Abstract
A number of prokaryotic proteins have been shown to contain nuclear localization signals (NLSs), although its biological role remains sometimes unclear. Terminal proteins (TPs) of bacteriophages prime DNA replication and become covalently linked to the genome ends. We predicted NLSs within the TPs of bacteriophages from diverse families and hosts and, indeed, the TPs of Φ29, Nf, PRD1, Bam35, and Cp-1, out of seven TPs tested, were found to localize to the nucleus when expressed in mammalian cells. Detailed analysis of Φ29 TP led us to identify a bona fide NLS within residues 1–37. Importantly, gene delivery into the eukaryotic nucleus is enhanced by the presence of Φ29 TP attached to the 5′ DNA ends. These findings show a common feature of TPs from diverse bacteriophages targeting the eukaryotic nucleus and suggest a possible common function by facilitating the horizontal transfer of genes between prokaryotes and eukaryotes.
Nuclear localization signals (NLSs) allow proteins to be recognized by the importin/karyopherin pathway and internalized into the eukaryotic cell nucleus (1). A number of NLSs in proteins of prokaryotic origin has been reported. Some of these proteins are produced by pathogenic or endosymbiotic microorganisms to perform or affect some functions inside their hosts (2, 3). However, the biological significance of NLSs from noneukaryotic origin is not well understood, like in the case of Escherichia coli Tus protein (4), phage P1 Cre recombinase (5), and terminal proteins (TPs) of chromosomes and linear plasmids of Streptomyces bacteria genus (6). For these two latter cases, possible roles in horizontal gene transfer (HGT) between distant organisms were suggested (5, 6).
The use of TP-mediated, protein-primed DNA replication has been reported or suggested in replicons from all domains of life, such as archaebacterial viruses, bacteriophages of Gram-positive and Gram-negative eubacterial hosts, Streptomyces chromosomes and plasmids, yeast plasmids, mitochondrial plasmids of plants and fungi, transposable elements, adenoviruses, and the recently described Mavirus virophage (6–11). Besides priming DNA replication, TPs can perform other important roles in vivo. In the case of phage Φ29, the TP is divided into three structural domains (12): (i) the C-terminal domain, which contains the serine 232 priming residue; (ii) the intermediate domain that contributes to the surface of interaction with the DNA polymerase; and (iii) the N-terminal domain (Nt) that is required in vivo for the recruitment of the viral genome (TP-DNA) and DNA polymerase at the bacterial nucleoid (13). The latter biological function seems analogous to that of adenovirus TP, which contains an NLS that directs the nuclear localization of the viral DNA polymerase (14).
The HGT phenomenon has been extensively reported in prokaryotes (15, 16), fungi (17), plants (18), and also humans (19), and it is an accepted driving force of evolution, although with some controversy about its relative contribution (20, 21). Actual transfer events are only rarely observed with unambiguous evidence. Sequence analysis can underestimate the number of transferred genes because sequences acquired from organisms of similar base composition and codon use patterns, as well as ancient transfer events, will escape detection (22). Indeed, some authors have suggested that the current accepted transfers are only the tip of a large iceberg of major evolutionary phenomena (16, 23, 24).
In bacteria, cell transformation, transduction, and conjugation account for most of gene transference (25). However, in eukaryotes the mechanisms for the uptake of genetic material for gene transfer are poorly understood in many cases. Given that many protists are phagotrophs and subsist by eating bacteria, it has been suggested that the evolution of eukaryotic organisms can be explained under the “you are what you eat” hypothesis, by which prokaryotic genes from symbiotic or food bacteria could have replaced ancient eukaryotic genes over evolutionary time (24, 26). In agreement with this theory, the occurrence of HGT in eukaryotes is higher in unicellular organisms that do not require specific germ lines to be reproduced and to spread the newly acquired genes (27). However, inside the eukaryotic cell, the nuclear envelope constitutes a barrier that only small DNA molecules (below 300 bp) can overcome, although the attachment of NLSs has been shown to enhance both nuclear transport and subsequent expression of larger fragments (28).
Here we report a widespread occurrence of predicted NLSs in phage TPs and show that the TPs of Φ29, Nf, PRD1, Bam35, and Cp-1 localize to the mammalian cell nucleus. Moreover, using an in vitro TP-primed amplification system that generates DNA molecules containing the TP covalently linked to the 5′ DNA ends (29), we show that the attached Φ29 TP increases gene delivery with respect to that of a control linear DNA, indicating that the attached TP may facilitate the nuclear translocation step. Altogether, our results show that eukaryotic nuclear targeting of TPs of diverse bacteriophages is a widespread feature of these proteins, supporting a possible role in HGT by transferring genes between prokaryotes and eukaryotes.
Results and Discussion
TPs of Diverse Phages Contain Predicted NLSs and Target the Eukaryotic Nucleus.
