Abstract
The bias of αβ T cells for MHC ligands has been proposed to be intrinsic to the T-cell receptor (TCR). Equally, the CD4 and CD8 coreceptors contribute to ligand restriction by colocalizing Lck with the TCR when MHC ligands are engaged. To determine the importance of intrinsic ligand bias, the germ-line TCR complementarity determining regions were extensively diversified in vivo. We show that engagement with MHC ligands during thymocyte selection and peripheral T-cell activation imposes remarkably little constraint over TCR structure. Such versatility is more consistent with an opportunist, rather than a predetermined, mode of interface formation. This hypothesis was experimentally confirmed by expressing a hybrid TCR containing TCR-γ chain germ-line complementarity determining regions, which engaged efficiently with MHC ligands.
Keywords: MHC restriction, TCR
T cells expressing an αβ T-cell receptor (TCR) are MHC-restricted, recognizing self- and foreign peptide epitopes presented by MHC class I and II molecules during thymic development and peripheral activation, respectively. Two mechanisms are proposed to underlie this ligand bias. First, the CD8 and CD4 coreceptors have dual specificity for extracellular MHC and the intracellular proximal kinase Lck. Consequently, when MHC class I or II ligands are engaged, Lck is colocalized with the TCR/CD3 complex initiating signal transduction. The importance of this mechanism in disadvantaging non-MHC ligands is highlighted by the recovery of T-cell selection, where MHC and the coreceptors are both absent in comparison to the absence of MHC alone (1). In this setting, non-MHC ligands drive thymic positive selection and are recognized by peripheral T cells (2). The ability of αβ TCRs to recognize non-MHC ligands does not rule out an intrinsic bias of the TCR for MHC ligands. Indeed, evidence for such a hardwired bias is suggested by pairwise interactions between TCR-β germ-line complementarity determining regions 1 and 2 (CDR1/2) and the MHC α-helices observed in several structures (3–7). Recently, such recurrent interactions have been shown to be dependent on the partner TCR-α chain, which can impose distinct modes of TCR-β engagement, suggesting they may not drive MHC specificity (8). Although the relatively limited set of TCR/MHC-peptide structures reveals a semiconserved docking geometry, the angle of TCR engagement varies by more than 60° and the generally central docking position can shift toward the peptide amino- or carboxy-terminus (9). Likewise, conserved features of the MHC α-helices, including exposure of the polypeptide backbone and surface depressions, have been suggested to provide energetically favorable sites for CDR engagement (5). However, a crucial role for specific MHC residues in TCR docking has not emerged (10). The role of germ-line TCR structure in the bias to MHC ligands has thus been perplexing, especially given the structural variability of both components. To investigate this, we have applied a unique mutagenesis approach based on redirecting V(D)J recombination, allowing extensive in vivo remodeling of the germ-line CDR regions. We find thymic T-cell selection and peripheral T-cell activation are not dependent on germ-line CDR structure, suggesting the TCR can adopt a highly versatile, antibody-like strategy for engaging MHC-peptide ligands. To test this hypothesis directly, the TCR-β germ-line CDR1 and CDR2 regions were replaced with TCR-γ chain CDRs. MHC class I and II are not natural ligands for γδ T cells, and their germ-line CDR regions have not coevolved with MHC molecules. This hybrid γβ TCR chain paired with the endogenous TCR-α repertoire and facilitated efficient recognition of MHC and thymic T-cell selection. These data demonstrate TCR-intrinsic specificity for MHC ligands is not an essential determinant of MHC restriction; rather, analogous to antibodies, the αβ TCR can use generic chemical features of the germ-line loops to engage ligands.
Results
TCR Germ-line MHC Contact Regions Are Structurally Diverse.
Hardwired specificity for MHC class I and II might constrain germ-line TCR CDR composition in comparison to Igs, which use their germ-line CDRs to engage an immense variety of ligands. To examine this, human and mouse Ig heavy/light and TCR-α/β germ-line CDR1 and CDR2 sequences were aligned and amino acid frequencies were determined for each position. For each CDR, the most common length was analyzed (Fig. S1 A and B). The highest sequence diversity is present in TCR-β CDR2 and αCDR1 rather than Ig germ-line regions (Fig. S1C). Further, CDR positions exhibiting strong amino acid preferences in the human and mouse are mostly apparent within Ig CDRs. TCR-intrinsic MHC reactivity would require most combinations of these structurally varied α- and β-germline regions to maintain MHC specificity. This analysis does not support a simple model for TCR-intrinsic MHC specificity in which the germ-line CDRs of TCR are more structurally constrained than their Ig equivalents.
Positive Selection Does Not Constrain TCR-β Germ-line Structure.
To determine whether cryptic MHC recognition codes are nonetheless present in the TCR germ-line CDRs, they were extensively mutated. The Vα8.3 and Vβ11 chains of the H2-Kk/TENSGKDI–specific C6 TCR were used as the supporting frameworks because their variable (V) segments are selected efficiently (11, 12). To avoid the long C6 βCDR3 loop influencing MHC engagement, it was reduced to triple glycine inserted between framework residues in all constructs (∆βCDR3) (13).
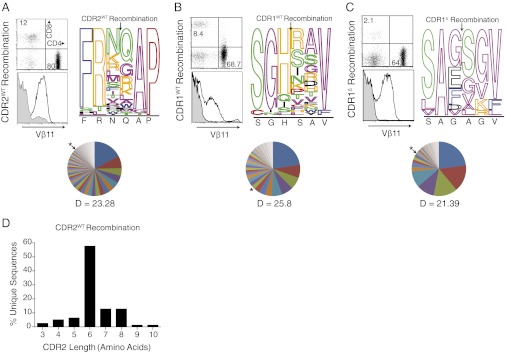
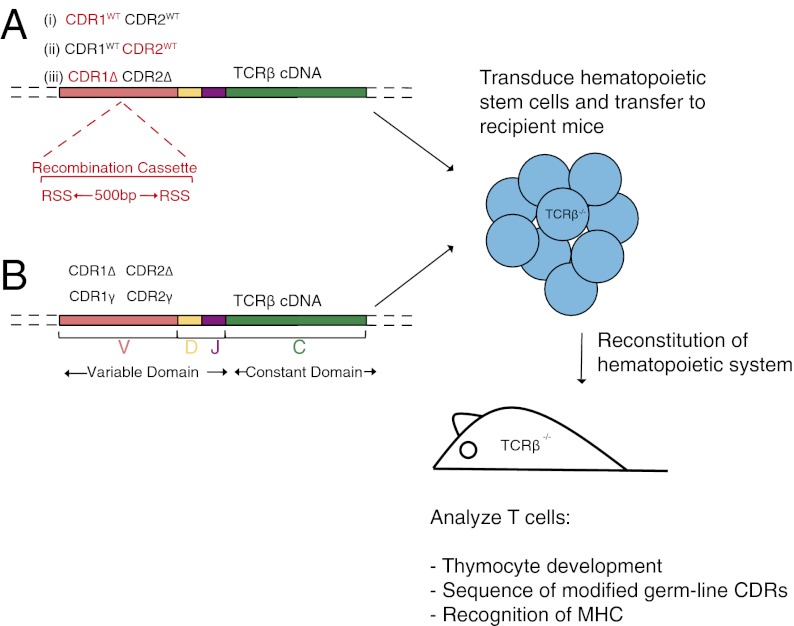
First, to introduce wide-ranging structural variation into the CDRs in vivo, our TCR-β chain retrogenic approach (14, 15) was modified to direct V(D)J recombination into the germ-line CDRs. This was achieved by appropriate positioning of recombination signal sequences (RSSs) and an intervening spacer into the center of the targeted germ-line CDR. Expression of the RAG genes during early T-cell development will initiate V(D)J recombination involving nucleotide deletion and TdT-mediated, nontemplated nucleotide insertion, leading to extensive diversification at the resulting join. This approach is particularly effective in generating diversity because each transduced early thymocyte progenitor will proliferate, generating many uniquely mutated TCR chains and increasing the total number of variants generated. This was applied to TCR-β by inserting RSSs derived from the mouse β-locus at the center of the WT βCDR1 or βCDR2 loop (Fig. 1 A and B). Constructs were transduced into TCR-βδ−/− deficient hematopoietic stem cells (HSCs) and transferred to