Abstract
The ability of proteins to locate specific targets among a vast excess of nonspecific DNA is a fundamental theme in biology. Basic principles governing these search mechanisms remain poorly understood, and no study has provided direct visualization of single proteins searching for and engaging target sites. Here we use the postreplicative mismatch repair proteins MutSα and MutLα as model systems for understanding diffusion-based target searches. Using single-molecule microscopy, we directly visualize MutSα as it searches for DNA lesions, MutLα as it searches for lesion-bound MutSα, and the MutSα/MutLα complex as it scans the flanking DNA. We also show that MutLα undergoes intersite transfer between juxtaposed DNA segments while searching for lesion-bound MutSα, but this activity is suppressed upon association with MutSα, ensuring that MutS/MutL remains associated with the damage-bearing strand while scanning the flanking DNA. Our findings highlight a hierarchy of lesion- and ATP-dependent transitions involving both MutSα and MutLα, and help establish how different modes of diffusion can be used during recognition and repair of damaged DNA.
Postreplicative mismatch repair (MMR) corrects errors in DNA synthesis before they lead to genomic instability (1–3). MMR increases the fidelity of DNA replication up to 1,000-fold, and MMR defects in humans cause hereditary nonpolyposis colorectal cancer and may influence the onset of other tumors (1). MutSα and MutLα are conserved eukaryotic protein complexes necessary for MMR. MutSα is responsible for recognition of mismatches and small insertion/deletion loops (1–3), whereas MutLα harbors an endonuclease activity necessary for cleavage of the lesion-bearing DNA strand (4, 5).
The challenges faced during MMR can be illustrated by considering that Saccharomyces cerevisiae should incur only approximately two mismatches per cell cycle (6). MutSα must find these rare lesions, MutLα must search for lesion-bound MutSα, and the lesion-bound MutSα/MutLα complex must search the flanking DNA for signals that distinguish the parental and daughter strands (1–3). Models describing how DNA-binding proteins search for specific targets include 3D diffusion (i.e., jumping), 1D hopping, 1D sliding, and intersegmental transfer; the latter three are categorized as facilitated diffusion because they allow target association rates exceeding limits imposed by 3D diffusion (7–10). New single-molecule and NMR techniques have led to resurgent interest in understanding how proteins locate targets (11–13), and using single-molecule imaging we previously demonstrated that MutSα and MutLα can undergo facilitated diffusion on undamaged DNA through 1Dsliding and 1D hopping, respectively (14, 15). However, no single-molecule study has directly revealed proteins searching for and subsequently engaging a target site through 1D diffusion (i.e., 1D sliding or 1D hopping) (7), and the inability to visualize target capture also prevents investigation of questions regarding downstream MMR events.
Here we used nanofabricated DNA curtains and total internal reflection fluorescence microscopy (TIRFM) to watch MutSα and MutLα as they interact with mismatch-containing substrates, and we asked how these proteins conduct their respective target searches throughout the early stages of MMR. We show that MutSα can be targeted to mismatched bases through either 1D sliding or 3D diffusion, that MutLα locates mismatch-bound MutSα through 1D hopping and 3D intersite transfer, and that mismatch-bound MutSα and MutSα/MutLα are released upon binding ATP and scan the flanking DNA for strand-discrimination signals by 1D diffusion. While searching for lesions, the movement of MutSα is consistent with a model wherein the protein rotates to maintain constant register with the helical contour of the DNA (14). However, once released from a mismatch, MutSα is altered so that mismatches no longer are recognized as targets, and the protein slides much more rapidly, suggesting its motion no longer is coupled to rotation around the DNA. Finally, we demonstrate that the mismatch-bound MutSα/MutLα complex undergoes an ATP-dependent functional transition rendering it resistant to dissociation from damaged DNA. These data provide a detailed view of how diffusion can contribute to the early stages of MMR.
Results
Visualization of Mismatch Recognition by MutSα on DNA Curtains.
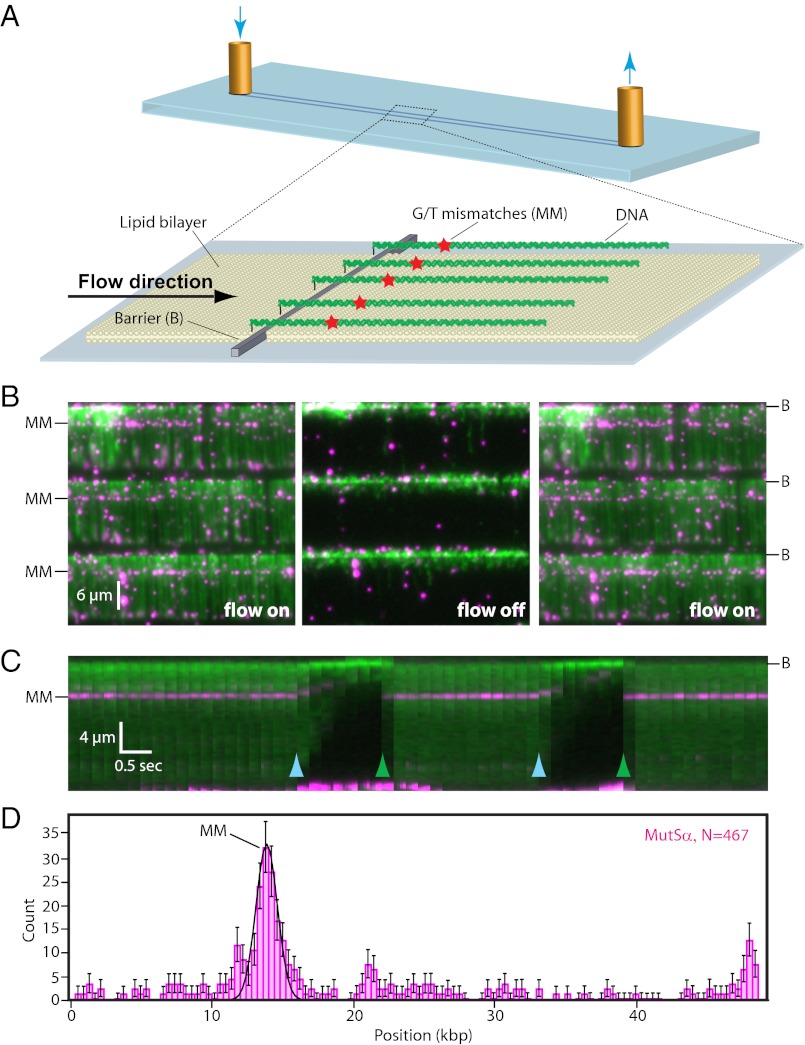
We have used DNA curtains previously to investigate the behavior of MutSα and MutLα on undamaged DNA (14, 15). Here we sought to determine how MutSα and MutLα behave on substrates with defined mismatches. For these experiments, we engineered a λ-DNA (47,467 bp) harboring three tandem G/T mismatches separated from one another by 38 bp (SI Appendix, Fig. S1; three mismatches were used to enhance efficiency of the assay). To make single-tethered DNA curtains, the DNA was anchored to a lipid bilayer on the surface of a microfluidic sample chamber, and hydrodynamic force was used to push the DNA into nanofabricated barriers (Fig. 1A) (16). The DNA was aligned along the barriers, enabling visualization of hundreds of molecules by TIRFM (Fig. 1 B and C and Movie S1). At 150 mM NaCl and 1 mM ADP MutSα showed preferential binding to the mismatches, as evidenced by the “lines” of QD-MutSα that spanned the DNA curtains at the mismatches (Fig. 1B and Movie S1) and as also was evident from histograms of the MutSα binding distributions (Fig. 1D). MutSα disappeared when flow was interrupted and reappeared when flow was resumed, verifying that the proteins were bound to the DNA and were not stuck to the surface of the sample chamber (Fig. 1 B and C and Movie S1). MutSα exhibited a half-life of 9.6 ± 1.5 min while bound to the mismatches in the presence of 1 mM ADP (n = 60; SI Appendix, Fig. S2).
Fig. 1.
Mismatch recognition by MutSα. (A) Schematic of single-tethered DNA curtains. DNA substrates are anchored to the bilayer and aligned along nanofabricated barriers. (B) Images of a three-tiered DNA curtain with flow on (Left), during a transient pause in flow (Center), and after flow has been resumed (Right). Flow is from top to bottom; DNA is green, and proteins are magenta. The location of the three tandem G/T mismatches (MM) is indicated. (C) Kymogram generated from a single DNA molecule subjected to transient pauses in buffer flow (light blue arrowheads) followed by quickly resuming flow (green arrowheads). (D) Distribution of MutSα bound to mismatch-containing DNA. Error bars in this and subsequent figures represent the SD from N bootstrap samples (44).