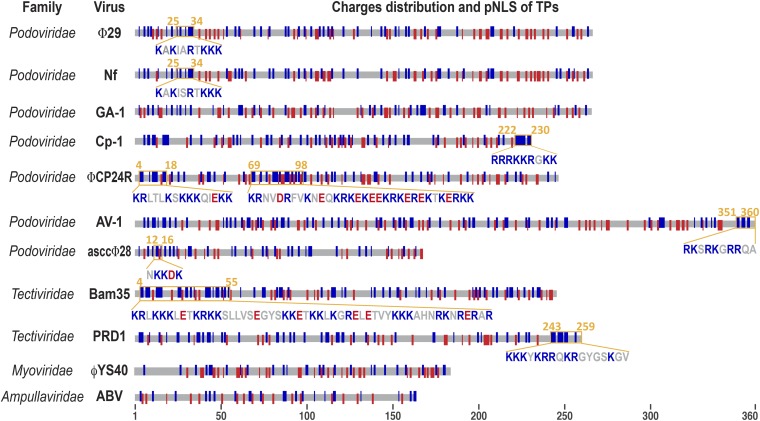
We analyzed annotated TP sequences from all representative types of bacteriophages for which a TP-primed replication mechanism has been proposed (Fig. 1). Except for the case of Φ29, Nf, and GA-1, which belong to the same virus genus, TP sequences do not show any clear sequence similarity, although they share a high proportion of basic residues, some of them grouped in positively charged clusters that, except for the case of ΦYS40 and Acidianus bottle-shaped virus (ABV), are predominant respect to negative charges in the Nt third of the protein. It is well known that both NLSs and DNA binding domains contain a high proportion of basic residues. In this respect, it has been hypothesized that evolution may have used part of the existing DNA binding regions as signals to compartmentalize DNA binding proteins into the nucleus, although different types of NLSs could have had alternative origins (30). However, the number of predicted NLSs in open reading frames (ORFs) from prokaryotes (1–2% respect to total ORFs vs. 8–10% in eukaryotes) (3, 30) seems to be underrepresented in relation to the DNA binding domains [about 10% of total proteins for both prokaryotes and eukaryotes, according to gene ontology annotations (GO:0003677) (31)]. Indeed, some prokaryotic DNA binding proteins such as E. coli LexA or the phage T7 RNA polymerase do not localize to the nucleus by themselves when expressed in eukaryotic cells (32, 33). This indicates that a particular prokaryotic DNA binding domain has clearly less likelihood of containing a predicted NLS than the eukaryotic ones.
Fig. 1.
Schematic representation of the primary structure and pNLS from diverse phage TPs. Protein sequences of Φ29 (11), GA-1 (45), Nf (46), Cp-1 (47), ΦCP24R (48), AV-1 (49), asccΦ28 (50), Bam35 (51), PRD1 (52), ABV (8), and ΦYS40 (53) are represented as gray lines of proportional length. Blue and red bars stand for positive and negative charged amino acid residues, respectively, in their equivalent positions. For reference, the viral family is also indicated. The sequences of the putative NLSs predicted with the NLStradamus server are highlighted in orange and detailed using the blue/red color code for charged residues.
We sought putative NLSs in sequences from representative annotated TPs using the NLStradamus Web server, which predicts NLSs in proteins based upon hidden Markov models (HMMs) statistical approaches and provides a high detection sensitivity with a low false positive rate (34). Predicted NLSs (pNLSs) were found in eight out of 11 TP sequences screened (Fig. 1). The PSORT server (35) found similar patterns, although it could not find any pNLS in the TPs of Φ29-like phages (Table S1).
Although GA-1 and Φ29 TPs share 40% of sequence identity, no NLSs were predicted in GA-1 TP, likely because a “KKK” cluster present within the Φ29 TP pNLS is restricted to a single “K” in GA-1 TP (see next section and Fig. S1). This suggests that the evolutionary role of NLSs in phage TPs is not essential for the phage life cycle. It is also interesting to note that ΦYS40 and ABV phages exist in natural extreme habitats where less eukaryotic biomass is usually found, which might explain the lack of NLSs in their putative TPs. Instead, one or more NLSs were detected in all but one of the analyzed mesophile phages, suggesting that the presence of NLSs may constitute a common feature in TPs.
To validate the in silico predictions, we analyzed the subcellular localization of double YFP fusions to Φ29, Nf, GA-1, PRD1, Bam35, Cp-1, and ABV TPs when expressed in COS-7 mammalian cells (Fig. 2). Except for the case of GA-1 and ABV, all TPs tested conferred a nuclear localization to the fluorescent fusion proteins. Because all of these 2YFP-TP fusions (75–85 kDa) are largely above the accepted nuclear pore exclusion limit (around 50 kDa; ref. 1), the exclusive nuclear localization of the TPs suggests that they are transported by the nuclear active import pathway. Altogether, in silico predictions and experimental results showed that eukaryotic nuclear targeting is widespread in phage TPs as an evolutionary common feature, likely due to the presence of functional NLSs.
Fig. 2.
