Abstract
Epigenetic regulation underlies the robust changes in gene expression that occur during development. How precisely epigenetic enzymes contribute to development and differentiation processes is largely unclear. Here we show that one of the enzymes that removes the activating epigenetic mark of trimethylated lysine 4 on histone H3, lysine (K)-specific demethylase 5A (KDM5A), reinforces the effects of the retinoblastoma (RB) family of transcriptional repressors on differentiation. Global location analysis showed that KDM5A cooccupies a substantial portion of target genes with the E2F4 transcription factor. During ES cell differentiation, knockout of KDM5A resulted in derepression of multiple genomic loci that are targets of KDM5A, denoting a direct regulatory function. In terminally differentiated cells, common KDM5A and E2F4 gene targets were bound by the pRB-related protein p130, a DREAM complex component. KDM5A was recruited to the transcription start site regions independently of E2F4; however, it cooperated with E2F4 to promote a state of deepened repression at cell cycle genes during differentiation. These findings reveal a critical role of H3K4 demethylation by KDM5A in the transcriptional silencing of genes that are suppressed by RB family members in differentiated cells.
Keywords: chromatin, histone demethylase, whole-genome sequencing, histone methylation
Regulation of gene expression is accomplished by transcription factors, histone-modifying enzymes, and chromatin remodeling machinery. The combined action of these three has been implicated in a number of biological processes, including cell cycle control, development, reprogramming, differentiation, and aging. Deregulation of chromatin-modifying enzymes has been strongly linked to the development of cancer. For example, two enzymes that regulate methylation at the histone H3 lysine 4 (H3K4) residue, mixed lineage leukemia-1 (MLL1) and lysine (K)-specific demethylase 5A (KDM5A), have been identified in translocations associated with human leukemia. H3K4 histone methylation states exhibit a highly distinct distribution pattern in the genome. Specifically, H3K4 trimethylation (H3K4me3) is strongly associated with transcriptional activation, with the highest levels observed near transcriptional start sites (TSS). In vitro studies suggest that the four KDM5 enzymes (KDM5A, KDM5B, KDM5C, and KDM5D) are able to remove methylation at lysine 4 of histone H3, and in vivo KDM5A and KDM5B may be recruited to common gene regions (1). This finding leads to multiple questions: (i) Do KDM5 proteins play a role in gene regulation by transcription factors? (ii) Are KDM5 enzymes nonredundant H3K4 demethylases?
During differentiation, cells exhibit two novel properties: repression of cell cycle genes associated with permanent cell cycle exit and activation of cell type-specific genes. Histone modifications are thought to be important epigenetic events intimately linked to initiation and maintenance of transcriptional changes for both of these processes. Cell cycle exit is associated with repression of a large group of genes, many of which are targets of the transcription factor complex between the retinoblastoma protein (pRB) and E2F. Derepression of pRB/E2F target genes plays an important role in human tumors, most of which carry mutations in the RB pathway. During cell cycle progression, pRB and the related proteins p107 and p130 (collectively called “pocket” proteins) are periodically and reversibly recruited to E2F target genes, accompanied by recruitment of histone deacetylase Sin3/HDAC and chromatin-remodeling activities (2). In the Go/quiescent state, the repression of cell cycle genes is mediated by the DP, RB, E2F, and MuvB (DREAM) complex that contains p130 and E2F4 (3).
Here we show that the histone demethylase KDM5A is recruited to genes experiencing strong repression during the course of differentiation, and that these genes are also occupied by components of the DREAM complex. Using high-resolution ChIP-seq analysis, we found that KDM5A preferentially binds to the TSS regions. Using cells where KDM5A expression level was decreased either by RNA inhibition or by using null and conditional (floxed) KDM5A alleles, we show that KDM5A is critical for H3K4 demethylation and repression of cell cycle genes. Differentiation time-course experiments showed a prominent recruitment of both KDM5A and E2F4 during later stages to specific genomic loci. Importantly, the recruitment of KDM5A and E2F4 was independent of one another. Specifically, E2F4 knockdown did not abolish KDM5 recruitment and H3K4 demethylation, and KDM5A knockdown did not affect E2F4 recruitment and histone acetylation. Nevertheless, KDM5A and E2F4 cooperated at cell cycle genes to enforce transcriptional repression.
