Abstract
“Sputnik” is a dsDNA virus, referred to as a virophage, that is coassembled with Mimivirus in the host amoeba. We have used cryo-EM to produce an electron density map of the icosahedral Sputnik virus at 3.5-Å resolution, sufficient to verify the identity of most amino acids in the capsid proteins and to establish the identity of the pentameric protein forming the fivefold vertices. It was also shown that the virus lacks an internal membrane. The capsid is organized into a T = 27 lattice in which there are 260 trimeric capsomers and 12 pentameric capsomers. The trimeric capsomers consist of three double “jelly-roll” major capsid proteins creating pseudohexameric capsomer symmetry. The pentameric capsomers consist of five single jelly-roll proteins. The release of the genome by displacing one or more of the pentameric capsomers may be the result of a low-pH environment. These results suggest a mechanism of Sputnik DNA ejection that probably also occurs in other big icosahedral double jelly-roll viruses such as Adenovirus. In this study, the near-atomic resolution structure of a virus has been established where crystallization for X-ray crystallography was not feasible.
Keywords: dsDNA egress, virus assembly, Mimivirus satellite, polypeptide tracing, cryo-electron microscopy
Mimivirus is one of the largest known viruses, first discovered in a water tower during a search for the cause of a hospital-acquired pneumonia outbreak in 2003 (1). Mimivirus has a dsDNA genome and uses Acanthamoeba as its natural host (1). Sputnik virus was discovered to be associated with Mimivirus when isolated from amoeba (2). Sputnik can only be propagated when coinfected with Mimivirus. The amoeba host implies that both Mimivirus and Sputnik are frequently present in aquatic food, resulting in disease when humans eat raw food (3). Unlike most satellite viruses, coinfection decreases the production of Mimivirus by approximately 70% (2). The requirement of Mimivirus for the propagation of Sputnik gave rise to the term “virophage” (4). EM images suggest that Sputnik virus matures in viral factories together with Mimivirus itself (5). This observation is supported by in situ hybridization experiments (6), which showed that Sputnik replicates its genome in viral factories. Other virophages such as Mavirus and Organic Lake virus, which are associated with giant dsDNA viruses, have been discovered recently (7, 8). These virophages might influence the ecosystem of aquatic environments (8).
Sputnik virions have an AT-rich, circular 18-kb dsDNA genome with 21 ORFs (1). Genes V20, V19, and V8 were tentatively associated with the major and two minor capsid proteins, respectively (1). Sequences homologous to the major capsid protein (MCP) of Sputnik have been found in Mavirus (19% sequence identity) and Organic Lake virus (28% sequence identity). Another eight ORFs of Spunik’s genome have similarity to eukaryal, archaeal, and bacterial virus genes.
A 10.7-Å resolution cryo-EM study of the Sputnik virus (9) had shown that the capsid protein assembled into an icosahedral shell with a diameter of approximately 750 Å and T = 27 quasi-symmetry (SI Materials and Methods). Thus, the capsid is composed of 260 quasi-hexomeric “hexons” and 12 pentomeric “pentons.” The double “jelly-roll” hexon structure of the Paramecium bursaria Chlorella virus 1 (PBCV-1) MCP (Vp54) fitted well into the 10.7-Å resolution cryo-EM density map of Sputnik (9). A mushroom-like fiber was found associated with the center of each hexon, although the icosahedrally averaged density suggested that the fiber had only partial occupancy. The capsid protein appeared to surround a membrane that enclosed the genome, consistent with the presence of lipid in the virus.
The capsid structure of numerous icosahedral viruses consists of several jelly-roll folds organized into pseudohexameric capsomers. The jelly-roll structure is a β-barrel formed by two opposing antiparallel-stranded β-sheets. If the strands along the polypeptide are identified by B, C, D, E, F, G, H, and I, then the strands in the two sheets are BIDG and CHEF (10). The jelly-roll structure was first recognized in the capsid of small icosahedral plant viruses (11, 12). The structure of small RNA animal viruses such as picornaviruses were then discovered to have similar tertiary structures for their capsid proteins as well as similar quaternary structure for the organization of the viral capsid (13, 14). Some ssDNA viruses were also found to have a jelly-roll structure for their capsid proteins (15, 16). The structure of the dsDNA adenovirus capsid protein was found to have two consecutive jelly-rolls in its MCP (17), giving the base of the capsomer a hexagonal shape. A similar double jelly-roll was found in the plant RNA cowpea mosaic virus (18) and later in PBCV-1 (19). Other viruses with double jelly-roll MCP structures include the dsDNA PRD1 bacterial phage (20), the marine phage PM2 (21, 22), the archaeal Sulfolobus turreted icosahedral virus (23, 24), and vaccinia virus (25, 26). Furthermore, there are numerous other viruses that most likely also have double jelly-roll MCPs according to sequence comparisons, as for instance Mimivirus (1) and African swine fever virus. Where the structure is known, the double jelly-roll forms a trimeric “capsomer” that has quasi-sixfold symmetry. In double jelly-roll structures, large insertions often occur between β-strands B and D and between F and G. These insertions form surface features of the virus. The term “tower” has been used for especially large (approximately 300 amino acids) insertions between strands DE and FG in the first jelly-roll of the adenovirus structure (17), giving the external part of the capsomer a triangular appearance.