Nuclear targeting of TPs from diverse phages. Confocal images of representative COS-7 cells expressing the indicated 2YFP fusions to wild-type Φ29, Nf, GA-1, PRD1, Bam35, Cp-1, and ABV TPs, selected among at least five pictures. Shown are YFP (green), DAPI (red), and YFP merged with DAPI images. The 2YFP–ABV construct gave rise to a disperse signal throughout the cell and a large focus of strong signal that might correspond to the aggresome formation reported for similar chimeric fusion proteins (54), although no significant degradation is observed in a Western blot to monitor the expression and stability of YFP fusions (Fig. S5).
Fragment 1–37 Nt of the Φ29 TP Constitutes a Functional Eukaryotic NLS.
A genuine NLS should be transposable and required to confer nuclear localization by an energy-dependent mechanism (1). We verified experimentally that nuclear localization of Φ29 TP in mammalian cells is intrinsic, independently of the type of construction or cells used (Fig. S2 A–C). Importantly, the YFP-TP signal was dispersed throughout the cell by an energy-depletion treatment, showing that an active mechanism is required for its nuclear localization in control conditions (Fig. S2D).
To further estimate the contribution of pNLSs, we mapped the NLS within the phage Φ29 TP. The Φ29 TP Nt domain that contains the pNLS and most of the positively charged residues (Fig. 1) was found to be required for nuclear localization (Fig. S3). Furthermore, the first 37 amino acids of the Φ29 TP confer nuclear localization to a 2YFP fusion protein, whereas the first 24 residues are not sufficient by themselves (Fig. 3B). On the other hand, the Nt (14–37) fusion, which contains the pNLS, shows an intermediate pattern, although with a clear accumulation of fluorescence in the nucleus. In addition, we performed site-directed mutagenesis to study the role of specific residues within the Nt (14–37) region. The mutation R19A had no effect on the subcellular localization of the TP (Fig. 3C), ruling out the role of this residue in the NLS function. The single mutant K27A showed an intermediate phenotype, with only partial accumulation in the nucleus, whereas the double K25A/K27A and the triple K32A/K33A/K34A mutants showed an impaired NLS function. This is in agreement with the fact that the GA-1 TP, which lacks the KKK motif, did not localize to the nucleus. These results indicate that the 25KAK27 and 32KKK34 motifs are required for the NLS function. Thus, it seems likely that the pNLS contributes to the major interaction with the carrier protein, whereas the 1–14 amino acids may constitute a required protein context, as has been found for other proteins (36, 37) and also shown for the specificity of importin-α isoforms (38). Importantly, a TP mutant lacking the first 37 Nt amino acids did not accumulate in the nucleus (Fig. 3B). Altogether, our results show that Φ29 TP contains a bona fide eukaryotic NLS, with residues 1–37 being necessary and sufficient for this function.
Fig. 3.
Identification of a minimal functional NLS in Φ29 TP. (A) Schematic representation of double YFP fusions to Φ29 TP domains and fragments. The first 37 amino acids that contain most (76%) of the positive charges were used as a reference for the NLS mapping. The NLS predicted by NLStradamus is boxed in red. The residues mutated in C are highlighted in navy blue. Confocal images of representative COS-7 cells expressing the indicated fusions to TP-fragments (B) or full-length point mutants (C), selected among at least five different images. Shown are YFP (green), DAPI (red), and YFP merged with DAPI images.
Φ29 TP Enhances Delivery of Linear DNA.
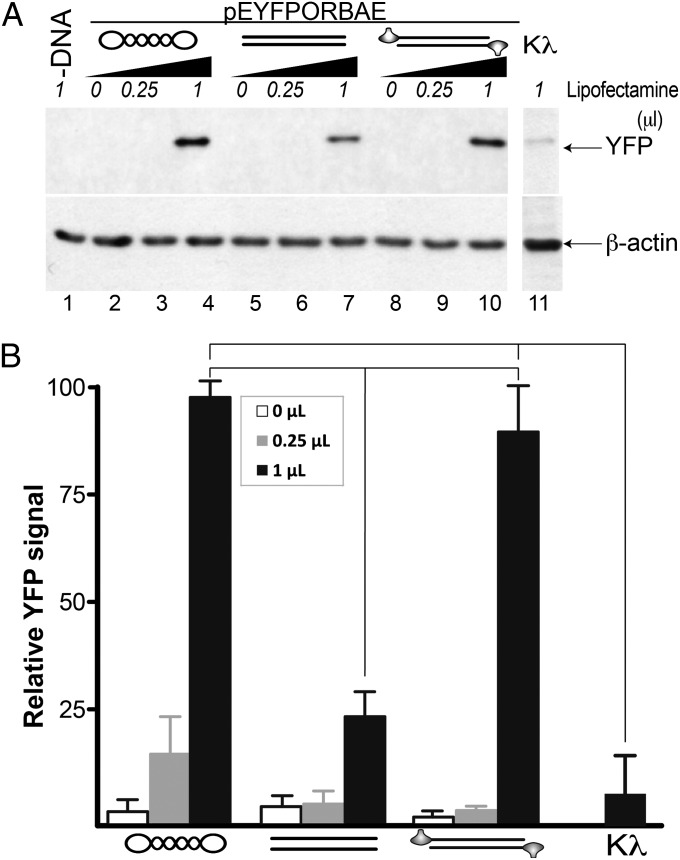