Results
ChIP-Seq Analysis for KDM5A Identifies Common Targets with E2F4.
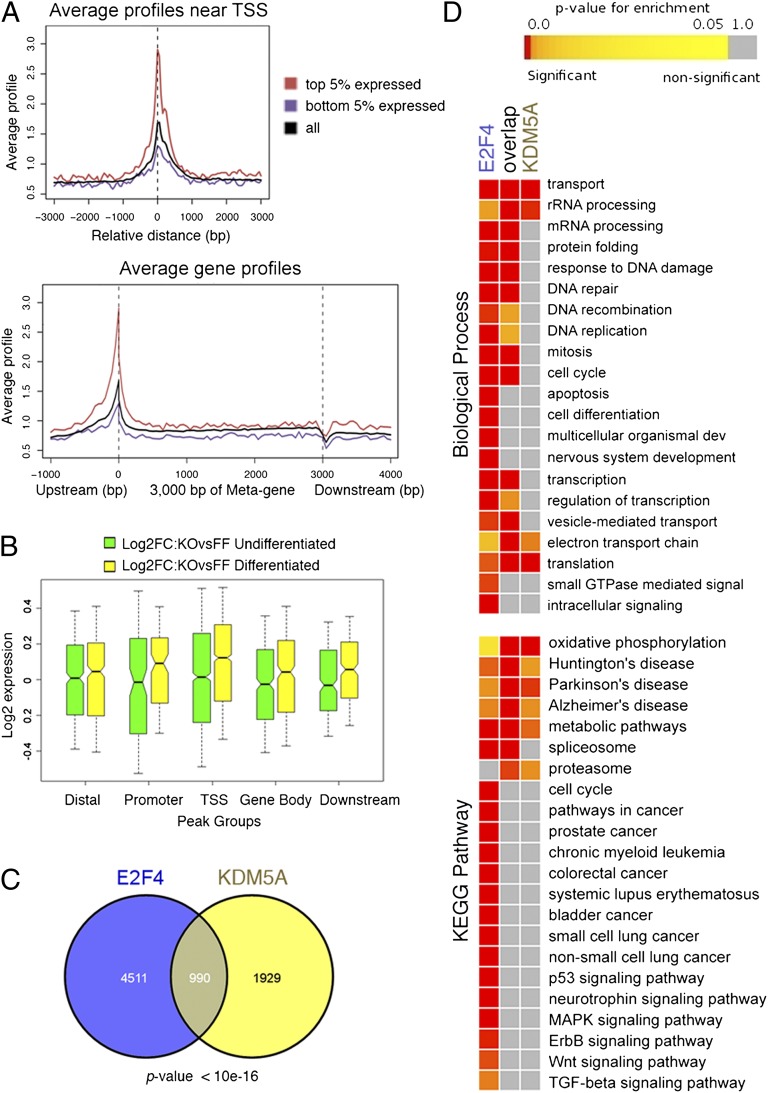
We previously showed that mouse ES cells lacking KDM5A are significantly impaired with respect to differentiation (4). To determine KDM5A targets genome-wide, we performed ChIP-seq experiments in two different clones of mouse ES cells with a conditional (floxed) KDM5A allele (Kdm5af/f) and a clone where the KDM5A allele was deleted (Kdm5a−/−). As expected, we were unable to detect a significant enrichment of bound DNA in ChIP-seq samples from Kdm5a−/− cells, confirming high specificity of the KDM5A antibody. We detected 3,093 regions bound by KDM5A in Kdm5af/f cells (Datasets S1 and S2). Relating KDM5A-bound regions to known mouse genes showed that KDM5A is preferentially bound at TSS regions, especially among the top 5% most-highly expressed genes (Fig. 1A). To determine whether direct KDM5A targets are changed in expression in cells lacking KDM5A, we further analyzed previously acquired gene-expression data (4). We found differences in expression of multiple KDM5A-bound genes (Dataset S3). KDM5A loss did not affect the overall level of KDM5A targets, irrespective of location of KDM5A peaks, in the undifferentiated condition (green boxes in Fig. 1B). Interestingly, in the differentiated condition a much higher number of genes were up-regulated (346 genes) than down-regulated (148 genes) where KDM5A was bound at the TSS (Fig. 1B, yellow boxes, and Dataset S4). Therefore, genes bound by KDM5A at TSS are significantly activated (P < 10−16), specifically when the cells are prone to differentiate.
Fig. 1.
KDM5A ChIP-seq analysis in mouse ES cells reveals corecruitment of KDM5A and E2F4. (A) Average KDM5A enrichment near TSS and in genic region. Genic regions are presented as a 3-kb-long metagene surrounded by a 1-kb region upstream TSS and 1-kb downstream region from transcription termination site (TTS). Dotted lines show TSS and TTS. Most KDM5A binding occurs at the TSS. High TSS ChIP signal is observed for KDM5A reads in the top 5% expressed genes compared with the bottom 5% expressed genes (expression data from GEO GSE5914). A drop in the signal occurs near the TTS, probably because of nucleosome depletion. (B) KDM5A targets in Kdm5a−/− ES cells are differentially expressed. Gene expression microarray data were generated from ES cells that were induced to differentiate by leukemia inhibitory factor (LIF) withdrawal (4). Change in expression for each gene was determined in Kdm5a−/− (KO) versus normal Kdm5af/f (FF) cells (Dataset S3). Box-and-whisker plots show distribution of Log2 fold-change values for genes associated with the classified KDM5A peaks (in relation to the closest gene as distal, promoter, TSS, gene body, or downstream regions). Solid bars of boxes display the 25–75% of ranked genes, with the median indicated as an intersection. The yellow bars depicting the expression changes that occur in differentiated cells in the group with KDM5A bound at the TSS are shifted toward higher values, indicating activation of this group of genes, which is statistically significant as determined by Wilcoxon rank sum test (Dataset S4). (C) KDM5A and E2F4 bind to many common genes. The Venn diagram shows the overlap between KDM5A targets (present study) and E2F4 targets (5). (D) Enrichment analysis for common and distinct target genes shows relations to GO terms and KEGG pathways. Corrected (false-discovery rate) P values are delineated in a colored heatmap, where red signifies overrepresentation of targets in a particular term. All large GO and KEGG groups with overrepresentation are shown.
Next, we compared our KDM5A ChIP-seq data with other genome-wide data obtained in mouse ES cells. Strikingly, our KDM5A data highly overlapped with E2F4 ChIP-on-chip data (5). Almost one-third of KDM5A targets (990 of 2,919) were also direct E2F4 targets (P < 10−16) (Fig. 1C and Dataset S5). KDM5A and E2F4 cooccupied a high proportion of genes with functions in transport and in the mitochondrion (e.g., rRNA processing, oxidative phosphorylation, electron transport chain) (center column “overlap” in Fig. 1D). KDM5A also bound multiple genes that were not direct targets of E2F4, yet these genes still participated in functions linked to genes regulated by both factors. In contrast, genes bound by E2F4 without KDM5A displayed a distinctive set of functions, including apoptosis [120 of 416 genes in this gene ontology (GO) are E2F4 targets], cell differentiation (115 of 498), as well as several cancer pathways [Kyoto Encyclopedia of Genes and Genomes (KEGG)]. Therefore, we were able to distinguish the functions of KDM5A/E2F4 cooccupied genes versus the functions of genes bound solely by E2F4.
KDM5A Is Corecruited with Components of the DREAM Complex.
