Abstract
The dispersal of modern humans across the globe began ∼65,000 y ago when people first left Africa and culminated with the settlement of East Polynesia, which occurred in the last 1,000 y. With the arrival of Polynesian canoes only 750 y ago, Aotearoa/New Zealand became the last major landmass to be permanently settled by humans. We present here complete mitochondrial genome sequences of the likely founding population of Aotearoa/New Zealand recovered from the archaeological site of Wairau Bar. These data represent complete mitochondrial genome sequences from ancient Polynesian voyagers and provide insights into the genetic diversity of human populations in the Pacific at the time of the settlement of East Polynesia.
Keywords: ancient DNA, colonization, origins, next generation sequencing, hybridization capture
The arrival of Polynesian colonists in Aotearoa/New Zealand only 750 y ago represents the end of the last major human migration event: the settlement of East Polynesia (1). The question of Polynesian origins has been debated since Europeans first arrived in the Pacific, and the genetic evidence for their origins and migration pathway continues to be debated today (2–6). Identification of a combination of mitochondrial DNA (mtDNA) mutations initially referred to as the “Polynesian motif” found at high frequency in Polynesian populations (7) has provided a marker for tracking the origins of this last major human expansion.
Despite the interest in identifying Polynesian origins, there has been little research on mtDNA variation within Polynesia. Studies focusing in the hypervariable region (HVR) of the mitochondrial genome have reported a near-ubiquitous distribution of the so-called Polynesian motif, now recognized as defining haplogroup B4a1a1a, within the Polynesian Triangle (3, 8). The lack of genetic diversity generally, including in the HVR, of modern Polynesians (ref. 9 and references cited therein) is typically hypothesized to be a reflection of a general lack of diversity in the founding population. This limited diversity has been explained as the combined result of multiple successive population bottlenecks that occurred during colonizing voyages and of the speed and recency of colonization (9). It is well known, however, that Polynesian populations—and East Polynesian populations in particular—were decimated by the introduction of European diseases at the time of, and following, initial European contact (10, 11). Thus, the lack of modern genetic variation in Polynesia may not necessarily reflect that of the colonizing populations.
Recent analyses of complete mitochondrial genomes have identified numerous new variants within the B4a1a1a haplogroup (2, 12), including a “Malagasy” motif. Madagascar, like East Polynesia, was settled within the last millennium or so by peoples speaking Austronesian languages (13). Significant mtDNA variation was also indicated in a recent study of both ancient and modern mtDNA in the Gambier Islands, in French Polynesia, where new variants of the B4a1a1a haplotype were reported as well as several ancient and modern individuals with the Q1 haplotype (14). Although a recent study by Benton et al. (15) found reduced mtDNA variation in a sample of 20 modern Maori compared with other worldwide populations, they still identified 11 individual haplotypes, including three previously unidentified lineages within the B4a1a1 haplotype. Furthermore they identified what they suggest are three unique Maori mutations (1185T, 4769G, and 16126C; sequence data not available from GenBank at the time of submission). These combined results suggest that we may be underestimating the mtDNA variation in the founding populations of Polynesia. Such variation could be critical for reconstructing the process of settlement of the Polynesian Triangle, for better understanding the genetic history of the region including the impact of infectious disease and other aspects of natural selection and ultimately for identifying the origins of the Polynesians. It might also provide key information regarding social organization and kinship structure in the founding populations of East Polynesia.
Here we present complete mitochondrial genome sequences from Polynesian voyagers that lived at the time of the settlement of East Polynesia. The ancient DNA (aDNA) evidence was obtained from the human remains excavated from the Wairau Bar archaeological site. The Wairau Bar site, located on the northeast coast of the South Island of New Zealand, is one of the earliest and best-dated sites in New Zealand and contains the earliest known burial grounds (Fig. S1). Excavations of the site undertaken from the 1940s through the 1960s identified three distinct burial groups, from which 42 individual burials were identified. These human remains and many of the artifacts recovered from the site were held at the Canterbury Museum as part of its permanent collection until 2009, when they were repatriated to Wairau Bar and Te Runanga a Rangitane o Wairau (Rangitane), the tribal group who have ancestral ties to and guardianship status over Wairau Bar (16, 17).
The Wairau Bar site is a large village site securely dated to A.D. 1285–1300 (17, 18). The rich artifactual assemblages of distinctive Archaic East Polynesian form are directly associated with several of the burials, particularly those in burial group 1, and are typical of the early colonization phase of Aotearoa/New Zealand (19). A large number of moa bones and eggs were also associated with some of these burials. It has been argued that moa (Aves: Dinornithiformes), the large, flightless birds endemic to New Zealand, became extinct within about a century of human arrival (20). Therefore, their presence in the burials also strongly suggests that these burials represent either a founding population or are at most one that was only a few generations removed from the founding population of Aotearoa/New Zealand. Similarly, a recently excavated oven at Wairau Bar contained the remains of moa, sea lion, fur seal, elephant seal, and bones of the extinct Haast’s Eagle (17), which were all consistent with the very early prehistoric/founding period occupation of the site. The aDNA recovered from the Wairau Bar individuals is therefore likely to represent that of the founding colonists of Aotearoa/New Zealand and also provides key evidence regarding the minimum variation present in the immediate ancestral population in central East Polynesia.