irradiated T cell-deficient recipients. Analogous to normal V(D)J recombination-mediated diversification of CDR3, immature retrogenic thymocytes in which the target germ-line region has undergone recombination, producing an in-frame β-chain, will express the pre-TCR and progress to the CD4+CD8+ double-positive (DP) stage. At the DP stage, the pre–TCR-α will be replaced with an endogenously rearranged TCR-α chain, forming a surface αβ TCR, which will audition for positive selection. This system thus provides a powerful screen for identifying mandatory elements of the germ-line CDRs that are required for engagement with MHC ligands. Following reconstitution, secondary lymphoid tissue of retrogenic mice expressing either construct contained high proportions of retrogenic CD4+ and CD8+GFP+Vβ11+ T cells (Fig. 2 A and B). A total of 51 and 101 unique variants were identified among 138 βCDR1 and 307 βCDR2 recombination events respectively analyzed within the peripheral CD4 T-cell repertoires (Table S1). More than 70% of the germ-line CDR variants displayed substantial change from the WT sequence with at least two amino acid substitutions and/or altered length (Fig. 2 A, B, and D and Table S1). Strikingly, for both CDRs, several variants were present at higher frequency than the WT, demonstrating the natural structures are not markedly advantaged in engaging MHC ligands (Fig. 2 and Table S1). Among variants that maintained germ-line length, all residues except proline at P6 of CDR2 were diversified, with the loop apices being extensively mutated with largely nonconservative substitutions, which were not biased to the corresponding residues of the germ-line CDR repertoires (Fig. 2 A and B). Because germ-line CDRs exhibit limited rigid body movement, remodeling is unlikely to significantly shift apex position (16). Given the diverse character and/or length of the modified loops, many are likely to engage with MHC-peptide rather than playing a passive role. Histidine, which can carry a positive charge, is present at P3 in >90% of the CDR1 repertoire, suggesting a key functional role (Fig. S1A). Consistent with this, P3 histidine was retained by most CDR1 variants (Fig. 2B). However, because this residue rarely contributes to bonding within the TCR/MHC interface, it may be principally involved in long-range electrostatic guidance rather than engaging in a conserved germ-line TCR contact with MHC (5, 17).
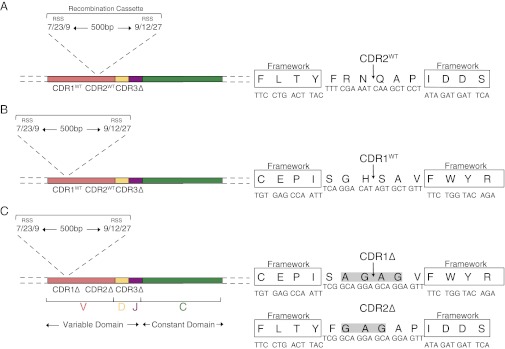
Fig. 1.
Design of recombination cassettes. The schematic represents the design of the ΔβCDR3 vector incorporating a recombination cassette in the germ-line CDR2 (A) or CDR1 (B). The constructs also contain a minimal triple-glycine CDR3 region flanked by framework V- and J-segment residues (13). The general design (Left) and details of the protein and nucleotide sequence (Right) are shown. The arrow represents the point of recombination. (C) Insertion of a recombination cassette into the CDR1 of a TCR-β chain with all three CDRs replaced with glycine-alanine linkers. Schematics are presented as for B. Highlighted residues represent those mutated from the original template.
Fig. 2.
Diverse “germ-line” CDR structures mediate thymic selection. V(D)J recombination was used to diversify germ-line βCDR2 (A), βCDR1 (B), and modified βCDR1 (C; also carrying mutated βCDR2). Flow cytometric analyses of splenic GFP+ T-cell populations (Upper) and Vβ11 expression on GFP+ CD4 T cells (Lower) are shown; filled plots are control GFP+CD4−CD8− cells (Upper Left). Stacked bar charts summarize amino acid frequencies within variant CDRs of peripheral GFP+ CD4 T cells. Only CDRs of the original length are shown. Arrows show the point of RSS insertion. Amino acid classification: blue, large hydrophobic; pink, small hydrophobic; green, polar; orange, positively charged; black, negatively charged; red, unique. The pie chart represents frequencies of recombinant CDR with unique amino acid composition [Shannon Diversity Index (D) is indicated]. The asterisk corresponds to the frequency of the total population occupied by regeneration of the original template. (D) Summary of CDR2 length of the data presented in A and Table S1.
These data show that TCR-β CDR structure is highly accommodating of structural change without loss of MHC engagement but do not exclude autonomous roles for each TCR-β germ-line CDR in MHC restriction. To examine this, the central regions of both βCDRs were replaced with flexible glycine-alanine linkers introducing seven substitutions. In vivo recombination was directed into the artificial βCDR1 of the template (Fig. 1C). Following reconstitution, retrogenic mice contained high proportions of GFP+Vβ11+ T cells in secondary lymphoid tissue (Fig. 2C). Again, the central regions of CDR1 variants from CD4 T cells were diversified without bias to the corresponding residues of the germ-line CDR1 repertoire (Fig. 2C and Table S1). These data further demonstrate that recognition of MHC imposes remarkably little constraint over TCR-β germ-line CDR structure, suggesting MHC recognition can be achieved in its absence.
TCRs Containing Artificial CDRs Lacking Germ-line Structure Direct Thymic Positive Selection.
To assess the impact of removing germ-line structure quantitatively, we next produced conventional retrogenic mice expressing TCR-β chains with WT germ-line regions or artificial loops lacking all germ-line structure. The mutant βCDR1 and βCDR2 template used above was mutated further by deleting framework residue Y54 positioned at the start of βCDR2, which makes contacts with MHC in several structures (5). This construct, ∆βCDR1/2/3, has eight mutations in the germ-line CDRs (four substitutions in CDR1 and three substitutions/one deletion in CDR2) (Fig. S2 A and B). The control construct, ∆βCDR3, has the germ-line CDRs in WT configuration. Both constructs contained the minimal CDR3 region as used in the V(D)J recombination vectors. In retrogenic mice, the ∆βCDR1/2/3 and the control chains directed development of similar numbers of peripheral T cells (Fig. S2C), although a higher proportion of ∆βCDR1/2/3 T cells were CD44hi (Fig. S2D). To determine whether the germ-line TCR-α CDRs are similarly accommodating of total loss of germ-line structure, the central five residues of both Vα8.3 CDRs were replaced with flexible glycine-alanine linkers (∆αCDR1/2, Fig. S3A). The WT and ∆αCDR1/2 constructs were transduced into TCR-α−/− HSCs and transferred to T cell-deficient recipients. Following reconstitution, retrogenic mice contained similar numbers of peripheral GFP+ T cells (Fig. S3B), demonstrating that T-cell development and MHC engagement are also not dependent on germ-line TCR-α CDR structure.
