Abstract
Antibody discovery platforms have become an important source of both therapeutic biomolecules and research reagents. Massively parallel DNA sequencing can be used to assist antibody selection by comprehensively monitoring libraries during selection, thus greatly expanding the power of these systems. We have therefore constructed a rationally designed, fully defined single-chain variable fragment (scFv) library and analysis platform optimized for analysis with short-read deep sequencing. Sequence-defined oligonucleotide libraries encoding three complementarity-determining regions (L3 from the light chain, H2 and H3 from the heavy chain) were synthesized on a programmable microarray and combinatorially cloned into a single scFv framework for molecular display. Our unique complementarity-determining region sequence design optimizes for protein binding by utilizing a hidden Markov model that was trained on all antibody-antigen cocrystal structures in the Protein Data Bank. The resultant ∼1012-member library was produced in ribosome-display format, and comprehensively analyzed over four rounds of antigen selections by multiplex paired-end Illumina sequencing. The hidden Markov model scFv library generated multiple binders against an emerging cancer antigen and is the basis for a next-generation antibody production platform.
Keywords: antibody display, synthetic antibody library, single framework antibody library
Antibodies are useful for their ability to bind molecular surfaces with high affinity and specificity. The genetic basis for their structural diversity is partially encoded in the germ line, but is also the result of stochastic genetic events, including chromosomal rearrangements, nontemplated nucleotide insertions, and somatic hypermutation. The majority of this diversity is localized to the complementarity-determining regions (CDRs), which are the six-peptide loops that protrude from the variable domain framework to form the antigen-combining surface of the antibody molecule. Three CDR loops are contributed by the heavy chain (H1, H2, and H3) and three by the light chain (L1, L2, and L3). CDRs 1 and 2 are encoded in the germ line, and are thus more constrained in their diversity. L3 is characterized by “junctional diversity,” formed during the recombination of two gene segments (V and J). Finally, H3 is formed by two consecutive genetic rearrangements (first between D and J, and then between V and DJ), and is additionally accompanied by nontemplated “N” nucleotides, making this CDR the source of most naturally occurring antibody diversity.
Our goal was to develop a synthetic antibody production platform inspired by nature, which could be seamlessly integrated with massively parallel, short-read DNA sequencing analysis (Fig. 1A) (1, 2). For maximum convenience, we required that library amplification and sequencing reactions should depend upon a single set of primers, rather than the complex mixture necessary for natural repertoire amplification and analysis. Like others before, we therefore constructed a highly diverse antibody library within a single variable-domain framework (3, 4). However, because it is well known that the natural diversity of variable-domain frameworks contributes to a naive repertoire’s functional shape space (5, 6), we sought to maximize the functional diversity in our library’s CDR repertoire by rationally designing sequences based on a mathematical model of antibody–antigen interaction.
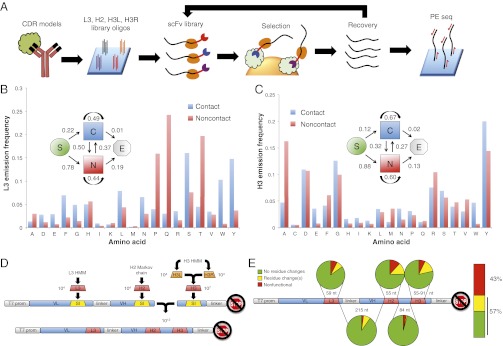
Fig. 1.
HMM antibody library design and synthesis. (A) Strategy for design and application of the rationally designed scFv library. Antigen–antibody crystal structures are used to design CDR-encoding DNA sequences, which are then synthesized on a programmable microarray. After ribosome display and enrichment for antigen binding clones, library recovery, and analysis by paired-end sequencing can be performed. (B) Model-defining parameters for the L3 HMM. Emission probability for each amino acid corresponding to the two possible states. State transition probabilities are inset: “S” denotes start of a chain, “C” denotes the contact state, “N” denotes the noncontact state, “E” denotes the end of the chain. (C) Model-defining parameters for the H3 HMM. Definitions are the same as for B. (D) Overview of the scFv ribosome display vector and library assembly strategy. “VL” and “VH” are the light and heavy variable domains, respectively. “T7 prom” is the T7 promoter, and the crossed stop sign denotes lack of a stop codon. L3, H2, and H3 are the CDR libraries designed to replace the “SI” suicide inserts. H3L and H3R sublibraries are brought together by combinatorial ligation to create H3. Similarly, the L3-H2 fragment is brought together with the H3 fragment in a combinatorial ligation. (E) Clonal Sanger sequencing analysis of 93 HMM scFv library members.
A single-chain variable fragment (scFv) is the simplest functional representation of an antibody molecule, and has become the platform of choice for most antibody engineers. Our first step was thus to identify the most suitable scFv framework to house libraries of rationally designed CDRs. Lloyd et al. screened a very large preimmune human scFv library against a panel of 28 different antigens, and after sequencing >5,000 postselection clones, they observed strong enrichment of a small subset of heavy- and light-chain variable domains (7). Among these domains, the most highly enriched were the heavy chain VH1–69 and the λ-light chain VL1–44. The authors attributed these framework enrichments to increased expression and optimal folding within the periplasm of the Escherichia coli host cells. These findings were further corroborated by the work of Glanville and colleagues (8). We therefore housed our CDR libraries within an scFv framework composed of VH1–69 and VL1–44.
