Abstract
Analyses of compounds in clinical development have shown that ligand efficient-molecules with privileged physical properties and low dose are less likely to fail in the various stages of clinical testing, have fewer postapproval withdrawals, and are less likely to receive black box safety warnings. However, detailed side-by-side examination of molecular interactions and properties within single drug classes are lacking. As a class, VEGF receptor tyrosine kinase inhibitors (VEGFR TKIs) have changed the landscape of how cancer is treated, particularly in clear cell renal cell carcinoma, which is molecularly linked to the VEGF signaling axis. Despite the clear role of the molecular target, member molecules of this validated drug class exhibit distinct clinical efficacy and safety profiles in comparable renal cell carcinoma clinical studies. The first head-to-head randomized phase III comparative study between active VEGFR TKIs has confirmed significant differences in clinical performance [Rini BI, et al. (2011) Lancet 378:193–1939]. To elucidate how fundamental drug potency–efficiency is achieved and impacts differentiation within the VEGFR TKI class, we determined potencies, time dependence, selectivities, and X-ray structures of the drug–kinase complexes using a VEGFR2 TK construct inclusive of the important juxtamembrane domain. Collectively, the studies elucidate unique drug–kinase interactions that are dependent on distinct juxtamembrane domain conformations, resulting in significant potency and ligand efficiency differences. The identified structural trends are consistent with in vitro measurements, which translate well to clinical performance, underscoring a principle that may be broadly applicable to prospective drug design for optimal in vivo performance.
Keywords: axitinib, pazopanib, sorafenib, sunitinib, tivozanib
The VEGF receptor (VEGFR) tyrosine kinases (TKs) are the clinically validated drug target of four structurally diverse TK inhibitors (TKIs) that have been approved in the renal cell carcinoma (RCC) setting (1, 2). The clear cell histology subtype of RCC is molecularly linked to the VEGF signaling axis by virtue of the ∼90% incidence of Von Hippel-Lindau (VHL) dysregulation. With VHL inactivation, hypoxia-inducible factor-α accumulates, leading to overproduction of the angiogenic factor VEGF among others. It is, therefore, generally accepted that on-target VEGFR TK inhibition accounts for the RCC efficacy seen within this class of TKIs. In addition to efficacy in RCC, VEGF signaling inhibition has been linked to side effects, with the most prominent being hypertension, which is consistently seen within the TKI class and the related monoclonal antibody to VEGF, bevacizumab (3).
Despite the clear role of VEGF signaling on both hypertension and efficacy in RCC, these on-target pharmacologic effects differ in frequency and degree between approved VEGFR TKI drugs, indicating that the extent of VEGF signal blockade may not be equivalent. Recent reports have analyzed similar RCC clinical studies across leading VEGFR TKIs for comparison purposes (1, 2). Clear distinction in both efficacy and safety across the class is apparent in observations of hypertension and progression-free survival (PFS), endpoints specifically linked to VEGF blockade. Additionally, the first head-to-head randomized phase III comparative study between active VEGFR TKIs has confirmed significant differences in clinical performance (4). Differences in hypertension and PFS suggest that distinct drug-specific factors limit the maximal exposure of each drug and consequently, the extent of VEGFR kinase blockade in patients. Principally, the greater the overall selectivity of a drug, the greater that the pharmacological effects will be driven by on- vs. off-target mechanisms. At the extreme, complete selectivity would ensure that the therapeutic dose is limited only by target-associated side effects and that maximal VEGF signal blockade would be realized. The VEGFR TKI cohort of molecules provides a unique opportunity to study the impact of molecular interactions and physical properties on potency and selectivity and how these in vitro measures translate to differentiated clinical profiles.
In a broad analysis of drug attrition, increased development halts and market withdrawals are associated with unfavorable molecular physical properties and dose burden to the liver, particularly when toxicophores are present (5–11). Indeed, modern programs increasingly use physical property-based drug design strategies in conjunction with high-resolution inhibitor–protein structures to discover low-dose, highly efficient drugs (12). Notwithstanding the convincing conclusions from general analysis of attrition, detailed side-by-side studies of the relationship between molecular interactions/properties and differences in clinical performance within single drug classes are needed. The receptor tyrosine kinases (RTKs) and their inhibitors have been a significant focus of research, supported at a molecular level with solved structures of TKI–kinase complexes and well-accepted classifications of inhibitor binding modes. Inhibitor classifications include type I for binders of the ATP pocket, type II for inhibitors that block the aspartate-phenylalanine-glycine (DFG) activation loop segment from docking in an active DFGin conformation, and type III inhibitors that are allosteric (13). Although typical protein studies focus on the kinase domain of RTKs, within the PDGF receptor (PDGFR) super family of kinases, inclusion of the juxtamembrane (JM) domain in constructs detects potency differences that distinguish some type I from type II TKIs (14).
To elucidate how fundamental drug potency–efficiency is achieved and impacts differentiation within the VEGFR TKI class, a JM domain-inclusive VEGFR2 TK construct was used to determine potencies, time dependencies, selectivities, and X-ray structures of drug–kinase complexes across a panel of TKIs (15). Distinct TKI molecular interactions and kinase conformations, which are not captured by type I, II, and III designations, shed light on in vitro results and corresponding clinical performance across the panel, revealing a drug design principle that may have general applicability for achieving optimal in vivo performance.
Results
Crystal Structures Reveal Distinct JM Conformations.
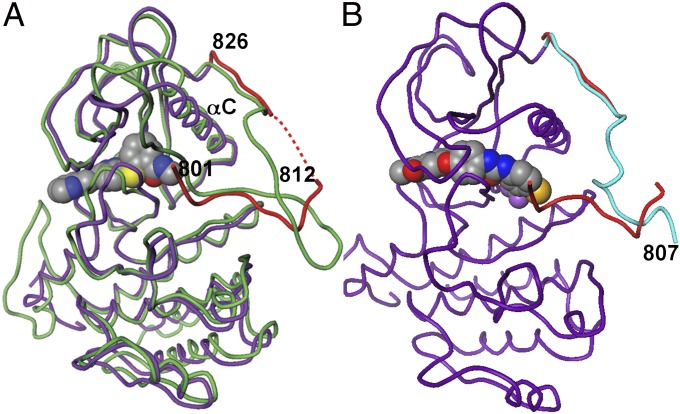
Structures were obtained of a nonphosphorylated VEGFR2 construct comprising the catalytic and JM domains (plus-JM) (15) in complex with five representative molecules: axitinib, pazopanib, sorafenib, sunitinib, and tivozanib. These complexes all show the activation loop adopting a DFGout position, similar to previously described inhibitor complexes of the nonphosphorylated VEGFR2 catalytic domain (minus-JM), including an axitinib complex (12). The plus-JM complexes with inhibitors axitinib and sunitinib show the JM to fold into the portion of the kinase interdomain space adjacent to α-helix C(JMin), analogous to autoinhibitory JMin conformations reported for other PDGFR kinase family members (16–19) and very similar to apo cKit (20) (Fig. 1A). This cleft will be referred to here as the regulatory domain pocket (RDP), because it is also where DFG resides in the active conformation (DFGin) and therefore, precludes a DFGin/JMin arrangement (21). Just before the classical kinase domain, the plus-JM residues 820–826 extend over the α-helix C. The density for residues 813–819 was not well-ordered, and therefore, positions for these residues are not included in the crystal structure coordinates. Residues 801–812 were well-ordered, packing into the RDP to participate in many interactions that stabilize an autoinhibitory conformation, analogous to the JM segment of FMS-related tyrosine kinase 3 (FLT3) (18) and cKit (17, 19, 20). The crystal structure of pazopanib shows insufficient electron density to support assignment of specific JM residues, but partial density is consistent with a mobile JM that transiently adopts a JMin conformation similar to the conformation seen with axitinib and sunitinib. Comparing the plus-JM complex of axitinib with the minus-JM counterpart (12) showed no significant differences in the catalytic domain, suggesting that JMin acts to preclude DFG rearrangement to active kinase but does not perturb any DFGout conformational features.