To mediate in HGT, TPs should be able to transport their attached DNA to the eukaryotic cell nucleus. To confirm this, we used a circular DNA (pEYFPORBAE, 29) that contains the yfp gene expression cassette and the Φ29 genome origins. Once linearized, the DNA was amplified in vitro by the phage replication system to incorporate the TP covalently linked at both 5′ DNA ends (Fig. S4). Thus, we transfected COS-7 cells with the circular DNA, the linear DNA amplified by a standard polymerase chain reaction, or the TP-DNA amplified by the Φ29 TP replication system. Fig. 4 shows that bacteria-purified plasmid DNA yielded a higher YFP signal than linear DNA did, probably due to a higher resistance to degradation and to the more compact form of supercoiled DNA (39). Importantly, the YFP expression in cells transfected with the TP-containing DNA was three- to fourfold higher than that of the linear DNA, indicating a stimulation in the presence of Φ29 TP covalently linked to the 5′ DNA ends, which most likely facilitates the nuclear translocation step and may also protect the DNA from degradation (39, 40). To further confirm the role of TP NLS in gene delivery, we treated the TP-DNA with proteinase K, which gives rise to a small acidic peptide attached to the DNA (41). This peptide does not contain the NLS but still can protect from exonuclease degradation (Fig. S4). As shown in Fig. 4, gene delivery was strongly impaired, which further supports a TP-mediated stimulation of nuclear transport by means of its NLS. The lower YFP signal in the proteinase K–treated DNA with respect to the naked linear DNA might be due to the interference of the peptide with a possible recircularization of the linear DNA. This result could provide a hint on the biological role of phage TPs in the ecosystems when infected bacteria interact with eukaryotic cells. In addition, in vitro generation of DNAs with improved gene delivery efficiency may constitute a powerful tool for a number of downstream applications, including transient expression of proteins or gene therapy (29, 42).
Fig. 4.
Φ29 TP linked to the DNA ends enhances gene delivery into eukaryotic cells. (A) Western blot of COS-7 cell extracts transfected with bacteria-purified plasmid pEYFPORBAE (29), linear form of the plasmid, and linear DNA with TP covalently linked at the 5′ ends, represented by simplified sketches. Amplified TP-DNA subsequently treated with proteinase K and λ exonuclease (Kλ) was also used. An anti-β-actin antibody was used as the loading control. The presence of TP does not modify the requirement of transfection reagent (lipofectamine), ruling out an enhancement of the cell internalization step. (B) Normalized YFP signal, obtained from the band densitometry of three independent experiments. As indicated, pairwise statistical tests showed that the relative YFP signal of supercoiled DNA and TP-DNA were significantly different from that of linear DNA as well as from the peptide containing DNA (P < 0.001).
Model of Phage TP-Mediated Prokaryotes–Eukaryotes Gene Transfer.
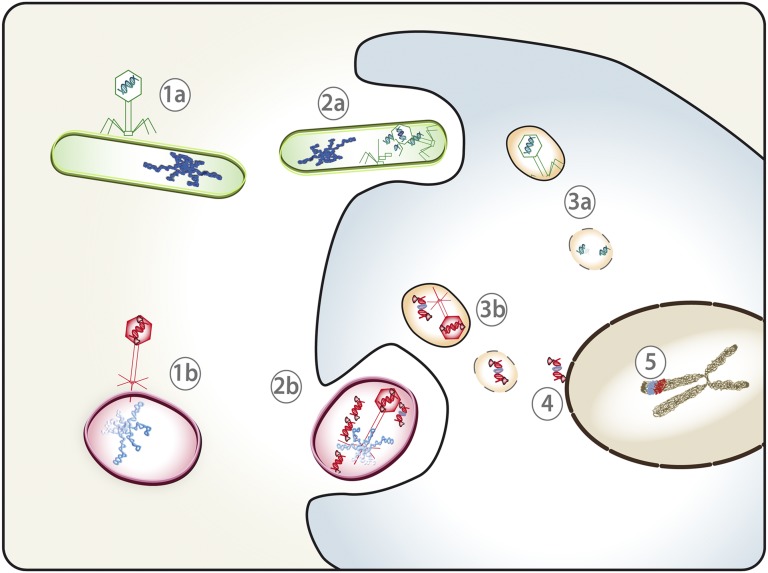
There are known specific mechanisms of prokaryotic–eukaryotic gene transfer, like Agrobacterium T-elements transformation, which also have NLS-containing linked proteins (2), yet they require a specific cellular internalization process. Genome sequencing has undoubtedly identified bacteriophages as substantial forces of bacterial adaptation and genomic divergence because they are highly abundant in all ecosystems and they have important roles in the transference of new biological traits (16). We propose a role of phage TPs in HGT in the ecosystems, by ferrying DNAs between evolutionarily distant organisms (Fig. 5). In a hypothetical simplified scenario, bacteria are infected by bacteriophages whose genome is constituted by DNA (1a) or TP-DNA (1b). Eventually, phages can mediate gene exchanges within different bacteria, thus incorporating foreign DNA into their genomes. As mentioned in the beginning of this article, phagocytosis has been suggested as a mechanism of cell uptake of foreign genes by many protists that are phagotrophs and subsist by eating bacteria (24, 26). In consequence, infected bacteria containing large amounts of viral particles and genomes can be internalized into a eukaryotic cell (2a and 2b). If the phage DNA contains 5′-linked TPs, it can be resistant to exonucleolytic degradation (3b) and, by means of the TP NLS, transported inside the cell nucleus (4). In this way, phage TP-mediated HGT could be a possible mechanism of interkingdom (prokaryotes–eukaryotes) genetic exchanges that, when conferring an advantage, would be fixed in a new lineage (5).