As a source of inspiration for CDR design features, we looked to the international ImMunoGeneTics’ (IMGT’s) annotated database of all antibody–antigen cocrystal structures present within Protein Data Bank (IMGT/3Dstructure-DB) as of May 2009 (9, 10). Amino acid residues within CDRs can contribute to antigen binding in two distinct ways: (i) direct, via contribution of a side group that makes contacts with the antigen, and (ii) indirect, affecting the conformation of the peptide backbone in a way that permits the direct interaction of neighboring amino acid side groups. This behavior of CDR amino acid sequences can be captured in a two-state hidden Markov model (HMM). The “contact” state should be enriched for amino acids capable of sharing/exchanging electrons or burying hydrophobic surfaces, whereas the “noncontact” state should be enriched for residues capable of appropriately constraining or relaxing the CDR polypeptide backbone. An important feature of HMMs is that the state of each position depends upon its nearest neighbor. It is thus important to note that traditional approaches to synthetic CDR construction typically use degenerate nucleotides or codons, and so cannot link the identity of a particular residue to that of its neighbors. To implement our HMM design, we took a different approach and synthesized complete CDR sequences as releasable oligonucleotides on a programmable DNA microarray. Importantly, this approach permits the filtering of deleterious sequences, such as restriction sites and undesirable peptide motifs (e.g., glycosylation signals and good HLA class II substrates), thus maximizing the functional utility of the library.
The transformation efficiency of bacterial cells with plasmid DNA is a significant barrier to construction of molecular libraries with a complexity greater than ∼1010. Because the utility of an scFv library scales with its diversity, we took advantage of the in vitro ribosome display technique, which has been used to generate antibodies with picomolar affinities (11). In this approach, mRNA molecules are tethered to the proteins they encode via noncovalent interactions with a ribosome. The mRNA is made to lack a stop codon necessary for peptide release, and so a population of ternary complexes composed of mRNA, encoded scFvs, and ribosomes is thus formed. Ribosome display libraries can be constructed and transcribed entirely in vitro, thus bypassing transformation bottlenecks.
After characterizing the quality of the HMM scFv library, we tested it by sequencing the library as it evolved over multiple rounds of selection on a protein antigen. We also developed robust methods to specifically recover desirable clones for expression and analysis in a simple two-step process. Our platform successfully produced antibodies against the emerging cancer antigen poliovirus receptor-related 4 (PVRL4) and sets the stage for a new paradigm in sequencing-assisted selection of rationally designed human antibodies.
Results
Library Design, Assembly, and Characterization.
We set out to diversify the three CDR loops most relevant to antigen binding. By examining the IMGT/3Dstructure-DB, we determined the average number of contacts per structure contributed by each CDR. Of contacts reported in this database, 76% are contributed by residues contained within CDRs. As expected, L3 and H3 contribute the most contacts, with H2 providing the third-most. In sum, 71% of CDR contacts are made by amino acids in these three CDRs (Fig. S1A).
To estimate the HMM-defining parameters for L3 and H3, we identified 236 unique L3 and 241 unique H3 sequences within IMGT/3Dstructure-DB. Each residue was classified as either making contact or not with the protein antigen, as determined by the corresponding 3D cocrystal structure. The resulting HMM state transition rates and amino acid emission probabilities for L3 and H3 are illustrated in Fig. 1 B and C. Notable features of these models are: (i) enrichment for the noncontact state at positions closer to the framework [i.e., probability of S (start) → N (noncontact) and N → E (end) transitions are much greater than S → C (contact) and C → E, respectively]; (ii) in H3, a tendency for blocks of contact/noncontact states (i.e., probability of staying in the same state is higher than transitioning between states); (iii) a strong enrichment in both L3 and H3 for contacts consisting of tyrosine and tryptophan [previously observed by Ofran et al. (12)]; and (iv) L3- or H3-specific enrichments for certain amino acids in each state (e.g., noncontact proline in L3, and contact glutamic acid in H3).
We used our HMM to generate >10,000 unique sequences for each of L3 and H3 (13). Whereas the length of L3 sequences was fixed at 13 residues, 1,000 H3 sequences were randomly chosen for each length from 9 to 21 amino acids long. As an analog to VJ recombination, we further expanded the diversity of H3 by separating each sequence into two halves: “H3L” and “H3R,” for subsequent combinatorial ligation to form full-length H3 sequences (Fig. 1D). This was accomplished by placing a type IIS restriction site downstream of H3L and upstream of H3R on their encoding oligos. After PCR and restriction digest with the SapI restriction endonuclease, the H3L and H3R fragments were combinatorially ligated together. The 3-nt 5′ SapI overhang on each sublibrary ensured that the H3 reading frames would remain intact.
The germ-line–encoded H2 CDR is characterized by structural features not present in L3 or H3 chains, and this is reflected in its heterogeneous contact profile (Fig. S1B). It has been suggested that H2 contributes to the stability of the variable domain of the heavy chain through interactions among its hydrophobic residues (14, 15). To avoid disrupting framework stability, we created a first-order Markov chain to generate H2 sequences that was trained on the 176 unique H2 chains in the IMGT database. This model was used to generate >10,000 H2 sequences.
Finally, all CDR sequences were passed through a series of three filters to maximize their utility. First, all restriction sites to be used during library construction were eliminated by introducing silent codon changes. Second, we sought to minimize the potential immunogenicity of the scFvs by discarding peptides with a high potential for loading onto HLA class II molecules during antigen presentation. We used the ProPred online server to filter our CDR sequences against the four most common HLA-DRB1 alleles (101, 301, 701, and 1501) with a stringency of 45% of the best substrate (16). This process resulted in replacement of about 18% of all H3 sequences by less immunogenic peptides. The third filter replaced sequences with the potential to interfere with industrial scale production (e.g., methionine oxidation, asparagine deamidation/cyclization), as well as glycosylation motifs.