MutSα Is Targeted to Mismatches Through a Combination of 1D Sliding and 3D Diffusion.
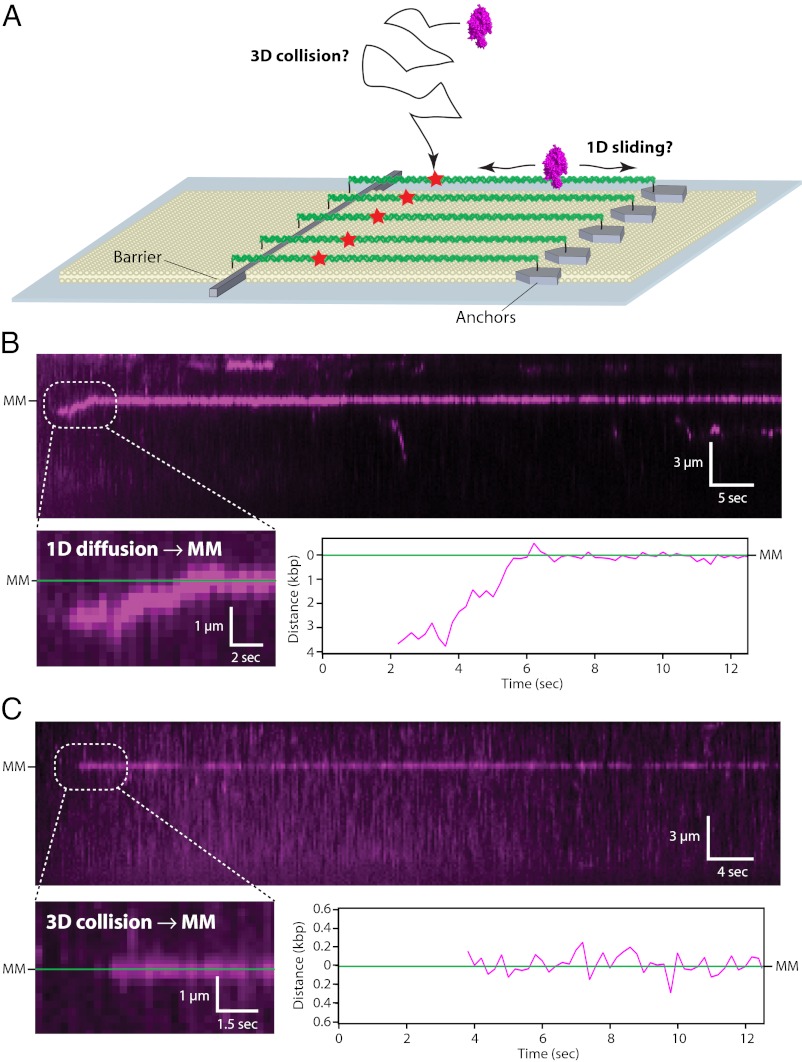
Next, to determine how MutSα located the mismatches, we used double-tethered DNA curtains where the DNA was aligned and anchored by both ends, allowing the molecules to be viewed in the absence of buffer flow (Fig. 2A) (17). MutSα was injected into the sample chamber, flow was terminated, and the proteins were observed in real time as they searched the DNA. At physiological ionic strength, MutSα located the mismatches either through 1D sliding (42.5% of observed events; n = 17/40) (Fig. 2B and SI Appendix, Fig. S3), with sliding observed over distances up to 3.7 µm (∼14.6 kbp), or through apparent 3D diffusion (57.5% of observed events; n = 23/40) (Fig. 2C). We defined target binding as MutSα being within three SDs of the target site for five consecutive frames; any submicroscopic 1D sliding events below this resolution were scored as apparent 3D diffusion. Therefore, the 42.5% of events attributed to 1D sliding represents the minimal fraction that can be described by this mechanism (SI Appendix).
Fig. 2.
Mechanisms of mismatch targeting by MutSα. (A) Schematic of the double-tethered DNA curtains. DNA substrates are anchored by one end to the lipid bilayer, are aligned along the nanofabricated barriers, and then are anchored at their downstream ends through a digoxigenin–antibody linkage. (B) Example of MutSα undergoing 1D diffusion until encountering the lesion. MutSα is magenta, the DNA is not labeled, and gaps in the trajectories reflect QD blinking. The lower panels highlight the first few seconds of the trajectory. (C) Example of MutSα capturing the mismatch through a direct 3D diffusion. Experiments in B and C were conducted with double-tethered curtains, and flow was terminated after MutSα entered the sample chamber.
MutSα Scans DNA Flanking the Mismatch by 1D Diffusion.
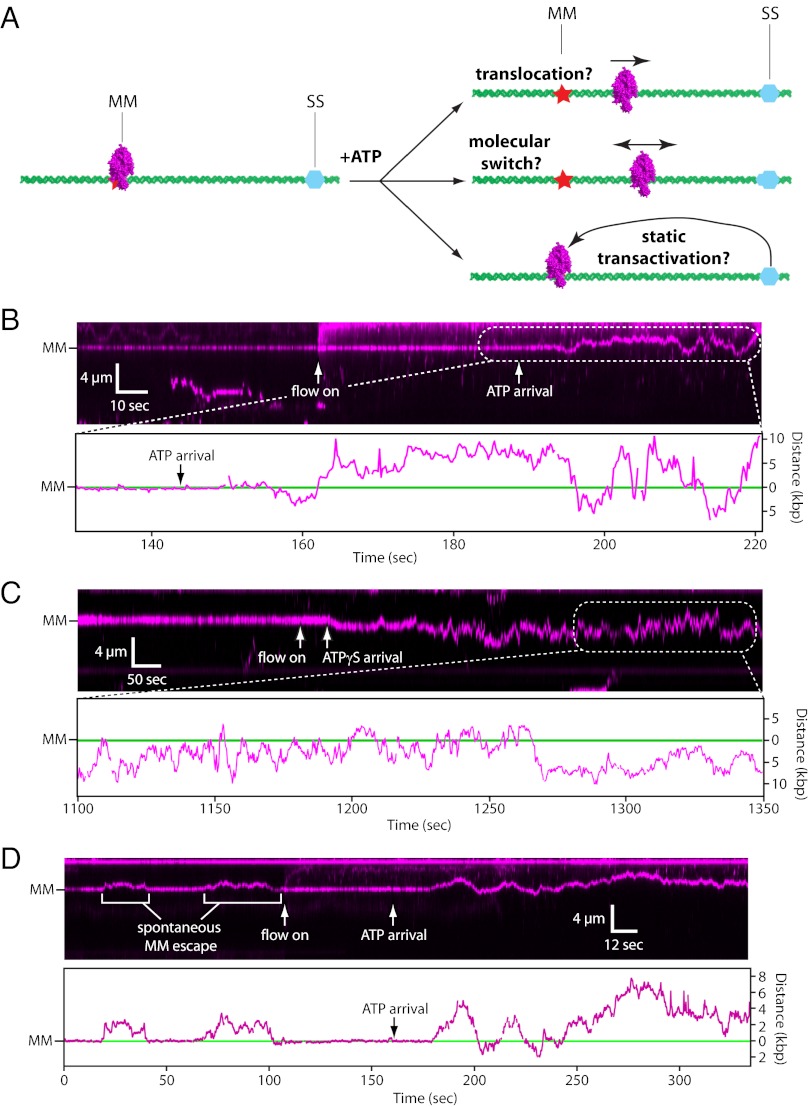
The mechanism by which MMR proteins search for strand-discrimination signals remains controversial (1–3, 18). Three proposed models are (i) translocation, in which MutSα uses the free energy released by ATP hydrolysis to move along DNA (19, 20); (ii) the molecular-switch model, in which ATP binding triggers a conformational change enabling MutSα to scan DNA by 1D diffusion (21–23); and (iii) static transactivation, in which ATP-binding allows stationary MutSα to search for distal strand-discrimination signals through DNA looping (Fig. 3A) (24–26). Each model makes unique predictions as to how MutSα should behave in the DNA curtain assay: Translocation predicts that MutSα should undergo ATP hydrolysis-dependent unidirectional motion; the molecular-switch model predicts that MutSα should exhibit ATP-binding–dependent 1D diffusion; and static transactivation predicts that MutSα should remain at the mismatch while awaiting looping-mediated interactions with flanking DNA.