Results
DNA Preservation and Sequence Recovery.
Of the 19 burials screened for DNA preservation, 4 provided sufficient sequence data for downstream analyses. These included burials 1, 2.1, 16A, and 18. Burial 1 represents the remains of a young to middle-aged female; burial 2.1 was a young adult male; burial 16A was a young adult female; and Burial 18 was a young to middle-aged female (16). Complete or nearly complete mitochondrial genomes were obtained from burials 1 (2,903 total mapped reads, 98.4% covered at 11.4-fold mean coverage) and 2.1 (1,199 total mapped reads, 99.3% covered at 6.9-fold mean coverage), and partial mitochondrial genome sequences could be recovered from burials 16A (128 total mapped reads, 29.3% covered at 0.8-fold mean coverage) and 18 (261 total mapped reads, 56.5% covered at 1.2-fold mean coverage) (Fig. S2). Sequence data have been deposited in the GenBank database.
Haplotype Assignments.
For burials 1, 2.1, and 16A, we were able to identify the mtDNA haplotype to the highest resolution. Burial 1 and 16A could be unambiguously identified as belonging to haplogroup B4a1a1a3, a haplogroup present in 7 of the 20 individuals studied by Benton et al. (15). Burial 2.1 was identified as belonging to B4a1a1a, although with two unique mutations at positions 4,917 and 8,790. The B4a1a1a haplogroup was only found in three individuals by Benton et al. (15). Burial 18 could be identified to the level of B4a1a1, but single nucleotide polymorphisms (SNPs) required for more fine-scale identification were not covered. Burials 1 and 2.1 differ by seven mutations, and burials 1 and 16A differ by at least one mutation (Table S1 and Table S2). All burials showed specific SNPs identified by Benton et al. (15) as unique to Maori. Burials 1 and 16A display the 1185T mutation; burials 2.1 and 18 display 4769G. Although two conflicting reads with 4769A were identified in burial 2.1 (Table S1), this result is consistent with expected aDNA deamination damage.
Sequence Authenticity.
Although strict precautions to avoid contamination of the laboratory experiments with present-day human DNA were taken, the teeth from which DNA was extracted were part of a museum collection for >50 y and may have been contaminated before entering our cleanroom facility. To estimate the level of contamination, we used five approaches.
First, throughout all experiments, the samples were handled alongside three no-template controls. None of these controls contained any reads mapping to the human mitochondrial genome, indicating no human contamination in our reagents.
Second, after identifying specific haplotypes, we evaluated the consistency of all haplotype-defining SNPs in the consensus sequences to determine whether more than one biological source could have contributed to the obtained consensus sequence (Table S1). None of the 68 haplotype-defining SNPs covered across all four analyzed burials was in unambiguous conflict with what was expected for the respective haplotype. However, we did identify two ambiguities. Position 263 in burial 2.1 was covered by two reads, one with the expected G, the other one with an A. The same combination could be observed for position 750 in burial 16A (Table S1). Both ambiguities are consistent with deamination damage.
Third, we evaluated the number of individual sequence reads that conflicted with the consensus sequences to identify potential contaminating molecules (Table S1). At 8 of the 68 informative sites (covered by a total of 438 reads), we found 15 reads that conflicted with the respective consensus sequence. All of these conflicting reads were either C to T or G to A mutations and are therefore consistent with deamination damage.
Fourth, we investigated the presence of increased C–T and G–A mutations at the 5′ and 3′ ends of all sequenced molecules. These mutations are characteristic for deamination-damaged ancient molecules and can be used to distinguish them from modern contamination. All four burials showed characteristic aDNA damage patterns at the sequence ends (Fig. S3). The observed C–T misincorporation frequency of ∼0.28–0.32 on the first base of the 5′ ends of all four samples is in good accordance with a rate of 0.1–0.3 (maximum of 0.4) “predicted” by Sawyer et al. (figure 4 in ref. 21) for samples between 500 and 2,000 y of age.
