by classification of a heterogeneous cryo-EM dataset. J Struct Biol. 2004;147(3):283–290. doi: 10.1016/j.jsb.2004.02.008. [DOI] [PubMed] [Google Scholar]
- 51.Spahn CM, Penczek PA. Exploring conformational modes of macromolecular assemblies by multiparticle cryo-EM. Curr Opin Struct Biol. 2009;19(5):623–631. doi: 10.1016/j.sbi.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Villa E, et al. Ribosome-induced changes in elongation factor Tu conformation control GTP hydrolysis. Proc Natl Acad Sci USA. 2009;106(4):1063–1068. doi: 10.1073/pnas.0811370106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.