Abstract
Natural products represent valuable lead structures for drug discovery. However, for most bioactive compounds no cellular target is yet identified and many substances predicted from genome analysis are inaccessible due to their life stage-dependent biosynthesis, which is not reflected in common isolation procedures. In response to these issues, an NMR-based and target-directed protease assay for inhibitor detection of the proteasome was developed. The methodology is suitable for one-shot identification of inhibitors in conglomerates and crude culture broths. The technique was applied for analysis of the different life stages of the bacterium Photorhabdus luminescens, which resulted in the isolation and characterization of cepafungin I (CepI), the strongest proteasome inhibitor described to date. Its biosynthesis is strictly regulated and solely induced by the specific environmental conditions determined by our methodology. The transferability of the developed technique to other drug targets may disclose an abundance of novel compounds applicable for drug development.
Keywords: NMR assay, UPS, natural compound, screening, virulence
The eukaryotic proteasome core particle (CP) is a 720-kDa multicatalytic protein degradation machinery and constitutes the main nonlysosomal protein degradation system in cells (1, 2). Its structure is composed of four stacked heptameric rings which contain the active subunits β1, β2, and β5 with an electrophilic Thr-1Oγ at their N-termini (3–5). They exhibit the proteolytic activities denominated caspase-like (CL), trypsin-like (TL), and chymotrypsin-like (ChTL) according to the chemical nature of their specificity pockets (6). Being involved in many cellular processes such as cell proliferation and antigen presentation, the CP is a potential target for the treatment of diseases as diverse as cancer or autoimmunity (7). The blockbuster drug Velcade, a synthetic dipeptide boronic acid, was the first proteasome inhibitor approved for cancer therapy (8). However, because of its severe side effects (9) and the vast potential of proteasome inhibitors with regard to other diseases (7), there is a high demand for the development of novel agents. Lead structures for new drugs are often deduced from natural compounds, as they are evolutionarily optimized (10). A current and highly prominent example is carfilzomib, a proteasome inhibitor in clinical phase III trials (11), which was derived from the natural product inhibitor epoxomicin (12, 13). Although other natural compounds have been elucidated in past decades (10), a persistent challenge for the identification of potent proteasome inhibitors still remains. However, in our days, natural product research suffers generally from two major limitations. First, for most compounds described in the literature, no cellular target has been identified so far, which generally complicates the rational application of bioactive compounds. Second, the life stage-dependent metabolite pattern of many organisms is not reflected by commonly applied growth conditions, thus leading to a severely limited accessibility within the huge spectrum of natural substances.
Results
Development of an NMR-Based Proteasome Assay.
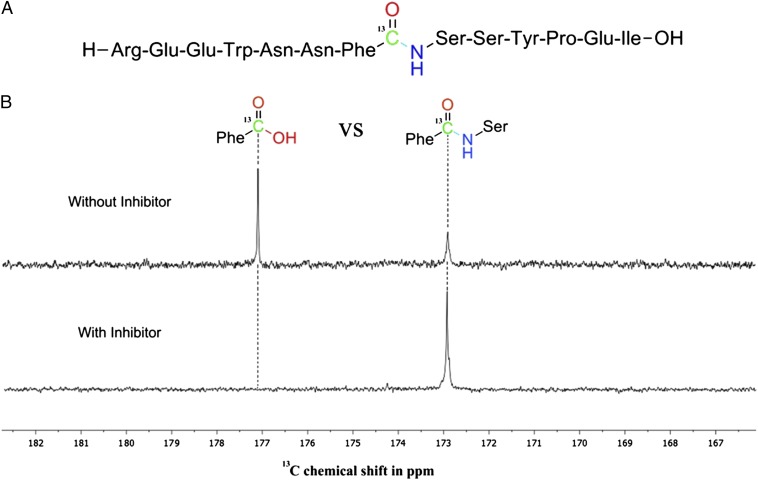
Addressing both issues, we have developed a proteasome assay for screening of inhibitor secretion under various growth conditions. It uses a peptide substrate of the CP, which was atom-selectively labeled for NMR spectroscopy and derived from the digestion pattern of the murine JAK1 tyrosine kinase, a well characterized natural substrate of the 20S proteasome (14). The peptide sequence is adapted toward specificity for the ChTL activity as inhibitors against the corresponding β5 subunit exert high cytotoxicity and strongly influence adaptive immune response by MHC I presentation. By introducing a carbonyl 13C probe at the scissile bond, hydrolysis can be qualitatively and quantitatively evaluated by a peak shift in the 13C-NMR spectrum from 173 to 177 ppm, corresponding to the amide educt and the carboxylic acid product, respectively (Fig. 1). Synthesis of the substrate was achieved by standard Boc-based solid-phase peptide chemistry, whereas the peptide cleavage site was determined by liquid chromatography–mass spectrometry (LC-MS) analysis of the digestion products in an in vitro assay. Confirming the specificity of the probe molecule toward the β5 subunit, the hydrolysis was prevented in the same assay after addition of the synthetic boronic acid inhibitor MG262 (15), which exhibits inhibition merely of the ChTL activity.