Fig. 5.
Model of TP-mediated HGT. In natural ecosystems, HGT within prokaryotic or archaea organisms is often mediated by phages that might contain (1b, red) or not contain (1a, green) TPs in their genome ends. Infected bacteria contain a large amount of viral particles and genomes, might encounter with eukaryotic cells, and eventually might be internalized by phagocytosis or other processes (2a and 2b). Normally, viral DNA would be degraded inside the cell (3a), but if the phage DNA contains 5′-linked TPs, it may go through lysosomes and endocytosis pathways (3b), subsequently be driven across the nuclear envelope by the NLS of the TP (4) and eventually be incorporated into the cell genome (5). Because phages mediate HGT between different bacteria, they may contribute to the incorporation of genetic information from themselves (red) or from their prokaryotic hosts (blue) to the eukaryotic genome.
Materials and Methods
Constructs Design.
All DNA manipulations and cloning were carried out by standard molecular biology methods (43). Oligonucleotides used for PCR amplifications (Sigma) are listed in Table S2. All DNA plasmid constructs were purified from XL-1 Blue E. coli strain with Promega Wizard Minipreps DNA Purification System and were sequence-verified prior to using.
Φ29, Nf, and GA-1 genomes were from the laboratory stock. Cp-1, PRD1, and Bam35 phage DNA were gifts from Drs. P. García (Centro de Investigaciones Biológicas, CSIC, Madrid, Spain), D. Bamford (Finnish Center of Excellence in Virus Research, Helsinki, Finland), and J. K. Bamford (University of Jyväskyla, Finland), respectively. The codon-optimized sequence of the ORF coding for the ABV putative TP was purchased from GeneScript. Single YFP-TP fusions to be expressed in transient transfection of COS-7 cells were generated by cloning the PCR-amplified TP ORFs into the EcoRI/BamHI sites of pEYFP-C1 or pEYFP-N1 vectors (Clontech). Plasmids expressing the double YFP fusions of TP constructs were generated by cloning the yfp gene into the XhoI/EcoRI sites of previously constructed single YFP plasmids.
Transient Expression and Confocal Analysis of YFP and YFP-TP Fusions.
COS-7 cells were obtained from the American Type Culture Collection and grown in Dulbecco’s modified Eagle medium (DMEM) containing 10% (vol/vol) fetal calf serum (FCS). For transient expression experiments, cells were grown over glass coverslips in multiwell 24 at 20,000 cells/cm2. Transfections were performed with 250 ng of DNA per well and Lipofectamine and Plus Reagent (Invitrogen) according to the manufacturer’s instructions. In the experiments to block active transport, cells expressing fusion proteins (16 h posttransfection) were energy-depleted by incubation for 2 h with 10 mM sodium azide and 10 mM 2-deoxyglucose in DMEM without glucose. Finally, cells were washed with phosphate-buffered saline (PBS) and doubly fixed with 4% paraformaldehyde (15 min at room temperature) plus methanol (5 min at –20 °C) and mounted with Mowiol-DABCO Mounting medium (containing 1 μg/mL 4′,6-diamidino-2-phenylindole, DAPI) on glass slides. Preparations were examined with a LSM510 confocal laser microscope (Zeiss). Images were processed using ImageJ software (44). Samples to monitor protein expression were washed with PBS and harvested directly in SDS/PAGE sample buffer. Western blots were performed with an anti-GFP antibody that cross-reacts with YFP (Invitrogen A-6455).
Effect of the Covalently Linked TP on Gene Delivery.