Fig. 3.
ATP binding provokes 1D diffusion of mismatch-bound MutSα. (A) Models showing how MutSα might search for strand-discrimination signals (SS). (B) Kymogram and tracking showing the response of mismatch-bound MutSα upon injection of 1 mM ATP. Experiments were conducted with double-tethered curtains, MutSα was prebound to the mismatch, ATP was injected at 0.1 mL min−1, and flow was terminated after ATP entered the sample chamber. The DNA was not labeled. “Flow on” indicates when ATP injection was initiated, and “ATP arrival” indicates when ATP entered the sample chamber. The difference between these time points corresponds to the dead volume of the microfluidics. (C) Response of mismatch-bound MutSα upon injection of 1 mM ATPγS. (D) Example of spontaneous, ATP-independent release of MutSα, followed by ATP-dependent release.
To distinguish among the models, we used double-tethered DNA curtains to investigate what happened when mismatch-bound MutSα was chased with ATP. When mismatch-bound MutSα was chased with ATP at physiological ionic strength, most proteins (85%; n = 60/71) were released from the mismatches after a brief delay (t1/2 = 14.6 s; n = 60), consistent with the 8.0 ± 2.7 s half-life reported for ATP-triggered release from G/T mismatches in biochemical studies (23), and the remaining 15% (n = 11/71) remained stationary and did not respond to ATP. Of those that were released upon injection of ATP, 15% (n = 9/60) directly dissociated from the DNA with no evident sliding, whereas the remaining 85% (n = 51/60) were released from the mismatch and scanned the flanking DNA through 1D diffusion (Fig. 3B, SI Appendix, Fig. S4, and Movie S2). Analysis of the mean squared displacement revealed a mean 1D diffusion coefficient (D1D) of 0.057 ± 0.064 µm2 s−1 (n = 25) after ATP-triggered mismatch release. Experiments conducted at 50 mM NaCl revealed significantly less ATP-dependent release of MutSα from the lesions: 78% of the proteins remained stationary upon ATP injection, and the remaining proteins either diffused (18%) or directly dissociated (4%) from the lesions (n = 78; SI Appendix, Fig. S5), indicating that ATP-triggered release and 1D diffusion were favored at physiological ionic strength. Our results also revealed changes in the lifetime of the complexes, as has been reported for Taq MutS (27). As demonstrated above, MutSα can scan DNA for lesions by 1D diffusion, and we have shown previously that at 150 mM NaCl the lifetime of Mutsα while scanning DNA before lesion recognition is 20 ± 4 s (14). In contrast, quantitation of the MutSα diffusion trajectories after lesion release yielded a lower bound for the lifetime, t1/2 ≥ 198 ± 23.4 s (Fig. 3 and SI Appendix, Fig. S4). MutSα also diffused along the DNA when chased with ATPγS (62% diffused, 23% dissociated, and 15% remained stationary; n = 26), indicating that nucleotide binding was sufficient to trigger mismatch release (Fig. 3C and SI Appendix, Fig. S4). These findings support the molecular-switch model in which MutSα scans the flanking DNA by 1D diffusion (21).
MutSα Must Remember Whether It Has Encountered a Mismatch.
The highly redundant nature of diffusion poses a conceptually important problem: Once MutSα is released from a mismatch and starts scanning the flanking DNA by 1D diffusion, it must not reengage the mismatch; otherwise it could become nonproductively trapped while undergoing reiterative cycles of mismatch binding and release. This problem can be illustrated by considering that when MutSα takes a single diffusive step away from the mismatch, it has a 50% probability of re-encountering the mismatch on the very next step, and the average number of times MutSα would re-encounter the mismatch is equal to N−1, where N is the distance in 1-bp diffusion steps between the mismatch and the nearest strand discrimination signal (SI Appendix, Fig. S6). These considerations suggest that MutSα must be functionally distinct after ATP-triggered release from a mismatch to avoid redundant lesion recognition.
To evaluate this hypothesis, we assessed the efficiency of lesion recognition by MutSα before and after ATP-triggered release from the mismatches. Of the MutSα molecules that recognized the lesions through a 1D search, none diffused past the lesions (n = 0/17) (Fig. 2B and SI Appendix, Fig. S3), indicating that initial target recognition must be efficient. Moreover, when MutS spontaneously escaped from the mismatches (i.e., ATP-independent release), the proteins typically diffused a short distance along the DNA and then quickly rebound to the lesions (n = 101 escapes, of which 97 resulted in rebinding to the lesions without bypass) (Fig. 3D, SI Appendix, Fig. S7, and Movie S3). Considered together, these data show that before the addition of ATP, MutSα stopped moving upon encountering the lesions during 1D searches in 97% of all observed cases (n = 114/118), with only 3% of the observed encounters leading to diffusion past the lesions. In contrast, after ATP- (or ATPγS)-triggered mismatch release, we observed a total of 325 independent, microscopically observed bypass events (n = 51 proteins, corresponding to an average of approximately six bypasses per protein), none of which led to detectable rebinding; these values represent the lower bounds for the number of potential bypass events, because the proteins often continued diffusing on the DNA beyond the duration of our observations. Notably, each microscopically observed bypass reflects ∼1,000 submicroscopic encounters with the lesions; these encounters are undetectable as independent events given current resolution limits (SI Appendix). These results indicate MutSα no longer recognizes mismatches as viable targets after ATP-triggered release.
MutSα Diffuses More Rapidly After Mismatch Recognition.
The mean D1D of MutSα before lesion recognition was 0.009 ± 0.011 µm2 s−1 (at 150 mM NaCl; n = 25) (14), but there was a 6.3-fold increase (Student t test, P = 1.5 × 10−9) in this value to 0.057 ± 0.064 µm2 s−1 (n = 25) after ATP-mediated release from the mismatches. Before lesion recognition, the diffusion coefficient of MutSα is consistent with 1D sliding wherein lateral motion of the protein is coupled to obligatory rotation as it tracks the helical pitch of the DNA (14). However, after lesion recognition, the mean diffusion coefficient of MutSα exceeded the theoretical threshold for rotation-coupled 1D diffusion (Drot,theor = 0.024 µm2 s−1) (14) and was physically incompatible with motion involving an obligatory rotational component (12, 28–30). Structures of MutS and MutSα reveal the proteins are in intimate contact with DNA along an interface that completely encircles the duplex (24, 31–33). This configuration could accommodate 1D sliding or could allow MutSα to make very small hops on the DNA as a closed ring, provided there was sufficient space between the protein and DNA surfaces to allow transient penetration of ions that could screen the charged surfaces; we cannot yet distinguish between these two possibilities experimentally. However, we can conclude that the rapid movement of MutSα after mismatch release is most consistent with 1D diffusion (hopping or sliding) in the absence of an obligatory rotational component. A similar conclusion was obtained recently from single-molecule measurements of Taq MutS bound to mismatch-containing DNA (34), suggesting that transitions from rotation-coupled to rotation-uncoupled diffusion upon lesion recognition and ATP-binding may be a common feature of the MutS family of proteins.