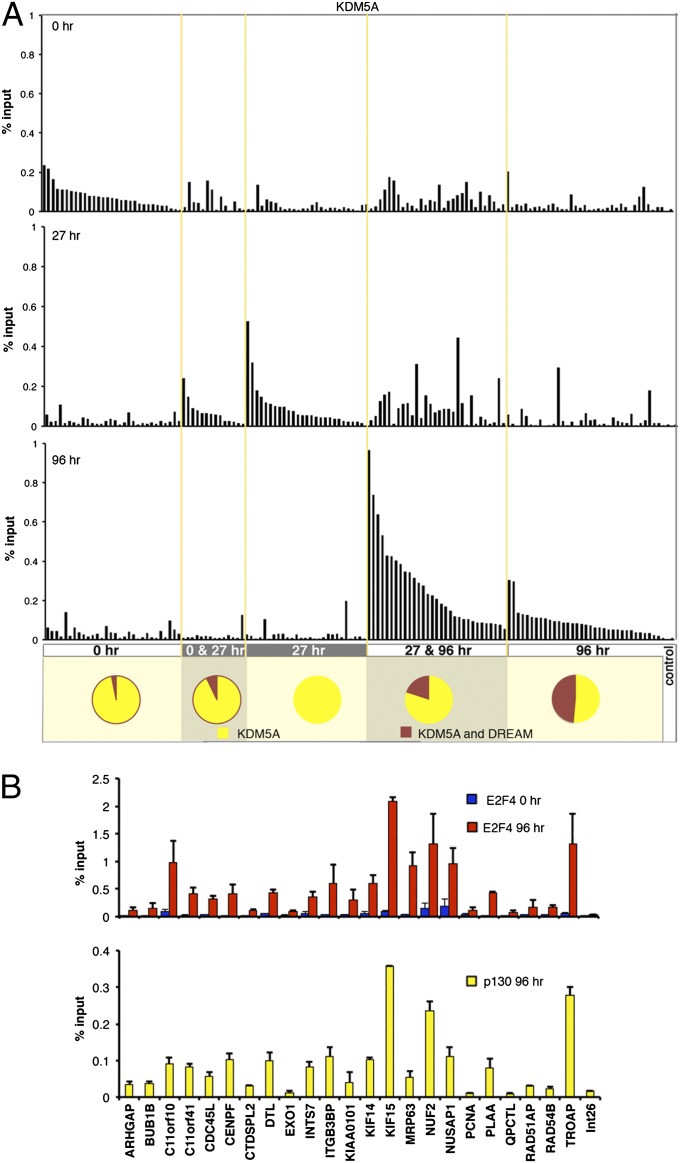
In proliferating cells pRB is inactive (phosphorylated) and E2F1–3 function as transcriptional activators (2). To guide cells out of the cell cycle, pRB is dephosphorylated and interacts with E2F1–3; these RB/E2F complexes bind to E2F target genes, repressing their activity. In contrast to E2F1–3, the E2F4–8 proteins seem to function only as repressors. In quiescent cells, p130/E2F4 is the most prominent pocket protein complex bound to E2F-regulated promoters (3, 6–8). We previously mapped human KDM5A/RBP2 binding regions during differentiation using ChIP-on-chip experiments in diffuse histiocytic lymphoma U937 cells (9). From these two ChIP-on-chip studies, we identified the groups of genes that mapped as condition-specific KDM5A targets (0-h-specific targets, common targets at 0 and 27 h, 27-h-specific targets, common targets at 27 and 96 h, and finally 96-h-specific targets) and DREAM complex targets (Fig. 2A and Dataset S6). We confirmed binding of KDM5A specifically at the relevant differentiation time points at 138 genes from these groups by using ChIP followed by real-time PCR analysis (ChIP-qPCR). At progressively later time points during differentiation, we found a proportional increase in the number of KDM5A targets that are also targets of the DREAM complex (Fig. 2A). Because of the generally high overlap between KDM5A and E2F4 target genes (Fig. 1C), we asked if condition-specific KDM5A targets that were predicted by our transcription factor binding analysis to contain E2F binding sites (9) are bound by E2F4 in the same condition. When these regions were subjected to ChIP-qPCR, all regions displayed high E2F4 binding, with p130 bound to most of these KDM5A targets after 96 h of differentiation (Fig. 2B). We conclude that during the course of differentiation KDM5A may be corecruited with E2F/pocket proteins components at DREAM complex targets.
Fig. 2.
KDM5A is recruited to DREAM complex targets. (A) Quantitative analysis of KDM5A enrichment at target genes during differentiation. Promoter regions bound by KDM5A only at 0, 27, or 96 h, as well as those bound at 0 and 27 h, or at 27 and 96 h were selected from KDM5A ChIP-on-chip analysis (P < 0.002). Primers were designed to the gene-promoter regions (Dataset S6) and the ChIP-qPCR data for each gene was generated in three different conditions, 0, 27, and 96 h after U937 cell treatment with 12-O-tetradecanoylphorbol-13-acetate (TPA). Int26E primer set was used as as control (Dataset S7). Enrichment is expressed relative to input chromatin. The comparison between different time points shows that all genes display differential enrichment for KDM5A, consistent with ChIP-on-chip data. A few genes from the KDM5A targets at 27 and 96 h after induction of differentiation are also enriched at 0 h, which may be because of the high enrichment of KDM5A at these genes in general. Pie charts show the proportion of KDM5A-bound genes and common KDM5A and DREAM target genes. (B) ChIP assays, using E2F4 and p130 antibodies, of 24 gene targets of KDM5A in differentiating cells containing predicted E2F-binding sites. Increased enrichment at 96 h indicates targets of the p130/E2F4 complex during differentiation. Error bars: means ± SEM, n = 2.
KDM5A Plays a Major Role in H3K4 Demethylation.