Fifth, we evaluated whether bait DNA could have been sequenced. We identified 19 positions at which the bait (haplotype T2b) differs from the identified sample haplotypes (Table S3). Of the 365 reads covering these informative positions, seven reads were consistent with the bait rather than the target. Two of those reads can be explained by DNA damage (burial 18 positions 1,888 and burial 2.1 position 14,905). However, position 4,917 in burial 2.1 is covered by five reads, all of which are consistent with T2b (G) rather than B4a1a1a (A). The mutation cannot be explained by deamination damage. Given the virtual absence of any other bait contamination, we are confident that this is a genuine mutation in burial 2.1.
Further measures of postmortem DNA degradation including the fragment length distribution and the base frequency at strand breaks were analyzed and were consistent with highly degraded DNA (Fig. S4 and Fig. S5).
Discussion
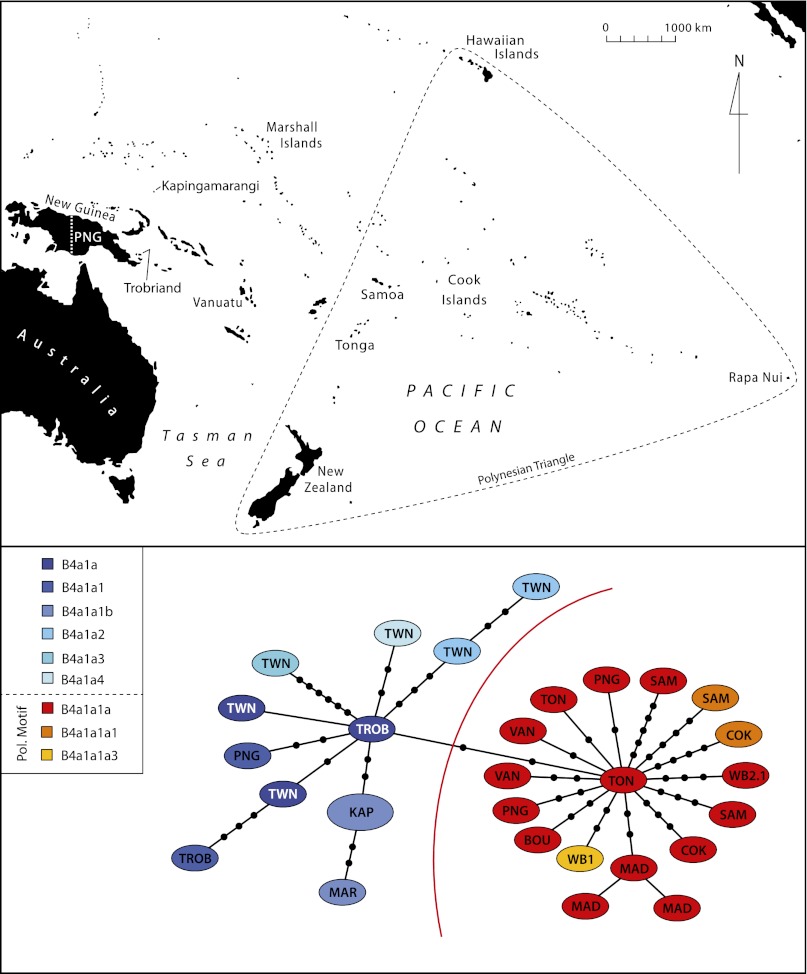
A recent reevaluation of the dates for the colonization of East Polynesia suggests that, contrary to earlier studies positing a relatively long (2,000 y) chronology for the region, the settlement of most of East Polynesia occurred rapidly, in the period from A.D. ∼1190–1290 (22). The authors determined that the expansion event occurred from the Society Islands, which were only settled 70–265 y previously. This rapid and recent expansion event, they argue, explains the “remarkable uniformity of East Polynesian culture, human biology and language” (22). We report here complete ancient mitochondrial genomes from Polynesia, which likely represent the founding population of New Zealand and which date almost directly to this period of rapid expansion and settlement. Far from being uniform, we identified three distinct mtDNA haplotypes in the four burials from which we obtained sequence data. When combined with the increasing number of complete mitochondrial genomes from the Pacific and related populations (e.g., Madagascar), a more complex picture is emerging.
Although all published mtDNA genomes sequenced to date from Maori belong to some branch on the B4a1a lineage, a recent analysis of ancient and modern mtDNA from the Gambier Islands in French Polynesia identified individuals with haplotype Q1, including one ancient sample dated to 1400–1450 cal A.D. (14). This haplotype has also been reported from modern Cook Island samples (6, 23, 24) and thus was likely to have existed in the source population for the East Polynesian expansion. It is possible that Q1 haplotypes were brought to New Zealand as well. Complete mtDNA analyses of these haplotypes in East Polynesia may indicate even more variation in the region.