We report here a 3.5-Å near-atomic resolution structure of Sputnik determined by cryo-EM. The MCP was confirmed as being the product of gene V20 and was shown, as previously suggested (9), to have a double jelly-roll structure. The protein that constitutes the penton was identified as being a combination of gene products V18 and V19, which had wrongly been assigned to have a frameshift relative to each other. The resultant V18/19 was shown to be a single jelly-roll structure. A 3.8-Å resolution structure of the emptied Sputnik virus was calculated and compared with the full virus structure, which suggests a possible mechanism of Sputnik DNA ejection that might apply to other large dsDNA viruses with double jelly-roll MCPs.
Results and Discussion
Overall Structure of the Sputnik Virus.
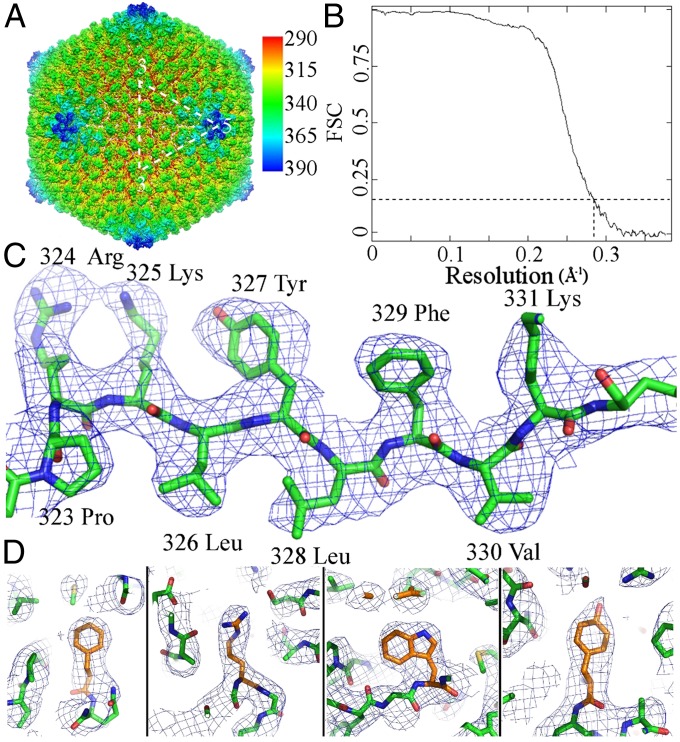
A 3.5-Å resolution, icosahedrally averaged map (Fig. 1 A–D) was determined by image reconstruction using approximately 12,000 cryo-EM particles. In addition, a 3.8-Å resolution map of empty capsids was calculated using approximately 13,000 empty particles. There are 4 and 1/3 hexon capsomers in each icosahedral asymmetric unit of the virus, where the 1/3 hexon is from a hexon sitting on the icosahedral threefold axis. In addition there is 1/5 of a penton capsomer adjacent to the fivefold vertex in the asymmetric unit. After the hexon and penton densities had been interpreted there remained three similar uninterpreted densities, each representing nine amino acids, presumably corresponding to the same sequence of a minor capsid protein. These densities were located on the inside of the capsid, at the boundary between pairs of hexons (Fig. S1), but are absent in the empty capsids and must have been associated with the DNA so as to be able to exit along with the DNA.
Fig. 1.
Reconstruction of Sputnik at 3.5-Å resolution. (A) Reconstruction of the whole virus, colored according to radius. (B) Fourier shell correlation (FSC) coefficient plot showing the resolution of Sputnik reconstruction to be 3.5 Å (FSC = 0.143) according to the criterion defined by Rosenthal and Henderson (42). (C) Cryo-EM density of a β-strand in the MCP. (D) Cryo-EM density of selected residues, Phe, Arg, Trp, and Tyr (from left to right), showing the quality of the reconstruction.
