Abstract
Interneurons are thought to be a primary pathogenic target for several behavioral disorders that arise during development, including schizophrenia and autism. It is not known, however, whether genetic lesions associated with these diseases disrupt established molecular mechanisms of interneuron development. We found that diminished 22q11.2 gene dosage—the primary genetic lesion in 22q11.2 deletion syndrome (22q11.2 DS)—specifically compromises the distribution of early-generated parvalbumin-expressing interneurons in the Large Deletion (LgDel) 22q11.2DS mouse model. This change reflects cell-autonomous disruption of interneuron migration caused by altered expression of the cytokine C-X-C chemokine receptor type 4 (Cxcr4), an established regulator of this process. Cxcr4 is specifically reduced in LgDel migrating interneurons, and genetic analysis confirms that diminished Cxcr4 alters interneuron migration in LgDel mice. Thus, diminished 22q11.2 gene dosage disrupts cortical circuit development by modifying a critical molecular signaling pathway via Cxcr4 that regulates cortical interneuron migration and placement.
Behavioral and psychiatric diseases, including autism and schizophrenia, are considered disorders of cortical circuitry (1–3). Despite variable onset and severity, the available evidence suggests that aberrant neural development leads to inappropriate circuit formation in these disorders. Cortical interneurons—essential regulators of cortical activity and synchrony for complex functions such as social interaction, learning, and memory (4)—are thought to be primary targets for this developmental pathogenesis (5–7). Indeed, the most consistent observations in postmortem cortex from patients with schizophrenia include altered interneuron number, distribution, or gene expression (8, 9). These studies cannot resolve whether such changes reflect developmental causes or pathologic consequences of cortical circuit dysfunction. Moreover, it is not clear whether pathogenic processes that lead to these disorders disrupt established or novel mechanisms that regulate interneuron development. Thus, we determined whether key cellular mechanisms and molecular pathways known to regulate interneuron differentiation are disrupted in the LgDel mouse model (10) of 22q11.2 deletion syndrome (22q11.2DS), a genetic syndrome highly associated with cortical circuit disorders.
The microdeletion at chromosome 22q11.2 that gives rise to 22q11.2DS is one of the most significant genetic risks for cortical circuit disorders (11–13). Postmortem studies of the cerebral cortex from 22q11.2DS patients implicate abnormal neuronal migration (14); imaging demonstrates related cortical anomalies including polymicrogyria and region-selective gray matter thinning (15, 16); and magnetic resonance spectroscopic studies show reduced GABA function (17). Thus, in 22q11.2DS, the available evidence indicates that cortical circuits are compromised and GABAergic interneurons are altered. We now report in the LgDel mouse—in which recent studies (18–20) suggest that cortical circuit assembly and function is compromised—a mechanistic relationship between diminished 22q11.2 dosage and a key molecular pathway that regulates interneuron migration during cortical development. LgDel interneuron phenotypes reflect interneuron-specific disruption of cytokine receptor Cxcr4, which has been shown to influence placement, migratory trajectory, and motility of these cells during cortical development (21–25). Thus, we define a relationship between 22q11.2DS, a compelling example of a copy number variant (CNV) syndrome thought to cause cortical circuit disorders, and Cxcr4-mediated signaling, an established mechanism that ensures normal cortical interneuron development.
Results
Disrupted Interneuron Placement in the LgDel Cortex Is Cell-Class Specific.
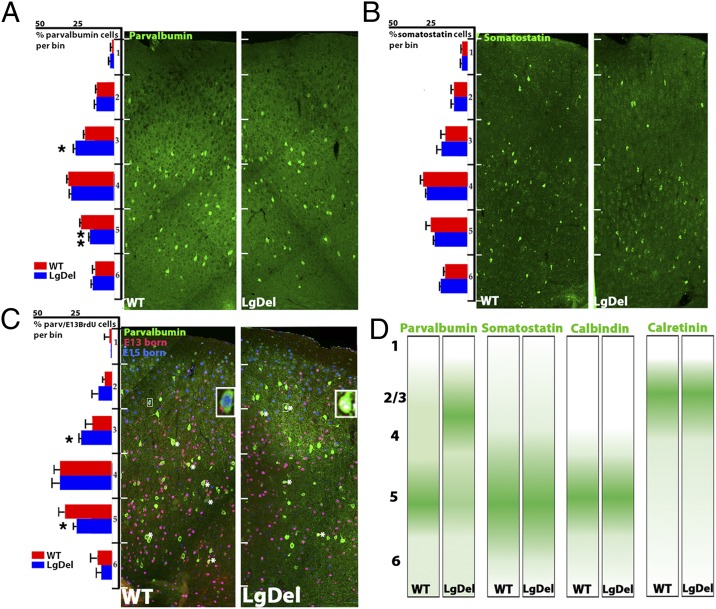
Our previous work indicates that interneuron distribution is altered in the LgDel cortex, with an apparent shift of parvalbumin cells toward more superficial layers at the expense of deeper layers (18). To determine whether this defect is similar to those associated with other mutations that alter laminar distribution of interneurons (22, 26, 27), we evaluated the frequency and laminar distribution of molecularly distinct interneuron classes in the LgDel mouse. Among four subtypes, we found specific disruption of parvalbumin interneurons (Fig. 1 A and B and Fig. S1). These changes are reminiscent of those reported for a small number of single-gene mutations that disrupt interneuron migration and placement, including Cxcr4 (22), Lhx6 (26), and Cxcr7 (27).
Fig. 1.
The laminar distribution of early-born parvalbumin interneurons is selectively altered in the LgDel cortex. Counting boxes (600- µm wide and divided into equidistant bins) spanning the cortex between the pial surface and white matter tract were analyzed in coronal brain sections as described previously (18). (A) Parvalbumin cells are significantly redistributed (ANOVA, n = 5 per genotype; significant differences comparing bins between genotypes determined by LSD test, P = 0.05 at bin 3 and 0.0017 at bin 5; error bars represent SEM) but not for the other primarily MGE-derived interneuron subtype, somatostatin (B). (C) Parvalbumin interneurons birth-dated at E13.5 by BrdU and then at E15.5 by IdU injection, were quantified using the same sampling strategy. Insets show parvalbumin interneurons with E13.5 (yellow nuclei) or E15.5 (blue nuclei) birthdates. A significant redistribution of E13.5-born parvalbumin cells was observed in the LgDel (ANOVA, n = 5 per genotype, P = 0.05 at bin 3 and 0.048 at bin 5). Asterisks mark E13.5-born BrdU-labeled parvalbumin cells. (D) Schematic of interneuron distribution in P21 cortex. Interneurons that are derived mainly from the caudal ganglionic eminence or are of broad origin are not altered in frequency or distribution in the LgDel (see Fig. S1 for calbindin and calretinin results), unlike the parvalbumin population derived from the MGE.