Fig. 1.
Crystal structures reveal a JMin and JMout in VEGFR2. (A) JMin position in the unactivated VEGFR2 axitinib complex and apoenzyme cKit. Coloring: VEGFR KD (purple), VEGFR JM (red), and cKit (green). The red dashed lines indicate a segment of the VEGFR JM domain that could not be unambiguously modeled from the electron density. (B) JMout position (cyan) in the VEGFR2 sorafenib complex. For comparison, the position of the JM in the VEGFR2 axitinib complex is also shown (red).
In the plus-JM complexes with type II inhibitors sorafenib and tivozanib, the JM is displaced from the RDP to a JMout position (Fig. 1B). Compared with sunitinib and axitinib, the type II TKIs have larger substituents extending into the RDP, preventing concurrent binding with JMin because of a steric clash, where sorafenib and tivozanib would have sub van der Waals distances to the JM I804 of 0.9 and 1.5 Å, respectively (Fig. 2 A and B). By contrast, the amide methyl of axitinib makes van der Waals contact with I804 (3.8 Å), defining the approximate boundary that an inhibitor substituent can approach without displacing the JMin (Fig. 2C).
Fig. 2.
A superposition of the autoinhibitory JMin (blue) in its axitinib complex position onto the (A) tivozanib and (B) sorafenib complexes illustrates how the larger inhibitor substituents clash with this position of I804. A van der Waals representation shows a favorable hydrophobic interaction between (C) axitinib and I804 of the JM.
The role of JM rearrangement in VEGFR2 activation can be inferred from the conformations of the structures reported here as well as the previously reported structure of the activated catalytic domain (21) (Fig. 3). In the autoinhibited state, the DFGout segment, particularly F1047, blocks ATP binding. The JMin position locks in the autoinhibitory state, because it prevents the DFGout to DFGin transition that allows ATP to bind through a clash between I804 and F1047 (Fig. 3 A–C). When the JM rearranges out of the RDP, then the DFG is unlocked and able to switch to the open activated state. Autophosphorylation, which activates the kinase presumably through stabilization of an active conformation, has been reported to occur on the JM and then the DFG-containing activation loop sequentially, consistent with the model (15).
Fig. 3.
Model of autoinhibitory to active conformation transitions for JM and DFG. The perspective in this graphic is from below with respect to Figs. 1 and 2 and rotated ∼160° from the top. Surface coloring: DFG (yellow), JM (red), ATP (cyan), and axitinib and tivozanib (gray). The ATP binding position was modeled from PDB ID code 1IR3. The DFG and JM positions are fixed in the vertical comparisons, where A and D are taken from the axitinib complex and B and E are taken from the tivozanib complex. (C) The activated VEGFR2 kinase domain including DFG was reported previously (PDB ID code 1VR2), and the JM is modeled from the tivozanib complex. The conformations from left to right correspond to TKI binding (D) type IV and (E) type II, type II, and type IV.
In Vitro Measurements.
Eight compounds served as the TKI panel of structurally diverse VEGFR2 TKIs for this study, including the four approved RCC VEGFR TKI drugs (axitinib, pazopanib, sorafenib, and sunitinib) and four drugs that have completed advanced clinical testing. Two previously described nonphosphorylated VEGFR2 tyrosine kinase constructs were used, the catalytic domain VEGFR2 construct (minus-JM) and the plus-JM construct (Ki values for axitinib have been previously reported) (15). The results of the side-by-side TKI studies determining potency, selectivity, and efficiency are shown in Table 1. The inhibitor names, along with structural features and clinical endpoints, are also shown (1, 2, 22).
Table 1.
VEGFR TKI panel summary of in vitro, structural, and clinical results
| Kinase Ki (nM) |
Indices |
HUVEC IC50 (nM) |
Binding |
Clinical results |
|||||||||
| TKI name | −JM | +JM | Ratio | KPI* | LipE | pVEGFR2 | Survival | Type | HA-RDP† | Dose | Cpavg-free | PFS1‡ | PFS2§ |
| Axitinib¶ | 1.1 | 0.020 | 55 | 0.95 | 7.4 | 0.013 | 0.18 | IVa | 1 | 10 | 0.5 | 12.1 | 13.7 |
| Pazopanib¶ | 8.20 | 0.29 | 28 | 0.83 | 5.8 | 0.63 | 12 | IVb | 0 | 800 | 42 | 7.4 | 9.2 |
| Cediranib | 8.10 | 0.46 | 18 | 0.61 | 4.0 | 0.088 | 7.4 | IVb | 0 | 45 | 6 | ||
| Sunitinib¶ | 60 | 3.9 | 15 | 0.02 | 5.4 | 2.7 | 14 | IVb | 0 | 50 | 16 | 8.8 | 11 |
| Brivanib|| | 44 | 3.9 | 11 | 0.77 | 5.0 | 27 | 50 | IVb | 0 | 800 | 300 | ||
| Tivozanib||** | 0.20 | 0.01 | 18 | 0.93 | 6.4 | 0.04 | 0.5 | II | 6 | 1.5 | 1.2 | 11.8 | 11.8 |
| Linifanib||** | 0.64 | 0.67 | 1 | 0.75 | 4.7 | 0.04 | 0.8 | II | 8 | 17.5 | 12 | ||
| Sorafenib¶** | 0.58 | 0.52 | 1 | 0.68 | 3.8 | 2.3 | 25 | II | 11 | 800 | 40 | 6.5 | 5.5 |
Selectivity and LipE are calculated from plus-JM determined Ki values.
*KPI for VEGFR TK vs. IC50 values for other kinases.
†Count of heavy atoms in regulatory domain pocket past E885 H-bond donor.
‡PFS1 taken from reported PFS in the cytokine refractory setting.