Major Capsid Protein.
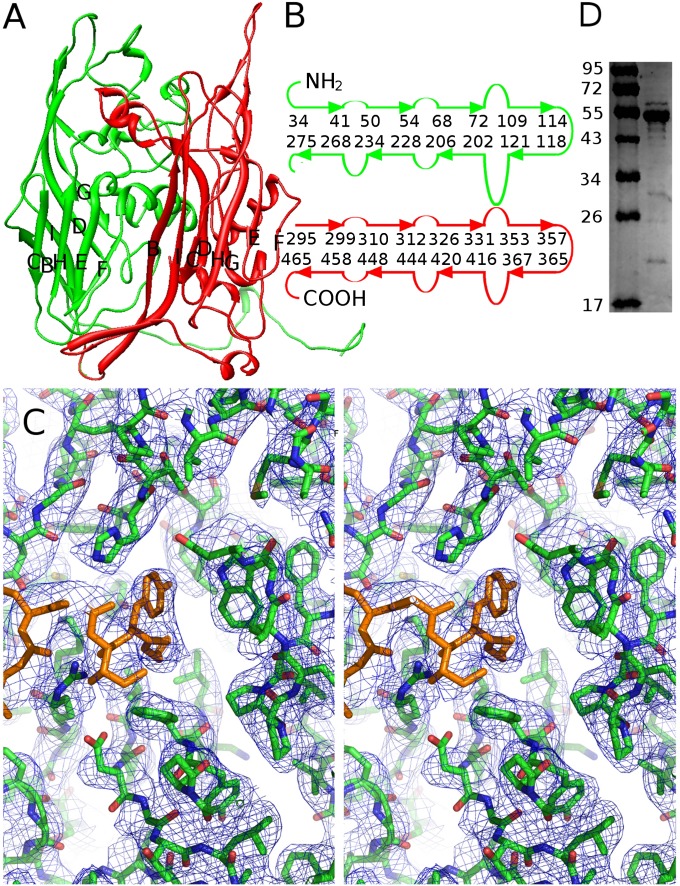
The structures of the 13 MCPs in the icosahedral asymmetric unit were built independently and then refined with the program CNS (27). The refined structure had an R-factor of 0.26 and 3.8% of residues in disallowed regions of the Ramachandran plot. Root-mean-square deviation of bond lengths from ideal values was 0.012 and that of angles was 1.7. The structure of the first 508 amino acids of each of the MCPs could be easily built using the amino acid sequence corresponding to gene V20 (Fig. 2 A and B). Residues 497–508 form an α-helix that is inserted into a hydrophobic pocket located in the center of each hexon. There is not enough space for amino acids that follow residue 508 to exit this pocket (Fig. 2C). Furthermore, an SDS/PAGE gel (Fig. 2D) shows that the molecular mass of the MCP is approximately 55 kDa, whereas the calculated molecular mass of the whole protein is 65.4 kDa. Therefore, the last 87 residues of the MCP must have been cleaved off probably during assembly. The overall structure of the MCP is a double jelly-roll fold with the BC, DE, FG, and HI loops pointing toward the quasi-sixfold axis of the capsomer, as in other jelly-roll–containing viral capsomers. Amino acids 1–279 of V20 form the first jelly-roll, and amino acids 280–507 form the second jelly-roll. As in the MCPs of PBCV-1 (Vp54), PRD1 (p3), and the adenovirus hexon, there is a “tower” structure created from the usual large insertions between strands D to E and F to G in both jelly-rolls (Fig. 2B). However, in Sputnik and PRD1 the insertion between strands H and I of the first jelly-roll also contributes to the tower structure. Furthermore, in Sputnik the insertion between strands F and G of the second jelly-roll contributes to the tower of the neighboring molecule.
Fig. 2.
Double jelly-roll structure of MCP. (A) Ribbon diagram of the Sputnik MCP. The first and second jelly-roll domains are colored green and red, respectively. (B) Diagrammatic representation of the arrangement of β-strands and residue numbers at the ends of the β-strands. (C) Stereo diagram showing the C-terminal region of the MCP. Residues 502–508 are colored orange. These residues bind into a hydrophobic pocket. There is no density or space for the remaining 87 C-terminal residues of the MCP sequence, suggesting that these residues had been cleaved. (D) Molecular mass of the MCP is estimated to be approximately 55 kDa by SDS/PAGE gel.