The four mitochondrial genomes presented here share two of the three unique “Maori” mutations identified by Benton et al. (15): 1185T in burials 1 and 16A and 4769G in burials 2.1 and 18. Although burial 1 has a haplotype already identified by Benton et al. (15), burials 2.1 and 16A have previously unidentified mutations and thus represent unique haplotypes, further increasing the known mtDNA variation within East Polynesia. Burials 1 and 2.1, for which complete mitochondrial genomes were sequenced, differ from the modern Polynesian mitochondrial genomes in GenBank by at least three positions (figure 2 in ref. 15). The Wairau Bar sequences are likely to represent a subset of the total mtDNA variation in the founding population of Aotearoa/New Zealand, which in itself is likely to be a subset of the variation present in the founding populations of East Polynesia.
At least three of the four individuals sequenced from the Wairau Bar site were not recently maternally related. Burials 1 and 2.1 were recovered in the same burial group (Group 1) with similar grave goods, presumed to be of high status (25), yet these two individuals belonged to two different haplotypes (B4a1a1a3 and B4a1a1a) (Fig. 1). This finding indicates that the founding populations were unlikely to be from a single matrilocal source. Furthermore, all four individuals displayed SNPs that have so far been identified only in Maori. It is highly unlikely that such unique SNPs evolved and gained dominance in a population within <50 y. Thus, these SNPs must have been present on the canoes of the New Zealand founding populations. They therefore provide a link to the immediate origins of the first New Zealanders. With more sequence data from contemporaneous Polynesian voyagers, these mutations may allow us to retrace the last major human migration that ended on the shores of Aotearoa/New Zealand.
Fig. 1.
Map of the Pacific with statistical parsimony network of Pacific B4a1a haplotypes. Haplotypes are represented by ellipses. Two haplotypes are connected by a line if they are separated by one mutation; each additional mutation is indicated by a small black circle. The red line represents the separation of Polynesian motif on the right and other B4a1a haplotypes on the left. BOU, Bougainville; COK, Cook Islands; KAP, Kapingamarangi; MAD, Madagascar; MAR, Marshall Islands; PNG, Papua New Guinea; SAM, Samoa; TON, Tonga; TWN, Taiwan; TROB, Trobriand Islands; VAN, Vanuatu; WB, Wairau Bar.
A particularly interesting SNP was identified in one of the Wairau Bar burials. Position 4917 in burial 2.1 was covered by five reads, all of which were consistent and indicate a G rather than the A typically found in the B4a1a1a haplotype. This mutation cannot be explained by deamination damage. While 4917G is a defining mutation for mitochondrial haplogroup T2b—the DNA used as bait in the hybridization enrichment of the Wairau Bar samples—given the virtual absence of any other bait contamination in any of the Wairau Bar samples, we assert that this is a genuine mutation in burial 2.1 (Table S3). This mutation is reported to be associated with insulin resistance (26). Type 2 diabetes (T2D) is a disease found at very high frequency in Maori and other Polynesian populations (27), and at least one other mtDNA mutation, which is a defining mutation for macrohaplogroup B (16189C) has also been associated with T2D in Asian populations (28). Interestingly, the recent analysis of mtDNA variation in modern Maori (15) did not report the 4917G variant, although their sample came from a single tribal group. It would therefore be particularly interesting to undertake further complete mtDNA genome sequencing of samples from Maori and other Polynesian populations to investigate the frequency of this mutation throughout Polynesia and to further assess its possible association with T2D and other metabolic disorders affecting Polynesians. In addition to having the mutations associated with insulin resistance, burial 2.1 showed skeletal evidence indicating that he also suffered from gout (16), another disease in the suite of metabolic disorders affecting modern Polynesians at remarkably high rates (29).
These complete ancient mitochondrial genomes obtained from the first New Zealanders are exciting for a number of reasons. First, the preservation conditions for aDNA in the Pacific region are generally poor, so recovery of reliable aDNA sequences, particularly for human remains, is rare. Our results indicate that the average length of DNA fragments obtained from the Wairau Bar burials was <70 bp and therefore below a reasonable fragment length for PCR amplification and Sanger sequencing (30, 31). The application of next generation sequencing technology and hybridization capture as demonstrated here, however, may mean that Pacific samples previously analyzed using older protocols for which reliable results could not be obtained might yield results in the future. We were very pleased to find that, despite storage and handling conditions that were far from ideal with regard to aDNA preservation and contamination, reasonable-quality DNA could be recovered from 700-y-old samples stored and studied in a museum context. More importantly, the consistency of haplotype-determining SNPs across the respective mitochondrial genomes or genome fragments suggests that each individual sequence derived from a single biological source of Polynesian ancestry. At least three of the four burials differed from each other, suggesting that the sequence data are authentic and that contamination was minimal to nonexistent; thus, the pre-extraction decontamination treatments applied to the Wairau Bar samples and our specific laboratory protocols to avoid contamination were successful. This finding has implications for future aDNA studies of museum samples should those be requested by recognized descendant groups or by museums—for example, as part of repatriation projects. Most importantly, the degree of variation seen in this small, colonizing population from Wairau Bar was beyond what we might have expected based on previous analyses of variation within the HVR of the mtDNA in Maori (8).