§PFS2 taken from a published summary table of similar RCC trials in mainly first-line patients. For axitinib, the time to progression (TTP) number of 15.7 was corrected to the PFS of 13.7.
¶Approved by the Food and Drug Administration for RCC.
||Not screened against all 283 kinases: tivozanib had Ki values determined against the 29 sorafenib kinase hits, and linifanib and brivanib had Ki values determined against the 17 shared kinase hits.
**Time-dependent TKIs as determined by 10 min preincubation studies.
When evaluated in the minus-JM assay, the measured Ki values ranged from 0.2 to 60 nM across the TKI panel. Using the plus-JM assay to test the same TKI panel, resulting potencies were equivalent to or greater than the minus-JM results, with Ki values ranging from 0.011 to 3.9 nM. The Ki ratio between the two constructs (minus-JM/plus-JM) ranged from 1- (for the two type II compounds with the largest RDP groups) to 55-fold for axitinib, which has an RDP group in closest contact to the JMin domain. The remaining inhibitors, which do not fully complement the binding pockets of either JM conformer, produced Ki ratios in between these two extremes. The binding modes were delineated by these ratios, where all inhibitors compatible with the JMin conformation (type IV, vide infra) showed a Ki ratio of at least 10-fold (Fig. 4). During experimentation with the two constructs, another divergence by binding-mode type was detected. Time dependence was observed only with the plus-JM construct and only for TKI binding modes that required the JM domain to rearrange from its autoinhibitory JMin conformation to JMout. This effect, which was limited to type II inhibitors sorafenib, linifanib, and tivozanib, necessitated a short preincubation time to observe a plateau of measured potency.
Fig. 4.
Graph of data generated from both constructs plotted on the y axis vs. cellular data. Coloring by binding mode: type II (red), type IVa (blue), and type IVb (green). Shape and shade by construct: plus-JM is darker and designated with +; minus-JM is lighter and designated with −. The minus-JM Ki values differ to the plus-JM for type IV TKIs and not for the two type II TKIs with the largest RDP groups. The type II exception is tivozanib. The minus-JM results fall above the 1:1 line vs. pVEGFR2 cell IC50, a rare circumstance for kinase inhibitors that bind the ATP site.
In the VEGF-induced VEGFR2 autophosphorylation (pVEGFR2) assay in human umbilical vein endothelial cells (HUVECs), the measured IC50 values for the TKI panel ranged from 0.013 to 27 nM. Anomalous to the typical experience for on-target kinase activity, the minus-JM biochemical Ki values were less potent than these cell-based pVEGFR2 IC50 values for all but one compound (Fig. 4). The TKIs that exhibited the largest discrepancy between minus-JM potency compared with the cell were those TKIs that bind the autoinhibitory JMin conformation. By contrast, biochemical Ki values generated from the more comprehensive plus-JM construct more closely matched pVEGFR2 cell potencies on the whole, with variations distributed equally across the two TKI binding modes (Fig. 4). The minus-JM construct result anomalies and the binding mode-specific deviations from the whole-cell results supported a focused use of the plus-JM results for additional translation analysis. The VEGF-dependent cell survival assay in HUVEC gave IC50 values that ranged from 0.18 to 50 nM across the TKI panel, an expected range for a functional response, averaging 10-fold higher than pVEGFR2 IC50 values.
To determine TKI selectivities, an initial extensive kinase screen was used to inform the selection of kinases for full Ki determinations. The initial kinase screen evaluated axitinib, cediranib, pazopanib, sunitinib, and sorafenib for inhibition against a panel of 283 kinases at two concentrations (23) (Dataset S1). The screen concentrations were normalized to the fixed multiples of 50- and 1,000-fold the determined plus-JM Ki values, with slight rounding to provide overlap of screen doses between a subset of drugs. Accordingly, the approximate 50-fold screen concentrations were set at 200 nM for sunitinib, 20 nM for pazopanib, cediranib, and sorafenib, and 1 nM for axitinib, and the 1,000-fold screen concentrations were 4,000 nM, 200 nM, and 20 nM for the above groupings, respectively. Other than the VEGFR1, -2, and -3 kinases, each TKI at the 50-fold concentration showed at least 50% inhibition of the following number of kinases: axitinib inhibited no other kinases; pazopanib inhibited 3; sorafenib inhibited 5; cediranib inhibited 5; and sunitinib inhibited 81 kinases.
Each TKI was further characterized by 10-point dose–response analysis for kinases that were >50% inhibited at the 1,000-fold screen concentration, with sunitinib being limited to the 87 kinases identified by the 50-fold concentration results and hits from other TKIs. To gain a broader perspective of the VEGFR TKI class, brivanib and linifanib were included in the dose–response studies for kinases that met the selection criteria for all five screen compounds (17 total kinases). Tivozanib was included in testing against kinases selected for sorafenib, a similar type II inhibitor (29 total); the understood caveat was that these add-on TKIs were not included in the initial screening protocol and therefore, may uniquely inhibit kinases that were not identified by the five screen TKIs. Findings from the initial screen were substantiated by the dose–response results. Follow-up dose–response Ki determinations for VEGFR1, -2, and -3 were triggered for all compounds. For each compound, the VEGFR1, -2, and -3 kinase Ki values were in good agreement with previously reported values (24) and within fourfold of each other, exhibiting little selectivity across these nearly identical kinases. Therefore, the below selectivity analysis was simplified for VEGFR kinases by using the plus-JM Ki results to represent the VEGFR TK family. Kinome selectivity trees were prepared for each of the approved TKIs using fold-selectivity values of plus-JM and kinase panel Ki data (Dataset S2). To generate a numerical representation of VEGFR kinase selectivity of each TKI, the kinase partition index (KPI) was derived from the plus-JM Ki and the kinase selectivity Ki data (Dataset S3), where VEGFR TK occupancy Z is described as ZVEGFR TK = 1/Ki and the KPI = ZVEGFR TK/(sum of Z values for all kinases).
Discussion
The ability to make robust conclusions about relative potency and selectivity across independently discovered TKIs is hampered by the multiple sources of data, often generated from nonidentical studies. Typical experimental conditions alone further complicate interpretation, because for practical reasons, they do not fully recapitulate the physiological conditions in the disease setting. For example, protein constructs of VEGFR2 TK that contain only the catalytic domain but not an important regulatory domain have been broadly used as practical surrogates of full-length protein to determine biochemical potency. Additionally, the reported inhibitor VEGFR2 TK crystal structures contain constructs comprised of only the catalytic domain (21, 25). Although these kinase catalytic domain constructs circumvent key technical problems inherent with full-length protein, the excised JM domain has been shown to impact both kinase regulation and measured inhibitor potency within the PDGF family of RTKs (14), including VEGFR2 TK (15). For meaningful comparative studies, it was deemed critical to incorporate the JM domain into the VEGFR kinase construct to better reflect whole-cell physiological conditions and the impact of important regulatory conformations on TKI potency and selectivity.