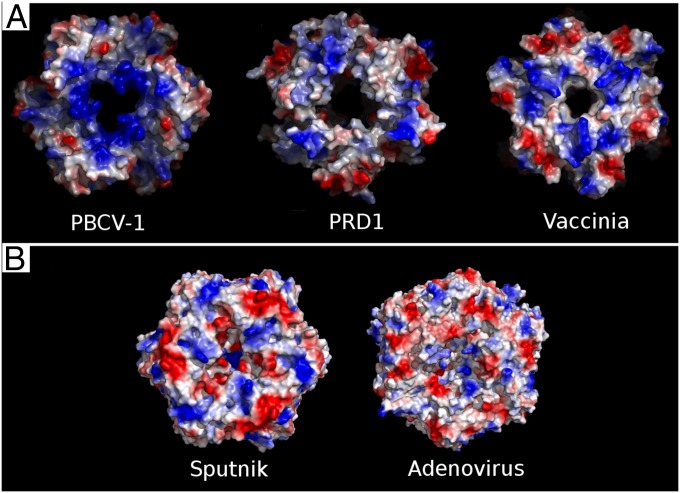
PBCV-1, PRD1, and vaccinia virus have a viral membrane, and their MCPs have highly positively charged surfaces facing the membrane (Fig. 3A). For Sputnik the surface of the MCP facing the inside of the virus has both negatively and positively charged patches (Fig. 3B) at neutral pH, similar to the charge distribution of the inside surface of the adenovirus hexon protein. Thus, the charge distribution of Sputnik’s MCP indicates that Sputnik probably does not have a viral membrane. The absence of a lipid membrane was further confirmed using paper spray mass spectroscopy (Figs. S2 and S3; SI Materials and Methods). The ease with which the genome can be persuaded to be ejected from the capsid by changes of pH (see below) also speaks to the absence of a barrier, were a membrane present.
Fig. 3.
Comparison of the inner surface charge distribution of dsDNA virus MCPs with a double jelly-roll fold. (A) Viruses that have a lipid membrane envelope. (B) Viruses that do not have a lipid membrane envelope.
The region immediately inside the capsid protein appears as three parallel layers, each approximately 25 Å thick, in a 20-Å resolution low-pass filtered map (Fig. S4A). The reconstruction of the empty virus does not have these layers (Fig. S4B). Therefore, the three consecutive density layers within the capsid probably represent ordered or semiordered dsDNA, as is observed for many dsDNA bacteriophages (28, 29). The outermost layer is more intense, presumably because the DNA structure is closely associated with the icosahedral capsid structure. This conclusion contradicts the earlier results (9), based on a 10.7-Å resolution map and mass spectroscopic results, that the density in this region corresponds to a membrane. Although the cause of this discrepancy is unclear, the present mass spectroscopic results have been confirmed by both positive and negative controls using viruses with and without a lipid membrane, respectively (Figs. S2 and S3).
The earlier 10.7-Å resolution study (9) had shown that the external surface of the capsid was decorated with occasional, approximately 100-Å-long, mushroom-like fibers. These features were not visible in the 3.5-Å resolution map, demonstrating that these features are essentially disordered at a resolution of better than 11 Å. It is unknown whether these features are proteins or glycans.
Penton Protein.
The Sputnik cryo-EM density representing the pentons and especially the insertion loops is slightly poorer than that of hexon densities, probably caused by greater flexibility of the penton loops. A Cα atom model was built into the electron density map, identifying the location of approximately 350 amino acids. Because the sequence of the Sputnik penton protein was unknown, a program was designed to align the protein sequences given by the Sputnik genome, expressed in terms of amino acid sizes, with the sizes of the amino acids in the cryo-EM density map (SI Materials and Methods). A satisfactory alignment was obtained for the approximately 150 N-terminal amino acids of gene product V18 and for the alignment of gene products V19 with the remaining C-terminal residues of the Cα atom penton structural model. In the gene sequencing results (2), there was a frame shift between the V18 and the initial bases of the V19 gene. However, according to the electron density map, the penton structure was only one polypeptide chain. Therefore, when the Sputnik genome was resequenced near the junction of these genes, it was found that the original sequence had one additional erroneous base that created a stop codon for V18. Thus, in the absence of the erroneous base, V18 and the whole V19 sequence were in the same frame without a stop codon between them, making V18 and V19 one V18/19 fusion gene, coding for 379 amino acids. The amino acid sequence was then built into the cryo-EM density except for 11 disordered amino acids at the N terminus and 5 disordered amino acids at the C terminus.