The final set of 43,803 CDR sequences (L3, H2, H3L, H3R) were flanked by the appropriate restriction site sequences, as well as sublibrary-specific PCR primer binding sequences, and then synthesized as releasable oligonucleotides on a silicon wafer (Agilent Technologies). The oligo libraries were PCR-amplified and separately cloned into the VH1–69 and VL1–44 human heavy- and light-chain variable fragments for combinatorial assembly (Fig. 1D and Methods). In vitro transcription was then performed to create the mRNA templates for ribosome display.
We characterized the HMM scFv library in two ways. First, we cloned a small sample of the library mRNA. This process allowed us to perform Sanger sequencing on individual colonies, and thereby estimate the overall fraction of the library expected to contain functional, full-length scFvs with no frameshift or nonsense mutations (57% functional, n = 93) (Fig. 1E). None of the colonies examined had retained their CDR “suicide insert,” and none had multiple CDR insertions. We found that the 43% nonfunctional clones derived mostly from oligo synthesis errors; ∼15% of each CDR was nonfunctional, resulting in 39% (1–0.853) nonfunctional clones. Second, we used our Illumina sequencing data to characterize the length distribution of the H3 loop (Fig. S2). Satisfied that our library was true to its design, we next performed selections against the cancer antigen PVRL4 (17, 18), and used Illumina sequencing to track the library during selection.
Selection and Analysis of HMM scFv Libraries on GST-PVRL4 Bait.
Four successive rounds of selection were performed with the HMM scFv library on GST-PVRL4. We quantified both enrichment of a spiked-in control scFv and the amount of RNA degradation during each round of selection to ensure these parameters met our quality-control thresholds (SI Methods). After three selection rounds, the library was split and selected on either GST-PVRL4 or GST-GCN4 (no PVRL4) in parallel, which allowed us to discriminate between PVRL4-specific scFvs and those that bind to GST or to some other component of the system.
The minimal region of the HMM scFvs that contains the three diversified CDRs is an appropriate size for analysis by paired-end Illumina sequencing. The sequencing libraries can thus be prepared conveniently by PCR. A small amount of each of the selected libraries, as well as the unselected HMM scFv library was amplified with Illumina sequencing adapters. These adapters include a 7-nt barcode to permit multiplexed analysis.
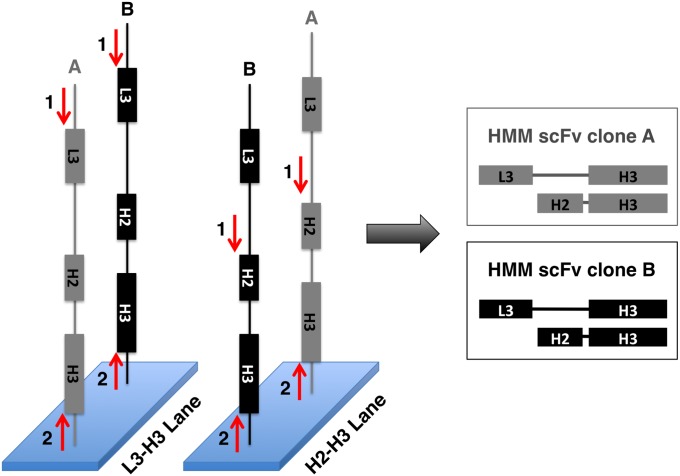
We performed paired-end sequencing in two separate Illumina HiSeq 2000 flow cell lanes. By obtaining L3-H3 mate pairs in one lane and H2-H3 mate pairs in the other lane, we could use the hyperdiversity of H3 sequences to match corresponding L3 and H2 sequences, thereby reconstructing the complete identity of each scFv clone (Fig. 2). After PCR amplification, however, we observed significant PCR chimerism, essentially resulting in CDR recombination. This complicated—but in most cases did not prevent—reconstruction of individual scFv clones. CDR recombination has been observed to significantly increase scFv affinity during ribosome display selection, suggesting that this process might actually improve the success rate of our platform (19).
Fig. 2.
Strategy for sequence reconstruction of HMM scFv clones. One-hundred nucleotide paired-end sequencing is performed on the same library in two independent lanes on an Illumina HiSeq 2000. In the L3-H3 lane, the first sequencing primer lands upstream of L3 (red “1” arrow). In the H2-H3 lane, the first sequencing primer lands upstream of H2 (red “1” arrow). The H3 sequence is then determined by reading from a common, second primer (red “2” arrow) in both lanes. L3 and H2 sequences are then paired using their unique H3 identifier to fully define the sequence of the scFv clone.