JM Domain Impact on in Vitro Results.
The initial evaluation using both kinase constructs provided useful information on the impact that the JM domain has on TKI binding across the panel. The addition of the JM domain as a component of the kinase construct causes two binding mode-dependent deviations from minus-JM results: (i) those TKIs that can bind a JMin conformation all become more potent, but two of three type II inhibitors do not (Fig. 4), and (ii) all three type II TKIs that displace the JM domain show a time dependence during routine potency measurements out to 10 min. Interestingly, tivozanib displayed a time dependence but showed an increase in affinity for the plus-JM construct. However, the crystal structure reveals only a JMout conformation where no direct contact is made between the JM domain and tivozanib, and a full JMin is precluded. Taken together, these data suggest that a mobile JM has an energetic preference for a JMin conformation bound to the RDP, displays a kinetic barrier to displacement from its resting state, and can adopt a state that clearly stabilizes at least one type II binding TKI. Of the type II TKIs, tivozanib does present the greatest opportunity for direct JM interactions, because it has the smallest RDP binding moiety; therefore, it has the largest residual RDP, where transient states between JMin and JMout could interact, although other explanations cannot be ruled out. The data consistently implicate the JM domain in binding interactions across multiple TKIs, underscoring the need to use the more complete plus-JM construct when characterizing highly optimized VEGFR TK inhibitors.
In assessing how the results translate to cells, measurements of HUVEC IC50 across the TKI panel substantiated the plus-JM results as physiologically relevant, with plus-JM–derived Ki values consistent with pVEGFR2 potencies in an absolute sense (plus-JM Ki/pVEGFR2 IC50 average ratio = 3) and high correlation across the panel to both pVEGFR2 (R = 0.77, P = 0.03) and survival IC50 endpoints (R = 0.81, P = 0.02). By comparison, minus-JM–generated Ki values showed a TKI-type divergence that showed particularly poor agreement of absolute potencies to pVEGFR2 values for JMin binding TKIs compared with type II TKIs (minus-JM Ki/pVEGFR2 IC50 average ratios = 41 and 7, respectively) (Fig. 4). The correlations of minus-JM Ki to pVEGFR2 (R = 0.65, P = 0.08) or survival IC50 values (R = 0.76, P = 0.03) are also weaker. Potencies derived from the plus-JM construct show significant correlation to selectivity (below) and average unbound plasma concentrations in the clinical (R = 0.83, P = 0.01) (Fig. S1), providing additional support for this construct as the most relevant.
Ligand Efficiency.
To facilitate analysis of drug–efficiency relationships to selectivities and in vivo properties, lipophilicity-based ligand efficiencies (LipEs) were calculated using LipE = pKi − clogP (10). The LipE value for a molecule normalizes target potency with lipophilicity, a physical property that strongly influences pharmaceutical properties. Increased molecular lipophilicity drives affinity for lipophilic environments, including binding affinity of the target protein, and also off-target proteins, eroding metabolic stability and selectivity among other pharmaceutical properties. Therefore, compounds of varying potencies and lipophilicities can be analyzed together using the LipE index to elucidate meaningful relationships between efficiency and selectivity measures or in vivo performance. The VEGFR TKI panel exhibits a broad LipE range from a high of 7.4 for axitinib to a low of 3.8 for sorafenib (Table 1), which is a useful spread to support analysis.
Selectivity.
Kinome screening at single drug concentrations (e.g., 1 μM) or views of IC50 values, without normalization to on-target potency, provides little knowledge of TKI selectivity, limiting the use of such representations for drug class comparisons. For evaluating selectivity, kinase tree representations that illustrate fold difference to target potency (26) were used (Fig. 5A). Because target potency defines these representations, the importance of using an on-target kinase construct that closely translates biochemical potency into the whole-cell context is amplified. The kinase tree representations provide a high-level view of selectivity, where the outlier sunitinib is most noticeable, and small selectivity differences between the other three can be discerned qualitatively on close inspection. In a quantitative analysis of selectivity, the calculated KPI was compared with the LipE index across TKIs. Because LipE is an expression of on-target efficiency that is blind to structural features of TKIs or any one receptor, a general association of LipE with overall selectivity is expected. Again, sunitinib is an understandable outlier, because it does not bind in the specificity region beyond the ATP pocket (Fig. 5B). With the exclusion of sunitinib, the correlation between LipE and overall kinase selectivity was very good (R = 0.96, P = 0.0008), indicating that the efficiency differences for these TKIs arise from interactions capable of delivering kinase specificity. Sunitinib is a powerful example that underscores the importance of deriving high efficiency from specific interactions outside the ATP binding site for exceptional selectivity in the kinase family.
Fig. 5.
(A) Kinome selectivity tree of Food and Drug Administration-approved VEGFR TK drugs in RCC. Circle sizes are inversely proportional to fold selectivity ratios calculated as (Ki for kinase)/(plus-JM Ki). (B) x axis broken after 0.1 and 0.8 and contracted. Coloring indicates channel occupancy: yes (blue) and no (red). The KPI measure of selectivity correlates with LipE for channel binders.
Molecular Interactions with JM Domain Conformations.
One structural feature that has defined type II kinase inhibitors is the presence of a large substituent to fill the RDP, forcing a DFG rearrangement. Therefore, type II inhibitors are characterized as incompatible with JM autoinhibition, which also docks JMin to this same RDP, an incompatibility that is corroborated by binding affinity loss against plus-JM relative to minus-JM constructs for cKIT, FLT3, and colony stimulating factor (CSF)-1R. The previous absence of TKIs with greater affinity for plus-JM constructs was specifically identified as a remaining medicinal chemistry challenge (14). Indeed, until now, this weaker binding of plus-JM constructs has been used as a signature of DFGout binders. To understand how some VEGFR TKIs achieve increased potency against plus-JM, challenging the JM incompatibility paradigm, the first cocrystal structures of plus-JM complexes were solved and analyzed for five TKIs: sorafenib, tivozanib, sunitinib, pazopanib, and axitinib. The results, which revealed that all TKIs bind the nonactive DFGout conformation, revealed two distinct JM conformations as the only significant protein conformational differences, enabling a focused dissection of drug–kinase interactions related to JM. The VEGFR TKIs that uniformly exhibit greater JM construct potency do not fill the RDP with a large inhibitor substituent, as type II TKIs do. The solved structures of these TKIs reveal a stabilized JMin acting as the physical lock in the RDP, precluding the DFG loop from adopting an active in position (21) (Fig. 3). This class is, therefore, termed type IV TKIs. The type IV mode of action extends the continuum moving from (Fig. 3, left to right) type IV binding of DGFoutJMin to type II binding of DGFoutJMout to type I binding of active DGFinJMout.
TKI Interactions with Plus-JM VEGFR2.