Colocalization of MutLα with Mismatch-Bound MutSα
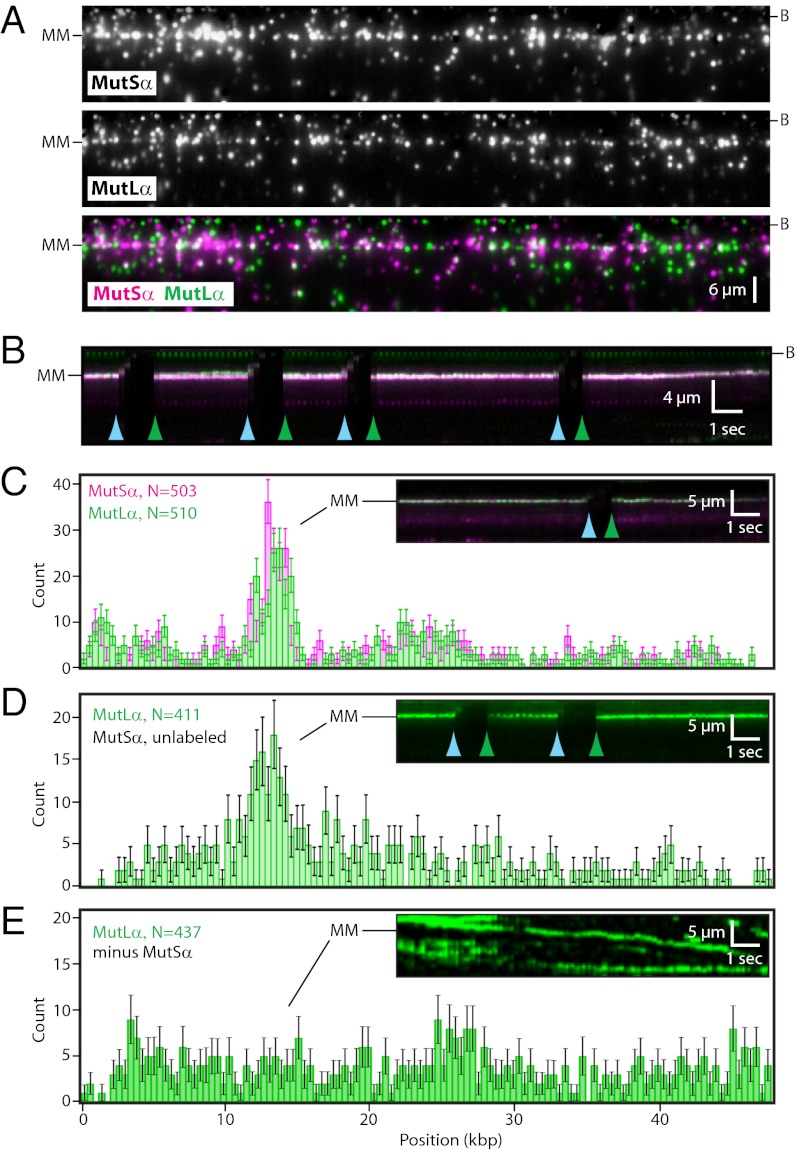
We next asked whether QD-tagged MutLα colocalized with mismatch-bound MutSα on the single-tethered DNA curtains (Fig. 4). We have shown previously that MutLα binds DNA, but rather than remaining stationary, most MutLα (≥95%) diffuses rapidly along the DNA by a 1D hopping mechanism (Movie S4) (15). We detected no colocalization of MutLα and MutSα on DNA that lacked mismatches (n ≥ 2,000; see below), and MutLα alone did not bind the G/T mismatches in the absence of MutSα but instead diffused past the lesions without stopping (Fig. 4E and Movie S5). However, when MutSα was bound to the mismatch, MutLα stopped diffusing at lesion-bound MutSα (Fig. 4 A and B). In the absence of ATP, both proteins remained at the lesions (Movies S6 and S7), with MutLα exhibiting a half-life of 7.8 ± 0.4 min (n = 65) when colocalized with mismatch-bound MutSα (SI Appendix, Fig. S2). Mismatch colocalization of MutLα was observed with both QD-tagged MutSα and untagged MutSα (Fig. 4 C and D). We conclude that MutLα was targeted specifically to mismatch-bound MutSα.
Fig. 4.
Colocalization of MutLα with mismatch-bound MutSα. (A) MutLα binding to mismatch-bound MutSα on single-tethered DNA curtains. MutSα was bound to the mismatch, followed by injection of MutLα into the sample chamber. MutLα and MutSα were labeled with different colored QDs. (Top) MutSα. (Middle) MutLα. (Bottom) Overlay with MutSα (magenta) and MutLα (green). The DNA was not labeled. Imperfect correspondence between all individual QD green/magenta pairs reflects the presence of “dark” proteins. (B) Kymogram generated from a single DNA molecule showing that MutLα remains stationary and colocalized with mismatch-bound MutSα over time; the green (MutLα) and magenta (MutSα) signals appear white in the overlay. Blue and green arrowheads indicate transient pauses in buffer flow, and the coincident disappearance of the QD signals verifies that neither protein was stuck to the sample chamber surface. (C) Distribution of QD-tagged MutLα in the presence of QD-tagged MutSα. (D) Distribution of QD-tagged MutLα with unlabeled MutSα. Insets in C and D show kymograms illustrating that MutSα/MutLα remains at the mismatch. (E) Distribution of QD-tagged MutLα on a single-tethered curtain in the absence of MutSα. MutLα diffuses on DNA continually in the absence of MutSα (Inset), so the distribution histogram in E represents the instantaneous distribution of mobile MutLα molecules, whereas the distribution peaks observed in C and D represent proteins that are stably bound to the lesions and are not moving along the DNA.
MutLα Is Targeted to Lesion-Bound MutSα by 1D Hopping and 3D Diffusion.
We next watched MutLα as it searched for mismatch-bound MutSα on double-tethered DNA curtains. MutLα could locate mismatch-bound MutSα by a 1D-hopping mechanism (55% of observed events; n = 33/60) or by apparent 3D diffusion (45% of observed events; n = 27/60) (Fig. 5A and SI Appendix, Fig. S8); the percentage of events attributed to 1D diffusion represents the minimal fraction that occurred through this mechanism, because the apparent 3D targeting events also could reflect submicroscopic 1D diffusion over distances less than our spatial resolution of ±30 nm. Control experiments verified that MutLα did not stop at mismatches in the absence of MutSα (n ≥ 2,000) (Fig. 5B and Movie S5). We conclude that MutLα can locate mismatch-bound MutSα through 1D hopping or 3D diffusion. Notably, when MutSα and MutLα collided while diffusing at sites other than a mismatch, they showed no evidence of establishing stable interactions (n ≥ 2,000) (Fig. 5C). This outcome is remarkable given that the local concentration of two proteins that encounter one another while undergoing a 1D search on the same DNA molecule is infinitely high. We conclude that the conformational context of MutSα is critical for controlling protein–protein interactions with MutLα and that the two complexes do not interact stably with one another while undergoing 1D diffusion in the absence of a mismatch despite being forced into close physical proximity through association with the same DNA molecule.
Fig. 5.
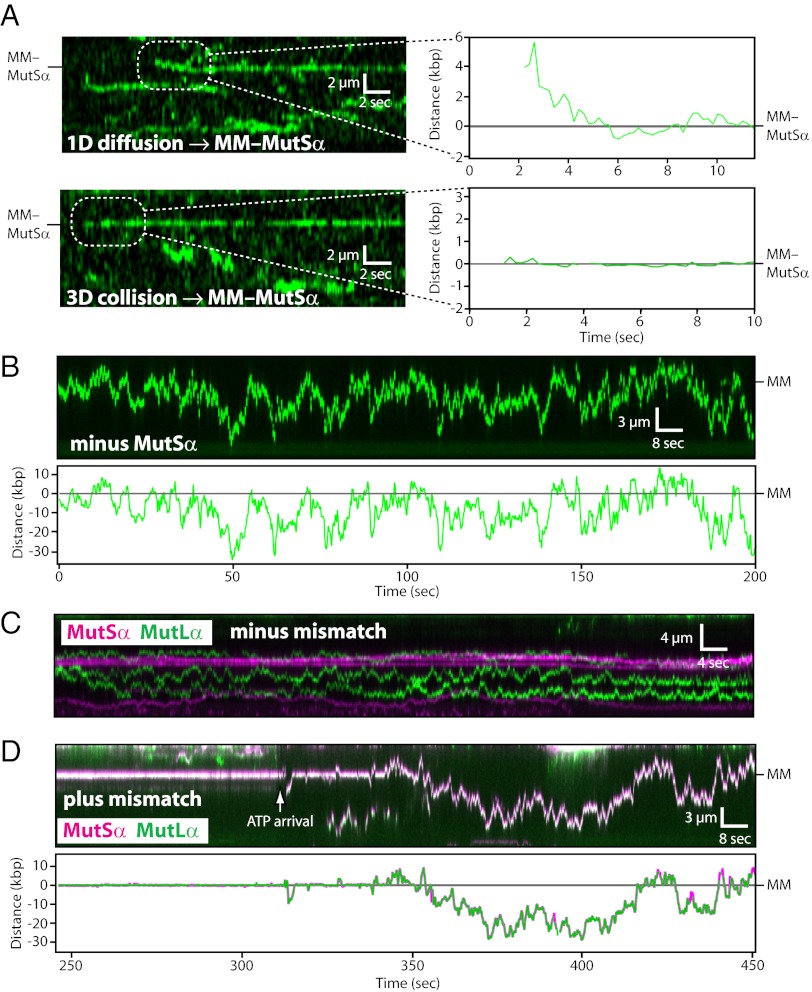
Target-search mechanisms of MutLα and the MutSα/MutLα complex. (A) Kymograms and tracking data showing examples of QD-tagged MutLα (green) engaging mismatch-bound MutSα (MM–MutSα; unlabeled) after undergoing a 1D or 3D target search. The DNA was not labeled. (B) Kymogram and tracking showing that MutLα does not stop at MutSα in the absence of a mismatch. (C) Kymogram showing that MutSα and MutLα do not establish stable interactions with one another on homoduplex DNA. (D) Kymogram and tracking showing ATP-triggered release of lesion-bound MutSα/MutLα and subsequent 1D diffusion along the flanking DNA. In the kymogram, the MutSα (magenta) and MutLα (green) signals appear white in the overlay. In the graph, MutSα (magenta) and MutLα (green) were tracked independently, and the tracking data were superimposed.