Fig. 1.
NMR peptide assay. (A) Primary sequence of the substrate with the scissile peptide bond and the 13C label. (B) NMR spectrum of substrate digestions with the uncleaved educt and the carboxylic acid product at 173 and 177 ppm, respectively (Upper). Product peak formation is suppressed upon addition of a CP-inhibiting culture broth (Lower).
The assay displays the superiority of NMR techniques over common UV-VIS and fluorescence-based methods by overcoming color quenching artifacts, which produce a plentitude of false-positive or -negative results in high-throughput arrays. In contrast, the developed approach yields unambiguous readout information and is applicable to extremely heterogeneous and colored conglomerates present, for example, in culture media. The technique is suitable for high-throughput analysis, as the recording time in a standard 500-MHz NMR machine, equipped with an autosampler, is only about 15 min per assay. Thus, a typical sample number of 96 experiments can be processed within 1 d. With a volume of ∼500 µL, the methodology allows to screen a large number of small cultures at different growth conditions in short periods.
Application of the Technique in a Real-Case Scenario.
For evaluation of the NMR assay by a positive control, we analyzed secretions of Pseudomonas syringae, which causes the brown spot disease in common bean plants and whose virulence is decisively determined by the proteasome inhibitor syringolin A (SylA), a member of the syrbactin family. The bacteria were grown in SRMAF medium, which contains the phenolic sugar arbutin present in plant leaves for induction of the pathogenic phase (16). The crude broth was then added to the assay mixture containing yeast CP. After an incubation period of only 10 min, 13C-labeled peptide substrate was added. NMR analysis of the digestion showed complete suppression of product formation, thus demonstrating the presence of proteasome-inhibiting substances. Despite the high chemical heterogeneity covering all types of biomolecules present in culture broth, the signal-to-noise ratio in the decisive spectral range from 173 to 177 ppm was unaffected by interference. Confirming the univocal results gained by our method, SylA was purified from the respective culture broth and analyzed by HPLC as described (16).
Identification of Candidate Organisms for Analysis.
Next, we aimed to identify other organisms producing inhibitors against the CP. SylA is produced in vivo by a nonribosomal peptide synthetase, which is constituted by an array of enzymes responsible for attachment of amino acid building blocks and introduction of chemical modifications. SylD, a 460-kDa multidomain enzyme, represents the major part of this assembly line and was chosen for BLAST alignment (17, 18). The search yielded hits among various Photorhabdus and Burkholderia species, which can therefore be suggested to produce analogous natural products. This group also comprises Burkholderia pseudomallei, the causative agent of melioidosis (19). Intriguingly, the related but less human pathogenic bacterium Burkholderia mallei, carries an analogous gene cluster, which is inactivated by transposon-mediated rearrangement, hence suggesting a significant contribution to virulence of the respective proteasome inhibiting compound (17). From this group of organisms, we chose the insect parasite bacterium Photorhabdus luminescens, as this S1 pathogen can be handled easily. The organism secretes intensely red-colored compounds (20, 21) not suitable for analysis with common assay types (Fig. S1), hence representing a perfect candidate for a proof of concept of our detection method.
Induction of the Pathogenic Phase in P. luminescens.
Photorhabdus was grown in SRMAF medium containing the phenolic sugar arbutin present in leaves, which induces secretion of the SylA virulence factor from the plant pathogen P. syringae (16). However, no inhibitor production was detected by using our NMR assay, thus implying a divergent evolution of the sensory system of both species to adapt to their respective host organisms. P. luminescens obviously requires a different molecular trigger present in insects to start its pathogenic phase. Screening of diverse growth media with our NMR-based methodology revealed that an osmotic shock is necessary to mimic the environmental changes upon intrusion into the insect body and in turn to initiate inhibitor secretion (SI Text, Table S1). Proteolytic processing of the substrate peptide was completely precluded under these conditions, indicating the presence of at least one highly potent CP inhibitor.