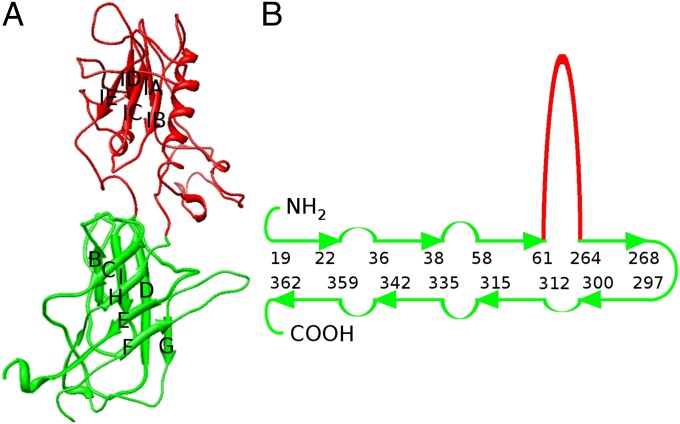
The penton protein folds into two domains: a “lower” domain that has a 184 amino acid jelly-roll structure located toward the interior of the virus, and an “upper” domain consisting of 195 amino acids (amino acids 66–261) located on the surface of the virus around the fivefold vertex (Fig. 4 A and B). The orientation of the five jelly-roll domains is roughly similar to the orientation of the VP1 subunits in picornaviruses. In Sputnik the β-strands are roughly radial, whereas in picornaviruses the β-strands are somewhat more tangential, but in both cases the BC, DE, FG, and HI loops are closest to the outside of the virus. The upper domain is primarily a β-barrel inserted between β-strands D and E of the lower jelly-roll domain. If the inserted β-strands along the polypeptide are named IA, IB, IC, ID, and IE, then the two β-sheets would consist of strands IB, IC, IE and of IA, ID. An α-helix, lying parallel to the β-strands, links β-strands IA and IB. The mean center position of the upper domain is rotated counterclockwise (seen from outside the virus) by approximately 60° around the fivefold axis, relative to the lower domain, placing the upper domain almost above the lower domain of the neighboring monomer. The penton has a pore along the fivefold axis with a diameter of 7–10 Å near the outside of the virus that is blocked near the base of the lower domain by residues 318–320. The closest structure found in a Dali search (30) was the penton protein of human adenovirus (31). The adenovirus penton structure has the same counterclockwise twist between the upper domain and lower domain as occurs in Sputnik. Like Sputnik, the upper domain of the adenovirus penton protein has the same fold as the upper domain of the Sputnik penton protein and is an insertion of 307 amino acids between β-strands D and E of the adenovirus lower domain. However, the adenovirus penton protein upper domain has an additional insertion of 53 amino acids between β-strands G and F. The adenovirus upper domain of the penton protein contains an Arg-Gly-Asp sequence that binds to the cellular αvβ3 receptor (32, 33). Only the nonenveloped viruses adenovirus and Sputnik have an insertion domain in their penton proteins. Sputnik, unlike adenovirus, does not have an Arg-Gly-Asp sequence or a fiber and attachment protein emanating from the fivefold vertices.
Fig. 4.
Single jelly-roll structure of sputnik penton protein. (A) Ribbon diagram of the Sputnik penton protein. The jelly-roll domain is colored green, and the insertion domain is colored red. (B) Diagrammatic representation of the arrangement of β-strands in the penton protein and the residue numbers at the ends of the β-strands.
Capsomer Interactions.
Any pseudohexameric capsomer formed by three double jelly-roll monomers has six edges (Fig. 5A). Each double jelly-roll monomer within a capsomer contributes to two edges, with each edge having an interaction with a neighboring capsomer. The interaction between neighboring capsomers can be divided into three classes. The first two classes form a pseudo-twofold axis between the first (f) edge of one capsomer (dominated by the CHEF sheet of the first jelly-roll) and the first (f) edge of a neighboring capsomer (ff), or between the second (s) edge of a capsomer (dominated by the CHEF sheet of the second jelly-roll) with the second (s) edge of another capsomer (ss). A third class of interaction (fs) is between the first (f) edge of one capsomer and the second (s) edge of a neighboring capsomer. This classification has similarities to that used to describe contacts between capsomers of PRD1 (34).
Fig. 5.