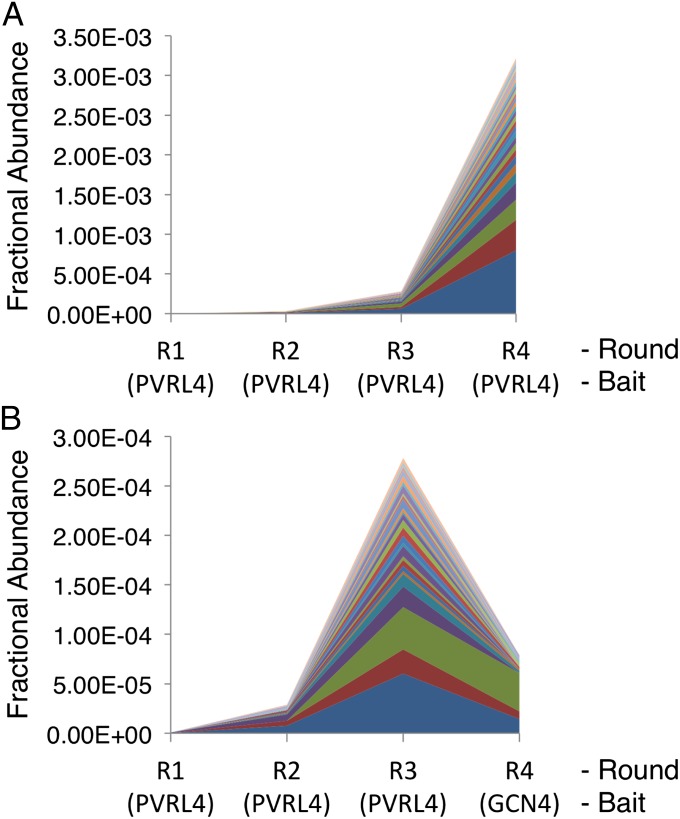
We next determined the relative abundance of each clone in the library over the course of four rounds of selection on GST-PVRL4, and compared this to the results from the round 3 PVRL4-selected library that was selected on GST-GCN4 (Fig. 3). Based on this analysis, a subset of the candidate PVRL4-specific clones was selected for further analysis.
Fig. 3.
Fractional abundance of top 30 PVRL4-specific HMM scFv clones during selection. (A) The fractional abundance of the top 30 PVRL4-specific HMM scFv clones shown over four rounds of enrichment on GST-PVRL4. Fraction is calculated as read number of a clone divided by the total number of reads from the corresponding library. (B) The fractional abundance of the same 30 PVRL4-specific HMM scFv clones from A. Data from rounds R1–R3 are the same in the two panels. Round 4 selection on the non-PVRL4 bait, GST-GCN4, results in a relative depletion of these clones from the population.
Recovery and Testing of Candidate Anti-PVRL4 HMM scFvs.
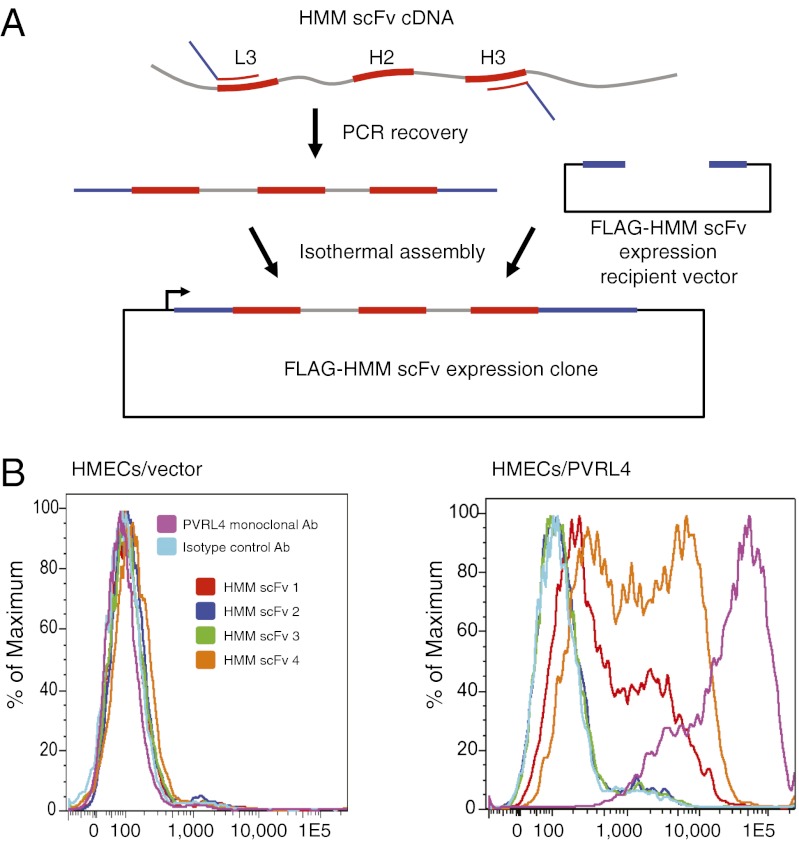
Before characterizing individual scFvs for their ability to bind antigen, they must first be isolated. This isolation can be accomplished either by resynthesizing the CDRs for cloning back into an expression framework, or by PCR-recovering the clones using primers specific for L3 and H3. We chose to recover candidate scFvs by performing PCR with L3/H3-specific primers that also contained 5′ homology arms for subsequent isothermal assembly into a FLAG/6His epitope-tag expression vector (Fig. 4A).
Fig. 4.
HMM scFv recovery strategy and FACS staining. (A) Candidate HMM scFv clones are recovered by PCR using primers specific for L3 and H3, and which also have 5′ homology arms for subsequent isothermal assembly into an expression vector with differing codon use. (B) Results of FACS staining to assess binding of candidate scFvs. HMECs infected with vector alone or vector-expressing PVRL4 antigen were stained with the indicated control antibodies or one of the top four most abundant postselection HMM scFv clones.
Recovered candidate anti-PVRL4 scFv clones were expressed in vitro as FLAG-tagged proteins. Of the top 25 most abundant postselection clones, four were found to specifically bind human mammary epithelial cell (HMEC)-expressed PVRL4 by FACS-staining analysis (Fig. 4B). The success rate of our method is likely underestimated by this analysis, however, as the selection bait was a bacterially-produced, unmodified GST-PVRL4 fusion, whereas HMECs display the glycosylated protein in the context of the cell surface.