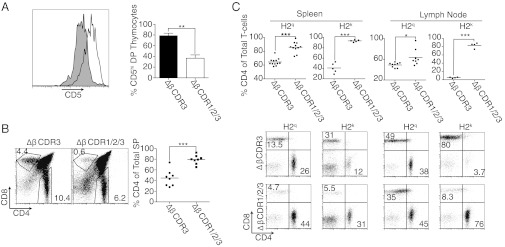
Increased CD44hi expression on ∆βCDR1/2/3 T cells may reflect homeostatic expansion associated with altered thymic selection. At the DP stage of thymocyte development, surface CD5 expression is positively correlated with the degree of MHC engagement by the repertoire of surface TCRs (6, 18). Although CD5 expression profiles overlapped, the proportion of ∆βCDR1/2/3 GFP+ DP thymocytes expressing high levels of CD5 is markedly reduced in the absence of the germ-line CDR structure (Fig. 3A), indicating diminished TCR engagement with MHC. This is also evident in the reduced single-positive (SP) 8 compartment (Fig. 3B). This skew may result from the ∆βCDR1/2/3 chain being most compromised in engaging MHC class I and/or preferential engagement of MHC class II by endogenous TCR-α chains in the absence of TCR-β germ-line structure. Deficient CD8 lineage selection was also manifest in the increased CD4/CD8 T-cell ratio ∆βCDR1/2/3 peripheral repertoires selected in both H2q and H2k hosts (Fig. 3C). The small thymic SP4 and SP8 populations of both TCR-α ∆αCDR1/2 and TCR-α WT retrogenic mice precluded similar analyses. This is due to nontransduced HSCs from TCR-α−/− donors progressing to the DP stage using endogenous TCR-β, thus competing with TCR-α transduced thymocytes, despite being unable to progress to the SP stage. Interestingly however, the absence of TCR-α germ-line CDRs introduced a small skew to the CD8 lineage in the periphery of both H2q and H2k hosts (Fig. S3C). Both TCR-α and TCR-β retrogenic WT and mutant mice contained CD4 T cells expressing regulatory markers FoxP3 and CD25 (Fig. S4). These data further highlight an unexpected adaptability of the TCR in engaging MHC.
Fig. 3.
T-cell populations in ΔβCDR3 and ΔβCDR1/2/3 retrogenic mice. (A) CD5 expression on GFP+ DP ΔβCDR3 and ΔβCDR1/2/3 thymocytes from H2q retrogenic mice (n = 3). A representative plot (Left) and summary bar chart (Right) are shown. (B) CD4 and CD8 expression on gated GFP+ thymocytes. Representative plots (Left) and summary graph (Right) show the percentage of SP4 of total SP. (C) GFP+ CD4 and CD8 T cells (spleen and lymph node) in ΔβCDR3 and ΔβCDR1/2/3 retrogenic mice of H2q and H2k haplotypes. Representative plots (Lower) and summary dot plots (Upper) show the percentage of GFP+ CD4 T cells of total T cells. Thymus, n = 8 (both groups); spleen (H2q), n = 12 (ΔβCDR3) and n = 11 (ΔβCDR1/2/3); spleen (H2k), n = 5 (both groups); lymph node (H2q), n = 9 (ΔβCDR3) and n = 8 (ΔβCDR1/2/3); lymph node (H2k), n = 4 (both groups). Data are collated from several batches of independent mice. *P < 0.05; **P < 0.005; ***P < 0.0001.
TCR Germ-line Structure Regulates Endogenous Partner Chain Diversity.
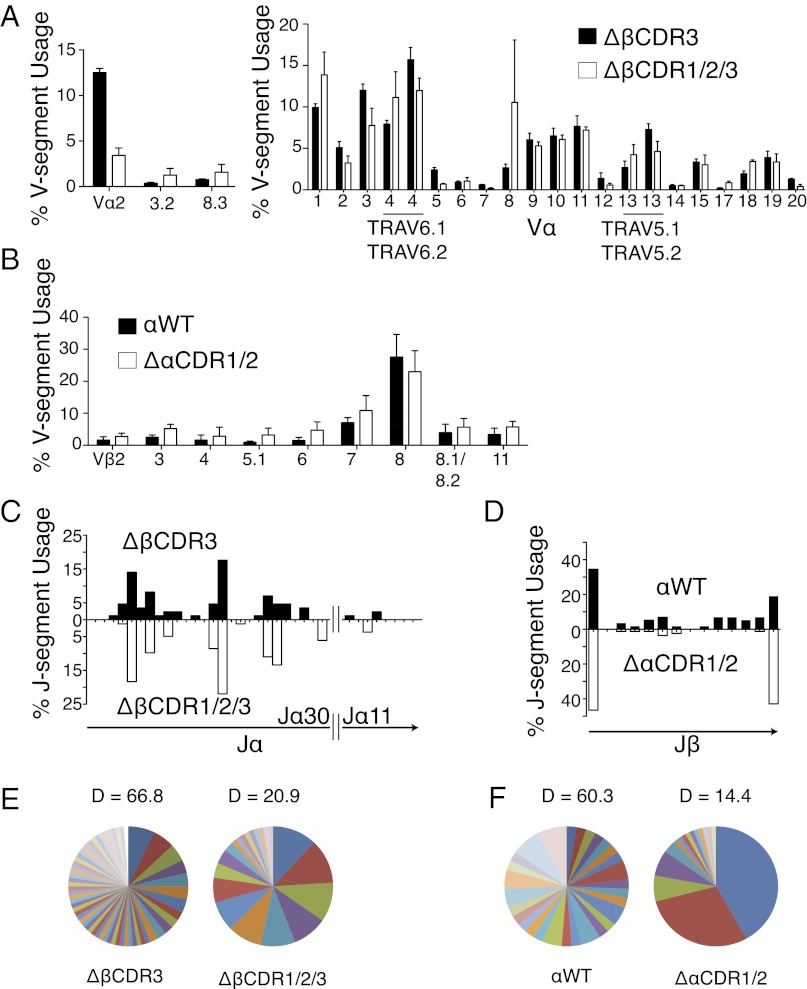
Reduced TCR engagement with MHC molecules in the absence of TCR germ-line CDRs clearly identifies an important role for these regions. In this setting, the WT germ-line regions of the endogenous partner chains are likely to play a dominant role in engaging MHC molecules. It was therefore of interest to determine whether the varied structures of the endogenous partner chains would have similar abilities to engage with MHC. Removal of germ-line TCR-β or TCR-α germ-line CDR structure had a limited impact on the coselected, endogenous variable segment repertoires, which were broadly similar in both cases (Fig. 4 A and B). Within the endogenous coselected repertoires, CDR3 rearrangements of the single-member Vα9 and Vβ7 gene segments were sequenced. Removal of the germ-line CDRs reduced CDR3 diversity and narrowed but did not distort joining (J)-segment use (Fig. 4 C–F). The set of α- and β-germline regions of the WT endogenous chains can thus function autonomously in engaging MHC ligands in the absence of germ-line structure on the retrogenic partner chains.