Within the adenine binding site, nonselective interactions are available across kinases, and they are accommodated by most ATP competitive inhibitors. Each TKI in the panel possesses a heterocycle that orients an H-bond acceptor to the backbone donor and the following variations of interactions with flanking backbone carbonyls (Fig. 6): H-bond donors (sunitinib), H-bond donor and a polarized C-H (axitinib, pazopanib, and sorafenib), or two polarized C-Hs (tivozanib). Across the TKI panel, other structural features with occupancy confined to the adenine site can also be found in nonspecific kinase inhibitors and are not further discussed.
Fig. 6.
The five TKI/plus-JM complexes are vertically aligned to depict the similarities of ATP site occupancy and differences in the interactions with the channel and RDP. The translucent skins are protein surfaces generated for each complex and colored by atom type. (A–E) The TKIs are ordered from top to bottom by the greatest fill of the channel and RDP for (A) sorafenib to the least for (E) sunitinib. Black dashed lines represent hydrogen bonds.
The rationale for harnessing selective interactions targeting DFGout is based on binding in the RDP, away from conserved regions of the ATP site (12, 27). However, the structurally diverse VEGFR TKIs, most of which do not occupy the RDP, all bind the DFGout conformation. The picture that emerges from in vitro data and structural analysis of TKI interactions beyond the conserved ATP site is that the most significant efficiency and selectivity differences arise from profoundly different fingerprints of interactions between the TKIs and the channel that links the ATP site to the RDP. Furthermore, good selectivity requires a rigid structural feature to complement the channel contours, because each TKI does except sunitinib; however, highly efficient interactions that stabilize polarity in the channel are required for superior levels of efficiency and selectivity (12). Both the E885 side chain and the D1046 backbone N-H present polarity that lines the kinase channel, providing significant opportunities for direct TKI interactions to impart distinct selectivity advantages. The importance of charge stabilization is magnified within the autoinhibitory JMin conformation, where the hydrophobic I804 of the JM caps the channel just beyond the charged E885. Indeed, the type IV TKIs with lowest LipE and selectivity do not form direct H-bonds to D1046 and E885, but the highest LipE TKI forms direct H-bonds to stabilize the internal polarity. Consequently, type IV inhibitors that are most efficient are both JM-complementary and form two direct H-bonds, providing additional classification as follows: IVa, JMin-complementary and H-bonds to both D1046 and E885; IVb, JMin-compatible. Detailed analysis of the TKI–kinase interactions by classification reveals subtle effects observed in the data.
VEGFR Type II TKI: JM Domain Displacers.
The type II TKIs within the panel are linifanib, sorafenib, and tivozanib. Solved cocrystal structures of plus-JM with the latter two TKIs reveal that they, indeed, bind the RDP, forcing a JMout conformation (Fig. 6 A and B). The type II TKIs fill the channel, making an important H-bond to D1046 and two H-bonds to the side chain carboxylate oxygen of E885. The type II TKIs have different-sized RDP groups ranging from 6 to 11 heavy atoms in the RDP (HA-RDP) for tivozanib and sorafenib, respectively, counting from the deepest H-bond donor. The increasing size of this group translates to a more complete fill of the RDP along with a fall in any JM domain contribution to binding affinity, which was reflected in the potency ratios of minus-JM/plus-JM. Despite being a defining component of type II TKIs, the increase in size of this structural feature is associated with weaker efficiency and selectivity. To support this observation, literature data were sought for methyl analogs of the type II TKIs, similar to the small RDP group found on axitinib, but none were found. Fortunately, VEGF-dependent cell survival assay in HUVEC IC50 values were reported for larger analogs of axitinib, which are structurally identical except for the increase in HA-RDP (12). These data fit with the trend observed for panel type II TKIs, supporting the conclusion that greater molecular size in the RDP leads to lost efficiency (Fig. S2).
VEGFR Type IVb TKI: JMin-Compatible.
Compared with type II, the depth of binding for type IVb TKIs spans a range from filling the contours of a channel up to but not including the RDP (pazopanib) (Fig. 6D) to little more depth than the ATP site (sunitinib) (Fig. 6E). Sunitinib extends only a few atoms beyond the ATP binding site, leaving the interior channel between the edge of the inhibitor and the JM domain unfilled. The unoccupied channel has volume to fit at least seven water molecules, two of which are observed crystallographically. The highly conserved K868–E885 salt bridge forms a wall of the channel adjacent to the inhibitor and is flanked on two sides by the observed water molecules, which serve to solvate and stabilize this interior charge. The salt bridge stabilization by waters is likely important to potency, because sunitinib does not make direct H-bonds to the charged groups. The JM domain in this structure makes no direct contact with the TKI, providing a view of an autoinhibitory JMin conformation that is likely unperturbed by TKI binding (Fig. 6E).
Pazopanib has a head group heterocycle that pushes into the channel within 4.7 Å of the JM position in the sunitinib complex, making a direct H-bond to the NH of D1046, a kinase backbone pivot point, and stabilizing the DFGout conformation (Fig. 6D). Pazopanib is unique among the TKIs by forming this one H-bond while not forming an H-bond to E885. Instead of stabilizing the buried charge of E885, the pazopanib head group displaces channel waters and positions two lipophilic methyls adjacent and nearly equidistant to one of its carboxylate oxygens (3.0 and 3.1 Å). A resting state JMin conformation, such as the conformation seen for sunitinib, would bring the lipophilic I804 adjacent to the E885, further insulating this charge. Not surprisingly, the JM domain exhibits only poor electron density, consistent with partial dynamic occupancy of the RDP. The greater JM mobility observed in this structure is likely caused by the need to orient polarity rather than an I804 side chain, adjacent to E885. The net result for pazopanib is greater selectivity arising from rigid TKI structural features that complement the contours of the channel and make the H-bond to D1046 but only slightly improved efficiency over sunitinib.
Brivanib and cediranib cocrystal structures were not solved. These inhibitors have a similar head group to pazopanib and published binding mode models that overlap with pazopanib, and for a similar indole head group, there is a solved TKI–kinase cocrystal structure in good agreement (28) with the pazopanib binding mode. In contrast to pazopanib, no direct H-bond to the backbone NH of D1046 is possible, because the indole head group lacks an acceptor. Furthermore, direct H-bonds to the side chain carboxylate oxygen of E885 are not observed in the model or related crystal structure because of the poor geometry and prohibitively long distance (3.8 Å) to make such an interaction. These two TKIs have slightly lower LipE values as a result of the more lipophilic structural features. Both TKIs fill the channel with rigid indole head groups, which translates to greater selectivity than sunitinib.
VEGFR Type IVa TKI: JM-Complementary Inhibitors and H-Bonds to D1046 and E885.