The settlement of the Pacific generally and East Polynesia specifically has been described as being the result of a series of successive bottleneck events. As a result, despite limited sampling in East Polynesia to support such a hypothesis, it has been suggested that little to no mtDNA variation would exist in the region—particularly in New Zealand, the last island group to be settled (9). Our results indicate that there was likely to be significant mtDNA variation within the founding population of Aotearoa/New Zealand, even if that variation may be within the B4a1a1a haplogroup. These first complete ancient mitochondrial genomes from Polynesia indicate that further complete mitochondrial genome sequencing from both modern and, where possible, ancient Polynesian populations will be valuable for testing current models of the settlement of Polynesia, which, we suggest, may require reassessment. We have now identified key mtDNA mutations in the founding populations that should allow us to locate the immediate origins of New Zealand Maori and better understand the history of disease susceptibility in Polynesian populations. These findings can perhaps even provide insight into the social and political structure of the founding communities of Aotearoa/New Zealand and of East Polynesia more generally.
Materials and Methods
Wairau Bar Site and Ancient Human Remains.
During repatriation discussions, Rangitane and the Canterbury Museum contacted researchers at the University of Otago regarding the possible archaeological and biological research that could be conducted before and in association with the reburial project. After a discussion regarding potential biological methods of analysis, Rangitane requested that a full macroscopic study of the human remains be undertaken (16) and that tooth and bone samples be taken and archived for aDNA and other biochemical analyses that might provide the tribal group with information about their ancestors.
Of the 42 burials recovered from the site, burials 1–7, or Group 1, were the most intact and best preserved. The remains from Group 1 represent a total of eight individuals, including five males, two females, and one individual of indeterminate sex. Group 2 consists of burials 8–11. Macroscopic analysis of these remains found their preservation to be generally poor. Group 3 includes burials 15–44, and the remains were of variable degrees of apparent preservation (Fig. S1) (16).
DNA Extraction.
All DNA extraction and 454 sequencing library preparation before PCR amplification were conducted in the purpose-built University of Otago aDNA facility (SI Text).
DNA was extracted from 19 individuals from Wairau Bar. These included burials 1, 2.1, 4, 5, 6, and 7 from burial group 1 and burials 12, 13, 14, 16A, 17, 18, 19, 25, 30, 31, 33, 35, and 37 from burial group 3. Because of poor quality, no samples from burial group 2 were processed. One tooth from each individual was made available for initial DNA extraction. Because the material was part of a museum collection and likely to have been extensively handled, the teeth were submerged in 5% bleach (wt/vol) for a minimum of 15 min (32) and rinsed in ultrapure water to reduce surface contamination before grinding for DNA extraction. Teeth were ground by using a sterile mortar and pestle, and 160–550 mg of dentin was used for DNA extraction following the silica based extraction protocol (33). Extractions were conducted in batches of five samples and one extraction blank (repeat extractions account for the total number of samples not being a multiple of five).
Preparation of 454 Sequencing Libraries.
The 454 sequencing libraries were produced directly from the aDNA extract by using truncated, barcoded, custom 454 shotgun adapters as described (34). Extraction blanks were treated as normal samples. Each sample was assigned a unique combination of eight-nucleotide A- and B-adapter barcodes that have never been used for any other study on human DNA in our laboratories. Barcodes differed by at least three nucleotides from any other barcode used. The total number of DNA molecules in the sequencing libraries and the number of PCR cycles to reach plateau were determined by quantitative PCR (qPCR) on a Stratagene MxPro 3000P system using SYBR Green dye (ABI). Products from the qPCR amplifications were run out on 2.5% agarose gels and checked for the presence of sequencing libraries and adapter dimers. Libraries characterized by strong adapter dimers and no visible library were excluded from further analyses because adapter dimers interfere with 454 sequencing and can severely reduce the reads on target. Good libraries with no or few visible adapter dimers included those for burials 1, 2.1, 16A, 18, 25, and 37. These 454 libraries and the respective extraction blanks were then “immortalized” by PCR amplification with tailed primers extending the truncated adapters to full length. To reduce PCR artifacts, the amplifications were stopped once plateau was reached. Immortalized libraries were purified by using Qiagen MinElute purification columns following the manufacturer’s protocol. Purified, immortalized libraries were then once again quantified on a Stratagene MxPro 3000P system to determine PCR cycles to plateau.
In-Solution Hybridization Capture.