Isolation and Structure Elucidation of the Inhibitors.
To identify the CP-inhibiting compounds from Photorhabdus, we have established an isolation procedure from liquid broth (SI Materials and Methods). Intriguingly, from 3-L culture broth we received two CP inhibitors with a final yield of 10 mg each, exhibiting molecular masses of 520 and 534 g·mol−1. Comparison of the LC-MS profile with a Photorhabdus culture exclusively grown at high salt condition revealed that the spectrum of secondary metabolites changed dramatically, thus reflecting a complete switch of life stages including the production of proteasome inhibitors (SI Text, Fig. S2 and Table S2).
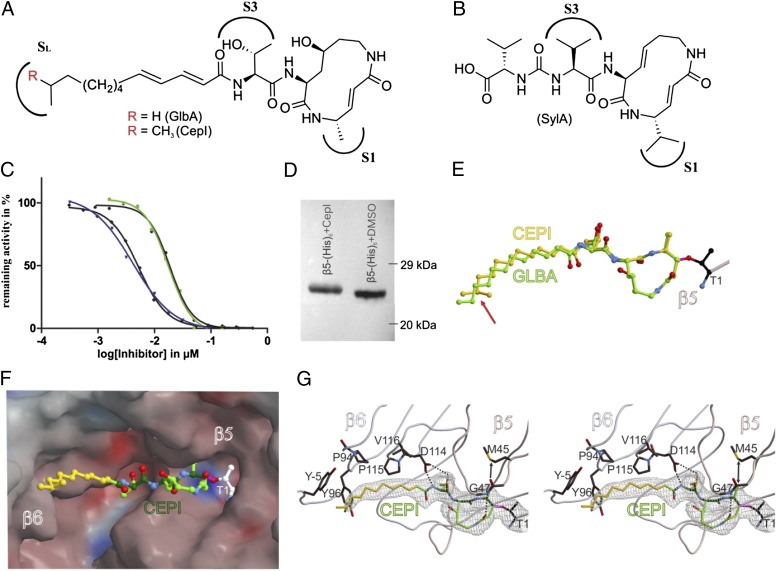
Structure elucidation of the two compounds by 2D-NMR analysis resulted in the molecules glidobactin A (GlbA), which had been known to be the strongest proteasome inhibitor identified so far (22, 23), as well as a molecule that has been named cepafungin I (CepI) in literature (24) (Fig. 2A and Fig. S3). Despite the high similarity of the isolated molecules, CepI has never been investigated with respect to its properties as a proteasome inhibitor. Both CepI and GlbA share a 12-membered macrolactam ring system that is linked to a fatty acid tail. As the only difference, this apolar chain is terminally branched and carries an additional methyl moiety in the case of CepI, whereas it is linear in GlbA.
Fig. 2.
In vitro functional and structural characterization of GlbA and CepI. (A) NMR-deduced structures of GlbA/CepI with the proteasome binding sites S1 and S3 as well as the hydrophobic patch SL. (B) SylA, featuring a hydrophilic carboxylic acid tail. (C) IC50 curves for the ChTL activity measured for GlbA (green) and CepI (blue) with 13C-labeled peptide substrate; black curves depict equivalent measurements with the fluorogenic Suc-Leu-Leu-Val-Tyr-AMC substrate (error bars are too small to be visible). (D) Western blot on a β5-(His)6 yeast CP mutant with or without treatment with CepI. The gel shift indicates an increased molecular mass by covalent and irreversible binding. (E) Structural superposition of GlbA and CepI bound to yeast CP; the differing methyl moiety is indicated by a red arrow. (F) Surface representation of CepI in the β5 substrate binding channel. Colors indicate positive and negative electrostatic potentials contoured from −30 kT/e (bright red) to +30 kT/e (bright blue). (G) Stereoview of the 2Fo − Fc omit electron density map (gray mesh, 1σ) for the β5 active site. Hydrogen bonds are indicated by dashed lines, whereas the arrow pronounces the accurate fitting of the P1 methyl side chain of the inhibitor. The lipophilic pocket of the SL site is formed by Tyr−5, Pro94, Tyr96, and Pro115 from β6 [amino acid numbering according to Löwe et al. (25)].