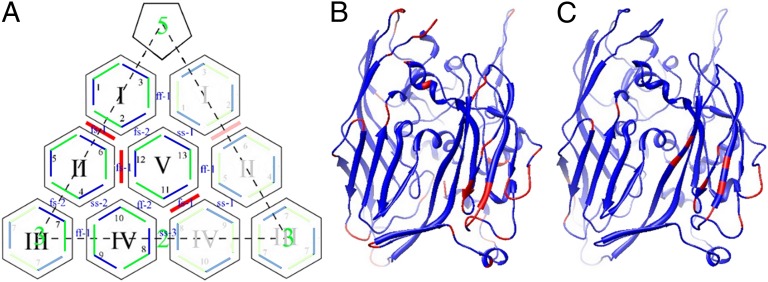
Asymmetric unit of Sputnik. (A) Capsomers are identified by Roman numerals I to V. Thirteen independent monomers are identified by Arabic numerals 1 to 13. Each MCP is shown by a green line (the first jelly-roll domain) followed by a blue line (the second jelly-roll domain). The independent minor proteins in the asymmetric unit are colored red. Icosahedrally related positions are indicated in the same manner but with corresponding faded colors. (B) The backbone of the MCP was colored according to the root mean square deviation of each Cα position from the mean of the superimposed 13 independent MCPs, ranging from blue (<0.6 Å) to red (>0.6 Å). (C) Number of times a given residue in the 13 independent MCPs is involved in capsomer to capsomer contacts ranging from blue (<8) to red (>8).
Each type of capsomer in one icosahedral asymmetric unit of Sputnik was identified by the Roman numerals I, II, III, IV, and V and the MCP monomers by Arabic numerals 1 to 13 (Fig. 5A). The rotational relationships between neighboring capsomers (Table S1) can be divided into a bend and twist component. The curvature of the viral capsid is determined by the bend angle. If all of the bend angles between neighboring capsomers were zero, then the capsomers would form a plane. The variation of bend angle was found to be limited to approximately 10°, 20°, and 40° degrees in the fs, ff, and ss classes, respectively.
Hydrogen bonds between capsomers were identified by visual inspection. The three classes of interactions, ff, ss and fs, could be subdivided into two or three subclasses depending on the conservation of hydrogen bonds (Table S1). Although there are five examples of class fs in the Sputnik structure, only three of these contact regions are associated with density probably representing the binding site of a minor capsid protein (see above). These three contact regions all belong to the same subclass fs-1, whereas the other two examples of fs contacts are associated with a different set of hydrogen bonds.
The root mean square deviation between equivalence Cα atoms is less than 0.43 Å on superimposing any pair of the 13 MCP structures on each other. However, there are specific regions where the MCP structure has greater variability (Fig. 5B), with displacements of the Cα atoms as much as 3.5 Å. These regions are mostly the loops at the top of each “tower” and in the CHEF β-sheets of the second jelly roll that make contacts with neighboring capsomers (Fig. 5C).
Empty Virus.
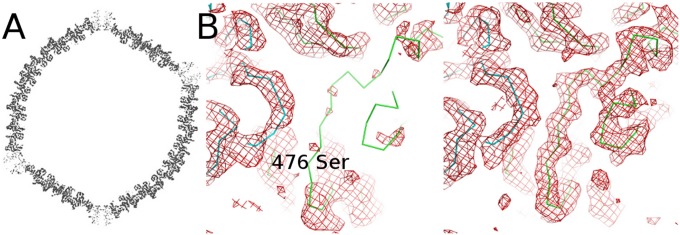
During the purification procedure of Sputnik (Materials and Methods), the virus separated into two bands while sedimenting through a cesium chloride gradient. The two bands corresponded to the full and empty virus. Subsequent work showed that empty particles can be produced from full particles either by raising the pH to 9 or lowering the pH to 5.5. It is therefore possible that the purified empty Sputnik particles were produced in a low-pH virus factory (35) during Mimivirus production. The purified empty particles were used in the reconstruction of a 3.8-Å resolution cryo-EM map (Materials and Methods and Fig. S5A). The empty Sputnik particles had a diameter that was approximately 5.2 Å larger than the diameter of the full infectious virus when measured along the threefold axes. However, there was no expansion along the fivefold axes. Presumably the expansion was made possible by the loss of interaction between the packaged DNA and the capsid structure.
The height of the penton density in the empty virus was approximately half of the hexon density, suggesting that some of the penton proteins were missing in the empty particles (Fig. 6A). Presumably the DNA had escaped from one of the vertices where there was no penton protein. Indeed, two particles that had been emptied by lowering the pH of the environment showed a feature that is probably DNA-like density spilling out from one of the vertices (Fig. S5B). A similar loss of the penton structure occurs for adenovirus, which can lose some pentons in the low-pH environment of an endosome (36). Similarly a mutant of the bacteriophage PRD1 can lose some pentons in the presence of a detergent, although PRD1, unlike Sputnik and Adenovirus, has a membrane (34). In the empty particle reconstruction of Sputnik, the electron density for residues 476–508 was missing in the MCP of monomer #1, the monomer adjacent to the penton (Fig. 6B). These residues are in the interface between the penton and hexon. However, it is not clear why these residues are completely disordered when only some of the pentons are missing, which would require disorder only in the #1 MCPs close to one of the missing penton sites. Thus, possibly the pH or other stress disorders these residues, resulting in only the loss of some pentons. The loss of pentons in double jelly-roll dsDNA viruses may be a general phenomenon for genome delivery.