Four PCRs of 1-µL immortalized library in a 100-µL reaction volume were performed for each sample to obtain 2 µg of 454 sequencing library per sample. Amplifications were stopped as soon as PCR plateau was reached. Libraries were purified and concentrated by using Qiagen MinElute purification columns following the manufacturer’s protocol. Human mtDNA was enriched in the 454 libraries by in-solution hybridization capture (35) with some modifications. Modern human DNA of a known European haplotype (T2b) was used to produce the hybridization capture probes. In contrast to the published protocol (35), each library was capture-enriched individually and subsequently quantified by qPCR on a Stratagene MxPro 3000P system using SYBR green dye (ABI). In addition, libraries were eluted from the streptavidin beads by heat treatment rather than sodium hydroxide melting (36). Libraries were pooled after capture enrichment according to the copy numbers identified by qPCR and sequenced on a Roche 454 GS Junior high-throughput sequencing platform.
Raw Data Analyses.
We used the program Geneious (37) to separate the sequencing reads by their barcodes and to remove the barcode and adapter sequences. The individual read collections were then assembled against the corrected Cambridge reference sequence (38) by using the Burrows–Wheeler Alignment Tool with the bwasw algorithm, which uses heuristic Smith–Waterman-like alignment to find high-scoring local hits. The Samtools software package (39) was used to remove clonal sequence reads and call the consensus sequence and SNPs. The filtered SNPs output of bcftools (part of the Samtools software package) was then transformed into an input file for the haplogroup assigning tool HaploGrep (http://haplogrep.uibk.ac.at). The HaploGrep output was used to find expected and unexpected haplogroup-defining SNPs as well as private SNPs. The assembly was checked by eye at each informative SNP position to identify sequencing reads conflicting with the consensus sequence. Local and global private mutations were only accepted if the respective position in the assembly was covered by at least three nonconflicting reads with different start and end positions (i.e., clearly molecules of unique origins). We further checked our assemblies for the presence of reads showing haplogroup-defining SNPs of the bait sequence (T2b).
aDNA is usually characterized by increase C–T mutations at the 5′ and increased G–A mutations at the 3′ ends of the molecules. These patterns are caused by deamination damage to the DNA after the death of an individual and can be used to distinguish aDNA from modern contamination (21, 40, 41). To confirm the authenticity of our sequence data, we therefore used the perl script mapdamage 0.3.5 (http://geogenetics.ku.dk/publications/mapdamage/) (42) to identify these characteristic aDNA damage patterns in all mapped sequencing reads. Because the bwasw algorithm ignores poorly matching sequence ends (“soft clipping”), it does not allow for subsequent analysis of such damage patterns. Thus, we used Bowtie 2 (http://bowtie-bio.sourceforge.net/bowtie2/index.shtml), which does not apply soft clipping if not run in the –local mode to reassemble the sequencing reads. To reduce the influence of actual sequence differences between the reference sequence and the target, we mapped the sequencing reads against a reference sequence of identical haplotype. The fragment length distribution plots presented in Fig. S4 were also produced from mapdamage output using the software package R.
Network Reconstruction.
We reconstructed a nonhierarchical statistical parsimony network including the two completely sequenced mitochondrial genomes from this study (burials 1 and 2.1) and the complete mitochondrial genomes of all Pacific B4a1a and derived haplotypes available on GenBank along with those from Madagascar and Taiwan (Fig. 1 and Table S4). Networks were constructed by using the freely available R script TempNet (43).
Supplementary Material
Acknowledgments
We thank Rangitane for support of the research, particularly Judith MacDonald and Richard Bradley; and Chris Jacomb and Emma Brooks (Southern Pacific Archaeological Research, Department of Anthropology and Archaeology, University of Otago) and Roger Fyfe (Canterbury Museum) for their work on the Wairau Bar project. The Wairau Bar research project has been funded by the Allan Wilson Centre for Molecular Ecology and Evolution; a grant-in-aid by the School of Medical Sciences, University of Otago; and a major grant from the Mason Foundation (to Canterbury Museum). The Wairau Bar Koiwi Research Project is a joint initiative between the University of Otago, the Canterbury Museum and Rangitane iwi. The kaitiakitanga (guardianship and protection) of Rangitane over the tupuna (ancestors) discussed in this document is acknowledged by the University of Otago and members of this research group.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequence data reported in this paper has been deposited in the GenBank database (accession no. JX893364–JX893367).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1209896109/-/DCSupplemental.