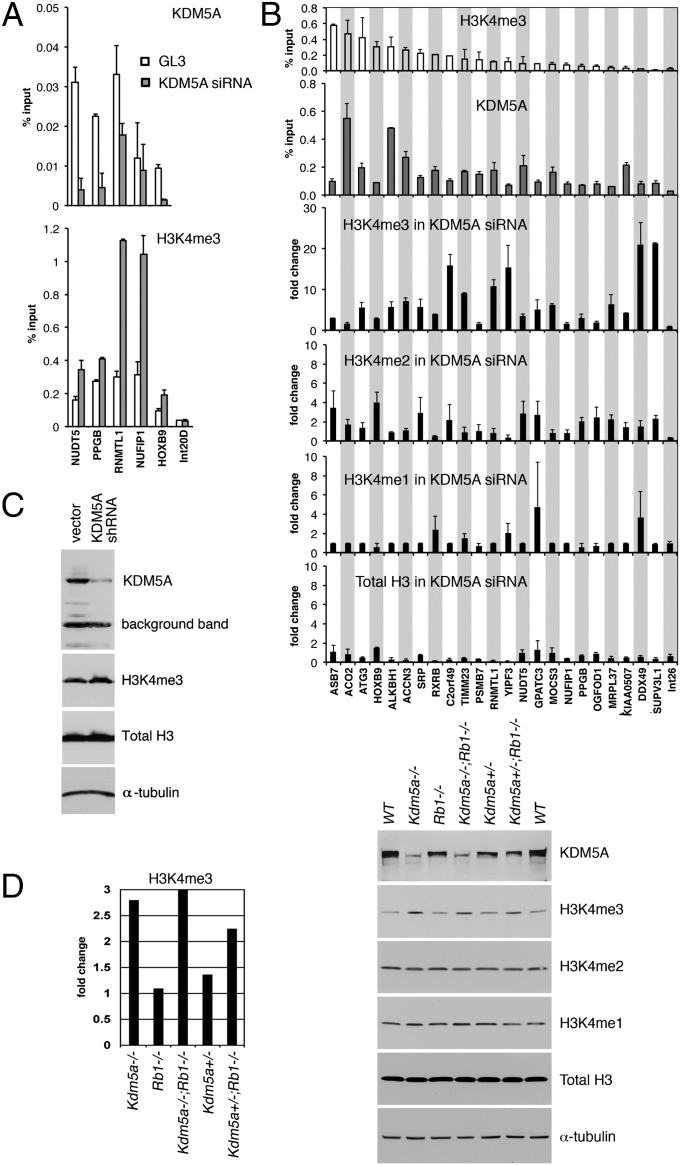
ChIP-on-chip analysis showed preferential association of KDM5A with promoter regions that are enriched with H3K4me3 (9). This finding is quite surprising, given that KDM5A has been shown to specifically demethylate H3K4me2 and me3 residues and forced expression of KDM5A resulted in a global loss of histone H3 methylation (10, 11). This result prompted us to study H3K4me3 in the regions bound by KDM5A by ChIP-qPCR and to ask if KDM5A binding to a promoter affects histone methylation. We performed ChIP-qPCR on 64 promoters displaying enrichment of KDM5A in two studied differentiation conditions, 0 and 96 h. Strikingly, all 64 promoters bound by KDM5A had a high level of H3K4 methylation in both conditions (Fig. S1 and Dataset S8).
Next, we tested H3K4me3 abundance in cells that are deficient for KDM5A. As previously shown, KDM5A protein levels can be efficiently decreased in SAOS-2 cells with KDM5A siRNA (12). Cells treated with a KDM5A siRNA displayed a quantifiable decrease in KDM5A binding in ChIP experiments compared with cells treated with control siRNA (Fig. 3A). The decrease in KDM5A binding at a promoter was always accompanied by increased H3K4me3 (Fig. 3A), suggesting that KDM5A affects histone methylation through direct binding to the promoter. To ask which H3K4 methylation state is most affected by the lack of KDM5A, we analyzed a set of 23 different KDM5A bound promoters (Fig. 3B). At 19 of these regions, the level of H3K4me3 increased 5–20 times in cells treated with KDM5A siRNA (Fig. 3B), whereas there was only a marginal increase in H3K4me2. Promoter regions normally lack H3K4me1, and we were unable to detect significant monomethylation after knockdown of KDM5A. The observed increase in H3K4 methylation was not because of an increase in the total histone H3 level. These data show that KDM5A is required for maintaining tri- and dimethylation of histone H3K4 at the promoter regions bound by KDM5A.
Fig. 3.
Cells deficient in KDM5A have global and gene-specific changes in H3K4 methylation. (A) SAOS-2 cells transfected with either KDM5A siRNA or control GL3 siRNA were analyzed by ChIP-qPCR for H3K4me3 and KDM5A binding at five random KDM5A target genes. The nucleosomes at the intergenic region (Int20D region), not bound by KDM5A, were not highly methylated at H3K4 in cells treated with either control siRNA or KDM5A siRNA. (B) The level of H3K4 methylation at KDM5A target genes is changed in KDM5A knockdown. ChIP-qPCR experiments for H3K4me3 and KDM5A were performed for the indicated genes in untreated SAOS-2 cells and enrichment is shown as a percentage of input. In KDM5A siRNA-treated SAOS-2 cells, enrichment in methylated histone H3K4 and total H3 are shown as a fold-change difference compared with cells treated with scrambled siRNA. Error bars in A and B: means ± SEM, n = 2. (C) Human U937 cells transduced with either KDM5A shRNA lentiviruses or control lentiviruses were analyzed for KDM5A, H3K4me3 and total H3 by immunoblotting. (D) Wild-type MEFs and MEFs with a deletion in the Kdm5a or Rb1 (RB) locus were analyzed for H3K4 methylation by immunoblotting. The KDM5A antibody gives a weak background band, which is evident in Kdm5a−/− cells. H3K4me3 was quantitated relative to α-tubulin that was used as a loading control in C and D. The levels of H3K4me3 in mutant MEFs are shown as fold-change differences compared with wild-type MEFs. Data are representative of three independent experiments.
We next asked if loss of KDM5A results in changes in the total level of H3K4 methylation. U937 cells, where we stably down-regulated KDM5A using a lentiviral shRNA vector, displayed a small but reproducible increase in the global level of H3K4me3 as revealed by immunoblotting analysis (Fig. 3C). This finding suggests that KDM5A is necessary for maintaining the total level of H3K4me3. Next, low-passage mouse embryonic fibroblasts (MEFs) were isolated from mice with a homozygous or heterozygous deletion of KDM5A. Consistent with this result in cells with shRNA, we found that the global level of H3K4me3 was again increased, almost threefold, in MEFs with genetic ablation of KDM5A (Fig. 3D). This result was not because of proliferation defect in Kdm5a−/− cells, which is pRB-dependent (4), because Kdm5a−/−;Rb1−/− cells still had high methylation. Strikingly, the H3K4me3 level still reproducibly increased (around 1.3-fold) in Kdm5a+/− cells compared with wild-type cells (Fig. 3D), indicating that deletion of only one copy of KDM5A is sufficient to affect methylation on a global scale. Therefore, by using two different systems of KDM5A depletion, RNA inhibition in human cells and deletion of the KDM5A allele in mouse cells, we showed that that the level of H3K4 trimethylation is dosage-sensitive to the amount of KDM5A, and is not compensated by KDM5B or other enzymes.
KDM5A and E2F4 Independently Contribute to Gene Repression.