Discussion
Synthetic biology has yet to deliver antibody production platforms that rival vertebrate immune systems in both product quality and manufacturing convenience (20). However, we anticipate that along with the maturation of gene-synthesis technologies and the affordability of high-throughput DNA sequencing will also come advances in antibody production pipelines that outperform animal immune systems in all regards (21).
To address the emerging need for a next-generation scFv production platform compatible with sequencing-assisted selection, we have created a rationally designed, single-framework scFv library. Single frameworks enable facile library amplification and sequencing using a single set of primers, but reduce framework-contributed functional diversity. To compensate for this potential loss of functionality, we used a mathematical model of structural data to capture subtle amino acid sequence biases that contribute to the formation of favorable antibody–antigen contacts. We additionally developed a method to mimic the junctional diversity of VJ recombination using type IIS restriction cleavage followed by shuffling ligation. Our combinatorial strategy significantly reduces the required size of each CDR sublibrary, making extreme diversification of the final scFv library a tractable problem. Finally, to accomplish the analysis of selected scFv libraries using short-read DNA sequencing, we have introduced a strategy for double mate pair-based reconstruction of full-length scFv clones.
One benefit of the three-CDR library presented here is the ease of clonal sequence reconstruction. We found that the hyperdiversity of our H3 CDR library permitted the near unambiguous pairing of L3 and H2 sequences with their shared H3, thus completely defining the repertoire at each round of selection. We went on to demonstrate an important advantage of single-framework libraries, which is the relative simplicity of recovering desirable clones. Our two-step protocol enables rapid isolation and assembly of clones into an expression vector for further functional characterization. In contrast, native heavy- and light-chain framework libraries are currently intractable to analysis with short-read sequencing, and require many more steps for clonal isolation. Although not demonstrated here, an additional design feature built into our platform is the ability to perform straightforward total synthesis of desirable clones from synthetic DNA oligos or oligo pools (Fig. 5). It is worth noting that the utility of the HMM library is not limited to the ribosome display format, and can be moved into traditional phage or yeast display vectors. Similarly, alternative heavy- and light-chain frameworks can be synthesized to house the HMM CDR libraries described in this work.
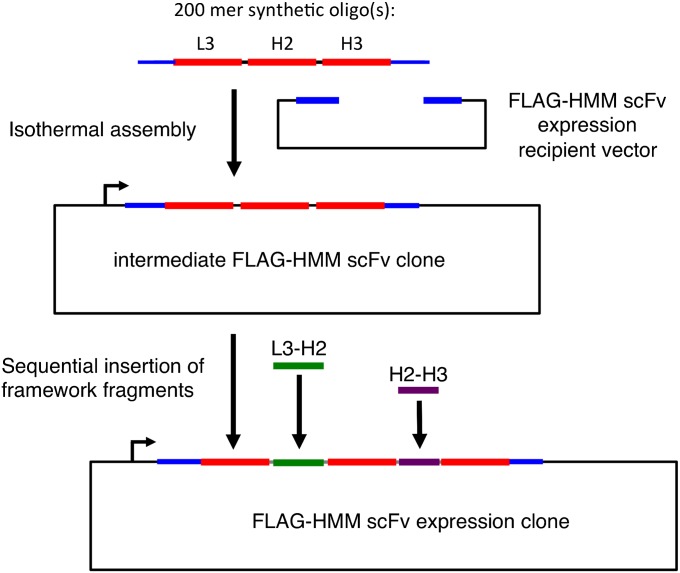
Fig. 5.
Total synthesis of HMM scFv clones. Strategy for reconstruction of desirable HMM scFv clones from synthetic oligos. After PCR amplification of the oligo or oligo library, the CDRs are assembled into an intermediate expression vector. The constant sequences of the framework regions between L3 and H2 (green) and between H2 and H3 (purple) are then sequentially inserted to form the complete HMM scFv expression clone or clone set.
An unrealized application of sequencing-assisted selection is the parallel production of antibody sets that target multiple antigens. For example, an “array” of antigens can be pooled in different configurations, screened, and the resulting antibodies deconvoluted so that scFvs specific to a single antigen can be deduced (22, 23). This strategy can reduce the number of selections to the square root of the number of antigens. Future single-pot, massively parallel selections will require the development of robust library-vs.-library deconvolution strategies, and preliminary progress has recently been reported (24).
As more sophisticated selection/deconvolution strategies emerge, and as immuno-PCR applications become more commonplace, we anticipate an increasing demand for efficient, low-cost production of high-quality synthetic antibody reagents. The methods presented here are a step toward this goal. As a first iteration, however, our platform is not without limitations. For example, not all of the scFvs predicted to bind PVRL4 were found to do so by FACS analysis. Indeed this finding could reflect the different antigenic context of cell surface PVRL4 or it could be the result of inaccurate scFv clone-enrichment quantification. Advances in DNA sequencing depth and read length will improve our ability to quantify clonal abundances, and will eliminate the need for the double mate-pair reconstruction of full-length sequences presented here. Read-length improvement will also facilitate analysis of libraries based on diversification of greater than three CDRs.
For this proof-of-principle work, we did not incorporate mutagenic PCR into the library-recovery protocol because it adds a layer of complexity to the analysis of enriched populations. However, the power of deep sequencing to map binding energy landscapes is now being realized (25) and will undoubtedly yield similar utility in the context of antibody selections. Finally, the profusion of additional antibody–antigen cocrystal structures will improve our ability to model CDR sequence biases that give rise to favorable antibody properties. With these considerations in mind, there is little doubt that sequencing-assisted selection of synthetic antibody libraries will play an increasingly important role in a wide variety of future biomolecular investigations.