In the experiments to evaluate the effect of TP on gene delivery, we used the Φ29 minimal amplification system of pEYFPORBAE linearized plasmid (29) to incorporate linked TP at both 5′ DNA ends. A map of plasmid pEYFPORBAE and the result of TP-mediated amplification are shown in Fig. S4. Control linear DNA was amplified by PCR with Herculase II DNA polymerase (Agilent) and diluted in the same reaction mixture but without magnesium to prevent exonuclease degradation. The PCR-amplified product is a linear DNA with 5′-phosphate groups that are required for subsequent Φ29 TP-mediated amplification (29). After amplification, samples were treated with EDTA followed by heat inactivation (10 min at 50 °C) and quantified in agarose gel by comparison with appropriated known standards. Transfections (50 ng DNA/well) were carried out as described above but in the absence of Plus Reagent, which according to manufacturer’s information may enhance the nuclear internalization step. Cells were harvested at 24 h posttransfection, and the YFP signal was monitored by Western blot as indicated above. An anti–β-actin antibody (AC-15, Sigma) was used as the loading control. Bands were quantified with ImageJ software (44) and the relative intensities from three independent experiments were subjected to a two-way analysis of variance statistical test and pairwise Bonferroni posttests with the GraphPad Prism 5 software.
Supplementary Material
Acknowledgments
We thank Dr. J. K. Bamford for DNA of Bam35 phage, Prof. María L. Salas for helpful discussions and Drs. M. de Vega and M. Serrano for critical reading of the manuscript. This work was supported by Grant BFU2011-23645 and Consolider-Ingenio Grant 2010 24717 from the Spanish Ministry of Economy and Competitiveness (to M.S.) and by an institutional grant from Fundación Ramón Areces to the Centro de Biología Molecular “Severo Ochoa.” I.H. is a holder of the Formación de Profesorado Universitario fellowship from the Spanish Ministry of Education.
Footnotes
Conflict of interest statement: A patent application related to this work has been filed for which the authors are inventors.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1216635109/-/DCSupplemental.
References
- 1.Görlich D, Mattaj IW. Nucleocytoplasmic transport. Science. 1996;271(5255):1513–1518. doi: 10.1126/science.271.5255.1513. [DOI] [PubMed] [Google Scholar]
- 2.Gelvin SB. Finding a way to the nucleus. Curr Opin Microbiol. 2010;13(1):53–58. doi: 10.1016/j.mib.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 3.Lee JC, et al. Prediction of bacterial proteins carrying a nuclear localization signal and nuclear targeting of HsdM from Klebsiella pneumoniae. J Microbiol. 2009;47(5):641–645. doi: 10.1007/s12275-009-0217-4. [DOI] [PubMed] [Google Scholar]
- 4.Kaczmarczyk SJ, Sitaraman K, Hill T, Hartley JL, Chatterjee DK. Tus, an E. coli protein, contains mammalian nuclear targeting and exporting signals. PLoS ONE. 2010;5(1):e8889. doi: 10.1371/journal.pone.0008889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Le Y, Gagneten S, Tombaccini D, Bethke B, Sauer B. Nuclear targeting determinants of the phage P1 cre DNA recombinase. Nucleic Acids Res. 1999;27(24):4703–4709. doi: 10.1093/nar/27.24.4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsai HH, Huang CH, Lin AM, Chen CW. Terminal proteins of Streptomyces chromosome can target DNA into eukaryotic nuclei. Nucleic Acids Res. 2008;36(10):e62. doi: 10.1093/nar/gkm1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fischer MG, Suttle CA. A virophage at the origin of large DNA transposons. Science. 2011;332(6026):231–234. doi: 10.1126/science.1199412. [DOI] [PubMed] [Google Scholar]
- 8.Peng X, Basta T, Häring M, Garrett RA, Prangishvili D. Genome of the Acidianus bottle-shaped virus and insights into the replication and packaging mechanisms. Virology. 2007;364(1):237–243. doi: 10.1016/j.virol.2007.03.005. [DOI] [PubMed] [Google Scholar]
- 9.Kapitonov VV, Jurka J. Self-synthesizing DNA transposons in eukaryotes. Proc Natl Acad Sci USA. 2006;103(12):4540–4545. doi: 10.1073/pnas.0600833103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim EK, Jeong JH, Youn HS, Koo YB, Roe JH. The terminal protein of a linear mitochondrial plasmid is encoded in the N-terminus of the DNA polymerase gene in white-rot fungus Pleurotus ostreatus. Curr Genet. 2000;38(5):283–290. doi: 10.1007/s002940000157. [DOI] [PubMed] [Google Scholar]
- 11.Salas M. Protein-priming of DNA replication. Annu Rev Biochem. 1991;60:39–71. doi: 10.1146/annurev.bi.60.070191.000351. [DOI] [PubMed] [Google Scholar]