Fig. 4.
Endogenous TCR repertoires coselected with retrogenic WT and mutant α and β TCR chains. (A) Flow cytometric (Left; n = 5) and quantitative RT-PCR (Right; n = 4) analysis of endogenous TCR Vα-segment use by splenic GFP+CD4+ ΔβCDR3 and ΔβCDR1/2/3 retrogenic T cells. TRAV, TCR alpha variable-segment. (B) Flow cytometric analysis of endogenous TCR Vβ-segment use by GFP+CD4+ splenic αWT and ΔαCDR1/2 retrogenic T cells (n = 3). Error bars represent SEM. (C) Analysis of endogenous TCR Jα-segment use by Vα9 recombination events from splenic ΔβCDR3 (n = 83) and ΔβCDR1/2/3 (n = 75) retrogenic CD4 T cells. (D) Analysis of endogenous TCR Jβ-segment use by Vβ7 recombination events from splenic αWT (n = 57) and ΔαCDR1/2 (n = 84) CD8 T cells. (E and F) Analysis of endogenous CDR3 repertoire diversity. Pie charts represent frequencies of CDR3 with unique regions with unique amino acid composition within the recombination events in C and D [Shannon Diversity Index (D) is indicated].
T Cells Containing Artificial CDRs Lacking Germ-line Structure Respond to MHC Ligands.
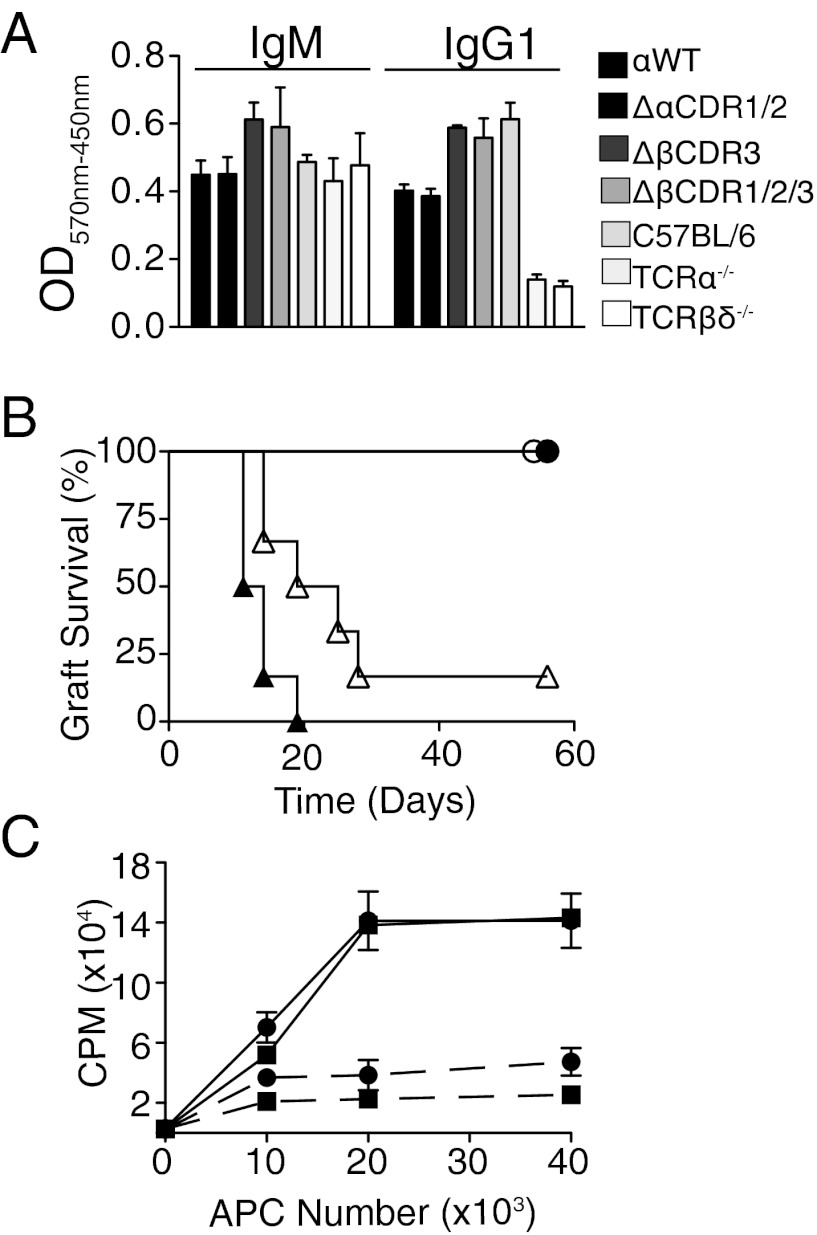
Activation of peripheral T cells requires engagement of MHC-peptide ligands with higher affinity than positive selection, and may therefore be more reliant than positive selection on intact germ-line CDR structure. The functional capacity of peripheral T cells lacking germ-line CDRs was assessed first by analyzing T cell-dependent B-cell Ig isotype switching. Sera from nonreconstituted TCR-βδ−/− and TCR-α−/− sera selectively lacked class-switched IgG1, which was recovered in retrogenic mice expressing either WT or artificial TCR-α and TCR-β germ-line CDRs (Fig. 5A), demonstrating peripheral T cells completely lacking germ-line structure on either chain can recognize and respond to MHC–peptide complexes presented by B cells. Reactivity to nonself-MHC molecules is a hallmark characteristic of T cells. To determine whether peripheral T cells lacking germ-line structure can recognize allogeneic MHC molecules and orchestrate a competent immune response, syngeneic T cell-deficient recipients were adoptively transferred with ∆βCDR3 or ∆βCDR1/2/3 retrogenic splenocytes and received adjacent allogeneic (H2b) and syngeneic (H2q) skin grafts. Both groups retained the syngeneic grafts and rejected the allogeneic grafts, although this was slightly retarded in the recipients receiving ∆βCDR1/2/3 retrogenic splenocytes (Fig. 5B). Primary in vitro proliferation assays confirmed these findings (Fig. 5C).
Fig. 5.
Functional analysis of T cells expressing mutated TCR chains. (A) Mutant germ-line CDR1/2 regions direct Ig class switching. Serum IgM and IgG1 levels of retrogenic mice expressing WT and mutant TCRs are shown. C57BL/6 and nonreconstituted TCR-α−/− TCR-βδ−/− mice were used as positive and negative controls, respectively. Bars represent triplicates from a single mouse. One representative experiment of two is shown. Lanes are identified in the key. (B) TCR-βδ−/− (H2q) mice were adoptively transferred with pooled T cells expressing ΔβCDR3 (filled symbols, n = 7) or ΔβCDR1/2/3 (open symbols, n = 7) and received adjacent allogeneic C57BL/6 (H2b, triangles) and syngeneic (H2q, circles) skin grafts. (C) Mixed lymphocyte response. Purified splenic retrogenic CD4 T cells expressing ΔβCDR3 (●) or ΔβCDR1/2/3 (■) were cultured with activated BMDCs from allogeneic C57BL/6 (H2b, solid lines, n = 3) and syngeneic (H2q, dashed line, n = 3; each in duplicate) donors in the presence of titrated thymidine. Error bars represent SEM. APC, Antigen Presenting Cell, CPM, counts per minute.