The laminar distribution of parvalbumin cells follows an inside–out gradient that reflects interneuron birthdates (28). To assess whether parvalbumin cell redistribution in LgDel mice is specific to interneurons born during particular embryonic epochs, we measured the cortical distribution of birth-dated interneuron subclasses born at embryonic day (E)13.5 and E15.5, respectively. We found that laminar redistribution is limited to E13.5-born parvalbumin cells. More of these cells are located superficially and fewer in deeper layers parallel to the overall redistribution of parvalbumin interneurons (Fig. 1C). Thus, interneuron placement is disrupted by diminished 22q11.2 dosage, similar to the disruption caused by single-gene mutations that compromise interneuron migration and placement.
Altered Interneuron Migration and Motility in the Embryonic LgDel Cortex.
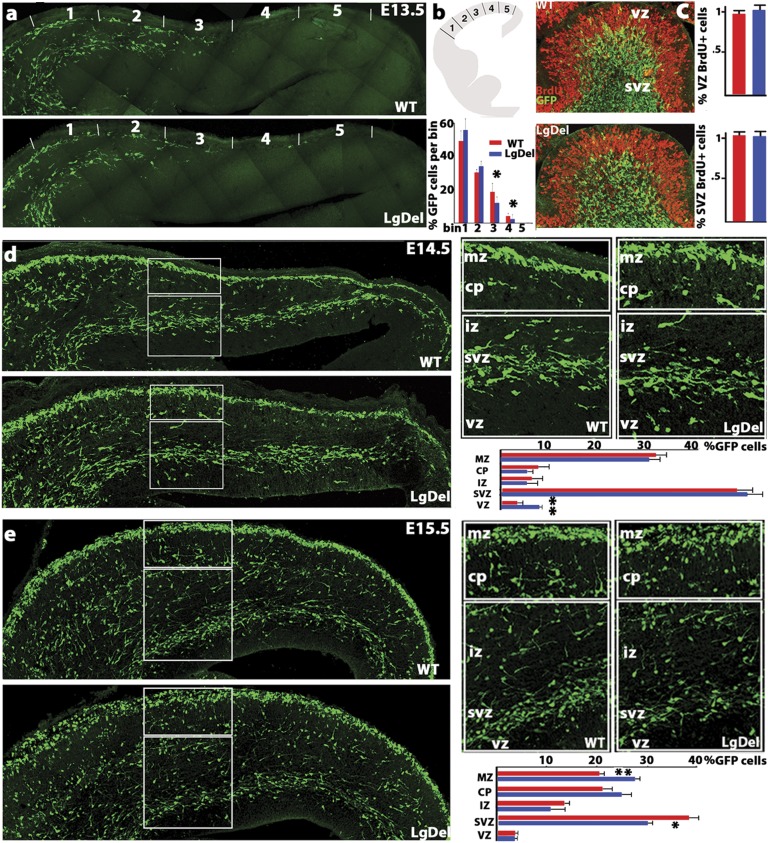
Changes in the distribution of cortical interneurons have been associated with genetic mutations that disrupt the migration of these cells during early development (22, 26, 27). To determine whether migration of basal forebrain-derived interneurons into the cortical rudiment is indeed altered in the LgDel 22q11.2DS model, we used a Dlx5/6 Cre-IRES-eGFP (CIE) reporter mouse line that labels postmitotic neurons generated in the basal forebrain (29). We found that interneurons advance less toward the dorsal aspect of the cortex in LgDel than in WT E13.5 embryos (Fig. 2 A and B) similar to migration deficits in homozygous Lhx6-mutant embryos (30).
Fig. 2.
Interneuron progress is disrupted in the embryonic LgDel cortex. LgDel embryonic cortical interneurons were labeled with GFP under the control of a Dlx5/6 promoter element that limits expression to ventral forebrain-derived cells (Dlx5/6-CIE line). (A) E13.5 LgDel interneurons (Lower) are less frequent in the LgDel dorsal cortex. (B) Tangential interneuron migration measured in five equidistant bins between the cortical/subcortical boundary (bin 1) and the cortical hem (bin 5) is less advanced in the LgDel (n = 5 per genotype, ANOVA with subsequent bin comparison by LSD test, *P ≤ 0.048 bin 3 and *P = 0.04 bin 4). (C) Images of E13.5 MGE showing Dlx5/6-CIE interneurons (green) and BrdU immunoreactivity (red). (BrdU was injected 45 min before animals were killed.). No significant proliferation differences were observed in apical (VZ; only BrdU single-labeled cells were counted) or basal (SVZ; only BrdU single-labeled cells were counted) MGE domains. n = 5 embryos per genotype; error bars represent SEM. (D) Cortical interneuron distribution between the ventricular and pial surfaces at E14.5 shows ectopic VZ interneurons (n = 5 per genotype, LSD test, **P = 0.007). Insets show MZ, CP, IZ, SVZ, and VZ. (E) The frequency of E15.5 LgDel migrating interneurons in the SVZ stream is reduced with a coincident increase in MZ stream cells (n = 5 per genotype, LSD test, **P = 0.003, *P = 0.02).
To determine whether these changes reflect reduced interneuron genesis in the basal forebrain, we administered a short-pulse of BrdU to E13.5 embryos and compared the numbers of cycling medial ganglionic eminence (MGE) apical and basal progenitors in LgDel and WT embryos (Fig. 2C). We did not observe proliferative changes in MGE apical or basal progenitor pools. Apparently, the capacity of MGE-derived interneurons to move into the cerebral cortex in the LgDel 22q11.2DS mouse model is selectively compromised, as seen in single-gene mutations that disrupt interneuron migration.