- 12.Kamtekar S, et al. The phi29 DNA polymerase:protein-primer structure suggests a model for the initiation to elongation transition. EMBO J. 2006;25(6):1335–1343. doi: 10.1038/sj.emboj.7601027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muñoz-Espín D, Holguera I, Ballesteros-Plaza D, Carballido-López R, Salas M. Viral terminal protein directs early organization of phage DNA replication at the bacterial nucleoid. Proc Natl Acad Sci USA. 2010;107(38):16548–16553. doi: 10.1073/pnas.1010530107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao LJ, Padmanabhan R. Nuclear transport of adenovirus DNA polymerase is facilitated by interaction with preterminal protein. Cell. 1988;55(6):1005–1015. doi: 10.1016/0092-8674(88)90245-0. [DOI] [PubMed] [Google Scholar]
- 15.Lang AS, Beatty JT. Importance of widespread gene transfer agent genes in alpha-proteobacteria. Trends Microbiol. 2007;15(2):54–62. doi: 10.1016/j.tim.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 16.Abby SS, Tannier E, Gouy M, Daubin V. Lateral gene transfer as a support for the tree of life. Proc Natl Acad Sci USA. 2012;109(13):4962–4967. doi: 10.1073/pnas.1116871109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marcet-Houben M, Gabaldón T. Acquisition of prokaryotic genes by fungal genomes. Trends Genet. 2010;26(1):5–8. doi: 10.1016/j.tig.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 18.Won H, Renner SS. Horizontal gene transfer from flowering plants to Gnetum. Proc Natl Acad Sci USA. 2003;100(19):10824–10829. doi: 10.1073/pnas.1833775100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lander ES, et al. International Human Genome Sequencing Consortium Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 20.Kurland CG, Canback B, Berg OG. Horizontal gene transfer: A critical view. Proc Natl Acad Sci USA. 2003;100(17):9658–9662. doi: 10.1073/pnas.1632870100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salzberg SL, White O, Peterson J, Eisen JA. Microbial genes in the human genome: Lateral transfer or gene loss? Science. 2001;292(5523):1903–1906. doi: 10.1126/science.1061036. [DOI] [PubMed] [Google Scholar]
- 22.Ochman H, Lawrence JG, Groisman EA. Lateral gene transfer and the nature of bacterial innovation. Nature. 2000;405(6784):299–304. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- 23.Zhaxybayeva O, Doolittle WF. Lateral gene transfer. Curr Biol. 2011;21(7):R242–R246. doi: 10.1016/j.cub.2011.01.045. [DOI] [PubMed] [Google Scholar]
- 24.Doolittle WF, et al. How big is the iceberg of which organellar genes in nuclear genomes are but the tip? Philos Trans R Soc Lond B Biol Sci. 2003;358(1429):39–57, discussion 57–58. doi: 10.1098/rstb.2002.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thomas CM, Nielsen KM. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat Rev Microbiol. 2005;3(9):711–721. doi: 10.1038/nrmicro1234. [DOI] [PubMed] [Google Scholar]
- 26.Doolittle WF. You are what you eat: A gene transfer ratchet could account for bacterial genes in eukaryotic nuclear genomes. Trends Genet. 1998;14(8):307–311. doi: 10.1016/s0168-9525(98)01494-2. [DOI] [PubMed] [Google Scholar]
- 27.Andersson JO. Lateral gene transfer in eukaryotes. Cell Mol Life Sci. 2005;62(11):1182–1197. doi: 10.1007/s00018-005-4539-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ludtke JJ, Zhang G, Sebestyén MG, Wolff JA. A nuclear localization signal can enhance both the nuclear transport and expression of 1 kb DNA. J Cell Sci. 1999;112(Pt 12):2033–2041. doi: 10.1242/jcs.112.12.2033. [DOI] [PubMed] [Google Scholar]
- 29.Mencía M, Gella P, Camacho A, de Vega M, Salas M. Terminal protein-primed amplification of heterologous DNA with a minimal replication system based on phage Φ29. Proc Natl Acad Sci USA. 2011;108(46):18655–18660. doi: 10.1073/pnas.1114397108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cokol M, Nair R, Rost B. Finding nuclear localization signals. EMBO Rep. 2000;1(5):411–415. doi: 10.1093/embo-reports/kvd092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carbon S, et al. AmiGO Hub Web Presence Working Group AmiGO: Online access to ontology and annotation data. Bioinformatics. 2009;25(2):288–289. doi: 10.1093/bioinformatics/btn615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dunn JJ, Krippl B, Bernstein KE, Westphal H, Studier FW. Targeting bacteriophage T7 RNA polymerase to the mammalian cell nucleus. Gene. 1988;68(2):259–266. doi: 10.1016/0378-1119(88)90028-5. [DOI] [PubMed] [Google Scholar]
- 33.Silver PA, Brent R, Ptashne M. DNA binding is not sufficient for nuclear localization of regulatory proteins in Saccharomyces cerevisiae. Mol Cell Biol. 1986;6(12):4763–4766. doi: 10.1128/mcb.6.12.4763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nguyen Ba AN, Pogoutse A, Provart N, Moses AM. NLStradamus: A simple hidden Markov model for nuclear localization signal prediction. BMC Bioinformatics. 2009;10:202. doi: 10.1186/1471-2105-10-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakai K, Horton P. PSORT: A program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem Sci. 1999;24(1):34–36. doi: 10.1016/s0968-0004(98)01336-x. [DOI] [PubMed] [Google Scholar]