Efficient Engagement of MHC by αβ TCRs Carrying Germ-line Regions from the Unconventional, γδ T-Cell Lineage.
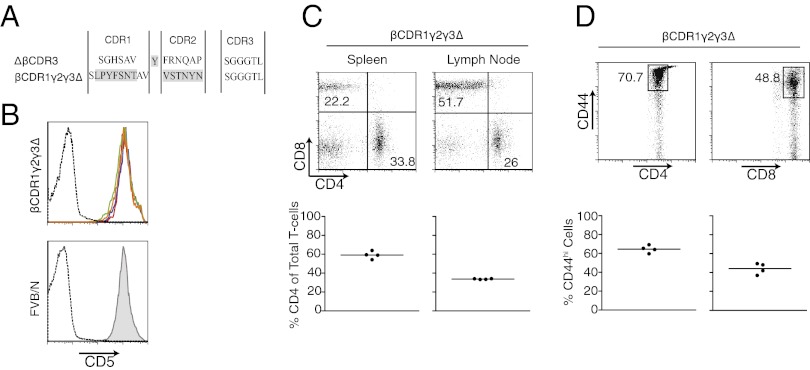
This unexpected degree of adaptability in MHC docking appears more compatible with a nonpredetermined, antibody-like strategy of ligand engagement by the TCR rather than an intrinsic hardwired bias. This hypothesis was explored by expressing a hybrid TCR-β chain in which both germ-line CDR loops derive from a TCR-γ chain (mouse γV1; construct βCDR1γ/2γ/3∆; Fig. 6A). The γV1 germ-line CDRs introduced 14 amino acid changes and increased CDR1 length by two residues. As for all TCR-β constructs, the CDR3 region was reduced to a triple glycine. The TCR-γ chain is highly unlikely to possess hardwired specificity for MHC ligands because the unconventional γδ T-cell lineage is not MHC-restricted. The hybrid (CDR1γ/2γ/3∆) chain behaved similar to the endogenous WT FVB/n TCR-β repertoire in directing development of uniformly high CD5hi DP thymocytes (Fig. 6B). Peripheral CD4/CD8 T-cell ratios and expression of CD44 resembled those of the ∆βCDR3 retrogenic mice, which have germ-line CDRs (Figs. 3 and 6 C and D and Fig. S2). These data demonstrate restoration of efficient recognition of MHC class I and II by TCRs containing combinations of TCR-α and γV1 germ-line regions, supporting the hypothesis that the αβ TCR can function similar to antibody in using generic chemical characteristics of the germ-line regions in a nonpredetermined fashion when forming interfaces with MHC-peptide ligands.
Fig. 6.
TCR-γ germ-line CDRs mediate αβ T-cell selection. (A) Schematic depiction of hybrid TCR chain containing germ-line CDR1/2 derived from TCR Vγ1 (construct βCDR1γ/2γ/3∆) grafted onto TCRVβ11 (framework Y54 deleted and minimal triple-glycine CDR3). (B) CD5 expression on GFP+ DP (solid line plots) and double-negative (dashed line plot) thymocytes from WT (TCR-β+δ+) FVB/n (Lower) and βCDR1γ/2γ/3∆ (Upper; n = 4). (C) Representative plots and summary charts of splenic and lymph node GFP+, CD4, and CD8 T-cell representation in βCDR1γ/2γ/3∆ retrogenic mice. (D) Representative plots and summary charts of CD44hi GFP+ CD4 and CD8 T cells from whole spleen in βCDR1γ/2γ/3∆ retrogenic mice. Compare with Fig. 3C and Fig. S3D.
Discussion
The functional specificity of αβ T cells for MHC class I and II molecules has been proposed to be underpinned by the CD4 and CD8 coreceptors that localize Lck with the TCR–CD3 complex only when MHC ligands are engaged (1). Additionally, the coreceptors have been proposed to contribute to the semiconserved docking geometry of the TCR, through constraints over their positioning for initiation of signal transduction (19, 20). Electrostatic complementarity-based guidance and shape complementarity have also been proposed to contribute to MHC bias and docking orientation. With ligand bias and docking orientation being conferred by these processes, the requirement for conventional ligand specificity based on predetermined interactions between receptor and ligand may be relaxed (9, 17, 21). To investigate this, we have used in vivo V(D)J recombination-based mutagenesis and extensive remodeling of the germ-line regions to investigate TCR-intrinsic structural requirements for engagement with MHC. We do not find evidence for such constraints. Rather, the αβ TCR is unexpectedly accommodating of structural change, with TCRs bearing >150 variant germ-line CDRs directing positive selection. Nucleotide bias of the enzymes mediating deletion/insertion will influence the composition of the diversified germ-line CDRs auditing for positive selection. To take account of this, it will be of interest to compare the preselection (DP-stage thymocytes) with the postselection (SP-stage thymocytes) repertoires to identify features that influence MHC engagement using deep sequencing and bioinformatic analyses. Removal of all germ-line structure on either chain by replacement with flexible glycine/alanine loops diminished but did not ablate T-cell selection or peripheral T-cell function. These processes depend on formation of TCR/MHC-peptide interfaces within distinct ranges of affinity, highlighting the extreme versatility of the TCR in productively engaging with MHC ligands.
Recently, the αβ TCR has been shown to engage the non-MHC ligand CD155 using antibody-like recognition (2). In this case, the T cells had developed in the absence of both the MHC molecules and the coreceptors. Our findings suggest recognition of the natural MHC ligands, occurring in the presence of the coreceptors, can also be achieved using an antibody-like strategy in which generic chemical features of the germ-line regions are used to form interfaces. Direct support for this model is provided by the efficient recognition of MHC class I and II molecules by hybrid TCR chains carrying structurally distinct germ-line CDRs derived from the unconventional γδ T-cell lineage, which is not MHC-restricted and cannot have intrinsic bias to MHC ligands (Fig. 6).
Relevant to this hypothesis, tyrosine is the most enriched amino acid within Ig germ-line CDRs and can dominate antigen recognition (22). Tyrosine also stands out within TCR germ-line CDRs for its frequent role in contacting the MHC α-helices (5, 23). The malleable Van der Waal interactions characteristic of tyrosines allow flexibility in forming interfaces with different MHC ligands through forming distinct molecular interactions (5, 24). This feature is further highlighted by such germ-line tyrosine residues contributing to engagement of the TCR with the non-MHC ligand CD155 (2). Both germ-line CDRs of γV1 contain a tyrosine residue, which may contribute to their enhanced ability to engage productively with MHC I and II molecules in comparison to artificial glycine/alanine loops (Figs. 3 and 6 C and D and Fig. S2). Further, because thymic positive selection proceeds at lower affinities than antigen recognition by primary antibodies, the contribution of the germ-line regions is likely to be correspondingly relaxed in comparison to antigen engagement by antibody (25, 26). Although this work demonstrates that TCR germ-line structure is dispensable for recognition of MHC, it is likely that evolution has optimized the germ-line regions to facilitate engagement across the spectrum of MHC alleles present within the species. Indeed, their conservation is evident between the mouse and human (Fig. S1), and an YXY motif is conserved in TCR-β CDR2 across jawed vertebrates (27). Our finding that TCR-γ germ-line CDRs can participate effectively in engagement with MHC class I and II suggests generic chemical features, independent of their context, can be used in forming interfaces with MHC. In some cases, the conserved TCR germ-line motifs may also function in this nonpredetermined antibody-like fashion in engaging MHC-peptide ligands.