MutSα/MutLα Complex Scans DNA Flanking the Mismatch by 1D Sliding.
We next asked whether the MutSα/MutLα complex also scanned the flanking DNA by 1D diffusion. As shown in Fig. 5D and SI Appendix, Fig. S9, in assays with double-tethered DNA curtains, ATP provoked release of MutSα/MutLα from the mismatches at physiological salt concentrations (150 mM NaCl). Most complexes then scanned flanking DNA by 1D diffusion (63% of observed events; n = 22/35), and all of those that scanned DNA by 1D diffusion remained intact as MutSα/MutLα complexes (n = 22/22), demonstrating that MutLα and MutSα remain associated with one another as they scan the flanking DNA by 1D diffusion, even though they do not interact while bound to duplex DNA before lesion recognition by MutSα. Smaller populations dissociated from the DNA upon injection of ATP (23%; n = 8/35) or remained at the ;mismatches (14%; n = 5/35). Following ATP-triggered mismatch release, the MutSα/MutLα complexes that underwent 1D diffusion remained on the DNA for up to several hundred seconds with a lower bound of t1/2 ≥ 267.6 ± 62.1 s (SI Appendix, Fig. S9). The complexes also repeatedly bypass the mismatches, whereas they remain stably bound to the mismatches in reactions containing only ADP (Fig. 5D and Movies S6 and S7). We conclude that the behavior of the MutSα/MutLα complex is consistent with the molecular-switch model (21). Furthermore, analysis of the postlesion diffusion trajectories revealed a mean D1D of 0.062 ± 0.095 µm2 s−1 (n = 22) for MutSα/MutLα, which was ∼6.9-fold larger than observed for MutSα alone before lesion recognition (D1D,MutSα = 0.009 ± 0.011 µm s−1 before mismatch release, at 150 mM NaCl; Student t test, P < 1 × 10−9), providing additional evidence that ATP-triggered release from lesions modifies the diffusive characteristics of the MMR proteins. Our results suggest that MutSα must be functionally distinct before and after lesion recognition and that these changes persist even after the proteins diffuse away from the mismatches.
Increased Stability of MutSα/MutLα After ATP-Triggered Release from Mismatches.
We next tested the relative resistance of the different MMR protein complexes to challenge with high-salt buffers. In the DNA curtain assays, all the DNA-bound MutSα dissociated when chased with moderately high salt (300 mM NaCl) before (14), during, or after lesion recognition (n ≥ 2,000). As previously shown, MutLα is more salt resistant than MutSα (15), but it also dissociated from DNA rapidly when challenged with higher salt (∼100% dissociation at 0.7 M NaCl; n ≥ 2,000). Mismatch-bound MutSα/MutLα also dissociated from DNA upon exposure to high salt, and in the presence of 1 mM ADP all the lesion-bound complexes (n = 40) dissociated from the DNA upon injection of 0.7 M NaCl. In contrast, after ATP-triggered release from the mismatch, MutSα/MutLα became resistant to increases in ionic strength, and 58% of the complexes (n = 18/31) remained bound to DNA and continued diffusing even after injection of buffer containing 0.7 M NaCl; the remaining 42% displayed a lifetime of 23.1 ± 8.3 s. We conclude that mismatch-bound MutSα/MutLα must undergo a structural change upon binding ATP, rendering the complex resistant to dissociation from the lesion-bearing DNA without altering its ability to scan the flanking duplex by 1D diffusion.
Intersite Transfer Between Juxtaposed DNA Molecules During MMR.
It is widely hypothesized that DNA-binding proteins can use some forms of facilitated diffusion (e.g., jumping or intersegmental transfer) to undergo intersite transfer between juxtaposed DNA segments that otherwise are separated by long regions of linear sequence (8, 9, 35). The potential for intersite transfer has profound implications for MMR. Before lesion recognition, either MutSα and/or MutLα might undergo intersite transfer, which in principle could assist in their respective target searches. However, if the proteins were to undergo intersite transfer while scanning the flanking DNA after lesion recognition, then in a best-case scenario repair would fail because the MMR machinery would lose track of the damaged DNA. In a worst-case scenario, intersite transfer after lesion recognition might lead to inappropriate cleavage of undamaged DNA by the MutLα endonuclease.
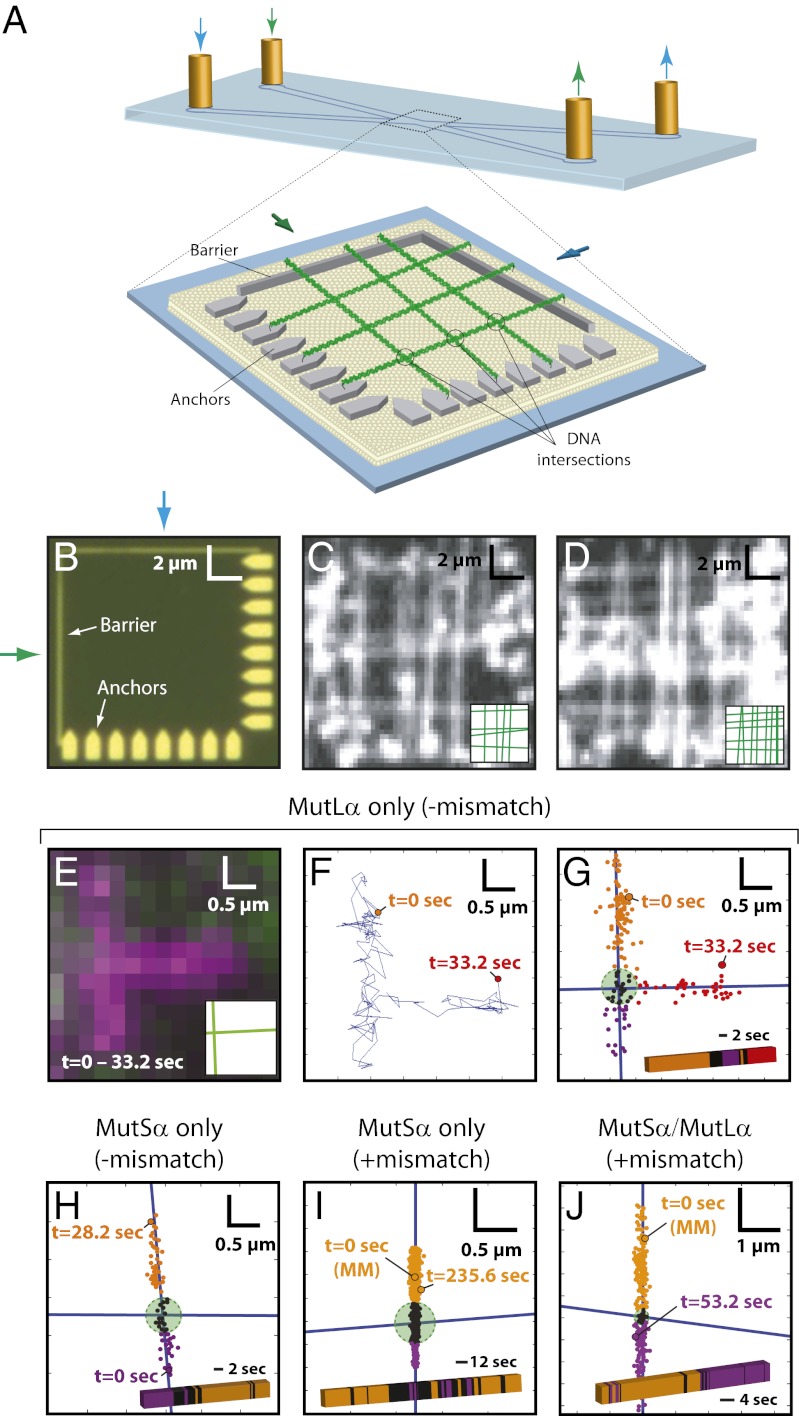
To assess intersite transfer during MMR, we used nanofabricated chromium patterns situated at the convergence of two buffer channels to arrange molecules into crisscrosses, where intersections between molecules represented regions of locally high DNA concentration (Fig. 6 A–D and SI Appendix, Fig. S10). The time-averaged distance between the DNA substrates at the crisscross was ∼106 nm, which was calculated by treating the DNA as two harmonic chains suspended above a surface at the height of the barriers (20 nm), and the probability that they approach within ≤20 nm of one another to during a 100-ms window is near unity (SI Appendix, Fig. S10). We reasoned that intersite transfer would be revealed as ∼90° turns in the protein diffusion trajectories at the DNA intersections. Accordingly, the diffusion trajectories of MutLα were punctuated by abrupt turns at the DNA intersections (Fig. 6 E–G). These results demonstrated that MutLα can undergo intersite transfer, with an observed probability of P = 0.188 (n = 32) for transferring from one DNA to another during each encounter with the intersections. This value represents a lower bound for the frequency of intersite transfer, because these events could be identified unambiguously only if the proteins diffused far enough away from the region encompassing the DNA intersection to verify whether they were bound to the first or second DNA molecule (Fig. 6G). This finding suggests that MutLα would be able to search for lesion-bound MutSα within the 3D volume of the eukaryotic nucleus through a combination of 1D hopping and intersite transfer. Our previous experiments suggest that MutLα travels while wrapped around DNA in a large ring-like configuration (15). If so, then this ring would have to open transiently to allow intersite transfer. In contrast, Mlh1 homodimers do not appear to form rings (15) and therefore would be expected to transfer more readily between two DNA molecules. In agreement with this hypothesis, Mlh1 alone also switched between DNA molecules and did so approximately twofold more efficiently (P = 0.333; n = 39) than MutLα. In contrast to MutLα, MutSα did not transfer between molecules readily before lesion binding (P = 0.067; n = 30) (Fig. 6H) or after ATP-triggered lesion release (P = 0.038; n = 130) (Fig. 6I). The MutSα/MutLα complex also remained confined to the same DNA after ATP-triggered lesion release (P = 0.052; n = 97) (Fig. 6J), indicating that the ability of MutLα to undergo intersite transfer was suppressed upon association with MutSα. These results, together with the finding that MutSα/MutLα is resistant to NaCl-induced dissociation after lesion release, indicate that MutLα is functionally altered within the context of the MutSα/MutLα complex, ensuring that the complex remains confined to the damaged DNA while scanning the flanking sequences.