- 36.Lischka P, Sorg G, Kann M, Winkler M, Stamminger T. A nonconventional nuclear localization signal within the UL84 protein of human cytomegalovirus mediates nuclear import via the importin α/β pathway. J Virol. 2003;77(6):3734–3748. doi: 10.1128/JVI.77.6.3734-3748.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nunes-Correia I, et al. African swine fever virus p10 protein exhibits nuclear import capacity and accumulates in the nucleus during viral infection. Vet Microbiol. 2008;130(1–2):47–59. doi: 10.1016/j.vetmic.2007.12.010. [DOI] [PubMed] [Google Scholar]
- 38.Friedrich B, Quensel C, Sommer T, Hartmann E, Köhler M. Nuclear localization signal and protein context both mediate importin alpha specificity of nuclear import substrates. Mol Cell Biol. 2006;26(23):8697–8709. doi: 10.1128/MCB.00708-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lechardeur D, Lukacs GL. Nucleocytoplasmic transport of plasmid DNA: A perilous journey from the cytoplasm to the nucleus. Hum Gene Ther. 2006;17(9):882–889. doi: 10.1089/hum.2006.17.882. [DOI] [PubMed] [Google Scholar]
- 40.Xavier J, Singh S, Dean DA, Rao NM, Gopal V. Designed multi-domain protein as a carrier of nucleic acids into cells. J Control Release. 2009;133(2):154–160. doi: 10.1016/j.jconrel.2008.09.090. [DOI] [PubMed] [Google Scholar]
- 41.Hermoso JM, Méndez E, Soriano F, Salas M. Location of the serine residue involved in the linkage between the terminal protein and the DNA of phage phi 29. Nucleic Acids Res. 1985;13(21):7715–7728. doi: 10.1093/nar/13.21.7715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tolmachov O, Coutelle C. Covalent attachment of multifunctional chimeric terminal proteins to 5′ DNA ends: A potential new strategy for assembly of synthetic therapeutic gene vectors. Med Hypotheses. 2007;68(2):328–331. doi: 10.1016/j.mehy.2006.06.055. [DOI] [PubMed] [Google Scholar]
- 43.Sambrook J, Russell D. Molecular Cloning: A Laboratory Manual. 4th Ed. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 2001. [Google Scholar]
- 44.Rasband WS. ImageJ. Bethesda, MD: National Institutes of Health; 1997–2011. [Google Scholar]
- 45.Illana B, Blanco L, Salas M. Functional characterization of the genes coding for the terminal protein and DNA polymerase from bacteriophage GA-1. Evidence for a sliding-back mechanism during protein-primed GA-1 DNA replication. J Mol Biol. 1996;264(3):453–464. doi: 10.1006/jmbi.1996.0653. [DOI] [PubMed] [Google Scholar]
- 46.Longás E, Villar L, Lázaro JM, de Vega M, Salas M. Phage phi29 and Nf terminal protein-priming domain specifies the internal template nucleotide to initiate DNA replication. Proc Natl Acad Sci USA. 2008;105(47):18290–18295. doi: 10.1073/pnas.0809882105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.García P, et al. Formation of a covalent complex between the terminal protein of pneumococcal bacteriophage Cp-1 and 5′-dAMP. J Virol. 1986;58(1):31–35. doi: 10.1128/jvi.58.1.31-35.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morales CA, et al. Complete genome sequence of the podoviral bacteriophage ΦCP24R, which is virulent for Clostridium perfringens. Arch Virol. 2012;157(4):769–772. doi: 10.1007/s00705-011-1218-2. [DOI] [PubMed] [Google Scholar]
- 49.Delisle AL, Barcak GJ, Guo M. Isolation and expression of the lysis genes of Actinomyces naeslundii phage Av-1. Appl Environ Microbiol. 2006;72(2):1110–1117. doi: 10.1128/AEM.72.2.1110-1117.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kotsonis SE, et al. Characterization and genomic analysis of phage asccphi28, a phage of the family Podoviridae infecting Lactococcus lactis. Appl Environ Microbiol. 2008;74(11):3453–3460. doi: 10.1128/AEM.02379-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ravantti JJ, Gaidelyte A, Bamford DH, Bamford JK. Comparative analysis of bacterial viruses Bam35, infecting a gram-positive host, and PRD1, infecting gram-negative hosts, demonstrates a viral lineage. Virology. 2003;313(2):401–414. doi: 10.1016/s0042-6822(03)00295-2. [DOI] [PubMed] [Google Scholar]
- 52.Hsieh JC, Jung GH, Leavitt MC, Ito J. Primary structure of the DNA terminal protein of bacteriophage PRD1. Nucleic Acids Res. 1987;15(21):8999–9009. doi: 10.1093/nar/15.21.8999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Naryshkina T, et al. Thermus thermophilus bacteriophage phiYS40 genome and proteomic characterization of virions. J Mol Biol. 2006;364(4):667–677. doi: 10.1016/j.jmb.2006.08.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.García-Mata R, Bebök Z, Sorscher EJ, Sztul ES. Characterization and dynamics of aggresome formation by a cytosolic GFP-chimera. J Cell Biol. 1999;146(6):1239–1254. doi: 10.1083/jcb.146.6.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.