References
- 1.Kirch PV. On the Road of the Winds: An Archaeological History of the Pacific Islands Before European Contact. Berkeley: Univ of California Press; 2000. [Google Scholar]
- 2.Pierson MJ, et al. Deciphering past human population movements in Oceania: Provably optimal trees of 127 mtDNA genomes. Mol Biol Evol. 2006;23(10):1966–1975. doi: 10.1093/molbev/msl063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Soares P, et al. Ancient voyaging and Polynesian origins. Am J Hum Genet. 2011;88(2):239–247. doi: 10.1016/j.ajhg.2011.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Richards M, Oppenheimer S, Sykes B. mtDNA suggests Polynesian origins in Eastern Indonesia. Am J Hum Genet. 1998;63(4):1234–1236. doi: 10.1086/302043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oppenheimer S, Richards M. Fast trains, slow boats, and the ancestry of the Polynesian islanders. Sci Prog. 2001;84(Pt 3):157–181. doi: 10.3184/003685001783238989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kayser M, et al. Melanesian and Asian origins of Polynesians: mtDNA and Y chromosome gradients across the Pacific. Mol Biol Evol. 2006;23(11):2234–2244. doi: 10.1093/molbev/msl093. [DOI] [PubMed] [Google Scholar]
- 7.Melton T, et al. Polynesian genetic affinities with Southeast Asian populations as identified by mtDNA analysis. Am J Hum Genet. 1995;57(2):403–414. [PMC free article] [PubMed] [Google Scholar]
- 8.Whyte ALH, Marshall SJ, Chambers GK. Human evolution in Polynesia. Hum Biol. 2005;77(2):157–177. doi: 10.1353/hub.2005.0045. [DOI] [PubMed] [Google Scholar]
- 9.Murray-McIntosh RP, Scrimshaw BJ, Hatfield PJ, Penny D. Testing migration patterns and estimating founding population size in Polynesia by using human mtDNA sequences. Proc Natl Acad Sci USA. 1998;95(15):9047–9052. doi: 10.1073/pnas.95.15.9047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McArthur N. Island Populations of the Pacific. Canberra, Australia: Australian National Univ Press; 1967. [Google Scholar]
- 11.Kirch PV, Rallu J-L, editors. The Growth and Collapse of Pacific Island Societies: Archaeological and Demographic Perspectives. Honolulu: Univ of Hawai'i Press; 2007. [Google Scholar]
- 12.Friedlaender JS, et al. Melanesian mtDNA complexity. PLoS ONE. 2007;2(2):e248. doi: 10.1371/journal.pone.0000248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Razafindrazaka H, et al. Complete mitochondrial DNA sequences provide new insights into the Polynesian motif and the peopling of Madagascar. Eur J Hum Genet. 2010;18(5):575–581. doi: 10.1038/ejhg.2009.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deguilloux M-F, et al. Human ancient and extant mtDNA from the Gambier Islands (French polynesia): Evidence for an early Melanesian maternal contribution and new perspectives into the settlement of easternmost Polynesia. Am J Phys Anthropol. 2011;144(2):248–257. doi: 10.1002/ajpa.21398. [DOI] [PubMed] [Google Scholar]
- 15.Benton M, et al. Complete mitochondrial genome sequencing reveals novel haplotypes in a Polynesian population. PLoS ONE. 2012;7(4):e35026. doi: 10.1371/journal.pone.0035026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Buckley H, Tayles N, Halcrow SE, Robb K, Fyfe R. The people of Wariau Bar: A re-examination. Journal of Pacific Archaeology. 2010;1(1):1–20. [Google Scholar]
- 17.Brooks E, Jacomb C, Walter R. Archaeological investigations at Wairau Bar. Archaeology in New Zealand. 2009;52(4):259–268. [Google Scholar]
- 18.Higham T, Anderson A, Jacomb C. Dating the first New Zealanders: The chronology of Wairau Bar. Antiquity. 1999;73(280):420–427. [Google Scholar]
- 19.Wilmshurst JM, Anderson AJ, Higham TFG, Worthy TH. Dating the late prehistoric dispersal of Polynesians to New Zealand using the commensal Pacific rat. Proc Natl Acad Sci USA. 2008;105(22):7676–7680. doi: 10.1073/pnas.0801507105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holdaway RN, Jacomb C. Rapid extinction of the moas (Aves: Dinornithiformes): model, test, and implications. Science. 2000;287(5461):2250–2254. doi: 10.1126/science.287.5461.2250. [DOI] [PubMed] [Google Scholar]