Fig. 6.
Intersite transfer of MMR proteins between juxtaposed DNA molecules. (A) Schematic of a crisscrossed DNA curtain. (B and C) Optical images showing pattern elements and TIRFM images showing examples of crisscrossed DNA molecules. Insets illustrate positions of each DNA molecule. (E–G) Behavior of MutLα (magenta) upon encountering a crisscrossed DNA junction. (E) Integrated trajectory. (F) Tracking data. (G) Tracking data superimposed on the DNA axes. In G, DNA molecules are shown as blue lines; the green circle identifies the DNA junction within a 90% confidence interval. Tracking data are color-coded according location relative to the crisscross. The color-coded bar shows the relative location of protein over time. (H) MutSα before encountering a lesion. (I) MutSα after ATP-triggered release from a mismatch. (J) MutSα/MutLα after ATP-triggered mismatch release; MutLα was QD-tagged, and MutS was untagged. In I and J the zero time points correspond to the location of the lesions, and the longer time trajectories for these datasets reflect the longer DNA-binding lifetimes of MutSα and MutSα/MutLα after ATP-triggered release from mismatches.
Discussion
Here we provide direct visual observation of proteins searching for and subsequently engaging target sites through facilitated diffusion mechanisms on single molecules of DNA. Our work also illustrates how transitions between different modes of diffusion are regulated during the early stages of MMR through a combination of lesion recognition, protein–protein association, and nucleotide cofactors. This work also suggests how facilitated diffusion might contribute to mismatch repair in vivo and yields insights into the structural changes necessary to accommodate the distinct behaviors of MutSα, MutLα, and the MutSα/MutLα complex at different stages of MMR.
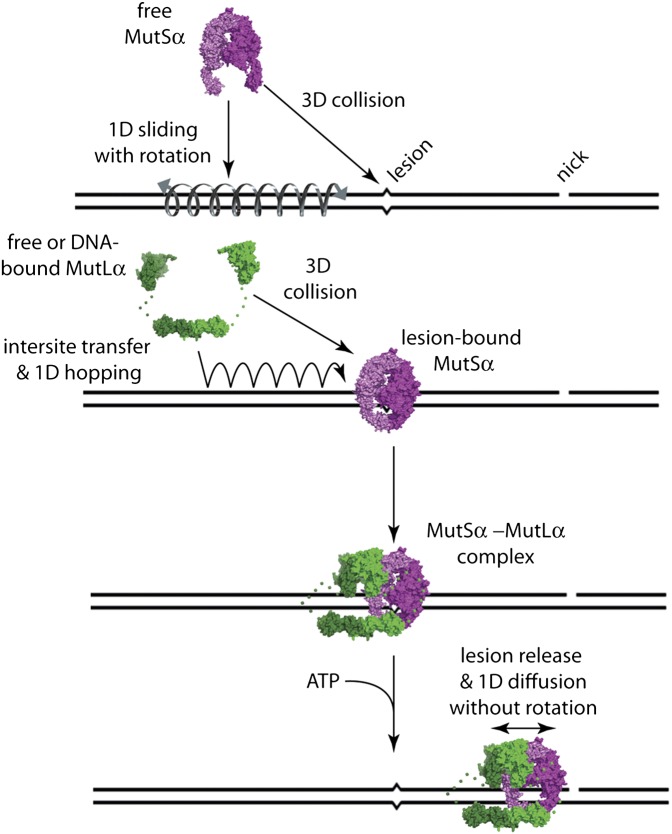
We have shown that MutSα can be targeted to mismatches in vitro by 1D sliding or through apparent 3D diffusion (Fig. 7A). Importantly, we previously demonstrated that sliding of MutSα is obstructed by nucleosomes (15), consistent with the notion that 1D sliding would be problematic for searches in crowded environments (8, 10, 36–38). We also anticipate that mismatch binding through 3D diffusion would be difficult if the mismatch were occluded by a nucleosome (39). These observations imply that any DNA searched by MutSα must be kept free of obstructions. This requirement could be accomplished if the MMR proteins were coupled to the DNA replication machinery. In support of this model, recent work has demonstrated that MutSα is physically associated with replication factories and that 10–15% of mismatch repair can be attributed to replication fork-associated MutSα (40). Together, these results suggest the possibility that the replisome might clear DNA of any potential obstacles that otherwise could impair lesion targeting, perhaps enabling MutSα to slide along the newly synthesized naked DNA while surveying for lesions at the rear of the progressing fork. Our finding that MutSα also can be targeted to lesions through a 3D mechanism (or submicroscopic 1D sliding over distances less than 30 nm) might explain how lesions are located for the 85–90% of repair events that do not involve direct association of MutSα with the replisomes (40).
Fig. 7.
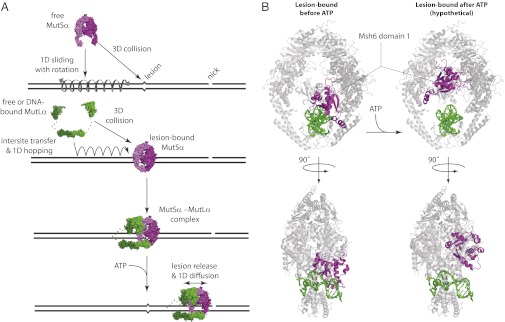
Model for early stages of MMR. (A) Model summarizing how MutSα finds lesions, how MutLα locates lesion-bound MutSα, and how the MutSα/MutLα scans the flanking DNA by 1D diffusion after ATP-triggered release from a lesion. (B) MutSα structural changes predicted upon ATP-triggered release from a lesion. (Left) The structures represent front and side views of mismatch-bound human MutSα (Protein Data Bank ID code 2O8B) in which the protein complex (gray) is wrapped around the DNA (green) with domain I of Msh6 (magenta) engaged with the mismatched base (33). (Right) Hypothetical structures were obtained by rigid-body rotation of Msh6 domain I out of the major groove to illustrate how retraction of Msh6 domain I out of the DNA major groove might allow the release of MutSα from the mismatch and still allow the protein to remain tightly wrapped around the DNA while enabling 1D diffusion in the absence of an obligatory rotational component.