Methods
HMM scFv Library Assembly.
We wished to use the J chains most commonly associated with the VH1–69 and VL1–44 segments. In a sequenced heavy-chain repertoire from an individual, IGHJ4 was the J chain most often recombined with VH1–69. We used work by Schofield et al. to determine that in a large pool of selected phage, IGLJ2 was the J chain that most often recombined with VL1–44 (26). These components were assembled and reverse translated into an E. coli codon preference (Dataset S1). We introduced silent mutations into the framework regions flanking L3, H2, and H3, for the purpose of cloning in the CDR libraries. We required that at least one of each of these pairs be nonpalindromic so as to minimize multiple CDR insertions during library cloning. To this end, we introduced a BbsI site 5′ and an Acc65I site 3′ of L3, a PflMI site 5′ and an ApoI site 3′ of H2, an AccI site 5′ and a BstEII site 3′ of H3. These pairs of cloning sites flanked replaceable suicide inserts, which contain a stop codon in all reading frames and a XhoI restriction site. The CDR libraries were released from the microarray as 10 pmol of single-stranded DNA and resuspended in 200 μL water. Next, 1 μL of each sublibrary was used as input for library-specific PCR using 1 μL Taq polymerase (TaKaRa) according to the manufacturer’s instructions (2 μM each primer). The thermal profile was: (i) 95 °C 5 m, [(ii) 94 °C 15 s, (iii) 55 °C 30 s, (iv) 68 °C 15 s] × 24. At this point, the reaction was divided in two and primers were replenished. The thermal profile was then: (i) 95 °C 5 m, (ii) 94 °C 45 s, (iii) 67 °C 7 m.
The L3 sublibrary was cloned into the scFv vector at the BbsI and Acc65I sites, electrotransformed into DH10B cells, and grown overnight on 15-cm carbenicillin plates. We harvested >107 transformants by scraping, and purified their plasmid DNA. Starting with this HMMscFv-L3 library, the same procedure was then used to replace the H2 suicide insert with the H2 library PCR product by using the engineered PflMI and ApoI sites. We obtained >107 transformants (HMMscFv-L3-H2 library) and purified the plasmid DNA. The H3L library PCR product was first NheI/BssHII subcloned into the pPAO2 vector (27). About 5 × 106 transformants were obtained and plasmid DNA collected. In parallel, H3R library PCR product was prepared. From the pPAO2-H3L plasmid pool, ∼300 bp of upstream sequence was PCR-amplified for subsequent size discrimination of H3L-H3R ligation product. Both pPAO2-H3L and H3R PCR products were digested with SapI for subsequent combinatorial ligation by their 5′ overhanging codons. High-concentration T4 ligation was carried out at 15 °C overnight, a condition that permits mismatched ligation at a relatively high frequency. Indeed, upon sequencing a large number of H3 clones, we observed many examples of library members with unmatched codons that were ligated together, and importantly, without disrupting the reading frame. After H3 ligation, the correct size product was gel-purified and PCR-amplified. This PCR product and the HMM scFv vector were then digested with AccI and BstEII, so that the final H3 library could replace the vector’s H3 suicide insert. If only complementary codons were able to ligate together, the theoretical diversity of the H3 sublibrary would be 1.2 × 107. However, we frequently observed noncomplementary ligation, thus increasing the expected diversity of H3. About 107 H3 clones were obtained.
To bring together HMMscFv-L3-H2 and HMMscFv-H3 in a final ligation (Fig. 1D), 60 μg of each of library was first digested with AccI and BbsI and the desired fragment gel-purified. In a high-concentration T4 ligation at 37 °C, the two fragments were ligated to form concatamers. Finally, the product was digested with both NotI (to release the desired in vitro transcription template) and XhoI (to destroy clones retaining a suicide insert) and gel-purified. We recovered 2.44 μg of HMMscFv-L3-H2-H3 library DNA at the correct size, which corresponds to 3.07 pmol or 1.85 × 1012, theoretically unique DNA molecules. This material was used as a template for in vitro transcription (RiboMAX Large Scale RNA Production System T7; Promega) to produce mRNA, which was subsequently isolated with TRI reagent (Ambion).
Ribosome Display.
Before immobilization of antigen-GST fusion protein, MagneGST beads (Promega) were washed 3× in 1× TBST. Five-microliter beads were used per immunoprecipitation, and beads were coated with 100 μL of bacterial lysate containing GST fusion protein mixed 1:1 with TBST. 2 μL of 1M dTT were included. Binding occurred overnight by rotating at 4 °C. RD Buffer, 1 L: 50 mM Tris Acetate (6.07 g), 150 mM NaCl (8.77 g), pH to 7.5 with acetic acid; autoclaved. Beads were washed 5× with buffer “RDWB+T” (RD Buffer plus 50 mM Mg Acetate and 0.5% Tween 20) and tubes were changed after every other wash. Beads were blocked in 50 μL “Selection Buffer” (RDWB+T plus 2.5 mg/mL heparin and 1% BSA and 83.3 μg/mL tRNA) plus 1 μL RNasin (Promega) at 4 °C for 2 h.