Overall, these findings suggest the TCR can use an antibody-like strategy to recognize MHC class I and II, which may confer several advantages. First, the two ligand types, MHC class I and II, are structurally distinct and highly polymorphic, making conventional receptor-ligand specificity, based on predetermined interactions, difficult to achieve. This is especially true for a highly diverse repertoire using many combinations of the TCR-α and TCR-β germ-line regions. Conversely, antibody-like recognition may facilitate low-affinity recognition across the plethora of MHC alleles and unique combinations of alleles inherited by each individual. Further, cross-reactivity, a key feature of the TCR, is likely to be increased by the broadened range of docking options provided by antibody-like recognition (28, 29). The parallel strategies of antigen recognition by T and B cells suggested by this work have interesting implications for the evolution of the two branches of adaptive immunity.
Materials and Methods
Mice.
TCR-α−/− mice [CBA (H2k)] and TCR-βδ−/− mice [FVB/n (H2q)] were bred in-house (30, 31). [C57BL/6 (H2b)], WT CBA, and FVB/n mice were from Harlan. Mice were maintained in specific pathogen free (SPF) conditions. All experiments were carried out with Home Office and Imperial College Ethical Review Process Committee approval.
C6 TCR and Generation of TCR Constructs.
The C6 TCR recognizes the Smcy TENSGKDI/H2-Kk complex (11). The C6 αβ TCR transgenic strain shows a skew to the CD8 lineage (32). Mutated C6α and C6β TCR genes were generated by overlap PCR. Custom DNA synthesis (Mr Gene) was used for constructs, including RSSs. TCR genes were cloned into the pMigR1 retroviral vector. The nomenclature of Arden et al. (33) is used throughout, except for the designation of additional TCR Vα-segments from Bosc and LeFranc (34), analyzed by real-time PCR.
Real-Time PCR.
Real-time PCR was performed using LightCycler FastStart DNA Master SYBR Green I (Roche Diagnostics). One microliter of cDNA template was mixed with 500 nM appropriate primers, 2 mM MgCl2, and 2 μL of LightCycler FastStart DNA Master SYBR Green I in a total volume of 20 μL. Amplification was performed after an initial denaturation step at 95 °C for 10 min using the following protocol: 40 cycles of 5 s at 95 °C, 5 s at the annealing temperature (determined for each primer pair), and 20 s at 72 °C, followed by data acquisition. Dissociation curves were assessed in the range of 60–95 °C. Threshold cycle (Ct) values were determined with the Fitpoint algorithm of the Lightcycler software (version 3.5; Roche Applied Science). The slope (efficiency) of each amplification was calculated by linear regression of four different Ct values taken in the linear amplification phase (0.2, 0.4, 0.8, and 1.6 units of fluorescence).
Media.
Iscove’s modified Dulbecco’s medium (IMDM) and RPMI (Gibco) were supplemented with 2 mM glutamine, 10% (vol/vol) FCS, 100 U/mL penicillin, and 100 μg/mL streptomycin unless otherwise stated. GM-CSF was produced by X63-Ag8 plasmacytoma cells transfected with the mouse GM-CSF gene maintained on 1 mg/mL G148. Cells were washed twice and cultured for a further 2–3 d in medium lacking antibiotics before harvest of supernatant (35).
Generation of Retrovirus.
Phoenix ecotropic packaging cells were plated (2 × 105 cells per well) in 2 mL of antibiotic-free IMDM in six-well plates (BD Biosciences). After 24 h, medium was replaced and cells were transfected using Lipofectamine-2000 (Invitrogen) with 3 μg of vector and 1 μg of pCL-Eco helper vector (Imgenex) in 500 μL of Opti-MEM medium (Invitrogen). The medium was changed 24 h after transfection, and the supernatant was harvested after a further 24 h. Transfection efficiency, typically 30–80%, was determined by flow cytometric detection of GFP.
Retroviral Transduction of Con A Blasts.
WT CBA spleen cell suspensions (3 × 106 cells/mL) were plated in 2 mL of RPMI in 12-well plates (Nunc), supplemented with 2 ng/mL recombinant mouse IL-7 (Santa Cruz Biotechnology), and stimulated with 4 ng/mL Con A (Sigma–Aldrich). After 24 h, 1 mL of medium containing IL-7 was added to each well. After a further 24 h, 4 × 106 cells were resuspended in 2 mL of viral supernatant (SN) containing 2 μg/mL polybrene (Sigma–Aldrich) and plated in 12-well plates. Plates were centrifuged at 1,200 × g for 90 min, and 1 mL each of preconditioned medium and fresh supplemented medium was added. Cells were harvested after 48 h and analyzed by FACS for expression of GFP and TCR.
Retroviral Transduction and Adoptive Transfer of HSCs.
Donor mice received 5-flurouracil (150 mg/kg; Invivogen) 72 h before flushing the long bones with PBS. Bone marrow cells were filtered through a 70-μm cell strainer and RBC-lysed by incubation in 2 mL of lysis buffer (Stem Cell Technologies) for 10 min on ice. A total of 2 × 106 cells were plated in 2 mL of IMDM containing 20 ng/mL recombinant mouse IL-3, 10 ng/mL IL-6, 10 ng/mL stem cell factor (SCF), and 10 ng/mL recombinant human FLT-3 ligand (all from Invitrogen). Transduction was carried out as for Con A blasts. Recipient mice were irradiated at 400 rad (syngeneic transfer) or 900 rad (allogeneic transfer). Transduction efficiencies was measured using flow cytometric analysis of GFP expression, and 0.1–1 × 106 GFP+ cells in 200 μL of PBS were injected (i.v.) per recipient.
Flow Cytometry.
Flow cytometric analyses were performed on a FACSCalibur (BD Bioscience) or Cyan-ADP (Dako) instrument and analyzed using FlowJo software (version 8.8.6; TreeStar). The following antibodies were used: CD4-FITC (RM4-5), CD8-APC (53-6.7), CD5-phycoerythrin (PE) (53-7.3), and TCR-PE (H57-597) (all from eBiosciences) and CD44-PE (IM7), CD4-PerCP (RM4-5), Vα2-PE (B20.1), Vα3.2-PE (8-8.96), Vα8.3-PE (B21.14), Vα12-PE (B20.1), Vβ2-PE (B20.6), Vβ3-PE (KJ25), Vβ4-PE (KT4), Vβ5.1-PE (MR9-4), Vβ6-PE (RR4-7), Vβ7-PE (TR310), Vβ8-PE (F23.1), Vβ8.1/8.2-PE (MR5-2), and Vβ11-PE (RR3.15) (all from BD Biosciences PharMingen).
Isolation of T Cells.
CD4+ or CD8+ T cells were isolated from spleen and inguinal lymph nodes using microbeads coated with anti-CD4 or anti-CD8 (Miltenyi Biotech) using manual magnetic activated cell sorting (MACS) columns following the manufacturer’s instructions. Typically, T cells were enriched to ≥97%.
Endogenous TCR Repertoire Analysis.