The channel interactions that define a type IVa TKI include two specific H-bonds and an overall fit of contours as deep as the JMin (axitinib). Compared with the other type IV inhibitors, the axitinib head group is positioned slightly higher and deeper into the channel (Fig. 6C). The head group amide substituent reaches to the JM, forming one direct H-bond to the key pivot point NH backbone of D1046 and a second direct H-bond to the carboxylate side chain of E885. Only one heavy atom extends past the last H-bond into the RDP within van der Waals distance of the JMin I804 (3.8 Å). The head group substantially complements the full length of the channel, contributing to affinity with polar charge stabilization as well as hydrophobic interactions. Together, the five TKI/plus-JM complexes and associated efficiency data reveal the importance of inhibitor–protein interactions that result in fully filling the contours of the channel, forming direct H-bonds to both D1046 and E885 and complementing the JMin conformation through favorable interactions rather than displacing it with large groups in the RDP. The high LipE values achieved in this manner produced high degrees of selectivity.
Relationships of in Vitro Results to Reported Clinical Endpoints.
Some drug-specific attributes become fixed in a given clinical setting, such as the selected daily administered dose or dose range, with related PK measurements expected to be comparable between clinical trials, such as average unbound plasma concentrations (Cpavg,free) for a given TKI (Table 1). However, comparing efficacy endpoints from different clinical trials comes with many caveats, where differences in trial design, such as patient selection criteria, can override differences attributable to test drug. Indeed, the need for biomarker-based patient selection of the most likely responders to a given therapy is receiving intense investigation as a key lever to potentially alter clinical outcomes for the better. The focus of this analysis on drug efficiency addresses the orthogonal key lever for impact on clinical results, maximizing target modulation. For this reason, trial designs that similarly selected patients with RCC of predominantly clear cell histology were evaluated to connect efficiency with mechanism-associated PFS and incidence of observed hypertension. This effort was aided greatly by recent summaries that compiled results from similar advanced RCC first-line or mostly first-line clinical trials (1, 2, 29). Another source of PFS and hypertension results comes from reports of similar clinical trials conducted in the very specific cytokine refractory RCC patient population (22, 30, 31). Confidence in comparisons is greatly strengthened in this latter patient population by a recent controlled phase III comparison study of axitinib and sorafenib (4). As an indication that different levels of VEGFR TK inhibition in the clinic are detected across TKIs, PFS in both patient settings was found to positively correlate with hypertension incidence (Fig. S3). Indeed, PFS has been shown previously to correlate with hypertension within studies of single VEGFR TKIs, where varying levels of VEGF blockade are believed to cause both endpoints to move in concert, prompting dose titration trial designs [e.g., tivozanib (32) and axitinib (33)]. To probe the link between drug efficiency and clinical effects associated with the target, the LipE indices were analyzed against the PFS endpoints, revealing convincing correlations in the cytokine refractory RCC setting (R = 0.86, P = 0.06) and the mostly first-line RCC setting (R = 0.97, P = 0.007) (Fig. 7). These results strongly implicate the extent of VEGF blockade as the determining factor in the differences observed for PFS and hypertension across the TKIs.
Fig. 7.
A shows the PFS results from cytokine refractory RCC trials (PFS1) plotted against plus-JM LipE. B shows PFS results from mostly first-line RCC studies (PFS2) plotted against plus-JM LipE.
Our findings highlight the importance of identifying and using the most physiologically relevant construct of a target kinase, including phosphorylation state and regulatory domains, to fully elucidate important binding modes and the consequential on-target potency and selectivity. The nonphosphorylated plus-JM VEGFR2 TK construct provided data consistent with other measurements, from in vitro potencies that mirrored results obtained in whole-cell assays to crystal structures that provided the basis for JM domain effects on TKI binding, to the magnitude of clinical effect derived from VEGF blockade. Understandably, the minus-JM construct was exposed to have deficiencies. Binding mode analysis across the TKI panel revealed that the greatest potency and drug efficiency was garnered from interactions that worked with a natural autoinhibitory state of the enzyme. Applying this learning to other target proteins prospectively in drug discovery will require the early identification of regulatory conformations so that drug mechanism of action can be aligned with inherent regulation. Directing drug design to potent interactions with such a conformation could garner significant efficiency advantages, which was shown in the VEGFR TKI class; it is an important principle to consider strategically in drug discovery efforts.
Materials and Methods
The following reagents were purchased from Sigma Chemical: poly(Glu4Tyr), lactate dehydrogenase, pyruvate kinase, phosphoenolpyruvate, Hepes, DTT, polyethylene glycol, sodium citrate, ammonium sulfate, 2-methyl-1,3-propanediol, MgCl2, ATP, tris(2-carboxyethyl)phosphine (TCEP), and NADH.
Expression and Purification of Proteins.
The expression and purification of the VEGFR2 protein encompassing the catalytic domain (residues 806–1171; minus-JM) and the catalytic domain with the JM domain (residues 786–1171; plus-JM) have been previously described (15). Both the minus-JM and plus-JM constructs have a deletion of the kinase insert domain (residues 940–989) and one point mutation (E990V). The kinases used for selectivity screening were fusion proteins (GST and hexahistidine) of kinase proteins, and they typically encompass the sequence from catalytic domain to the C-terminal end. The proteins were typically baculovirally expressed in insect cells.
Enzymatic Assays.
The spectrophotometric-coupled enzymatic assay used to measure VEGFR2 enzymatic activity was as described (15). Ki determinations were made from a plot of the fractional velocity as a function of inhibitor concentration fit to the Morrison equation with the enzyme concentration as a variable (34, 35). Selectivity screening was done by Carna Biosciences with the Caliper LabChip3000 assay (Caliper Life Science). Compounds were prepared in 100% (vol/vol) DMSO, diluted to 25% (vol/vol) DMSO with 20 mM Hepes, pH 7.5, and added to the reaction for a final DMSO concentration of 6%. Inhibitor concentrations varied from 1.0 to 0.00003 μM. Typical reactions were 20 μL and contained 120 ng/mL (1.24 nM) KDR, 75 μM ATP (Km,ATP = 74 μM), 1.0 μM CSKtide peptide (KKKKEEIYFFFG), 5 mM MgCl2, 2 mM DTT, 0.01% Triton X-100, and 6.25% (vol/vol) DMSO in 20 mM Hepes, pH 7.5. The mixture was incubated in a 384-well polypropylene plate at room temperature for 1 h and terminated by the addition of 60 μL QuickScout Screening Assist MSA Buffer (Carna Biosciences). The reaction mixture was applied to a LabChip3000 system, and the product/substrate peptide peaks were separated. The kinase reaction was quantified by the product ratio peak heights of product (P) and substrate (S) peptides [P/(P + S)]. The selectivity screening assays were primarily mobility shift assays, with a smaller subset using either ELISA or immobilized metal ion affinity-based fluorescence polarization assay (36) (Molecular Devices). ELISAs were BMPR1A, BRAF, BRAF(V600E), COT, DLK, LIMK1, LKB1, MAP2K1/2/3/5/6/7, MAP3K1/2/3/4/5, MLK1/2/3, MOS, PLK4, RAF1, TAK1, TTK, and WEE1. The following kinase assays were in the ion affinity-based fluorescence polarization assay format: PEK, PKN1, PKR, PLK2, and SRPK1. Screening was performed at a Km concentration of ATP for between 1 and 5 h, dependent on the individual kinase. Each assay had a positive control, staurosporine, or other. All assays contained between 5 and 10 mM Mg2+, with a small subset also including Mn2+, Ca2+, or K+.