Next, 6.37 μg RNA (1 × 1013 RNA molecules) per 14-μL translation reaction were used. Translations were performed using the RTS 100 E. coli Disulfide kit (5 PRIME) according the manufacturer’s instructions, except that the feeding solution was not used. Translation was allowed to proceed for 13 min 45 s at 30 °C. Each 14-μL reaction was immediately diluted with 96 μL ice-cold Selection Buffer and 3 μL RNasin. Reactions were centrifuged 14,000 × g for 5 min at 4 °C. Supernatant was then moved to a new, cold tube. Fifty-microliter beads in Selection Buffer was added to the ribosome-displayed HMM scFv library and rotated 4 h at 4 °C. Beads were washed six times with 500 μL ice-cold RDWB+T. Tubes were changed after every other wash. Ribosomal complexes were disrupted after the final wash by resuspending beads in 50 μL “EB20” (RD Buffer plus 20 mM EDTA) plus 1 μL RNasin and incubated at 37 °C for 10 min. Released RNA was then purified on Qiagen RNeasy column and eluted into 33 μL nuclease-free H2O.
Superscript III kit (Invitrogen) was used to reverse transcribe the selected RNA library from the preTolA primer. Next, 1 μL (5 U) of E. coli RNase H (New England Biolabs) was incubated with the RT product at 37 °C for 20 min. Recovered cDNA was first PCR-amplified using primers that flank an insert region containing the CDRs (LLF2 and LLR2). PCR amplification was performed with the GC-RICH PCR kit (Roche) using the following the conditions: 1× GC-RICH Buffer, 0.2 mM of dNTP, 0.2 μM LLF2 primer, 0.2 μM of LLR2 primer, 0.5 μM of Resolution Solution, 1 μL of enzyme per 50 μL reaction. The thermal profile was: (i) 95 °C for 3 min, [(ii) 95 °C for 15 s, (iii) 55 °C for 30 s, (iv) 72 °C for 1 min] × 40 cycles, (v) 72 °C for 7 min. The resulting PCR product was then double-digested with BbsI and BamHI (New England Biolabs), gel-extracted, and ligated using T4 Ligase into the pRDscFv2 vector. The ligation product was then PCR amplified using primers specific for the T7 promoter and the TolA linker (T7B2 and TolA). PCR amplification was performed as above but with the T7B2 and TolA primers. The final PCR product was digested with XhoI (New England Biolabs) and gel-purified for either Illumina sequencing or use in a subsequent round of selection.
Recovery of HMM scFv Clones from a Selected Library.
Single HMM scFv clones were recovered from the selected library by PCR with CDR-specific primers followed by assembly into a protein expression vector. Forward and reverse primers were designed to amplify target clone’s L3-H2-H3 insert and contained a 20-bp adapter sequence for assembly into the protein expression vector. PCR amplification was performed with the following conditions: 1× Phusion High-Fidelity PCR Master Mix with HF Buffer, 0.2 μM each of the forward and reverse primers, 1 μL of cDNA recovered after library selection per 50-μL reaction. The thermal profile was: (i) 98 °C for 30 s, [(ii) 98 °C for 10 s, (iii) 72 °C for 1 min] × 30 cycles, (iv) 72 °C for 10 min.
PCR products were subsequently gel-purified and assembled into a protein expression vector using an isothermal assembly method. The protein expression vector contains the scFv framework followed by a FLAG tag and two in-frame stop codons. The isothermal assembly reaction was performed as previously described (28). Each reaction contained 100 ng of linear vector DNA lacking the L3-H2-H3 insert and 20 ng of the recovery PCR product, and was incubated at 50 °C for 1 h. One-microliter of the assembly reaction product was transformed into DH5α E. coli cells and colonies were picked for sequence verification. Plasmids were expressed using the RTS 100 Disulfide Kit (5 PRIME) according the manufacturer’s instructions, except that the feeding solution was not used. The resulting product was used directly in subsequent experiments.
Please refer to the SI Methods to find further details regarding the methods used to construct the ribosome display vector, the selection quality control measures, the Illumina sequencing and analysis pipeline, and the FACS confirmation procedure.
Supplementary Material
Acknowledgments
We thank Uri Laserson for sharing IgG heavy-chain sequencing data, François Ehrenmann and Marie-Paule Lefranc for providing crystal structure data, and Andreas Plückthun for providing p4c11L34Ser. S.J.E. is an investigator with the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1215549109/-/DCSupplemental.
See Commentary on page 18245.