MutLα can search for lesion-bound MutSα through a combination of 1D hopping, 3D diffusion, and intersite transfer (Fig. 7A), and we anticipate that this search could occur on chromatin because MutLα can diffuse readily past nucleosomes (15). After assembling at a lesion, the MutSα/MutLα complex is released upon binding ATP and scans the flanking DNA by 1D diffusion. During this search, MutSα/MutLα is rendered incapable of intersite transfer and becomes highly resistant to dissociation, which could ensure that the MutSα/MutLα complex remained confined to the damaged DNA. These properties are established through a sequence of events including lesion recognition by MutSα and establishment of mismatch-dependent protein–protein interactions between MutSα and MutLα followed by ATP-triggered release of MutSα/MutLα from the lesion. This strict hierarchy would enforce tight regulatory control over the formation of higher-order MMR protein intermediates, thereby preventing inappropriate assembly of MutSα/MutLα complexes at sites other than DNA lesions. Bacterial MutL and eukaryotic MutLα both undergo ATP-driven conformational changes consistent with the formation of closed-ring architectures mediated through dimerization of the N-terminal domains (41, 42). Therefore, we hypothesize that MutLα within the context of the MutSα/MutLα complex engages the DNA in a closed-ring configuration after ATP-triggered mismatch release, rendering the complex resistant to dissociation from damaged DNA (Fig. 7A). The marked resistance of the MutSα/MutLα complex to dissociation from the DNA after ATP-triggered release from the mismatches also is consistent with the recent finding that Pms1–4GFP foci do not turn over when the downstream stages of MMR are compromised (40).
MutLα form oligomers comprised of ∼11 ± 5 proteins at sites of repair in vivo, as evidenced by the presence of Pms1–4GFP foci (40). In our assays, ∼79% of all observed MutLα appeared consistent with single proteins based on quantum dot (QD) blinking (SI Appendix). The predominance of single MutLα molecules in our study can be attributed to the fact that we were probing the early stages of MMR involving initial lesion recognition and assembly of the first MutSα/MutLα complex. In contrast, MutLα foci observed in vivo reflect later stages of the reaction (40). Taken together these results suggest that MutLα oligomerization on MutSα occurs only after the first MutSα/MutLα complex is released from the lesion. This hypothesis also is supported by the observation that the msh6-G114D mutant of MutSα, which is capable of forming a ternary complex with MutLα at mismatches but is defective for ATP-triggered release, does not support formation of detectable Pms1–4GFP foci in vivo. Therefore, ATP-triggered release of the initial MutSα/MutLα complex from the lesions may represent an intermediate step preceding the assembly of higher-order MutLα oligomers.
MutSα alone or within the context of the MutSα/MutLα complex displays dramatically altered diffusive characteristics before and after lesion recognition, likely reflecting distinct functional and structural states necessary to accommodate the different stages of MMR. Before mismatch recognition, MutSα diffuses through a mechanism consistent with 1D sliding while tracking the helical pitch of the DNA (14, 15), but after ATP-triggered release from the mismatch, MutSα diffuses much more rapidly and no longer recognizes mismatches as binding targets. Inspection of available MutS and MutSα structures provides a potential explanation for these differences (Fig. 7B) (24, 31, 33). MutSα completely encircles DNA, and domain I of Msh6 lies within the major groove, allowing a conserved phenylalanine and glutamic acid to engage the mismatch; all remaining contacts with the DNA lie along the phosphate backbone (24, 31, 33). This configuration of Msh6 domain I would impose steric constraints requiring MutSα to track the helical pitch of the DNA during any 1D diffusion (i.e., just as a bolt tracks the helical threads of a screw). Retraction of domain I from the major groove would be necessary and sufficient to allow MutSα to diffuse as a closed ring on DNA without obligatory rotation (Fig. 7B) and also is consistent with the recent observation that domain I of Taq MutS undergoes large structural changes upon being released from mismatches based upon single-pair fluorescence resonance measurements of energy transfer (43). Therefore, we hypothesize that domain I of Msh6 is inserted into the major groove before lesion recognition (as necessary to engage a mismatch and consistent with a rotation-coupled 1D diffusion) and remains within the major groove upon binding the lesion (as shown in the crystal structures) but then is retracted from the major groove after ATP-triggered release from the mismatch (consistent with more rapid 1D diffusion observed after lesion recognition). Retraction of Msh6 domain I from the major groove also would explain how MutSα and MutSα/MutLα are released from the mismatch upon binding ATP and how they avoid rebinding the mismatch while searching for strand-discrimination signals.
Materials and Methods
Experiments were performed with a custom-built TIRF microscope and nanofabricated DNA curtains, as previously described (14–17). Images were acquired at 5–10 Hz using NIS-Elements software (Nikon) and were saved as uncompressed, 16-bit TIFF files. Experiments requiring two-color detection used a Dual-View image-splitting device (Optical Insights) equipped with a dichroic mirror (630 DCXR; Chroma Technologies). Image alignment of the two channels was performed during postprocessing [ImageJ software (National Institutes of Health) with the “Align RGB Planes” plug-in] using the dark signal from the nanofabricated DNA barriers as a reference, and aligned images were pseudocolored and digitally recombined in ImageJ. Before use, MutSα was affinity purified after being labeled with QDs, thus eliminating any QDs not bound by active MutSα before injection of the sample for single-molecule imaging. Unless otherwise stated, reactions were performed as previously described (14, 15), except that all buffers contained either 100 or 150 mM NaCl. In brief, all buffers contained 20 mM Tris (pH 7.8), 1 mM MgCl2, 1 mM DTT, and 4 mg/mL BSA, along with the indicated concentration of NaCl. Unless otherwise stated, standard reaction conditions for looking at lesion binding all contained 1 mM ADP. In the nucleotide chase experiments, ADP was replaced by injecting 1 mM ATP or 1 mM ATPγS, as specified. Finally, YOYO1 was omitted in most reactions because its presence inhibited ATP-triggered release of MutSα from the mismatches. Additional details are provided in SI Appendix.
Supplementary Material
Acknowledgments
We apologize to colleagues whose work we were unable to cite because of space limitations. This work was supported by a National Science Foundation (NSF) Presidential Early Career Award for Scientists and EngineersAward and by National Institutes of Health (NIH) Grants GM082848 (to E.C.G.) and GM53085 (to E.E.A.), and by the Howard Hughes Medical Institute. J.G. was supported by NIH Training Grant T32GM00879807. A.J.P. was supported by a State University of New York fellowship. The work was supported in part by the Initiatives in Science and Engineering program through Columbia University, the NSF Nanoscale Science and Engineering Initiative CHE-0641523; and by the New York State Office of Science, Technology, and Academic Research.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
See Commentary on page 18243.
See Author Summary on page 18251 (volume 109, number 45).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1211364109/-/DCSupplemental.