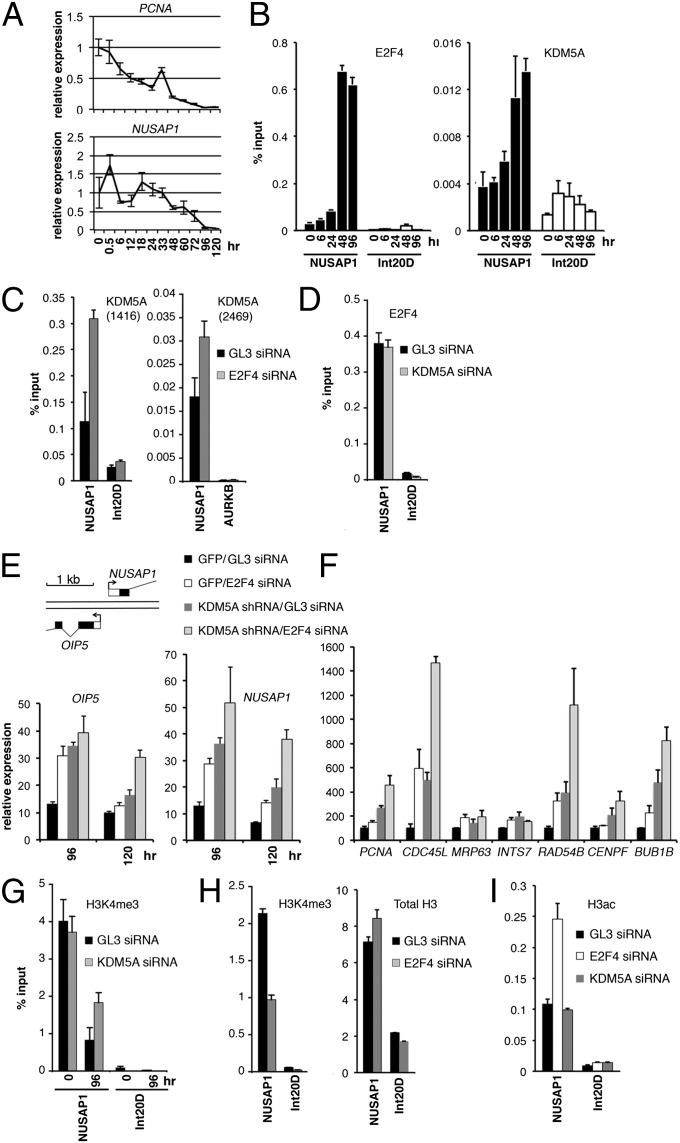
In the asynchronous cell population that we used in ChIP-seq, E2F4 occupied a high number of cell cycle genes (218 of 470 cell cycle genes, P < 1e-16) (Fig. 1D). We detected a significant enrichment of genes involved in cell cycle (i.e., GOs: DNA repair, DNA recombination, DNA replication, mitosis, cell cycle) bound by both E2F4 and KDM5A (Fig. 1D), raising the possibility that E2F4 and KDM5A cooperate with one another in the regulation of cell cycle genes. PCNA (proliferating cell nuclear antigen) and NUSAP1 (nucleolar spindle-associated protein) are targets of both E2F and KDM5A, and have specific roles in proliferating cells. PCNA acts as a processivity factor for DNA polymerase δ, and NUSAP1 plays a role in spindle microtubule organization during mitosis. mRNA and protein levels of NUSAP1 peak at the G2-M cell cycle transition, associated with microtubule formation, and abruptly decline after cell division, localizing to nucleoli during interphase (13). After induction of differentiation in U937 cells by 12-O-tetradecanoylphorbol-13-acetate (TPA) treatment, the steady-state mRNA levels of PCNA and NUSAP1 gradually decline (Fig. 4A). We performed ChIP-qPCR for E2F4 and KDM5A at the times of no/low repression of PCNA and NUSAP1 (0, 6, and 24 h), intermediate repression (48 h), and substantial repression (96 h). From 24 to 48 h NUSAP1 expression dropped about 2-fold, and between 48 h and 96 h it dropped 10-fold (Fig. 4A). This drop paralleled a two- to threefold increase in KDM5A binding and an almost sevenfold increase in E2F4 binding (Fig. 4B). The difference in KDM5A recruitment is unlikely because of changes in its cellular protein level, as the KDM5A protein level does not change significantly during the cell cycle (Fig. S2) or differentiation (12). Therefore, the binding of both E2F4 and KDM5A simultaneously increased at the point when the NUSAP1 promoter was experiencing repression.
Fig. 4.
KDM5A-mediated H3K4 demethylation represses cell cycle genes during U937 cell differentiation independent of E2F4. (A) Relative transcript level of NUSAP1 and PCNA at timed intervals after TPA addition, normalized to the level of B2M. (B) The difference in occupancy of the E2F4- and KDM5A-dependent promoter at different time points of differentiation as determined by ChIP. Intergenic unbound region Int20D is shown as a control. (C) E2F4 knockdown results in increased occupancy of KDM5A at the NUSAP1 promoter. The cells treated with the indicated siRNAs were induced with TPA for 48 h and ChIP experiments were performed using the KDM5A antibody 1416. To confirm the obtained result, ChIP experiments were done using the KDM5A antibody 2469, a distinct control region, AURKB. (D) E2F4 binding to the NUSAP1 promoter is unchanged in cells with KDM5A knockdown. ChIP E2F4 experiments were performed in parallel with the experiments in C. (E) Combined E2F4 and KDM5A knockdown result in derepression of the NUSAP1 promoter. The siRNA treatments did not change the expression level of unrelated genes (Fig. S3C). Gene map of the bidirectional NUSAP1/OIP5 promoter is shown. (F) Combined E2F4 and KDM5A knockdown results in additive and cooperative effects. The cells were induced for differentiation with TPA for 120 h. (G) KDM5A knockdown results in increased methylation in differentiating cells. (H) E2F4 knockdown results in decreased methylation in differentiating cells. (I) Histone acetylation changes at the NUSAP1 promoter occur after E2F4 loss but not after KDM5A loss. In H and I, U937 cells were treated with the indicated siRNAs, induced with TPA for 72 h and the ChIP experiments were performed with the antibodies to H3K4me3, total H3 or acetylated H3. Error bars: means ± SEM, n = 3.