We next asked whether this disruption of cortical interneuron progress is transient or sustained. The delay in tangential progress visible at E13.5 was not noticeable at either E14.5 or E15.5. At E14.5, however, there are significantly more interneurons in the ventricular zone (VZ) in the LgDel cortex (Fig. 2D), again resembling phenotypes seen in homozygous Cxcr4 mutants (21, 27). At E15.5, interneurons have accumulated in the marginal zone (MZ) and have decreased in the subventricular zone (SVZ) stream (Fig. 2E). This dynamism in the distribution of migratory interneurons between the MZ, SVZ, and VZ streams in LgDel embryos parallels that reported for Cxcr4-homozygous mutants during this developmental epoch and prefigures altered distribution of interneurons in both genotypes. Apparently changes in interneuron migratory trajectories following 22q11.2 deletion resemble those associated with loss of function of genes that influence interneuron motility during midgestation.
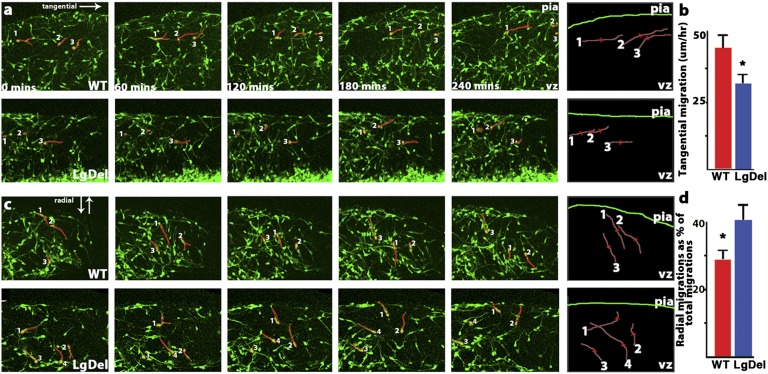
To determine directly whether these defects reflect altered motility of nascent LgDel interneurons, we imaged interneurons as they migrate into the cortex in live slices from E13.5 Dlx5/6-CIE embryonic forebrains. Interneurons migrate tangentially into the LgDel cortex at a significantly reduced rate (Fig. 3 A and B and Movies S1 and S2), consistent with diminished progress of cortical interneurons through the cortical hemisphere (Fig. 2). LgDel interneurons also deviate from tangential routes. Significantly more LgDel interneurons move radially between MZ and SVZ migratory streams (Fig. 3 C and D and Movies S3 and S4), consistent with the redistribution of cells in the MZ and SVZ in the LgDel that accumulate by E15.5 (Fig. 2). Together, our data demonstrate aberrant position, motility, and migratory route selection of LgDel interneurons in the developing cortex. These changes are similar to those reported for interneuron motility in Cxcr4-homozygous mutants (25). Thus, diminished 22q11.2 gene dosage compromises cellular mechanisms of interneuron migration that also are the target of single genes that define regulatory pathways for the movement of these cells in developing cortex.
Fig. 3.
Movement of embryonic interneurons is disrupted in the LgDel. (A) Time series of E13.5 live-imaged forebrain slices in which migrating interneurons (left to right) have been labeled genetically using the Dlx5/6-CIE transgene. (Upper) WT. (Lower) LgDel. The numerals and red outlines identify representative cells and their positions over the 4-h interval shown. (Far Right) Examples of migratory trajectories of individual cells imaged over 4 h. (B) Interneurons have a reduced velocity in the cortex relative to WT (n = 30 migratory neurons in five embryos per genotype, t test, *P = 0.032). (C) Time series of embryonic interneurons in transition from tangentially to radially directed migration (left to right). (Upper) WT. (Lower) LgDel. The numbers and red outlines indicate the migratory state and direction for representative interneurons over the 4-h interval. (Far Right) Representative examples of radial migratory trajectories of individual cells imaged over 4 h. (D) Radial migrations occur more frequently in LgDel interneurons (five embryos per genotype, t test, *P = 0.039).
Cell-Autonomous Defects in LgDel Cortical Interneurons.
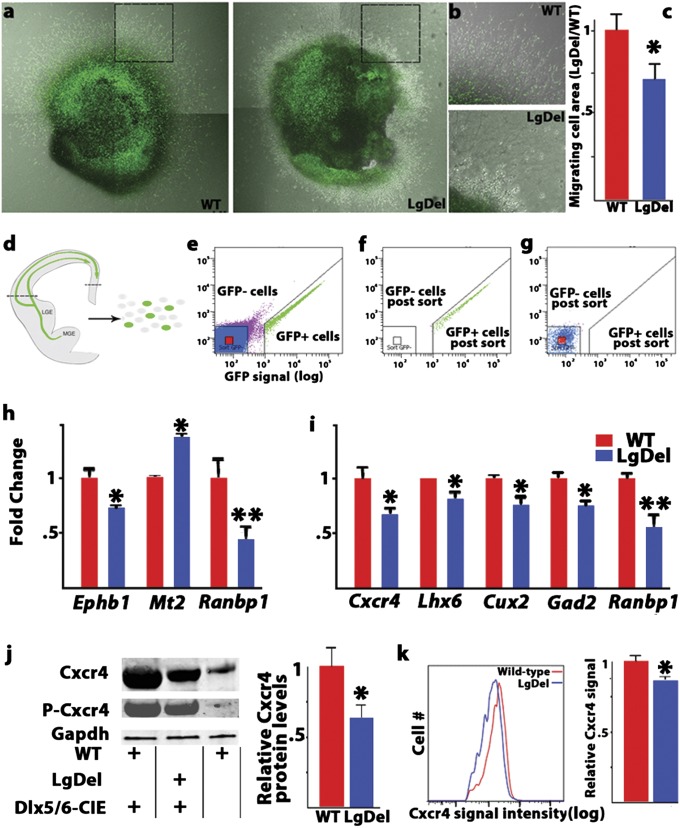
We next asked whether the altered position, progress, and movement of LgDel interneurons reflect altered motility and migratory capacity of MGE-generated interneurons themselves. We dissected E12.5 MGE from Dlx5/6-CIE WT and LgDel embryos and grew these explants in a 3D collagen matrix. After 3 d, we compared the degree of dispersion of interneurons migrating from the MGE tissue mass. We observed a reduced area of cells dispersed from LgDel MGE explants (Fig. 4 A–C). This reduction was observed in a minimally enriched environment consisting of 3D collagen substrate and serum-free medium. Accordingly, it is likely that cell-autonomous disruption of the migratory capacity of MGE-derived cells contributes to altered movement of interneurons into the cortical rudiment during early development in LgDel mice.