References
- 1.Modrich P. Mechanisms in eukaryotic mismatch repair. J Biol Chem. 2006;281:30305–30309. doi: 10.1074/jbc.R600022200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jiricny J. The multifaceted mismatch-repair system. Nat Rev Mol Cell Biol. 2006;7:335–346. doi: 10.1038/nrm1907. [DOI] [PubMed] [Google Scholar]
- 3.Kunkel TA, Erie DA. DNA mismatch repair. Annu Rev Biochem. 2005;74:681–710. doi: 10.1146/annurev.biochem.74.082803.133243. [DOI] [PubMed] [Google Scholar]
- 4.Kadyrov FA, Dzantiev L, Constantin N, Modrich P. Endonucleolytic function of MutLalpha in human mismatch repair. Cell. 2006;126:297–308. doi: 10.1016/j.cell.2006.05.039. [DOI] [PubMed] [Google Scholar]
- 5.Kadyrov FA, et al. Saccharomyces cerevisiae MutLalpha is a mismatch repair endonuclease. J Biol Chem. 2007;282:37181–37190. doi: 10.1074/jbc.M707617200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kunkel TA. DNA replication fidelity. J Biol Chem. 2004;279:16895–16898. doi: 10.1074/jbc.R400006200. [DOI] [PubMed] [Google Scholar]
- 7.Halford SE. An end to 40 years of mistakes in DNA-protein association kinetics? Biochem Soc Trans. 2009;37:343–348. doi: 10.1042/BST0370343. [DOI] [PubMed] [Google Scholar]
- 8.von Hippel PH, Berg OG. Facilitated target location in biological systems. J Biol Chem. 1989;264:675–678. [PubMed] [Google Scholar]
- 9.Halford SE, Marko JF. How do site-specific DNA-binding proteins find their targets? Nucleic Acids Res. 2004;32:3040–3052. doi: 10.1093/nar/gkh624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hager GL, McNally JG, Misteli T. Transcription dynamics. Mol Cell. 2009;35:741–753. doi: 10.1016/j.molcel.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tang C, Iwahara J, Clore GM. Visualization of transient encounter complexes in protein-protein association. Nature. 2006;444:383–386. doi: 10.1038/nature05201. [DOI] [PubMed] [Google Scholar]
- 12.Blainey PC, van Oijen AM, Banerjee A, Verdine GL, Xie XS. A base-excision DNA-repair protein finds intrahelical lesion bases by fast sliding in contact with DNA. Proc Natl Acad Sci USA. 2006;103:5752–5757. doi: 10.1073/pnas.0509723103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gorman J, Greene EC. Visualizing one-dimensional diffusion of proteins along DNA. Nat Struct Mol Biol. 2008;15:768–774. doi: 10.1038/nsmb.1441. [DOI] [PubMed] [Google Scholar]
- 14.Gorman J, et al. Dynamic basis for one-dimensional DNA scanning by the mismatch repair complex Msh2-Msh6. Mol Cell. 2007;28:359–370. doi: 10.1016/j.molcel.2007.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gorman J, Plys AJ, Visnapuu ML, Alani E, Greene EC. Visualizing one-dimensional diffusion of eukaryotic DNA repair factors along a chromatin lattice. Nat Struct Mol Biol. 2010;17:932–938. doi: 10.1038/nsmb.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fazio T, Visnapuu ML, Wind S, Greene EC. DNA curtains and nanoscale curtain rods: High-throughput tools for single molecule imaging. Langmuir. 2008;24:10524–10531. doi: 10.1021/la801762h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gorman J, Fazio T, Wang F, Wind S, Greene EC. Nanofabricated racks of aligned and anchored DNA substrates for single-molecule imaging. Langmuir. 2010;26:1372–1379. doi: 10.1021/la902443e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kolodner RD, Mendillo ML, Putnam CD. Coupling distant sites in DNA during DNA mismatch repair. Proc Natl Acad Sci USA. 2007;104:12953–12954. doi: 10.1073/pnas.0705698104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Allen DJ, et al. MutS mediates heteroduplex loop formation by a translocation mechanism. EMBO J. 1997;16:4467–4476. doi: 10.1093/emboj/16.14.4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blackwell LJ, Martik D, Bjornson KP, Bjornson ES, Modrich P. Nucleotide-promoted release of hMutSalpha from heteroduplex DNA is consistent with an ATP-dependent translocation mechanism. J Biol Chem. 1998;273:32055–32062. doi: 10.1074/jbc.273.48.32055. [DOI] [PubMed] [Google Scholar]
- 21.Gradia S, Acharya S, Fishel R. The human mismatch recognition complex hMSH2-hMSH6 functions as a novel molecular switch. Cell. 1997;91:995–1005. doi: 10.1016/s0092-8674(00)80490-0. [DOI] [PubMed] [Google Scholar]
- 22.Gradia S, et al. hMSH2-hMSH6 forms a hydrolysis-independent sliding clamp on mismatched DNA. Mol Cell. 1999;3:255–261. doi: 10.1016/s1097-2765(00)80316-0. [DOI] [PubMed] [Google Scholar]
- 23.Mendillo ML, Mazur DJ, Kolodner RD. Analysis of the interaction between the Saccharomyces cerevisiae MSH2-MSH6 and MLH1-PMS1 complexes with DNA using a reversible DNA end-blocking system. J Biol Chem. 2005;280:22245–22257. doi: 10.1074/jbc.M407545200. [DOI] [PubMed] [Google Scholar]
- 24.Obmolova G, Ban C, Hsieh P, Yang W. Crystal structures of mismatch repair protein MutS and its complex with a substrate DNA. Nature. 2000;407:703–710. doi: 10.1038/35037509. [DOI] [PubMed] [Google Scholar]
- 25.Junop MS, Obmolova G, Rausch K, Hsieh P, Yang W. Composite active site of an ABC ATPase: MutS uses ATP to verify mismatch recognition and authorize DNA repair. Mol Cell. 2001;7:1–12. doi: 10.1016/s1097-2765(01)00149-6. [DOI] [PubMed] [Google Scholar]
- 26.Wang H, et al. DNA bending and unbending by MutS govern mismatch recognition and specificity. Proc Natl Acad Sci USA. 2003;100:14822–14827. doi: 10.1073/pnas.2433654100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jeong C, et al. MutS switches between two fundamentally distinct clamps during mismatch repair. Nat Struct Mol Biol. 2011;18:379–385. doi: 10.1038/nsmb.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bagchi B, Blainey PC, Xie XS. Diffusion constant of a nonspecifically bound protein undergoing curvilinear motion along DNA. J Phys Chem B. 2008;112:6282–6284. doi: 10.1021/jp077568f. [DOI] [PubMed] [Google Scholar]
- 29.Blainey PC, et al. Nonspecifically bound proteins spin while diffusing along DNA. Nat Struct Mol Biol. 2009;16:1224–1229. doi: 10.1038/nsmb.1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schurr JM. The one-dimensional diffusion coefficient of proteins absorbed on DNA. Hydrodynamic considerations. Biophys Chem. 1979;9:413–414. [PubMed] [Google Scholar]
- 31.Lamers MH, et al. The crystal structure of DNA mismatch repair protein MutS binding to a G x T mismatch. Nature. 2000;407:711–717. doi: 10.1038/35037523. [DOI] [PubMed] [Google Scholar]
- 32.Mendillo ML, et al. Probing DNA- and ATP-mediated conformational changes in the MutS family of mispair recognition proteins using deuterium exchange mass spectrometry. J Biol Chem. 2010;285:13170–13182. doi: 10.1074/jbc.M110.108894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Warren JJ, et al. Structure of the human MutSalpha DNA lesion recognition complex. Mol Cell. 2007;26:579–592. doi: 10.1016/j.molcel.2007.04.018. [DOI] [PubMed] [Google Scholar]
- 34.Cho W-K, et al. ATP alters the diffusion mechanics of MutS on mismatched DNA. Structure. 2012;20:1264–1274. doi: 10.1016/j.str.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vuzman D, Polonsky M, Levy Y. Facilitated DNA search by multidomain transcription factors: Cross talk via a flexible linker. Biophys J. 2010;99:1202–1211. doi: 10.1016/j.bpj.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mirny L, et al. How a protein searches for its site on DNA: The mechanism of facilitated diffusion. J Phys A. 2009;42:434013. [Google Scholar]
- 37.Slutsky M, Mirny LA. Kinetics of protein-DNA interaction: Facilitated target location in sequence-dependent potential. Biophys J. 2004;87:4021–4035. doi: 10.1529/biophysj.104.050765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gorski SA, Dundr M, Misteli T. The road much traveled: Trafficking in the cell nucleus. Curr Opin Cell Biol. 2006;18:284–290. doi: 10.1016/j.ceb.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 39.Li F, Tian L, Gu L, Li GM. Evidence that nucleosomes inhibit mismatch repair in eukaryotic cells. J Biol Chem. 2009;284:33056–33061. doi: 10.1074/jbc.M109.049874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hombauer H, Campbell CS, Smith CE, Desai A, Kolodner RD. Visualization of eukaryotic DNA mismatch repair reveals distinct recognition and repair intermediates. Cell. 2011;147:1040–1053. doi: 10.1016/j.cell.2011.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ban C, Junop M, Yang W. Transformation of MutL by ATP binding and hydrolysis: A switch in DNA mismatch repair. Cell. 1999;97:85–97. doi: 10.1016/s0092-8674(00)80717-5. [DOI] [PubMed] [Google Scholar]
- 42.Sacho EJ, Kadyrov FA, Modrich P, Kunkel TA, Erie DA. Direct visualization of asymmetric adenine-nucleotide-induced conformational changes in MutL alpha. Mol Cell. 2008;29:112–121. doi: 10.1016/j.molcel.2007.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qiu R, et al. Large conformational changes in MutS during DNA scanning, mismatch recognition and repair signalling. EMBO J. 2012;31:2528–2540. doi: 10.1038/emboj.2012.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Efron B, Tibshirani R. An Introduction to the Bootstrap. New York: Chapman and Hall, Inc.; 1993. [Google Scholar]