The cooccurrence of KDM5A and E2F4 at genomic regions (Figs. 1C and 4B) raised the possibility that E2F4 or KDM5A may be recruiting one another to these regions. We studied KDM5A enrichment at the NUSAP1 promoter in cells treated with E2F4 siRNA. Using two different KDM5A antibodies in ChIP assays, we detected an increase rather than a decrease in KDM5A recruitment in cells lacking E2F4 (Fig. 4C). Conversely, we did not see any change in E2F4 occupancy in KDM5A siRNA-treated cells (Fig. 4D). These data imply that KDM5A and E2F4 are recruited independently of each other and are consistent with earlier studies that failed to detect a physical interaction between KDM5A and E2F4 (14–16).
To ask if KDM5A binding has functional impact on expression of E2F targets and whether E2F4 and KDM5A operate through the same or different mechanisms, we generated U937 cells with single and combined E2F4 and KDM5A knockdowns and measured the mRNA levels of NUSAP1, PCNA, and a panel of other common KDM5A/E2F4 targets during differentiation (Fig. 4 E and F, and Fig. S3A). The NUSAP1 promoter region is bidirectional, containing the TSS of another gene, OIP5, separated by only 107 nucleotides (human genome assembly NCBI36/hg18) from the NUSAP1 TSS. Loss of E2F4 led to increased expression of NUSAP1, OIP5, and other genes (Fig. 4 E and F), consistent with the repressive effects of E2F4 on its direct targets. Knockdown of LIN9, another component of the DREAM complex, resulted in derepression similar to E2F4 knockdown (Fig. S3 B and C). Loss of KDM5A also generally resulted in greater derepression than the loss of E2F4 alone (Fig. 4 E and F). Finally, the combined loss of E2F4 and KDM5A resulted in greater derepression of their shared targets than the loss of each individual factor, suggesting an additive effect. For example, CDC45L mRNA increased 15-fold in the double knockdown compared with about 5-fold in each single knockdown (Fig. 4F). Significantly, these experiments showed that KDM5A and the DREAM complex not only bind independently to the promoters of cell cycle-regulated genes in the repressed state, but also independently and actively repress these genes, resulting in the cumulative and synergistic effects.
KDM5A Represses Cell Cycle Genes Through H3K4 Demethylation.
Decreased H3K4 methylation has been detected during differentiation at the promoters of MFN2, BRD8/KIF20A, FGF4, OTX2, HOXA1, HOXA5, and HOXA7 genes bound by KDM5A (9, 11, 14, 17). The question of whether the regulation of a cell cycle gene involves removal of H3K4 trimethylation by KDM5A has not been studied. Treatment of U937 cells with TPA for 96 h decreased H3K4me3 at the NUSAP1 and PCNA promoters, but KDM5A knockdown resulted in twofold higher H3K4 methylation at the promoters of differentiated cells (Fig. 4G and Fig. S3D). If E2F4 is required for KDM5A function in gene repression, then removing E2F4 may increase H3K4 methylation similar to the removal of KDM5A. However, the H3K4me3 level at the NUSAP1 promoter was actually decreased in cells treated with E2F4 siRNA (Fig. 4H), which was consistent with the increased KDM5A enrichment at NUSAP1 promoter (Fig. 4C). This finding was not associated, however, with changes in nucleosome density because it was not accompanied by a decrease in total histone H3 (Fig. 4H). Consistent with E2F4 recruitment of Sin3/HDAC, knockdown of E2F4 resulted in increased acetylation at the NUSAP1 promoter, but not at a control genomic region that is not bound by E2F4 (Fig. 4I). In contrast, no effect on acetylation was observed in KDM5A knockdown. This finding suggests that KDM5A-mediated repression is critical for E2F target-gene repression and it involves the demethylation of H3K4me3 positioned close to the TSS. The combined regulation by both histone H3K4 demethylation and histone deacetylation results in cumulative effect (Fig. 4 E and F) that may be critical to achieving the dramatic drop in the level of gene expression during differentiation (Fig. 4A).
Discussion
The concerted action of transcription factors and histone-modifying enzymes is important for transcriptional regulation and epigenetic memory. We found that the deep transcriptional repression of cell cycle genes is achieved by targeting both KDM5A, a major H3K4 histone demethylase, and pocket domain proteins to their promoter regions. Importantly, recruitment of KDM5A to target genes was independent of E2F4 binding, although both regulators cooperated in repressing cell cycle genes. Previous studies have implicated KDM5A together with three other paralogs in histone H3K4 methylation. In this study, we used three different systems to decrease the level of the KDM5A protein and are unique in showing that the four KDM5 family members are not entirely redundant and KDM5A plays a prominent role in gene repression during differentiation.
We showed that KDM5A cooccupies a substantial portion of E2F4 gene targets. The monocyte/macrophage differentiation system recapitulated the dynamics of transcription factor binding and histone modifications at E2F promoters: coincident to the silencing of cell cycle genes, E2F4/p130 and KDM5A were recruited, and the promoter lost H3K4 methylation. Recent RNA-seq experiments, which are more quantitative than microarrays, showed that the cell cycle genes as a group experience the greatest decrease in expression, subsequent to the onset of differentiation (18). Previous genome-wide in silico computational analyses of promoters identified key regulators of human cell cycle-regulated genes, with significant enrichment of the E2F, NRF1, NF-Y, and cyclic AMP-responsive element binding motifs in their promoters (19). Interactions between such factors could have a cooperative effect at some promoters. Studies in Caenorhabditis elegans, Drosophila, and mammalian cells showed that the E2F4 protein is involved in the formation of several chromatin complexes. In association with p130 in the DREAM complex, E2F4 lacks the Sin3 component (3). Nonetheless, E2F4-mediated repression at KDM5A-bound genes still involves histone deacetylation, which is likely to occur through association of E2F4 with HDAC activity. The cooperative activity with KDM5A may lead to a higher level of repression than that which is achieved by pocket protein complexes alone. In human retinoblastoma, many chromosomal alterations include regions of genes involved in cell cycle regulation (20), suggesting that their additional loss is important for tumorigenesis. We propose that the pocket proteins are central regulators in cell growth and proliferation because they are able to regulate genes involved in cell cycle progression using distinct mechanisms (e.g., KDM5A and E2F).