Fig. 4.
Cell-autonomous defects in LgDel cortical interneurons. (A) Cell migration differs in explants of WT (Left) and LgDel (Right) Dlx5/6-CIE MGE. (B) Higher-magnification images show reduced migration from LgDelMGE explants. (C) There was a significant reduction in surface area covered by LgDel cells (n = 8 embryos per genotype, *P = 0.021, t test). (D) Schematic of dissected tissue (dotted lines) collected for FACS analysis of migrating interneurons and their cortical substrate cells. (E) A representative FACS plot of Dlx5/6-CIE–labeled and –unlabeled cortical cells (green and red, respectively). (F and G) Dlx5/6-CIE–labeled (F) and –unlabeled (G) cortical cells were reanalyzed after FACS to verify sorting efficiency. (H and I) RT-qPCR confirmation of altered gene expression between WT and LgDel Dlx5/6-CIE–unlabeled cortical cells (H) and WT and LgDel Dlx5/6-CIE–labeled cortical interneurons (I) (n = 3 pooled samples per genotype, t test, *P ≤ 0.05, **P ≤ 0.01). (J) Western blot for Cxcr4 protein in Dlx5/6-CIE WT (Left) and LgDel (Center) interneurons isolated by FACS shows reduced total Cxcr4 protein in LgDel versus WT (*P = 0.039, t test). Phosphorylated Cxcr4 (P-Cxcr4) was similarly reduced in LgDel samples. Both phosphorylated and total Cxcr4 protein levels are minimal in Dlx5/6-CIE–unlabeled cells (Right). (K) Representative flow cytometry histograms showing reduced signal for cell-surface Cxcr4 antibody labeling of E14.5 dissociated LgDel Dlx5/6-CIE cortical interneurons. The mean Cxcr4 cell-surface fluorescence in LgDel Dlx5/6-CIE+ Cxcr4+ cells was reduced relative to WT cells (Right) (n = 10 WT, 9 LgDel embryos; *P = 0.032, t test).
Aberrant migration of nascent LgDel interneurons, either isolated in explants or observed in the developing cortex (Figs. 2 and 3), suggests significant changes in gene expression in these cells or in their migratory substrates because of diminished 22q11.2 gene dosage. Thus, we separated cortical substrate cells from Dlx5/6-CIE–labeled interneurons in microdissected E14.5 LgDel and WT cortex using fluorescence-activated cell sorting (FACS) (Fig. 4 D–G) and compared gene expression using Affymetrix microarrays (Tables S1 and S2). These changes then were confirmed by quantitative PCR (qPCR) in cDNAs as well as quantitative protein measurements in parallel samples of FACS-purified cells.
In Dlx5/6-CIE–negative cortical substrate cells, some altered transcripts could modify the interneurons’ migratory environment. These alterations include the cell-adhesion molecule Ephb1, whose preferred ligand, Ephrin-b1 is expressed at comparable levels in WT and LgDel migrating interneurons. Cellular stress genes also were altered, including Metalliothionen2 (Mt2) (Fig. 4H), consistent with a previous report of elevated expression in cardiac primordia in the Df1 22q11.2DS model (31).
In the Dlx5/6-CIE–labeled interneuron population, expression levels of several genes that regulate interneuron migration and placement decline (Fig. 4I). These genes include Cux2 and Lhx6, transcription factors previously shown to regulate interneuron migration and differentiation (26, 30, 32, 33). Gad2 (Gad65), which encodes one of the key synthetic enzymes for the predominant cortical interneuron neurotransmitter GABA (34), is also diminished. Finally, expression of the chemokine receptor Cxcr4, which has a cell-autonomous role in guiding nascent interneurons in the cortex (23, 25), is decreased by ∼35%. In contrast, expression of an additional cytokine receptor implicated in migration, Cxcr7, and the primary ligand for Cxcr4, Cxcl12, are not altered in LgDel interneurons or substrate cells. This observation raises the possibility that 22q11.2 interneuron migratory phenotypes might be the consequence of cell-autonomous changes in Cxcr4 expression and activity.
To establish whether Cxcr4 is indeed diminished in FACS-purified Dlx5/6-CIE interneurons, we measured total Cxcr4 protein and found a reduction comparable to the Cxcr4 transcript (Fig. 4J). We also assessed phosphorylated Cxcr4, which was reduced by ∼34% compared with WT FACS-purified interneurons (Fig. 4J). Disrupted interneuron migration may reflect decreased cell-surface Cxcr4 capable of transducing extracellular signals; however, Cxcr4 is present both at the interneuron surface and in cytoplasmic compartments (27). Accordingly, we compared the abundance of cell-surface Cxcr4 receptors on LgDel and WT interneurons using flow cytometry. We found a significant reduction in the immunolabeling intensity for Cxcr4 on the cell surface of LgDel interneurons (Fig. 4K). Thus, interneurons in LgDel embryos have a reduction of Cxcr4 protein on their surfaces as they migrate in the developing cerebral cortex. These changes provide a framework for evaluating whether specific molecular mechanisms for cell-autonomous disruption of interneuron migration resulting from diminished 22q11.2 gene dosage engage the same pathways as those that depend on Cxcr4 function.
Diminished Cxcr4 Dosage Modifies Distribution of Migrating LgDel Interneurons.
Altered Cxcr4 expression and activity and the similarity of LgDel and Cxcr4−/− interneuron phenotypes (21–25) suggest that diminished 22q11.2 gene dosage and disrupted Cxcr4 signaling compromise similar aspects of interneuron migration, particularly the distribution of cells in the MZ, SVZ, and VZ migratory streams. If so, one would expect further diminished Cxcr4 expression to modify distinct aspects of the LgDel interneuron phenotype selectively and proportionately.