Fig. 6.
Structure of the empty Sputnik. (A) Cross-section through the empty Sputnik cryo-EM map, showing where the density is at a higher level. The density of the penton protein is lower than that of the MCPs. (B) Electron density (red) of the hexon capsomer, adjacent to the penton protein in the empty virus (Left) and full virus (Right). The Cα backbone structure (green) of MCP #1, closest to the penton. The electron density from residue 476 to residue 508 of MCP #1 is missing in the empty virus. The Cα backbone structure of penton protein is colored blue.
Evolution.
Single and double jelly-roll MCP structures have been observed in ssRNA, ssDNA, and dsDNA icosahedral viruses (19, 20, 34). In all these cases the jelly-roll is assembled into threefold symmetric capsomers that have quasi-sixfold symmetry. These capsomers are assembled into hexagonal arrays as predicted by Caspar and Klug (37), separated by approximately 75 Å center-to-center (19, 38). However, a double jelly-roll assembly is not feasible around the pentameric vertices. Hence, a single jelly-roll protein fold has been retained in all such viruses to complete the assembly around the pentameric vertices. For example, in picornaviruses the pentamer is formed by five copies of VP1 (13); in adenoviruses (39), PRD1 (40), PM2 (21, 22), and Sputnik the pentamer is formed by five copies of the penton molecule.
Many, but not all, dsDNA viruses that have a double jelly-roll capsid fold also have a lipid membrane envelope. Thus, the mode of genome delivery will be quite different for these viruses, even if the assembly process might be similar. Both Sputnik and adenovirus, neither of which have a membrane envelope, have a similarly structured inserted “upper” domain at the pentameric vertex that participates in viral entry for adenovirus (33, 41) and possibly therefore also in Sputnik. However, in contrast, PRD1 has a membrane and the penton is missing the “upper” insertion domain (22), suggesting a different genome delivery system.
Materials and Methods
Sputnik virus was grown and purified as previously described (9). After concentrating the virus to a concentration of approximately 3 mg/mL, a 3-μL sample was placed onto a glow discharged C-flat EM grid with 1.2-μm-diameter holes in a thin carbon layer. The grid was flash-frozen using a Gatan Cryoplunge3 device with a blotting time of approximately 12 s in liquid ethane at approximately 100 K. The particle images were collected on Kodak film (Kodak Electron Image Film SO-163) with an FEI Titan Krios electron microscope using a nominal magnification of 59,000×. The images were taken with a defocus range between 1.5 and 2.5 μm. The electron dose was approximately 22 electrons/Å2. The films were digitized with a Nikon scanner (Super Cool Scan 9000) using a 6.35-μm scan step size, giving a final pixel size of 1.1 Å.
A 3.5-Å resolution, icosahedrally averaged map (Fig. 1A) was determined by image reconstruction using approximately 12,000 cryo-EM particles. In addition, a 3.8-Å resolution map of empty capsids was calculated using approximately 13,000 empty particles. The overall resolution was calculated according to where the Fourier shell correlation coefficient (42) fell below 0.143 (Fig. 1B). The quality of the map was also judged by visual inspection, with particular reference to the ability to differentiate between large residues such as arginine, phenylalanine, tyrosine, and tryptophan (Fig. 1 C and D).
A total of approximately 18,000 virus particles were selected from approximately 250 micrographs, based on the Thon rings extending beyond 5 Å. These particles were boxed using the program e2boxer.py from the EMAN2 software package (43). The contrast transfer function parameters of the particles from each micrograph were determined with the CTFIT program in the EMAN software package (44). An initial model was generated, using a fourfold binned dataset (adjacent pixels in a 4 × 4 box were averaged), by selecting particle images with the best two-, three-, and fivefold rotational symmetry by the program starticos. The initial model was improved with the program refine (from the EMAN software package) (44) by refining the orientation and center of each boxed image. The refinement was stopped when the reconstruction showed no improvement in the Fourier shell correlation coefficient compared with the previous reconstruction cycle.