In Vitro Functional and Structural Characterization of GlbA and CepI.
Investigating the inhibitory potential of both compounds, we performed IC50 measurements with yeast CP using the established NMR assay for the ChTL activities. Remarkably, the IC50 value for CepI (4 nM) was five times lower compared with GlbA (19 nM; Fig. 2C). Evaluation of the stoichiometry between CepI and CP reveals that 100% inhibition is already observed at a ratio of 2:1, thus reaching the detection limit of the assay with regard to the twofold symmetry of the proteasomal subunits. These results could be verified by established fluorescence-based assays, as the isolated compounds were uncolored and therefore did not cause quenching effects compared with the crude culture broth. Moreover, the high consistency between the data gained from both recording techniques confirms the equivalence of the NMR approach, using a natural and active-site specific proteasome substrate, in contrast to common assay types. In addition, measurements were expanded on evaluation of the TL and CL sites. The TL activity was inhibited to 24 nM for CepI and 193 nM for GlbA, respectively, thus exhibiting the same tendency as observed for the β5 subunit (Fig. S4). The proteasomal CL activity was not affected by either compound up to concentrations as high as 100 µM. These results were surprising because only a minor change such as a methyl group at the distal end from the active proteasomal Thr-1Oγ residue has such an impact on the binding affinity.
Therefore, we performed crystal structure analysis of CepI and GlbA in complex with the yeast CP at 2.8-Å (Rfree = 24.3%) and 3.0-Å (Rfree = 22.7%) resolution for characterizing both inhibitors at the molecular level (SI Text, Table S3). Electron densities display covalent and irreversible binding of Thr-1Oγ to the macrolactam ring via ether bond formation, which was confirmed by a gel shift assay (Fig. 2 D and E). Intriguingly, the aliphatic fatty acid chains of GlbA and CepI point toward a hydrophobic patch formed by subunit β6 adjacent to the catalytically active β5 subunit. However, in contrast to the linear aliphatic chain of GlbA, the branched isopropyl side chain of CepI is further stabilized in a lipophilic pocket by strong van der Waals interactions (Fig. 2 E–G). In conclusion, structural evaluation of SylA, GlbA, and CepI suggests a directed evolution: starting from a charged and highly polar moiety adjacent to the macrolactam scaffold with impaired binding affinity, continuing with enhanced decoration as well as exploitation of the distant hydrophobic pocket in GlbA by an aliphatic linker, and eventually yielding an optimized solution with maximum inhibition values for CepI. Analysis of the other active proteasomal subunits reveals that the β2 active site is occupied, whereas β1 was found empty, hereby confirming the in vitro results. Equivalently to β5, the enhanced inhibitory strength of CepI versus GlbA against the TL activity can be explained by apolar interactions with a distal lipophilic patch via its branched fatty acid tail.
Cell Culture Experiments.
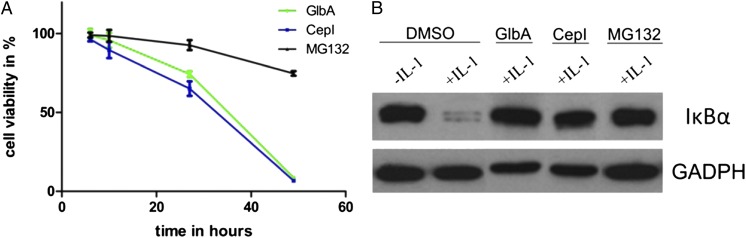
To investigate cytotoxic effects on HeLa cancer cells upon treatment with GlbA/CepI, we determined the time-dependent viability at a final compound concentration of 1 µM (Fig. 3A). Both compounds, GlbA and CepI, exhibit significantly increased cytotoxicity versus the standard proteasome inhibitor MG132, an artificial and cell-penetrating aldehyde peptide (26). This finding is also in agreement with their superior binding strength over MG132, which features an IC50 value of 270 nM against the ChTL activity in vitro. Although it has been shown that inhibition of β5 alone is sufficient for cytotoxicity of a compound, it is considerably increased if also a second subunit is affected (27). This furthermore explains the enhanced effects of the syrbactin compounds, which bind both to the β5 and the β2 subunits. Comparison between CepI and GlbA reveals that cancer cells treated with CepI displayed increased lethality versus GlbA, again reflecting the results received by the in vitro experiments.
Fig. 3.