We crossed LgDel:Dlx5/6-CIE mice with mice carrying one floxed Cxcr4 allele (Cxcr4wt/f) (35), resulting in heterozygous Cxcr4 deletion in Dlx5/6-CIE cells (Fig. 5 A–C). We confirmed that this manipulation results in decreased Cxcr4 cell-surface labeling by FACS analysis (Fig. S2). Disrupted tangential interneuron migration into the cortical rudiment in the LgDel is not altered further by additionally reduced Cxcr4 expression in E13.5 LgDel:Dlx5/6-CIE/Cxcr4wt/f embryos (Fig. 5D), in concert with observations in Cxcr4−/− at this stage (21). In contrast, the SVZ interneuron stream was altered in the LgDel:Dlx5/6-CIE/Cxcr4wt/f embryos, with substantially more interneurons displaced into the VZ, often displaying radially orientated processes (Fig. 5E), in accord with the ectopic distribution of interneurons in the VZ of E13.5 Cxcr4−/− embryos (21). Indeed, the severity of the VZ/SVZ phenotype is graded and proportional so that LgDel:Dlx5/6-CIE/Cxcr4wt/f > LgDel:Dlx5/6-CIE > WT:Dlx5/6-CIE/Cxcr4wt/f (Fig. 5E) with approximate doubling observed between genotypes.
Fig. 5.
Targeted heterozygous Cxcr4 mutation selectively modifies interneuron migration in the developing LgDel cortex. There is a visible difference in the distribution of Dlx5/6-CIE interneurons in the VZ/SVZ of (A) E13.5 WT/Dlx5/6-CIE/Cxcr4wt/f, (B) LgDel, and (C) LgDel/Dlx5/6-CIE/Cxcr4wt/f cortex. (D) Tangential interneuron progress into cortex was quantified between the cortical/subcortical boundary and the cortical hem. There was an equivalent, significant reduction of tangential migration in the LgDel and LgDel/Dlx5/6-CIE/Cxcr4wt/f embryos. (E) Interneuron distribution was quantified in six equidistant bins between the pial and ventricular surfaces. Migratory interneuron frequency in the VZ of the LgDel is increased relative to WT (WT/Dlx5/6-CIE/Cxcr4wt/f; LSD test, P = 0.045 at bin location 5), and this altered frequency is increased further in the LgDel/Dlx5/6-CIE/Cxcr4wt/f (LSD test; P = 0.003 at bin location 5). Furthermore, there is a significant increase in interneurons in the VZ of the LgDel/Dlx5/6-CIE/Cxcr4wt/f over the LgDel (LSD test; P = 0.05; error bars represent SEM).
Thus, 22q11.2 deletion disrupts an established molecular mechanism for interneuron migration—Cxcr4-mediated signaling—resulting in strikingly similar changes in interneuron movement and distribution.
Discussion
Interneuron phenotypes in the LgDel mouse model of 22q11.2DS—a genetic syndrome associated with increased risk for cortical circuit disorders—are substantially similar to those caused by loss-of-function mutations in single genes that regulate parallel aspects of interneuron development. We established interneuron cell autonomy for these shared phenotypes and found cellular and molecular deficits specific to migration. Diminished 22q11.2 dosage reduces the expression of a number of genes with significant influence on interneuron migration, including Cxcr4. Interneuron-specific mutation of Cxcr4 in the context of diminished 22q11.2 gene dosage selectively and proportionately enhances migratory phenotypes shared by 22q11.2-deleted LgDel and Cxcr4−/− mice. Accordingly, we suggest 22q11.2DS is in part a neuronal migration disorder in which diminished 22q11.2 gene dosage disrupts interneuron migration via a Cxcr4-dependent mechanism.
Diminished 22q11.2 gene dosage, like several single-gene mutations including Lhx6, Cxcr7, and Cxcr4, alters cortical circuitry by changing the placement and frequency of interneurons. Diminished 22q11.2 dosage selectively compromises a temporally distinct subset of parvalbumin interneurons most likely generated in the ventral MGE. Indeed, the migration and placement of calbindin (broad origin), calretinin [mainly caudal ganglionic eminence (36)], and somatostatin interneurons [dorsal MGE (37)] are not compromised in LgDel mice. The laminar redistribution of parvalbumin interneurons, without a reduction in frequency, likely reflects the disruption of two established migration mechanisms. First, additional MZ migratory interneurons result in increased upper-layer parvalbumin cells because of enhanced local radial ingression into layers 2/3 (28). Second, heterochronic, heterotopic VZ interneurons anomalously ascend radially to layers 2/3 rather than to layers 5/6. In 22q11.2DS, such changes may alter local inhibitory networks and oscillatory properties that depend upon parvalbumin interneuron integrity (20, 38, 39) and lead to cellular pathology parallel to that in the cortex of schizophrenic and autistic patients (8, 9).
There is significant cell autonomy and specificity for interneuron deficits caused by diminished 22q11.2 gene dosage. The transcriptional profile of cortical substrate cells yields few obvious candidates: genes for motogenic or chemotropic ligands and adhesion, trophic, and tropic molecules are not altered. In contrast, transcriptional differences in migratory interneurons include diminished expression of candidates known to influence interneuron genesis, migration, placement, and function: Lhx6, Cux2, Gad2, and, perhaps most compelling, the cytokine receptor gene Cxcr4. Accordingly, 22q11.2DS targets interneurons by disrupting a genetic network that regulates migration. Our results provide direct evidence that CNVs associated with cortical circuit disorders can modify cellular and molecular mechanisms essential for circuit development.