We showed that KDM5A is required for gene regulation during differentiation, because in differentiating Kdm5a−/− ES cells KDM5A TSS targets generally have higher expression compared with wild-type ES cells (Fig. 1B). Consistent with this observation, our previous microarray study showed that correlation between recruitment of KDM5A and decreased transcription was characteristic of the whole group of genes specifically bound by KDM5A in the differentiated condition (9). Besides cell cycle genes, during differentiation KDM5A is bound to genes that code for proteins involved in mitochondrial function and to developmental genes, such as the HOX genes (9, 11, 21). The coexistence of H3K27me3 and H3K4me3 epitomize the epigenetic state of HOX genes and other developmental genes in stem and progenitor cells. H3K27 methylation is accomplished by recruitment of PRC2, which contains the histone H3K9 and H3K27 methyltransferase EZH2 as the catalytic component (22). HOX are another group of genes besides cell cycle genes dramatically changed in expression during terminal differentiation. In particular, HOXA9 is expressed more than 100-fold higher in stem and progenitor cells than in differentiated cell populations (23). One study showed that the PRC2 complex recruits KDM5A to a large number of genes, which is required for repression of PRC2 target genes during ES cell differentiation (17). However, we and another group were unable to see a significant overlap between ChIP-seq KDM5A peaks and the PRC2 location in U937 cells and mouse ES cells (24), indicating that this is not a major mechanism for repression of KDM5A bound genes.
Previous studies of mammalian H3K4 methyltransferases showed that deletion of any one of the MLL proteins had only a minimal effect on the global levels of H3K4 methylation. MLL1 overexpression increases levels of histone H3K4me2/3 at specific genes, such as HOXA9, yet does not change the global level of H3K4me2/3 (25). This finding is in sharp contrast to the demethylases that are not wholly redundant because decreasing the level of KDM5A, either by shRNA in a human cancer cell line or using homo- and heterozygous mice, increased the global level of H3K4me3. The majority of histone H3 in proliferating cells is part of chromatin (26), and despite observations that KDM5 proteins occur in different chromatin complexes and bind to different genome locations (this study and refs. 27), KDM5A, based on our findings, plays a major role in global histone demethylation.
Materials and Methods
Cell Culture and Immunoblotting.
For induction of differentiation, TPA was added to cells at different times as described (12) and cells were collected simultaneously for RNA isolation or ChIP assays. Immunoblotting was performed using our rabbit KDM5A antiserum 2469 in Fig. 3C, a mixture of 2469 and the affinity purified antibody 1416 in Fig. 3D, and with the antibody 1416 in Fig. S2A, anti-H3K4me3 (07-473), anti-H3K4me2 (07-030), and anti-H3K4me1 (07-436) from Upstate, total histone H3 (ab-1791) antibodies from Abcam, mouse RB G3-245 (BD Biosciences), and α-tubulin (T9026) (Sigma) antibodies.
ChIP and Gene Expression Analyses.
Kdm5af/f (wild-type KDM5A) ES cells were isolated from mouse blastocysts of Kdm5af/f mice, which were maintained on a pure C57BL/6 background. Successful Cre-dependent recombination was performed as previously described (4). ChIP-seq experiments were performed with the KDM5A antibody 1416 following the previously described procedure (28). ChIP experiments were performed as described (9) using the following rabbit antibodies: KDM5A antibodies 1416 and 2469, H3K4me3 (ab-8580) and total histone H3 (ab-1791) from Abcam, H3K4me2 (07-030) and H3K4me1 (07-436) from Upstate, and E2F4 (sc-1082) and p130 (sc-317) from Santa Cruz. To determine transcript levels in U937 cells, RNA was isolated and reverse-transcribed. Real-time PCR was performed using the SYBR Green PCR master mix and iCycler MyiQ system or SsoFast EvaGreen Supermix and the CFX96 system (Bio-Rad).
Enriched probe genomic locations (29) were annotated to the closest TSS of EnsEMBL genes (v55) using Bioconductor (v2.7, R v2.12.1) (30) package ChIPpeakAnno (v1.60) (31). Target genes of KDM5A in U937 and expression data of genes in this cell line were obtained from our previous study (9). Genomic location of E2F4 targets was taken from GSE20551 [particularly GSM516408 (5)]. GiTools were used for enrichment analysis and heatmap generation (32). Expression data in differentiating Kdm5af/f and Kdm5a−/− ES cells were taken from one of our other studies (4). Expression data in ES cells from C57BL/6 mouse were used from the Gene Expression Omnibus (GEO) dataset GSE5914. ChIP-seq data are available from http://www.ncbi.nlm.nih.gov/geo/ under accession number GSE28343.
Supplementary Material
Acknowledgments
We thank Ms. Alexandra Vilkova for technical assistance. The project was supported by Grants R01 CA138631 (to E.V.B.) and R01 CA076120 (to W.G.K.) from the National Institutes of Health; Education Grant SAF2009-06954 (to N.L.-B.) from the Spanish Ministry of Science; a Chercheur boursier (junior 1) award from the Fonds de la recherche en santé du Québec (to N.G.); Clinical and Translational Science Award Grant UL1 RR024139 and a V Scholar Award (to Q.Y.); and a fellowship from the Autonomous Government of Catalonia, Spain (to A.B.M.M.K.I.). W.G.K. is a Howard Hughes Medical Institute Investigator and a Doris Duke Distinguished Scientist.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE28343).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1216724109/-/DCSupplemental.