Cell culture assays. (A) Cytotoxicity is measured by performing the Alamar Blue assay with HeLa cells after 6, 10, 24, and 48 h for MG132 (black), GlbA (green), and CepI (blue) at 1 µM concentration. (B) Western blot on IκB with or without IL-1 induction in HeLa cells, which were treated for 6 h with 50 µM MG132, GlbA, CepI, or DMSO. Proteasomal IκB degradation is initiated by IL-1 signaling, whereas it is prevented after addition of the cell-penetrating CP inhibitors. The GADPH housekeeping protein was costained for quantification.
Assessing intracellular proteasome inhibition upon treatment with both natural compounds, we analyzed degradation of the natural proteasome substrate IκB. In most human cells, the nuclear NF-κB transcription factor is inactively sequestered in the cytosol by the inhibitory IκB protein, whose ubiquitin-mediated degradation is initiated by the inflammatory cytokine IL-1 (28). By incubation with GlbA or CepI before addition of IL-1, however, the digestion of IκB is prevented, hence demonstrating the cell penetration and intracellular proteasome inhibition by the natural compounds (Fig. 3B).
Discussion
The results gained by our experiments are promising and make CepI a candidate for further drug development. Being the most powerful proteasome inhibitor described to date, its secretion is strictly controlled in Photorhabdus (SI Text). The bacterium, which lives in the gut of the nematode Heterorhabditis, presumably suspends production of GlbA and CepI until it is released into the hemocoel of an insect larva, whereupon they kill their mutual prey in a concerted action to reproduce new bacteria and nematodes (29). Hence, GlbA and CepI are proteasome inhibitors to which a biological function could be assigned. Despite the rigid environmental regulation, the discovery of the two natural compounds from Photorhabdus was feasible due to the approach established in this work. The determination of the appropriate stimuli for pathogenic organisms to enter the virulent life stages, when most secondary metabolites are produced, is of crucial importance, because the majority of all genes involved in production of natural compounds are usually found silent under common growth conditions. Because many other organisms, like the human pathogen Burkholderia pseudomallei, carry gene clusters for putative proteasome inhibitors, the application of the developed methodology for screening of such triggers will identify promising new natural products, which may lead to the development of new drugs for treatment of various diseases. Eventually, we anticipate that a transfer of the established screening procedure to other targets of pharmaceutical interest will facilitate the identification of natural inhibitory compounds for these enzymes.
Materials and Methods
NMR Proteasome Assay.
Purification of the 20S proteasome from Saccharomyces cerevisiae was performed as described previously (3). The assay mixture containing 10 µg/mL yeast CP, 100 mM Tris (pH 8.0), 0.01% SDS, and 300 µL of sterile filtrated culture broth was incubated for 10 min at room temperature. Labeled peptide was added to a final concentration of 2 mM and incubated for 6 h. The assay was quenched by ultrafiltration, completed with D2O, and recorded in a 500-MHz spectrometer (Bruker Topspin 13C-Cryo NMR). IC50 measurements were carried out accordingly with the purified compounds dissolved in DMSO. Remaining activity was calculated by scaling the ratio of the product to the educt peak and compared with a blank sample.
Secretion and Purification of GlbA/CepI.
A preculture of P. luminescens subsp. laumondii (DSM no. 15139) was grown at high osmolarity [0.2% (wt/vol) tryptone, 0.5% (wt/vol) yeast extract, 1% (wt/vol) NaCl] for 3 d at 28 °C, which was then used to inoculate medium without NaCl in a 1:100 ratio. During further incubation for 3 d, a hydrophobic resin (XAD-16) was added to the culture broth, which was afterward extracted with methanol. The extract was subjected to a silica column, which was washed with cyclohexane and eluted with ethyl acetate. Fractions containing inhibitors were identified by the proteasome assay and applied to RP-C18 HPLC chromatography. GlbA and CepI eluted at ∼55% (vol/vol) acetonitrile in a 10–70% (vol/vol) gradient over 1 h on a 250/10-mm column.
Cell Culture Assay.
Viability measurements with HeLa cells were performed at a final inhibitor concentration of 1 µM at 6, 10, 24, and 48 h by Alamar Blue assays as described previously (30); MG132 was purchased from Bachem. For Western blot analysis of IκB accumulation, HeLa cells were incubated with 40 µM inhibitors for 6 h and then stimulated by addition of IL-1 at a concentration of 30 ng/mL for 20 min before harvesting. After cell lysis, 10 µg of protein was applied to 15% (wt/vol) SDS/PAGE. Western blot analysis of IκB was standardized by comparison with the GADPH housekeeping protein.