Reduced Cxcr4 expression in LgDel migrating interneurons—based upon mRNA and protein measurements—places cytokine regulation of interneuron migration via Cxcr4 (21-25) squarely in the context of cortical circuit disorder pathogenesis. There are several similarities between interneuron defects in LgDel and Cxcr4−/− embryos. In both instances, pallial migration is slowed and disordered, and ectopic interneurons are seen in the VZ (21, 22, 25). In utero pharmacological blockade of Cxcr4 in the MGE as well as transplantation of Cxcr4−/− MGE cells into WT embryos results in altered distribution of presumed parvalbumin interneurons in the postnatal cortex (22). Thus, reduced Cxcr4 in migratory interneurons emerges as a likely contributor to disrupted transit and final distribution of parvalbumin interneurons caused by diminished 22q11.2 gene dosage. In LgDel interneurons, altered transcriptional regulation via Lhx6 and Cux2 may diminish Cxcr4 expression (30) in presumptive parvalbumin cells, whereas Cxcr4 levels in other classes of cells approximate WT levels (21, 40). Our results therefore outline a network of interacting genes, with Cxcr4 as a key node, perhaps modulated by Lhx6 and Cux2, by which diminished 22q11.2 gene expression selectively disrupts interneuron development.
Our targeted genetic inactivation of one copy of Cxcr4 in LgDel interneurons defines a molecular mechanism for 22q11.2 gene dosage-dependent regulation of interneuron migration. LgDel interneurons, with decreased surface Cxcr4, are relatively insensitive to cortical Cxcl12 signals. Reduced Cxcr4 signaling causes displacement of interneurons into the VZ. We suggest that this displacement may be followed by heterochronic migration into the cortex that terminates in layers 2/3, as is appropriate based on the time of arrival in the VZ, rather than in layers 5/6, as would be appropriate for the birthdates of these interneurons. Normally, at E13.5, cohorts of recently MGE-generated interneurons (41) enter the cortex via the SVZ stream (28). This population, delayed and misdirected as it enters the pallium, likely constitutes the ectopic migrating interneurons in the SVZ/VZ of E13.5 LgDel embryos. It is this subset of cells whose numbers likely are enhanced when Cxcr4 cell-surface availability is diminished further in the LgDel:Dlx5/6-CIE/Cxcr4wt/f cortex. These Cxcr4-compromised cells eventually could account for misplaced parvalbumin neurons in the LgDel model of 22q11.2DS.
Neurodevelopmental hypotheses of schizophrenia and autism, the cortical circuit disorders most frequently associated with 22q11.2DS (42, 43), have suggested that pathogenesis results from disrupting cellular and molecular mechanisms for cortical development. Nevertheless, there was little evidence that specific mechanisms are compromised by diminished 22q11.2 gene expression or any other mutation associated with cortical circuit disorders. Our evidence shows that dosage changes can alter key regulatory pathways for cortical development in 22q11.2DS. It remains to be determined if other genetic syndromes associated with cortical circuit disorders similarly disengage established cellular and molecular mechanisms, resulting in aberrant cortical circuit formation.
Materials and Methods
Mice.
Animal experimental procedures were approved by The George Washington University institutional animal care and use committee. The Dlx5/6-CIE (29), Cxcr4 f/f (35), and LgDel (10) lines were kept on a C57/BL6 background.
Real-Time Imaging.
Forebrain slices (250 µm) prepared on a vibratome from E13.5 brains were placed on nucleopore membrane filters (Whatman) over 35-mm imaging dishes containing slice culture medium (44). Slices were transferred to a 37 °C/5% CO2 chamber and visualized with an Olympus FV1000 confocal microscope. Slices were imaged every 6 min for 12 h. Migration was quantified using Imaris software (Bitplane).
Quantitative and Statistical Analysis.
To determine interneuron distribution at postnatal day (P)21, a counting box subdivided into six equidistant bins that spanned the pial surface-to-white matter tract was placed systematically in cortical locations at the level of the anterior commissure (area 6 of the mouse frontal association cortex). At E13.5, cells were counted in five equidistant bins between the cortical/subcortical boundary and the cortical hem. At E14.5 and E15.5, interneuron distribution between the pial and ventricular surfaces was determined for cytologically distinct zones [MZ, cortical plate (CP), intermediate zone (IZ), SVZ, and VZ] within a single counting box. Two independent observers, blinded to genotype, counted cells from the immunolabeled material. The data were analyzed using Student t test, ANOVA, and post hoc Fisher’s least significant difference test (LSD) (Graphpad, Inc.).
Tissue Dissociation and FACS.
Embryonic cortices were dissociated in papain (Worthington Biochemical). FACS then was performed on a FACSAria flow cytometer (BD Biosciences). Cells were gated using forward and orthogonal light scatter, and GFP fluorescence was captured in the detector with a 530/30 bandpass filter. After collection, cells were lysed in TRIzol for RNA isolation (Invitrogen). For Cxcr4 cell-surface labeling, an Alexa Fluor 647-nm–conjugated rat anti-Cxcr4 antibody (BioLegend) was added to dissociated embryonic cortical cells and measured by flow cytometry using a HeNe laser (633-nm excitation). Analysis was conducted using FlowJo software (Tree Star,Inc.).
Microarray and RT-qPCR.
RNA was extracted from TRIzol samples and purified using RNeasy spin columns (Qiagen). For cRNA synthesis and array hybridization, 100 ng of total RNA was used. Samples were hybridized to the mouse gene 1.0 ST array (Affymetrix). cDNA synthesis reactions were conducted as previously described (18). qPCR was conducted on a CFX384 thermal cycler using EvaGreen reagent (BioRad). Primer sequences used are listed in Table S3.
Immunohistochemistry.
Fixed embryonic and postnatal brains were processed and stained as described previously (18). Primary antibodies used are listed in Table S4. Species-specific fluorescently conjugated secondary antibodies (Invitrogen) were applied subsequently.
Nucleotide Analog Injections.
Timed pregnant mice (E13.5 for proliferation and E13.5/E15.5, for birth-dating) were injected i.p. with BrdU (50 mg/kg body weight) and iododeoxyuridine (IdU; 70 mg/kg). Standard BrdU/IdU immunolabeling techniques were used after antigen retrieval by steam treatment in sodium citrate buffer.
Explant Assay.
E12.5 MGE explants were grown in collagen matrix for 72 h at 37 °C/5% CO2 in neurobasal medium supplemented with B27 and l-glutamine (Invitrogen). Collagen (Rat Tail Collagen I; BD Biosciences) was prepared by NaHCO3 neutralization and diluted 1:3 before plating.