References
- 1.Ravn U, et al. By-passing in vitro screening—Next generation sequencing technologies applied to antibody display and in silico candidate selection. Nucleic Acids Res. 2010;38:e193. doi: 10.1093/nar/gkq789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang H, et al. Phenotype-information-phenotype cycle for deconvolution of combinatorial antibody libraries selected against complex systems. Proc Natl Acad Sci USA. 2011;108:13456–13461. doi: 10.1073/pnas.1111218108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barbas CF, 3rd, Bain JD, Hoekstra DM, Lerner RA. Semisynthetic combinatorial antibody libraries: A chemical solution to the diversity problem. Proc Natl Acad Sci USA. 1992;89:4457–4461. doi: 10.1073/pnas.89.10.4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barbas CF., 3rd Synthetic human antibodies. Nat Med. 1995;1:837–839. doi: 10.1038/nm0895-837. [DOI] [PubMed] [Google Scholar]
- 5.Vargas-Madrazo E, Lara-Ochoa F, Almagro JC. Canonical structure repertoire of the antigen-binding site of immunoglobulins suggests strong geometrical restrictions associated to the mechanism of immune recognition. J Mol Biol. 1995;254:497–504. doi: 10.1006/jmbi.1995.0633. [DOI] [PubMed] [Google Scholar]
- 6.Lee CV, et al. High-affinity human antibodies from phage-displayed synthetic Fab libraries with a single framework scaffold. J Mol Biol. 2004;340:1073–1093. doi: 10.1016/j.jmb.2004.05.051. [DOI] [PubMed] [Google Scholar]
- 7.Lloyd C, et al. Modelling the human immune response: Performance of a 1011 human antibody repertoire against a broad panel of therapeutically relevant antigens. Protein Eng Des Sel. 2009;22:159–168. doi: 10.1093/protein/gzn058. [DOI] [PubMed] [Google Scholar]
- 8.Zhai W, et al. Synthetic antibodies designed on natural sequence landscapes. J Mol Biol. 2011;412:55–71. doi: 10.1016/j.jmb.2011.07.018. [DOI] [PubMed] [Google Scholar]
- 9.Kaas Q, Ruiz M, Lefranc MP. IMGT/3Dstructure-DB and IMGT/StructuralQuery, a database and a tool for immunoglobulin, T cell receptor and MHC structural data. Nucleic Acids Res. 2004;32(Database issue):D208–D210. doi: 10.1093/nar/gkh042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ehrenmann F, Kaas Q, Lefranc MP. IMGT/3Dstructure-DB and IMGT/DomainGapAlign: A database and a tool for immunoglobulins or antibodies, T cell receptors, MHC, IgSF and MhcSF. Nucleic Acids Res. 2010;38(Database issue):D301–D307. doi: 10.1093/nar/gkp946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hanes J, Schaffitzel C, Knappik A, Plückthun A. Picomolar affinity antibodies from a fully synthetic naive library selected and evolved by ribosome display. Nat Biotechnol. 2000;18:1287–1292. doi: 10.1038/82407. [DOI] [PubMed] [Google Scholar]
- 12.Ofran Y, Schlessinger A, Rost B. Automated identification of complementarity determining regions (CDRs) reveals peculiar characteristics of CDRs and B cell epitopes. J Immunol. 2008;181:6230–6235. doi: 10.4049/jimmunol.181.9.6230. [DOI] [PubMed] [Google Scholar]
- 13.Schütz F, Delorenzi M. MAMOT: Hidden Markov modeling tool. Bioinformatics. 2008;24:1399–1400. doi: 10.1093/bioinformatics/btn201. [DOI] [PubMed] [Google Scholar]
- 14.Bond CJ, Wiesmann C, Marsters JC, Jr, Sidhu SS. A structure-based database of antibody variable domain diversity. J Mol Biol. 2005;348:699–709. doi: 10.1016/j.jmb.2005.02.063. [DOI] [PubMed] [Google Scholar]
- 15.Lara-Ochoa F, Vargas-Madrazo E, Jimenez-Montano MA, Almagro JC. Patterns in the complementary determining regions of immunoglobulins (CDRs) Biosystems. 1994;32:1–9. doi: 10.1016/0303-2647(94)90014-0. [DOI] [PubMed] [Google Scholar]
- 16.Singh H, Raghava GP. ProPred: Prediction of HLA-DR binding sites. Bioinformatics. 2001;17:1236–1237. doi: 10.1093/bioinformatics/17.12.1236. [DOI] [PubMed] [Google Scholar]
- 17.Fabre-Lafay S, et al. Nectin-4 is a new histological and serological tumor associated marker for breast cancer. BMC Cancer. 2007;7:73. doi: 10.1186/1471-2407-7-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Athanassiadou AM, Patsouris E, Tsipis A, Gonidi M, Athanassiadou P. The significance of Survivin and Nectin-4 expression in the prognosis of breast carcinoma. Folia Histochem Cytobiol. 2011;49:26–33. doi: 10.5603/fhc.2011.0005. [DOI] [PubMed] [Google Scholar]
- 19.Chodorge M, Fourage L, Ravot G, Jermutus L, Minter R. In vitro DNA recombination by L-Shuffling during ribosome display affinity maturation of an anti-Fas antibody increases the population of improved variants. Protein Eng Des Sel. 2008;21:343–351. doi: 10.1093/protein/gzn013. [DOI] [PubMed] [Google Scholar]
- 20.Hoogenboom HR. Selecting and screening recombinant antibody libraries. Nat Biotechnol. 2005;23:1105–1116. doi: 10.1038/nbt1126. [DOI] [PubMed] [Google Scholar]
- 21.Beck A, Wurch T, Bailly C, Corvaia N. Strategies and challenges for the next generation of therapeutic antibodies. Nat Rev Immunol. 2010;10:345–352. doi: 10.1038/nri2747. [DOI] [PubMed] [Google Scholar]
- 22.Erlich Y, et al. DNA Sudoku—Harnessing high-throughput sequencing for multiplexed specimen analysis. Genome Res. 2009;19:1243–1253. doi: 10.1101/gr.092957.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prabhu S, Pe’er I. Overlapping pools for high-throughput targeted resequencing. Genome Res. 2009;19:1254–1261. doi: 10.1101/gr.088559.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bowley DR, Jones TM, Burton DR, Lerner RA. Libraries against libraries for combinatorial selection of replicating antigen-antibody pairs. Proc Natl Acad Sci USA. 2009;106:1380–1385. doi: 10.1073/pnas.0812291106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Whitehead TA, et al. Optimization of affinity, specificity and function of designed influenza inhibitors using deep sequencing. Nat Biotechnol. 2012;30:543–548. doi: 10.1038/nbt.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schofield DJ, et al. Application of phage display to high throughput antibody generation and characterization. Genome Biol. 2007;8:R254. doi: 10.1186/gb-2007-8-11-r254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zacchi P, Sblattero D, Florian F, Marzari R, Bradbury AR. Selecting open reading frames from DNA. Genome Res. 2003;13:980–990. doi: 10.1101/gr.861503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gibson DG, et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.