Supplementary Material
Acknowledgments
We thank Richard Feicht for yeast proteasome purification, as well as Nerea Gallastegui, Haissi Cui, and Achim Krüger for assistance in cell culture assays. Katja Bäuml is acknowledged for help with peptide synthesis. We also thank the staff of the Beamlines X06SA and X06DA at the Paul Scherrer Institute, Swiss Light Source (Villingen, Switzerland), for their support during data collection. M.K. was funded by European Research Council Starting Grant 258413.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org [PDB ID codes 4FZC (20S yeast proteasome in complex with Cepafungin I) and 4FZG (20S yeast proteasome in complex with Glidobactin A)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1211423109/-/DCSupplemental.
References
- 1.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 2.Gallastegui N, Groll M. The 26S proteasome: Assembly and function of a destructive machine. Trends Biochem Sci. 2010;35(11):634–642. doi: 10.1016/j.tibs.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 3.Groll M, et al. Structure of 20S proteasome from yeast at 2.4 Å resolution. Nature. 1997;386(6624):463–471. doi: 10.1038/386463a0. [DOI] [PubMed] [Google Scholar]
- 4.Huber EM, et al. Immuno- and constitutive proteasome crystal structures reveal differences in substrate and inhibitor specificity. Cell. 2012;148(4):727–738. doi: 10.1016/j.cell.2011.12.030. [DOI] [PubMed] [Google Scholar]
- 5.Unno M, et al. Structure determination of the constitutive 20S proteasome from bovine liver at 2.75 Å resolution. J Biochem. 2002;131(2):171–173. doi: 10.1093/oxfordjournals.jbchem.a003084. [DOI] [PubMed] [Google Scholar]
- 6.Orlowski M. The multicatalytic proteinase complex, a major extralysosomal proteolytic system. Biochemistry. 1990;29(45):10289–10297. doi: 10.1021/bi00497a001. [DOI] [PubMed] [Google Scholar]
- 7.Huber EM, Groll M. Inhibitors for the immuno- and constitutive proteasome: Current and future trends in drug development. Angew Chem Int Ed Engl. 2012;51(35):8708–8720. doi: 10.1002/anie.201201616. [DOI] [PubMed] [Google Scholar]
- 8.Richardson PG, Hideshima T, Anderson KC. Bortezomib (PS-341): A novel, first-in-class proteasome inhibitor for the treatment of multiple myeloma and other cancers. Cancer Contr. 2003;10(5):361–369. doi: 10.1177/107327480301000502. [DOI] [PubMed] [Google Scholar]
- 9.Arastu-Kapur S, et al. Nonproteasomal targets of the proteasome inhibitors bortezomib and carfilzomib: A link to clinical adverse events. Clin Cancer Res. 2011;17(9):2734–2743. doi: 10.1158/1078-0432.CCR-10-1950. [DOI] [PubMed] [Google Scholar]
- 10.Gräwert MA, Groll M. Exploiting nature’s rich source of proteasome inhibitors as starting points in drug development. Chem Commun (Camb) 2012;48(10):1364–1378. doi: 10.1039/c1cc15273d. [DOI] [PubMed] [Google Scholar]
- 11.Khan ML, Stewart AK. Carfilzomib: A novel second-generation proteasome inhibitor. Future Oncol. 2011;7(5):607–612. doi: 10.2217/fon.11.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meng L, et al. Epoxomicin, a potent and selective proteasome inhibitor, exhibits in vivo antiinflammatory activity. Proc Natl Acad Sci USA. 1999;96(18):10403–10408. doi: 10.1073/pnas.96.18.10403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Groll M, Kim K, Kairies N, Huber R, Crews C. Crystal structure of epoxomicin: 20S proteasome reveals molecular basis for selectivity of α'β'-epoxyketone proteasome inhibitors. J Am Chem Soc. 2000;122(6):1237–1238. [Google Scholar]
- 14.Dick TP, et al. Contribution of proteasomal beta-subunits to the cleavage of peptide substrates analyzed with yeast mutants. J Biol Chem. 1998;273(40):25637–25646. doi: 10.1074/jbc.273.40.25637. [DOI] [PubMed] [Google Scholar]