Western Blot.
FACS samples were resolved using 4–12% SDS–PAGE and were transferred electrophoretically onto PVDF membrane (Amersham Biosciences). The membrane was incubated in primary antibody for 1 h at room temperature and then with species-appropriate fluorescently conjugated secondary antibody (Licor). Each sample was probed with a rabbit anti-Cxcr4 antibody (Abcam) or rabbit anti-phospho Cxcr4 antibody (Ecm Biosciences) and normalized to Gapdh. Fluorescence was measured on a Licor Odyssey system.
Supplementary Material
Acknowledgments
We thank Thomas Harrigan for technical support. We thank Teresa Hawley for FACS analysis. This work was supported by National Institutes of Health (NIH)National Institute of Child Health and Human Development Grant 042182 and NIH/National Institute of Mental Health Grant 64065 (to A-S.L.), Grant S10RR025565 for microscopic analysis, and National Institute of Neurological Disorders and Stroke Grant NS031768 for the University of North Carolina at Chapel Hill Neuroscience Center core facility. D.W.M. was funded by a National Alliance for Research on Schizophrenia and Depression Young Investigator Award from the Brain and Behavior Research Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1211507109/-/DCSupplemental.
References
- 1.Rubenstein JL, Merzenich MM. Model of autism: Increased ratio of excitation/inhibition in key neural systems. Genes Brain Behav. 2003;2(5):255–267. doi: 10.1034/j.1601-183x.2003.00037.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rippon G, Brock J, Brown C, Boucher J. Disordered connectivity in the autistic brain: Challenges for the “new psychophysiology”. Int J Psychophysiol. 2007;63(2):164–172. doi: 10.1016/j.ijpsycho.2006.03.012. [DOI] [PubMed] [Google Scholar]
- 3.Insel TR. Rethinking schizophrenia. Nature. 2010;468(7321):187–193. doi: 10.1038/nature09552. [DOI] [PubMed] [Google Scholar]
- 4.Fishell G, Rudy B. Mechanisms of inhibition within the telencephalon: “Where the wild things are”. Annu Rev Neurosci. 2011;34:535–567. doi: 10.1146/annurev-neuro-061010-113717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benes FM. Emerging principles of altered neural circuitry in schizophrenia. Brain Res Brain Res Rev. 2000;31(2-3):251–269. doi: 10.1016/s0165-0173(99)00041-7. [DOI] [PubMed] [Google Scholar]
- 6.Lewis DA, Hashimoto T, Volk DW. Cortical inhibitory neurons and schizophrenia. Nat Rev Neurosci. 2005;6(4):312–324. doi: 10.1038/nrn1648. [DOI] [PubMed] [Google Scholar]
- 7.Blatt GJ, Fatemi SH. Alterations in GABAergic biomarkers in the autism brain: Research findings and clinical implications. Anat Rec (Hoboken) 2011;294(10):1646–1652. doi: 10.1002/ar.21252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beasley CL, Zhang ZJ, Patten I, Reynolds GP. Selective deficits in prefrontal cortical GABAergic neurons in schizophrenia defined by the presence of calcium-binding proteins. Biol Psychiatry. 2002;52(7):708–715. doi: 10.1016/s0006-3223(02)01360-4. [DOI] [PubMed] [Google Scholar]
- 9.Hashimoto T, et al. Gene expression deficits in a subclass of GABA neurons in the prefrontal cortex of subjects with schizophrenia. J Neurosci. 2003;23(15):6315–6326. doi: 10.1523/JNEUROSCI.23-15-06315.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Merscher S, et al. TBX1 is responsible for cardiovascular defects in velo-cardio-facial/DiGeorge syndrome. Cell. 2001;104(4):619–629. doi: 10.1016/s0092-8674(01)00247-1. [DOI] [PubMed] [Google Scholar]
- 11.Murphy KC, Jones LA, Owen MJ. High rates of schizophrenia in adults with velo-cardio-facial syndrome. Arch Gen Psychiatry. 1999;56(10):940–945. doi: 10.1001/archpsyc.56.10.940. [DOI] [PubMed] [Google Scholar]
- 12.Niklasson L, Rasmussen P, Oskarsdottir S, Gillberg C. Autism, ADHD, mental retardation and behavior problems in 100 individuals with 22q11 deletion syndrome. Res Dev Disabil. 2009;30(4):763–773. doi: 10.1016/j.ridd.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 13.Fine SE, et al. Autism spectrum disorders and symptoms in children with molecularly confirmed 22q11.2 deletion syndrome. J Autism Dev Disord. 2005;35(4):461–470. doi: 10.1007/s10803-005-5036-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kiehl TR, Chow EW, Mikulis DJ, George SR, Bassett AS. Neuropathologic features in adults with 22q11.2 deletion syndrome. Cereb Cortex. 2009;19(1):153–164. doi: 10.1093/cercor/bhn066. [DOI] [PubMed] [Google Scholar]
- 15.Schaer M, et al. Abnormal patterns of cortical gyrification in velo-cardio-facial syndrome (deletion 22q11.2): An MRI study. Psychiatry Res. 2006;146(1):1–11. doi: 10.1016/j.pscychresns.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 16.van Amelsvoort T, et al. Brain anatomy in adults with velocardiofacial syndrome with and without schizophrenia: Preliminary results of a structural magnetic resonance imaging study. Arch Gen Psychiatry. 2004;61(11):1085–1096. doi: 10.1001/archpsyc.61.11.1085. [DOI] [PubMed] [Google Scholar]
- 17.Mori T, et al. Neuroradiological and neurofunctional examinations for patients with 22q11.2 deletion. Neuropediatrics. 2011;42(6):215–221. doi: 10.1055/s-0031-1295479. [DOI] [PubMed] [Google Scholar]
- 18.Meechan DW, Tucker ES, Maynard TM, LaMantia AS. Diminished dosage of 22q11 genes disrupts neurogenesis and cortical development in a mouse model of 22q11 deletion/DiGeorge syndrome. Proc Natl Acad Sci USA. 2009;106(38):16434–16445. doi: 10.1073/pnas.0905696106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Long JM, et al. Behavior of mice with mutations in the conserved region deleted in velocardiofacial/DiGeorge syndrome. Neurogenetics. 2006;7(4):247–257. doi: 10.1007/s10048-006-0054-0. [DOI] [PubMed] [Google Scholar]