References
- 1.Islam AB, Richter WF, Lopez-Bigas N, Benevolenskaya EV. Selective targeting of histone methylation. Cell Cycle. 2011;10(3):413–424. doi: 10.4161/cc.10.3.14705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Frolov MV, Dyson NJ. Molecular mechanisms of E2F-dependent activation and pRB-mediated repression. J Cell Sci. 2004;117(Pt 11):2173–2181. doi: 10.1242/jcs.01227. [DOI] [PubMed] [Google Scholar]
- 3.Litovchick L, et al. Evolutionarily conserved multisubunit RBL2/p130 and E2F4 protein complex represses human cell cycle-dependent genes in quiescence. Mol Cell. 2007;26(4):539–551. doi: 10.1016/j.molcel.2007.04.015. [DOI] [PubMed] [Google Scholar]
- 4.Lin W, et al. Loss of the retinoblastoma binding protein 2 (RBP2) histone demethylase suppresses tumorigenesis in mice lacking Rb1 or Men1. Proc Natl Acad Sci USA. 2011;108(33):13379–13386. doi: 10.1073/pnas.1110104108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim J, et al. A Myc network accounts for similarities between embryonic stem and cancer cell transcription programs. Cell. 2010;143(2):313–324. doi: 10.1016/j.cell.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rayman JB, et al. E2F mediates cell cycle-dependent transcriptional repression in vivo by recruitment of an HDAC1/mSin3B corepressor complex. Genes Dev. 2002;16(8):933–947. doi: 10.1101/gad.969202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Balciunaite E, et al. Pocket protein complexes are recruited to distinct targets in quiescent and proliferating cells. Mol Cell Biol. 2005;25(18):8166–8178. doi: 10.1128/MCB.25.18.8166-8178.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith EJ, Leone G, DeGregori J, Jakoi L, Nevins JR. The accumulation of an E2F-p130 transcriptional repressor distinguishes a G0 cell state from a G1 cell state. Mol Cell Biol. 1996;16(12):6965–6976. doi: 10.1128/mcb.16.12.6965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lopez-Bigas N, et al. Genome-wide analysis of the H3K4 histone demethylase RBP2 reveals a transcriptional program controlling differentiation. Mol Cell. 2008;31(4):520–530. doi: 10.1016/j.molcel.2008.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klose RJ, et al. The retinoblastoma binding protein RBP2 is an H3K4 demethylase. Cell. 2007;128(5):889–900. doi: 10.1016/j.cell.2007.02.013. [DOI] [PubMed] [Google Scholar]
- 11.Christensen J, et al. RBP2 belongs to a family of demethylases, specific for tri-and dimethylated lysine 4 on histone 3. Cell. 2007;128(6):1063–1076. doi: 10.1016/j.cell.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 12.Benevolenskaya EV, Murray HL, Branton P, Young RA, Kaelin WG., Jr Binding of pRB to the PHD protein RBP2 promotes cellular differentiation. Mol Cell. 2005;18(6):623–635. doi: 10.1016/j.molcel.2005.05.012. [DOI] [PubMed] [Google Scholar]
- 13.Raemaekers T, et al. NuSAP, a novel microtubule-associated protein involved in mitotic spindle organization. J Cell Biol. 2003;162(6):1017–1029. doi: 10.1083/jcb.200302129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Oevelen C, et al. A role for mammalian Sin3 in permanent gene silencing. Mol Cell. 2008;32(3):359–370. doi: 10.1016/j.molcel.2008.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hayakawa T, et al. RBP2 is an MRG15 complex component and down-regulates intragenic histone H3 lysine 4 methylation. Genes Cells. 2007;12(6):811–826. doi: 10.1111/j.1365-2443.2007.01089.x. [DOI] [PubMed] [Google Scholar]
- 16.Silverstein RA, Ekwall K. Sin3: A flexible regulator of global gene expression and genome stability. Curr Genet. 2005;47(1):1–17. doi: 10.1007/s00294-004-0541-5. [DOI] [PubMed] [Google Scholar]
- 17.Pasini D, et al. Coordinated regulation of transcriptional repression by the RBP2 H3K4 demethylase and polycomb-repressive complex 2. Genes Dev. 2008;22(10):1345–1355. doi: 10.1101/gad.470008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brunskill EW, Lai HL, Jamison DC, Potter SS, Patterson LT. Microarrays and RNA-Seq identify molecular mechanisms driving the end of nephron production. BMC Dev Biol. 2011;11:15. doi: 10.1186/1471-213X-11-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elkon R, Linhart C, Sharan R, Shamir R, Shiloh Y. Genome-wide in silico identification of transcriptional regulators controlling the cell cycle in human cells. Genome Res. 2003;13(5):773–780. doi: 10.1101/gr.947203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen D, Gallie BL, Squire JA. Minimal regions of chromosomal imbalance in retinoblastoma detected by comparative genomic hybridization. Cancer Genet Cytogenet. 2001;129(1):57–63. doi: 10.1016/s0165-4608(01)00427-7. [DOI] [PubMed] [Google Scholar]
- 21.Wang GG, et al. Haematopoietic malignancies caused by dysregulation of a chromatin-binding PHD finger. Nature. 2009;459(7248):847–851. doi: 10.1038/nature08036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cao R, Zhang Y. The functions of E(Z)/EZH2-mediated methylation of lysine 27 in histone H3. Curr Opin Genet Dev. 2004;14(2):155–164. doi: 10.1016/j.gde.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 23.Novershtern N, et al. Densely interconnected transcriptional circuits control cell states in human hematopoiesis. Cell. 2011;144(2):296–309. doi: 10.1016/j.cell.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peng JC, et al. Jarid2/Jumonji coordinates control of PRC2 enzymatic activity and target gene occupancy in pluripotent cells. Cell. 2009;139(7):1290–1302. doi: 10.1016/j.cell.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Milne TA, et al. Multiple interactions recruit MLL1 and MLL1 fusion proteins to the HOXA9 locus in leukemogenesis. Mol Cell. 2010;38(6):853–863. doi: 10.1016/j.molcel.2010.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loyola A, Bonaldi T, Roche D, Imhof A, Almouzni G. PTMs on H3 variants before chromatin assembly potentiate their final epigenetic state. Mol Cell. 2006;24(2):309–316. doi: 10.1016/j.molcel.2006.08.019. [DOI] [PubMed] [Google Scholar]
- 27.Xie L, et al. KDM5B regulates embryonic stem cell self-renewal and represses cryptic intragenic transcription. EMBO J. 2011;30(8):1473–1484. doi: 10.1038/emboj.2011.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beshiri ML, et al. Genome-wide analysis using ChIP to identify isoform-specific gene targets. J Vis Exp. 2010 doi: 10.3791/2101. (41) pii:2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lander ES, et al. International Human Genome Sequencing Consortium Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 30.Gentleman RC, et al. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 2004;5(10):R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu LJ, et al. ChIPpeakAnno: A Bioconductor package to annotate ChIP-seq and ChIP-chip data. BMC Bioinformatics. 2010;11:237. doi: 10.1186/1471-2105-11-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perez-Llamas C, Lopez-Bigas N. Gitools: Analysis and visualisation of genomic data using interactive heat-maps. PLoS ONE. 2011;6(5):e19541. doi: 10.1371/journal.pone.0019541. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.