Cellular Assays.
The VEGF-dependent cell survival assay in HUVEC has been previously described (15). VEGF-induced autophosphorylation (pVEGFR2) was measured by ELISA in HUVEC cells. Cells (passage < 6) were seeded at 20,000 per well in 96-well plates containing endothelial growth medium (Lonza) for 1 d. The cells were then serum-starved (endothelial growth medium plus 0.1% FBS and 0.2% BSA) overnight. The cells were treated with compounds formulated in 0.1% DMSO in the starvation medium at 1:3 serial dilution in duplicates for 1 h at 37 °C plus CO2. The cells were stimulated with 50 ng/mL rhVEGF (R & D Systems) for 5 min. The positive control cells received VEGF only without compounds, and the negative control cells received starvation medium only without VEGF or compounds. After VEGF stimulation, the cells were lysed in 100 μL/well in cell lysis buffer (Cell Signaling) for 25 min (at 4 °C cold room). The cell lysates were transferred to pVEGFR2 ELISA plates (Cell Signaling), and the procedure was followed by the pVEGFR2 ELISA protocol (Cell Signaling). The VEGF-induced autophosphorylation (pVEGFR2) was measured at 450 nm on EN Vision plate reader.
Crystal Structure Determinations.
Coordinates and methods for the crystal structures shown here have been deposited to the Protein Data Bank (PDB) with the accession ID codes listed in Table S1. Crystals were obtained by the hanging drop vapor diffusion method at 13 °C using precipitant solutions containing approximately (i) plus-JM complexes: 0.2 M sodium citrate (pH 6.0–6.5) and 14–21% (wt/vol) polyethylene glycol molecular weight 3,350 and (ii) minus-JM complex with axitinib: 0.1 M Hepes, pH 7.5, 0.2 M ammonium sulfate, 5% (vol/vol) 2-methyl-1,3-propanediol, 60 mM β-mercaptoethanol, and 15–20% (wt/vol) polyethylene glycol molecular weight 6,000. X-ray data and refinement statistics are listed in Table S1. X-ray data for the plus-JM complexes were collected using (i) sorafenib: a Rigaku rotating anode source; (ii) axitinib: advanced light source synchrotron beamline 5.0.2; (iii) sunitinib: Advanced Photon Source LS-CAT beamline; and (iv) tivozanib: Advanced Photon Source beamline 17-ID. Data for the minus-JM–axitinib complex were collected using a Rigaku rotating anode source. Crystallographic software packages used included (i) data processing: HKL2000 and autoPROC (37) and (ii) structure solution and refinement: X-plor (38), Refmac (39), CNX (40), Xtalview (41), and Coot (42).
Supplementary Material
Acknowledgments
We thank Robert A. Kumpf and Beth A. Lunney for editorial review of the manuscript. This study was sponsored by Pfizer Inc.
Footnotes
Conflict of interest statement: The authors are Pfizer employees and stockholders or were at the time that this study was conducted. This study was sponsored by Pfizer Inc.
This article is a PNAS Direct Submission.
Data deposition: The crystallography, atomic coordinates, and structure factors have been deposited in the Protein Data Bank, www.pdb.org [PDB ID codes 4AGC (inhibitor complexes axitinib/plus-JM), 4ASD (sorafenib/plus-JM), 4AGD (sunitinib/plus-JM), 4ASE (tivozanib/plus-JM), and 4AG8 (axitinib/minus-JM)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1207759109/-/DCSupplemental.
References
- 1.Albiges L, Salem M, Rini B, Escudier B. Vascular endothelial growth factor-targeted therapies in advanced renal cell carcinoma. Hematol Oncol Clin North Am. 2011;25:813–833. doi: 10.1016/j.hoc.2011.04.006. [DOI] [PubMed] [Google Scholar]
- 2.Coppin C, Kollmannsberger C, Le L, Porzsolt F, Wilt TJ. Targeted therapy for advanced renal cell cancer (RCC): A Cochrane systematic review of published randomised trials. BJU Int. 2011;108:1556–1563. doi: 10.1111/j.1464-410X.2011.10629.x. [DOI] [PubMed] [Google Scholar]
- 3.Roodhart JM, Langenberg MH, Witteveen E, Voest EE. The molecular basis of class side effects due to treatment with inhibitors of the VEGF/VEGFR pathway. Curr Clin Pharmacol. 2008;3:132–143. doi: 10.2174/157488408784293705. [DOI] [PubMed] [Google Scholar]
- 4.Rini BI, et al. Comparative effectiveness of axitinib versus sorafenib in advanced renal cell carcinoma (AXIS): A randomised phase 3 trial. Lancet. 2011;378:1931–1939. doi: 10.1016/S0140-6736(11)61613-9. [DOI] [PubMed] [Google Scholar]
- 5.Empfield JR, Leeson PD. Lessons learned from candidate drug attrition. IDrugs. 2010;13:869–873. [PubMed] [Google Scholar]
- 6.Meanwell NA. Improving drug candidates by design: A focus on physicochemical properties as a means of improving compound disposition and safety. Chem Res Toxicol. 2011;24:1420–1456. doi: 10.1021/tx200211v. [DOI] [PubMed] [Google Scholar]
- 7.Stepan AF, et al. Structural alert/reactive metabolite concept as applied in medicinal chemistry to mitigate the risk of idiosyncratic drug toxicity: A perspective based on the critical examination of trends in the top 200 drugs marketed in the United States. Chem Res Toxicol. 2011;24:1345–1410. doi: 10.1021/tx200168d. [DOI] [PubMed] [Google Scholar]
- 8.Price DA, Blagg J, Jones L, Greene N, Wager T. Physicochemical drug properties associated with in vivo toxicological outcomes: A review. Expert Opin Drug Metab Toxicol. 2009;5:921–931. doi: 10.1517/17425250903042318. [DOI] [PubMed] [Google Scholar]
- 9.Leeson PD, Empfield JR. Reducing the risk of drug attrition associated with physicochemical properties. Annu Rep Med Chem. 2010;45:393–407. [Google Scholar]
- 10.Edwards MP, Price DA. Role of physicochemical properties and ligand lipophilicity efficiency in addressing drug safety risks. Annu Rep Med Chem. 2010;45:381–391. [Google Scholar]
- 11.Wenlock MC, Austin RP, Barton P, Davis AM, Leeson PD. A comparison of physiochemical property profiles of development and marketed oral drugs. J Med Chem. 2003;46:1250–1256. doi: 10.1021/jm021053p. [DOI] [PubMed] [Google Scholar]