- 20.Sigurdsson T, Stark KL, Karayiorgou M, Gogos JA, Gordon JA. Impaired hippocampal-prefrontal synchrony in a genetic mouse model of schizophrenia. Nature. 2010;464(7289):763–767. doi: 10.1038/nature08855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li G, et al. Regional distribution of cortical interneurons and development of inhibitory tone are regulated by Cxcl12/Cxcr4 signaling. J Neurosci. 2008;28(5):1085–1098. doi: 10.1523/JNEUROSCI.4602-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.López-Bendito G, et al. Chemokine signaling controls intracortical migration and final distribution of GABAergic interneurons. J Neurosci. 2008;28(7):1613–1624. doi: 10.1523/JNEUROSCI.4651-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stumm RK, et al. CXCR4 regulates interneuron migration in the developing neocortex. J Neurosci. 2003;23(12):5123–5130. doi: 10.1523/JNEUROSCI.23-12-05123.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tiveron MC, Cremer H. CXCL12/CXCR4 signalling in neuronal cell migration. Curr Opin Neurobiol. 2008;18(3):237–244. doi: 10.1016/j.conb.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 25.Wang Y, et al. CXCR4 and CXCR7 have distinct functions in regulating interneuron migration. Neuron. 2011;69(1):61–76. doi: 10.1016/j.neuron.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liodis P, et al. Lhx6 activity is required for the normal migration and specification of cortical interneuron subtypes. J Neurosci. 2007;27(12):3078–3089. doi: 10.1523/JNEUROSCI.3055-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sánchez-Alcañiz JA, et al. Cxcr7 controls neuronal migration by regulating chemokine responsiveness. Neuron. 2011;69(1):77–90. doi: 10.1016/j.neuron.2010.12.006. [DOI] [PubMed] [Google Scholar]
- 28.Miyoshi G, Fishell G. GABAergic interneuron lineages selectively sort into specific cortical layers during early postnatal development. Cereb Cortex. 2011;21(4):845–852. doi: 10.1093/cercor/bhq155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stenman J, Toresson H, Campbell K. Identification of two distinct progenitor populations in the lateral ganglionic eminence: Implications for striatal and olfactory bulb neurogenesis. J Neurosci. 2003;23(1):167–174. doi: 10.1523/JNEUROSCI.23-01-00167.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao Y, et al. Distinct molecular pathways for development of telencephalic interneuron subtypes revealed through analysis of Lhx6 mutants. J Comp Neurol. 2008;510(1):79–99. doi: 10.1002/cne.21772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prescott K, et al. Microarray analysis of the Df1 mouse model of the 22q11 deletion syndrome. Hum Genet. 2005;116(6):486–496. doi: 10.1007/s00439-005-1274-3. [DOI] [PubMed] [Google Scholar]
- 32.Cobos I, Long JE, Thwin MT, Rubenstein JL. Cellular patterns of transcription factor expression in developing cortical interneurons. Cereb Cortex. 2006;16(Suppl 1):i82–i88. doi: 10.1093/cercor/bhk003. [DOI] [PubMed] [Google Scholar]
- 33.Cubelos B, et al. Cux-1 and Cux-2 control the development of Reelin expressing cortical interneurons. Dev Neurobiol. 2008;68(7):917–925. doi: 10.1002/dneu.20626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Erlander MG, Tillakaratne NJ, Feldblum S, Patel N, Tobin AJ. Two genes encode distinct glutamate decarboxylases. Neuron. 1991;7(1):91–100. doi: 10.1016/0896-6273(91)90077-d. [DOI] [PubMed] [Google Scholar]
- 35.Nie Y, et al. The role of CXCR4 in maintaining peripheral B cell compartments and humoral immunity. J Exp Med. 2004;200(9):1145–1156. doi: 10.1084/jem.20041185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu Q, Cobos I, De La Cruz E, Rubenstein JL, Anderson SA. Origins of cortical interneuron subtypes. J Neurosci. 2004;24(11):2612–2622. doi: 10.1523/JNEUROSCI.5667-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wonders CP, et al. A spatial bias for the origins of interneuron subgroups within the medial ganglionic eminence. Dev Biol. 2008;314(1):127–136. doi: 10.1016/j.ydbio.2007.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hensch TK. Critical period plasticity in local cortical circuits. Nat Rev Neurosci. 2005;6(11):877–888. doi: 10.1038/nrn1787. [DOI] [PubMed] [Google Scholar]
- 39.Sohal VS, Zhang F, Yizhar O, Deisseroth K. Parvalbumin neurons and gamma rhythms enhance cortical circuit performance. Nature. 2009;459(7247):698–702. doi: 10.1038/nature07991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tanaka DH, et al. CXCR4 is required for proper regional and laminar distribution of cortical somatostatin-, calretinin-, and neuropeptide Y-expressing GABAergic interneurons. Cereb Cortex. 2010;20(12):2810–2817. doi: 10.1093/cercor/bhq027. [DOI] [PubMed] [Google Scholar]
- 41.Rubin AN, et al. The germinal zones of the basal ganglia but not the septum generate GABAergic interneurons for the cortex. J Neurosci. 2010;30(36):12050–12062. doi: 10.1523/JNEUROSCI.6178-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Geschwind DH, Levitt P. Autism spectrum disorders: Developmental disconnection syndromes. Curr Opin Neurobiol. 2007;17(1):103–111. doi: 10.1016/j.conb.2007.01.009. [DOI] [PubMed] [Google Scholar]
- 43.Weinberger DR. Implications of normal brain development for the pathogenesis of schizophrenia. Arch Gen Psychiatry. 1987;44(7):660–669. doi: 10.1001/archpsyc.1987.01800190080012. [DOI] [PubMed] [Google Scholar]
- 44.Polleux F, Ghosh A. The slice overlay assay: A versatile tool to study the influence of extracellular signals on neuronal development. Sci STKE. 2002;2002(136):pl9. doi: 10.1126/stke.2002.136.pl9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.