- 21.Sawyer S, Krause J, Guschanski K, Savolainen V, Pääbo S. Temporal patterns of nucleotide misincorporations and DNA fragmentation in ancient DNA. PLoS ONE. 2012;7(3):e34131. doi: 10.1371/journal.pone.0034131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilmshurst JM, Hunt TL, Lipo CP, Anderson AJ. High-precision radiocarbon dating shows recent and rapid initial human colonization of East Polynesia. Proc Natl Acad Sci USA. 2011;108(5):1815–1820. doi: 10.1073/pnas.1015876108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lum JK, Cann RL. mtDNA lineage analyses: origins and migrations of Micronesians and Polynesians. Am J Phys Anthropol. 2000;113(2):151–168. doi: 10.1002/1096-8644(200010)113:2<151::AID-AJPA2>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 24.Sykes B, Leiboff A, Low-Beer J, Tetzner S, Richards M. The origins of the Polynesians: an interpretation from mitochondrial lineage analysis. Am J Hum Genet. 1995;57(6):1463–1475. [PMC free article] [PubMed] [Google Scholar]
- 25.Duff R. The Moa-Hunter Period of Maori Culture. 3rd Ed. Wellington, New Zealand: Government Printer; 1977. [Google Scholar]
- 26.Crispim D, et al. The European-specific mitochondrial cluster J/T could confer an increased risk of insulin-resistance and type 2 diabetes: An analysis of the m.4216T > C and m.4917A > G variants. Ann Hum Genet. 2006;70(Pt 4):488–495. doi: 10.1111/j.1469-1809.2005.00249.x. [DOI] [PubMed] [Google Scholar]
- 27.Elliott RB. Epidemiology of diabetes in Polynesia and New Zealand. In: Levymarchal C, Czernichow P, editors. Epidemiology and Etiology of Insulin-Dependent Diabetes in the Young. Basel: Karger; 1992. pp. 66–71. [Google Scholar]
- 28.Park KS, et al. Study Group of Molecular Diabetology in Asia A mitochondrial DNA variant at position 16189 is associated with type 2 diabetes mellitus in Asians. Diabetologia. 2008;51(4):602–608. doi: 10.1007/s00125-008-0933-z. [DOI] [PubMed] [Google Scholar]
- 29.Buckley HR. Epidemiology of gout: Perspectives from the past. Current Rheumatology Reviews. 2011;7:106–113. [Google Scholar]
- 30.Knapp M, Hofreiter M. Next generation sequencing of ancient DNA: Requirements, strategies and perspectives. Genes. 2010;1:227–243. doi: 10.3390/genes1020227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prüfer K, et al. Computational challenges in the analysis of ancient DNA. Genome Biol. 2010;11(5):R47. doi: 10.1186/gb-2010-11-5-r47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kemp BM, Smith DG. Use of bleach to eliminate contaminating DNA from the surface of bones and teeth. Forensic Sci Int. 2005;154(1):53–61. doi: 10.1016/j.forsciint.2004.11.017. [DOI] [PubMed] [Google Scholar]
- 33.Rohland N, Hofreiter M. Ancient DNA extraction from bones and teeth. Nat Protoc. 2007;2(7):1756–1762. doi: 10.1038/nprot.2007.247. [DOI] [PubMed] [Google Scholar]
- 34.Knapp M, Stiller M, Meyer M. In: Generating Barcoded Libraries for Multiplex High-Throughput Sequencing. Ancient DNA: Methods and Protocols, Methods in Molecular Biology. Shapiro B, Hofreiter M, editors. New York: Humana, Springer; 2012. pp. 155–170. [DOI] [PubMed] [Google Scholar]
- 35.Maricic T, Whitten M, Pääbo S. Multiplexed DNA sequence capture of mitochondrial genomes using PCR products. PLoS ONE. 2010;5(11):e14004. doi: 10.1371/journal.pone.0014004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maricic T, Pääbo S. Optimization of 454 sequencing library preparation from small amounts of DNA permits sequence determination of both DNA strands. Biotechniques. 2009;46(1):51–52, 54–57. doi: 10.2144/000113042. [DOI] [PubMed] [Google Scholar]
- 37.Drummond AJ, et al. 2011. Geneious (Biomatters, Auckland), Version 5.4.
- 38.Andrews RM, et al. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet. 1999;23(2):147. doi: 10.1038/13779. [DOI] [PubMed] [Google Scholar]
- 39.Li H, et al. 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Briggs AW, et al. Patterns of damage in genomic DNA sequences from a Neandertal. Proc Natl Acad Sci USA. 2007;104(37):14616–14621. doi: 10.1073/pnas.0704665104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krause J, et al. A complete mtDNA genome of an early modern human from Kostenki, Russia. Curr Biol. 2010;20(3):231–236. doi: 10.1016/j.cub.2009.11.068. [DOI] [PubMed] [Google Scholar]
- 42.Ginolhac A, Rasmussen M, Gilbert MT, Willerslev E, Orlando L. mapDamage: Testing for damage patterns in ancient DNA sequences. Bioinformatics. 2011;27(15):2153–2155. doi: 10.1093/bioinformatics/btr347. [DOI] [PubMed] [Google Scholar]
- 43.Prost S, Anderson CNK. TempNet: A method to display statistical parsimony networks for heterochronous DNA sequence data. Methods in Ecology and Evolution. 2011;2(6):663–667. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.