- 12.Kania RS. In: Structure-Based Design and Characterization of Axitinib. Kinase Inhibitor Drugs, Drug Discovery and Development. Li R, Stafford JA, editors. New York: Wiley; 2009. pp. 167–201. [Google Scholar]
- 13.Dar ACSK, Shokat KM. The evolution of protein kinase inhibitors from antagonists to agonists of cellular signaling. Annu Rev Biochem. 2011;80:769–795. doi: 10.1146/annurev-biochem-090308-173656. [DOI] [PubMed] [Google Scholar]
- 14.Wodicka LM, et al. Activation state-dependent binding of small molecule kinase inhibitors: Structural insights from biochemistry. Chem Biol. 2010;17:1241–1249. doi: 10.1016/j.chembiol.2010.09.010. [DOI] [PubMed] [Google Scholar]
- 15.Solowiej J, et al. Characterizing the effects of the juxtamembrane domain on vascular endothelial growth factor receptor-2 enzymatic activity, autophosphorylation, and inhibition by axitinib. Biochemistry. 2009;48:7019–7031. doi: 10.1021/bi900522y. [DOI] [PubMed] [Google Scholar]
- 16.Walter M, et al. The 2.7 A crystal structure of the autoinhibited human c-Fms kinase domain. J Mol Biol. 2007;367:839–847. doi: 10.1016/j.jmb.2007.01.036. [DOI] [PubMed] [Google Scholar]
- 17.Mol CD, et al. Structural basis for the autoinhibition and STI-571 inhibition of c-Kit tyrosine kinase. J Biol Chem. 2004;279:31655–31663. doi: 10.1074/jbc.M403319200. [DOI] [PubMed] [Google Scholar]
- 18.Griffith J, et al. The structural basis for autoinhibition of FLT3 by the juxtamembrane domain. Mol Cell. 2004;13:169–178. doi: 10.1016/s1097-2765(03)00505-7. [DOI] [PubMed] [Google Scholar]
- 19.Gajiwala KS, et al. KIT kinase mutants show unique mechanisms of drug resistance to imatinib and sunitinib in gastrointestinal stromal tumor patients. Proc Natl Acad Sci USA. 2009;106:1542–1547. doi: 10.1073/pnas.0812413106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chien E, et al. 2007. US Patent 7,303,893.
- 21.McTigue MA, et al. Crystal structure of the kinase domain of human vascular endothelial growth factor receptor 2: A key enzyme in angiogenesis. Structure. 1999;7:319–330. doi: 10.1016/s0969-2126(99)80042-2. [DOI] [PubMed] [Google Scholar]
- 22.Motzer RJ, et al. Sunitinib efficacy against advanced renal cell carcinoma. J Urol. 2007;178:1883–1887. doi: 10.1016/j.juro.2007.07.030. [DOI] [PubMed] [Google Scholar]
- 23.Card A, et al. High-throughput biochemical kinase selectivity assays: Panel development and screening applications. J Biomol Screen. 2009;14:31–42. doi: 10.1177/1087057108326663. [DOI] [PubMed] [Google Scholar]
- 24.Davis MI, et al. Comprehensive analysis of kinase inhibitor selectivity. Nat Biotechnol. 2011;29:1046–1051. doi: 10.1038/nbt.1990. [DOI] [PubMed] [Google Scholar]
- 25.Bender SL, et al. 2004. International Patent WO 2004/092217.
- 26.Karaman MW, et al. A quantitative analysis of kinase inhibitor selectivity. Nat Biotechnol. 2008;26:127–132. doi: 10.1038/nbt1358. [DOI] [PubMed] [Google Scholar]
- 27.Alton GR, Lunney EA. Targeting the unactivated conformations of protein kinases for small molecule drug discovery. Expert Opin Drug Discov. 2008;3:595–605. doi: 10.1517/17460441.3.6.595. [DOI] [PubMed] [Google Scholar]
- 28.Harmange J-C, et al. Naphthamides as novel and potent vascular endothelial growth factor receptor tyrosine kinase inhibitors: Design, synthesis, and evaluation. J Med Chem. 2008;51:1649–1667. doi: 10.1021/jm701097z. [DOI] [PubMed] [Google Scholar]
- 29.Motzer RJ, et al. Axitinib second-line therapy for metastatic renal cell carcinoma (mRCC): Five-year (yr) overall survival (OS) data from a phase II trial. J Clin Oncol. 2011;29(Suppl):4547. [Google Scholar]
- 30.Sternberg CN, et al. Pazopanib in locally advanced or metastatic renal cell carcinoma: Results of a randomized phase III trial. J Clin Oncol. 2010;28:1061–1068. doi: 10.1200/JCO.2009.23.9764. [DOI] [PubMed] [Google Scholar]
- 31.Escudier B, et al. Sorafenib for treatment of renal cell carcinoma: Final efficacy and safety results of the phase III treatment approaches in renal cancer global evaluation trial. J Clin Oncol. 2009;27:3312–3318. doi: 10.1200/JCO.2008.19.5511. [DOI] [PubMed] [Google Scholar]
- 32.De Luca A, Normanno N. Tivozanib, a pan-VEGFR tyrosine kinase inhibitor for the potential treatment of solid tumors. IDrugs. 2010;13:636–645. [PubMed] [Google Scholar]
- 33.Rini BI, et al. Diastolic blood pressure as a biomarker of axitinib efficacy in solid tumors. Clin Cancer Res. 2011;17:3841–3849. doi: 10.1158/1078-0432.CCR-10-2806. [DOI] [PubMed] [Google Scholar]
- 34.Morrison JF. Kinetics of the reversible inhibition of enzyme-catalysed reactions by tight-binding inhibitors. Biochim Biophys Acta. 1969;185:269–286. doi: 10.1016/0005-2744(69)90420-3. [DOI] [PubMed] [Google Scholar]
- 35.Murphy DJ. Determination of accurate KI values for tight-binding enzyme inhibitors: An in silico study of experimental error and assay design. Anal Biochem. 2004;327:61–67. doi: 10.1016/j.ab.2003.12.018. [DOI] [PubMed] [Google Scholar]
- 36.Sportsman JR, Gaudet EA, Boge A. Immobilized metal ion affinity-based fluorescence polarization (IMAP): Advances in kinase screening. Assay Drug Dev Technol. 2004;2:205–214. doi: 10.1089/154065804323056549. [DOI] [PubMed] [Google Scholar]
- 37.Vonrhein C, et al. Data processing and analysis with the autoPROC toolbox. Acta Crystallogr D Biol Crystallogr. 2011;67:293–302. doi: 10.1107/S0907444911007773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brunger AT. X-PLOR Version 3.1. A System for X-Ray Crystallography and NMR. New Haven, CT: Yale Univ Press; 1992. [Google Scholar]
- 39.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 40.Brünger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 41.McRee DE. A visual protein crystallographic software system for X11/Xview. J Mol Graph. 1992;10:44–46. [Google Scholar]
- 42.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.