RNA was extracted from purified T cells using phenol-chloroform extraction (Invitrogen iProtocols) or RNAqueous RNA extraction kits (Applied Biosciences) and was converted to cDNA using SuperScript III kits (Invitrogen) following the manufacturer’s instructions. cDNA was amplified with reverse primers specific for the relevant constant region of the TCR-α or TCR-β chain and forward primers specific for Vα9 and Vβ7 for endogenous TCR-α and TCR-β diversity repertoire, respectively. Products were cloned into a TOPO vector, transformed into DH5α cells via One Shot transformation (Invitrogen), and sequenced from colony PCRs (Medical Research Council Genomic Services, Hammersmith Hospital, London). Sequences were analyzed with Four Peaks software Mekentosj (version 1.7.2). J-segment use was assigned using the ImMunoGeneTics database (33).
Recombination Cassette Repertoire Analysis.
GFP+CD4+ cells derived from spleen and lymph nodes of mice expressing retrogenic WT or mutated C6β TCR genes containing RSS were sorted on a FACSAria II (BD Biosciences). RNA extraction and repertoire analysis was carried out as described above using primers designed to amplify across the recombined Vβ11 CDR region.
Generation of Bone Marrow-Derived Dendritic Cells.
The method of Inaba et al. was followed (36). Briefly, bone marrow was flushed from the long bones of FVB/n TCR-βδ−/− and C57BL/6 mice using IMDM and 107 cells plated in 10 mL in 20-cm2 flasks (BD Falcon) with 10% (vol/vol) GM-CSF containing supernatant. On day 3, half of the medium was replaced and fresh GM-CSF containing supernatant was added to achieve 10% (vol/vol). Cells were harvested on day 6 by scraping and replated in IMDM plus 10% GM-CSF at the initial density. On day 7, the bone marrow-derived dendritic cells (BMDCs) were activated overnight with LPS (Sigma–Aldrich) at 100 ng/mL.
Mixed Lymphocyte Response.
A mixed lymphocyte response was carried out as described (37). Briefly, 105 CD4 T cells purified as described above were cocultured with 0, 1 × 104, 2 × 104, or 4 × 104 allogeneic or syngeneic irradiated BMDCs in 200 μL of RPMI in round-bottomed, 96-well plates (Nunc). After 72 h, wells were pulsed with 1 μCi [3H] thymidine and left for a further 18 h. Thymidine uptake was measured using a Wallac 1205 Betaplate liquid scintillation counter (PerkinElmer).
Skin Grafting.
FVB TCR-βδ−/− (H2q) mice were adoptively transferred with 3 × 106 splenocytes pooled from three TCR-βδ−/− (H2q) retrogenic mice expressing either the ΔβCDR3 or ΔβCDR1/2/3 TCR chain. Cells were allowed to expand for 4 wk. Mice then received adjacent syngeneic [FVB TCR-βδ−/− (H2q)] and allogeneic [C57BL/6 (H2b)] skin grafts from donor tail skin. Skin grafting was carried out as described (38). Plasters were removed after 8 d, and graft integrity was assessed every 2–3 d.
Ig Class Switch ELISA.
An ELISA was performed using the Mouse Ig Isotyping ELISA Kit (BD PharMingen) on serum following the manufacturer’s instructions. Plates were read at 570 nm subtracted from 450 nm (SpectramaxM2; Molecular Devices).
Additional Software.
The heat maps used in Fig. S1 were generated utilizing JColorGrid (39). The stack charts used in Fig. 2 were generated utilizing WebLogo (http://weblogo.berkeley.edu/logo.cgi).
Statistical Analysis.
Statistical significance between groups of mice was determined using an unpaired Student t test (Prism, version 5.0c for Mac OSX; GraphPad). Statistically significant results are represented by: *P < 0.05, **P < 0.005, ***P < 0.0001, and not significant. Statistical analysis of Ig and TCR diversity in Fig. S1C was carried out using the protein variability server with the Shannon’s entropy analysis output for position-by-position measurement of diversity (40). The Shannon’s diversity index used in Figs. 2 and 4 was calculated as described previously (41, 42).
Supplementary Material
Acknowledgments
This work was funded by the Wellcome Trust and the UK Medical Research Council.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
See Author Summary on page 18259 (volume 109, number 45).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1210882109/-/DCSupplemental.
References
- 1.Van Laethem F, et al. Deletion of CD4 and CD8 coreceptors permits generation of alphabetaT cells that recognize antigens independently of the MHC. Immunity. 2007;27(5):735–750. doi: 10.1016/j.immuni.2007.10.007. [DOI] [PubMed] [Google Scholar]
- 2.Tikhonova AN, et al. αβ T cell receptors that do not undergo major histocompatibility complex-specific thymic selection possess antibody-like recognition specificities. Immunity. 2012;36(1):79–91. doi: 10.1016/j.immuni.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feng D, Bond CJ, Ely LK, Maynard J, Garcia KC. Structural evidence for a germline-encoded T cell receptor-major histocompatibility complex interaction ‘codon.’. Nat Immunol. 2007;8(9):975–983. doi: 10.1038/ni1502. [DOI] [PubMed] [Google Scholar]
- 4.Dai S, et al. Crossreactive T Cells spotlight the germline rules for alphabeta T cell-receptor interactions with MHC molecules. Immunity. 2008;28(3):324–334. doi: 10.1016/j.immuni.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marrack P, Scott-Browne JP, Dai S, Gapin L, Kappler JW. Evolutionarily conserved amino acids that control TCR-MHC interaction. Annu Rev Immunol. 2008;26:171–203. doi: 10.1146/annurev.immunol.26.021607.090421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scott-Browne JP, White J, Kappler JW, Gapin L, Marrack P. Germline-encoded amino acids in the alphabeta T-cell receptor control thymic selection. Nature. 2009;458(7241):1043–1046. doi: 10.1038/nature07812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garcia KC, Adams JJ, Feng D, Ely LK. The molecular basis of TCR germline bias for MHC is surprisingly simple. Nat Immunol. 2009;10(2):143–147. doi: 10.1038/ni.f.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stadinski BD, et al. A role for differential variable gene pairing in creating T cell receptors specific for unique major histocompatibility ligands. Immunity. 2011;35(5):694–704. doi: 10.1016/j.immuni.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rudolph MG, Stanfield RL, Wilson IA. How TCRs bind MHCs, peptides, and coreceptors. Annu Rev Immunol. 2006;24:419–466. doi: 10.1146/annurev.immunol.23.021704.115658. [DOI] [PubMed] [Google Scholar]
- 10.Burrows SR, et al. Hard wiring of T cell receptor specificity for the major histocompatibility complex is underpinned by TCR adaptability. Proc Natl Acad Sci USA. 2010;107(23):10608–10613. doi: 10.1073/pnas.1004926107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scott DM, et al. Identification of a mouse male-specific transplantation antigen, H-Y. Nature. 1995;376(6542):695–698. doi: 10.1038/376695a0. [DOI] [PubMed] [Google Scholar]
- 12.Laouini D, et al. V beta T cell repertoire of CD8+ splenocytes selected on nonpolymorphic MHC class I molecules. J Immunol. 2000;165(11):6381–6386. doi: 10.4049/jimmunol.165.11.6381. [DOI] [PubMed] [Google Scholar]