- 15.Adams J, et al. Potent and selective inhibitors of the proteasome: Dipeptidyl boronic acids. Bioorg Med Chem Lett. 1998;8(4):333–338. doi: 10.1016/s0960-894x(98)00029-8. [DOI] [PubMed] [Google Scholar]
- 16.Wäspi U, Blanc D, Winkler T, Rüedi P, Dudler R. Syringolin, a novel peptide elicitor from Pseudomonas syringae pv. syringae that induces resistance to Pyricularia oryzae in rice. Mol Plant Microbe Interact. 1998;11(8):727–733. [Google Scholar]
- 17.Schellenberg B, Bigler L, Dudler R. Identification of genes involved in the biosynthesis of the cytotoxic compound glidobactin from a soil bacterium. Environ Microbiol. 2007;9(7):1640–1650. doi: 10.1111/j.1462-2920.2007.01278.x. [DOI] [PubMed] [Google Scholar]
- 18.Krahn D, Ottmann C, Kaiser M. The chemistry and biology of syringolins, glidobactins and cepafungins (syrbactins) Nat Prod Rep. 2011;28(11):1854–1867. doi: 10.1039/c1np00048a. [DOI] [PubMed] [Google Scholar]
- 19.Wiersinga WJ, van der Poll T, White NJ, Day NP, Peacock SJ. Melioidosis: Insights into the pathogenicity of Burkholderia pseudomallei. Nat Rev Microbiol. 2006;4(4):272–282. doi: 10.1038/nrmicro1385. [DOI] [PubMed] [Google Scholar]
- 20.Frackman S, Anhalt M, Nealson KH. Cloning, organization, and expression of the bioluminescence genes of Xenorhabdus luminescens. J Bacteriol. 1990;172(10):5767–5773. doi: 10.1128/jb.172.10.5767-5773.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brachmann AO, et al. Triggering the production of the cryptic blue pigment indigoidine from Photorhabdus luminescens. J Biotechnol. 2012;157(1):96–99. doi: 10.1016/j.jbiotec.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 22.Groll M, et al. A plant pathogen virulence factor inhibits the eukaryotic proteasome by a novel mechanism. Nature. 2008;452(7188):755–758. doi: 10.1038/nature06782. [DOI] [PubMed] [Google Scholar]
- 23.Oka M, et al. Glidobactins A, B and C, new antitumor antibiotics. I. Production, isolation, chemical properties and biological activity. J Antibiot (Tokyo) 1988;41(10):1331–1337. doi: 10.7164/antibiotics.41.1331. [DOI] [PubMed] [Google Scholar]
- 24.Shoji J, et al. Isolation of cepafungins I, II and III from Pseudomonas species. J Antibiot (Tokyo) 1990;43(7):783–787. doi: 10.7164/antibiotics.43.783. [DOI] [PubMed] [Google Scholar]
- 25.Löwe J, et al. Crystal structure of the 20S proteasome from the archaeon T. acidophilum at 3.4 Å resolution. Science. 1995;268(5210):533–539. doi: 10.1126/science.7725097. [DOI] [PubMed] [Google Scholar]
- 26.Bush KT, Goldberg AL, Nigam SK. Proteasome inhibition leads to a heat-shock response, induction of endoplasmic reticulum chaperones, and thermotolerance. J Biol Chem. 1997;272(14):9086–9092. doi: 10.1074/jbc.272.14.9086. [DOI] [PubMed] [Google Scholar]
- 27.Screen M, et al. Nature of pharmacophore influences active site specificity of proteasome inhibitors. J Biol Chem. 2010;285(51):40125–40134. doi: 10.1074/jbc.M110.160606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ortis F, et al. Differential usage of NF-κB activating signals by IL-1β and TNF-α in pancreatic beta cells. FEBS Lett. 2012;586(7):984–989. doi: 10.1016/j.febslet.2012.02.021. [DOI] [PubMed] [Google Scholar]
- 29.Waterfield NR, Ciche T, Clarke D. Photorhabdus and a host of hosts. Annu Rev Microbiol. 2009;63:557–574. doi: 10.1146/annurev.micro.091208.073507. [DOI] [PubMed] [Google Scholar]
- 30.Macherla VR, et al. Structure-activity relationship studies of salinosporamide A (NPI-0052), a novel marine derived proteasome inhibitor. J Med Chem. 2005;48(11):3684–3687. doi: 10.1021/jm048995+. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.