- 13.Bartok I, et al. T cell receptor CDR3 loops influence alphabeta pairing. Mol Immunol. 2010;47(7-8):1613–1618. doi: 10.1016/j.molimm.2010.01.012. [DOI] [PubMed] [Google Scholar]
- 14.Furmanski AL, et al. Public T cell receptor beta-chains are not advantaged during positive selection. J Immunol. 2008;180(2):1029–1039. doi: 10.4049/jimmunol.180.2.1029. [DOI] [PubMed] [Google Scholar]
- 15.Furmanski AL, et al. Peptide-specific, TCR-alpha-driven, coreceptor-independent negative selection in TCR alpha-chain transgenic mice. J Immunol. 2010;184(2):650–657. doi: 10.4049/jimmunol.0902291. [DOI] [PubMed] [Google Scholar]
- 16.Armstrong KM, Piepenbrink KH, Baker BM. Conformational changes and flexibility in T-cell receptor recognition of peptide-MHC complexes. Biochem J. 2008;415(2):183–196. doi: 10.1042/BJ20080850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khan JM, Ranganathan S. Understanding TR binding to pMHC complexes: How does a TR scan many pMHC complexes yet preferentially bind to one. PLoS ONE. 2011;6(2):e17194. doi: 10.1371/journal.pone.0017194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Azzam HS, et al. CD5 expression is developmentally regulated by T cell receptor (TCR) signals and TCR avidity. J Exp Med. 1998;188(12):2301–2311. doi: 10.1084/jem.188.12.2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yin Y, Wang XX, Mariuzza RA. Crystal structure of a complete ternary complex of T-cell receptor, peptide-MHC, and CD4. Proc Natl Acad Sci USA. 2012;109(14):5405–5410. doi: 10.1073/pnas.1118801109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adams JJ, et al. T cell receptor signaling is limited by docking geometry to peptide-major histocompatibility complex. Immunity. 2011;35(5):681–693. doi: 10.1016/j.immuni.2011.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stewart-Jones G, et al. Rational development of high-affinity T-cell receptor-like antibodies. Proc Natl Acad Sci USA. 2009;106(14):5784–5788. doi: 10.1073/pnas.0901425106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fellouse FA, Wiesmann C, Sidhu SS. Synthetic antibodies from a four-amino-acid code: A dominant role for tyrosine in antigen recognition. Proc Natl Acad Sci USA. 2004;101(34):12467–12472. doi: 10.1073/pnas.0401786101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ofran Y, Schlessinger A, Rost B. Automated identification of complementarity determining regions (CDRs) reveals peculiar characteristics of CDRs and B cell epitopes. J Immunol. 2008;181(9):6230–6235. doi: 10.4049/jimmunol.181.9.6230. [DOI] [PubMed] [Google Scholar]
- 24.Yin L, et al. A single T cell receptor bound to major histocompatibility complex class I and class II glycoproteins reveals switchable TCR conformers. Immunity. 2011;35(1):23–33. doi: 10.1016/j.immuni.2011.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Juang J, et al. Peptide-MHC heterodimers show that thymic positive selection requires a more restricted set of self-peptides than negative selection. J Exp Med. 2010;207(6):1223–1234. doi: 10.1084/jem.20092170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yin J, Beuscher AE, 4th, Andryski SE, Stevens RC, Schultz PG. Structural plasticity and the evolution of antibody affinity and specificity. J Mol Biol. 2003;330(4):651–656. doi: 10.1016/s0022-2836(03)00631-4. [DOI] [PubMed] [Google Scholar]
- 27.Scott-Browne JP, et al. Evolutionarily conserved features contribute to αβ T cell receptor specificity. Immunity. 2011;35(4):526–535. doi: 10.1016/j.immuni.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wooldridge L, et al. A single autoimmune T cell receptor recognizes more than a million different peptides. J Biol Chem. 2012;287(2):1168–1177. doi: 10.1074/jbc.M111.289488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maynard J, et al. Structure of an autoimmune T cell receptor complexed with class II peptide-MHC: Insights into MHC bias and antigen specificity. Immunity. 2005;22(1):81–92. doi: 10.1016/j.immuni.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 30.Philpott KL, et al. Lymphoid development in mice congenitally lacking T cell receptor alpha beta-expressing cells. Science. 1992;256(5062):1448–1452. doi: 10.1126/science.1604321. [DOI] [PubMed] [Google Scholar]
- 31.Mombaerts P, et al. Mutations in T-cell antigen receptor genes alpha and beta block thymocyte development at different stages. Nature. 1992;360(6401):225–231. doi: 10.1038/360225a0. [DOI] [PubMed] [Google Scholar]
- 32.Chai JG, et al. Critical role of costimulation in the activation of naive antigen-specific TCR transgenic CD8+ T cells in vitro. J Immunol. 1999;163(3):1298–1305. [PubMed] [Google Scholar]
- 33.Arden B, Clark SP, Kabelitz D, Mak TW. Mouse T-cell receptor variable gene segment families. Immunogenetics. 1995;42(6):501–530. doi: 10.1007/BF00172177. [DOI] [PubMed] [Google Scholar]
- 34.Bosc N, Lefranc MP. The mouse (Mus musculus) T cell receptor alpha (TRA) and delta (TRD) variable genes. Dev Comp Immunol. 2003;27(6-7):465–497. doi: 10.1016/s0145-305x(03)00027-2. [DOI] [PubMed] [Google Scholar]
- 35.Karasuyama H, Kudo A, Melchers F. The proteins encoded by the VpreB and lambda 5 pre-B cell-specific genes can associate with each other and with mu heavy chain. J Exp Med. 1990;172(3):969–972. doi: 10.1084/jem.172.3.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Inaba K, et al. Generation of large numbers of dendritic cells from mouse bone marrow cultures supplemented with granulocyte/macrophage colony-stimulating factor. J Exp Med. 1992;176(6):1693–1702. doi: 10.1084/jem.176.6.1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chai JG, Bartok I, Scott D, Dyson J, Lechler R. T:T antigen presentation by activated murine CD8+ T cells induces anergy and apoptosis. J Immunol. 1998;160(8):3655–3665. [PubMed] [Google Scholar]
- 38.Billingham R, Medawar PB. The technique of free skin grafting in mammals. J Exp Biol. 1951;28:385–402. [Google Scholar]
- 39.Joachimiak MP, Weisman JL, May BCh. JColorGrid: Software for the visualization of biological measurements. BMC Bioinformatics. 2006;7:225. doi: 10.1186/1471-2105-7-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Garcia-Boronat M, Diez-Rivero CM, Reinherz EL, Reche PA. PVS: A web server for protein sequence variability analysis tuned to facilitate conserved epitope discovery. Nucleic Acids Res. 2008;36(Web Server issue):W35–W41. doi: 10.1093/nar/gkn211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ferreira C, et al. Non-obese diabetic mice select a low-diversity repertoire of natural regulatory T cells. Proc Natl Acad Sci USA. 2009;106(20):8320–8325. doi: 10.1073/pnas.0808493106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Singh Y, Ferreira C, Chan AC, Dyson J, Garden OA. Restricted TCR-alpha CDR3 diversity disadvantages natural regulatory T cell development in the B6.2.16 beta-chain transgenic mouse. J Immunol. 2010;185(6):3408–3416. doi: 10.4049/jimmunol.1001088. [DOI] [PubMed] [Google Scholar]