A twofold binned dataset was used to determine the initial orientation and translation parameters of each particle by iterative refinement. The best match between the observed images and the projected model derived from the previous iteration was used to determine orientation and translation parameters. Particles that had a discrepancy of more than 2.5° in any of the angles among the best three projections were excluded from the next reconstruction. For the following refinement cycles only the density of the capsid in the improved reconstruction was retained as a model. The refinement was terminated when the resolution of the reconstruction showed no further improvement in the Fourier shell coefficient (FSC).
Because Sputnik particles were embedded in a layer of vitreous ice, the defocus of individual particles have variation within a single film, which would also affect the pixel size of the individual particle. A full-size (nonbinned) dataset was used to refine the defocus, pixel size, orientation, and translation of each particle. A combination of FREALIGN (45) and EMAN (44) was used to refine these parameters locally according to the initial parameters. Particles were not included in the reconstruction if any of the parameters had an unexpected large change in their angular identification compared with the previous cycle. The quality of the reconstruction was monitored by visual inspection of various features, such as the connectivity of the main chain density in the loop area and the integrity of the side chain density in the map. The improved reconstruction was then used as a model for next cycle until the quality of the map showed no further improvement in the FSC. Approximately 12,000 particles were used in the final reconstruction at a resolution of approximately 3.5 Å. The 3.8-Å resolution map of the empty virus was calculated using the same methods, starting with approximately 20,000 particles and a final selection of approximately 13,000 particles.
Initially Cα-only models of both MCP and penton protein were built into the EM density by using the “Cα baton mode” in Coot (46). The Cαs were then replaced by the corresponding amino acids in the sequence. The atomic positions were refined by using “real space refine zone” in the program Coot (46). The atomic model was refined using the CNS software package (47). The whole icosahedral asymmetric unit was refined with strict NCS constraints to take into account the icosahedral symmetry and the contacts between various subunits.
Calculation of the bend and twist angles was based on the direction of the quasi-threefold axes (normal) of neighboring capsomers and the vector connecting their centers. A plane A was defined by the normal of capsmer “a” and the vector connecting the center of this capsomer to the neighboring capsomer “b.” A similar plane B was defined for the neighboring capsomer “b.” The angle between planes A and B defined the twist angle. To determine the bend angle a vector was calculated that was the projection of the normal of capsomer “b” onto plane A. The angle between this vector and the normal of capsomer “a” defined the bend angle.
Supplementary Material
Acknowledgments
We thank Graham Cooks, Thomas Müller, and Christina Ferreira (Department of Chemistry, Purdue University) for providing expertise and assistance with paper spray mass spectroscopy (SI Materials and Methods), and Sheryl Kelly for help in the preparation of the manuscript. This work was supported by a National Institutes of Health (NIH) Grant AI011219 (to M.G.R.). The purchase of the FEI Titan microscope was made possible by NIH Grant RR028984.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. N.G. is a guest editor invited by the Editorial Board.
Data deposition: The atomic coordinates and structure factors have been deposited in the Electron Microscopy Data Bank, www.ebi.ac.uk/pdbe/emdb [EMDB ID codes 5495 (3.5-Å resolution cryo-EM map of Sputnik) and 5496 (3.8-Å resolution cryo-EM map of empty Sputnik)]; and the Protein Data Bank, www.pdb.org [PDB ID code 3J26 (coordinates of the near-atomic resolution structure of Sputnik)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1211702109/-/DCSupplemental.
References
- 1.Raoult D, et al. The 1.2-megabase genome sequence of Mimivirus. Science. 2004;306(5700):1344–1350. doi: 10.1126/science.1101485. [DOI] [PubMed] [Google Scholar]
- 2.La Scola B, et al. The virophage as a unique parasite of the giant mimivirus. Nature. 2008;455(7209):100–104. doi: 10.1038/nature07218. [DOI] [PubMed] [Google Scholar]
- 3.Parola P, et al. Acanthamoeba polyphaga mimivirus virophage seroconversion in travelers returning from Laos. Emerg Infect Dis. 2012;18(9):1500–1502. doi: 10.3201/eid1809.120099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krupovic M, Cvirkaite-Krupovic V. Virophages or satellite viruses? Nat Rev Microbiol. 2011;9(11):762–763. doi: 10.1038/nrmicro2676. [DOI] [PubMed] [Google Scholar]
- 5.Desnues C, Raoult D. Inside the lifestyle of the virophage. Intervirology. 2010;53(5):293–303. doi: 10.1159/000312914. [DOI] [PubMed] [Google Scholar]
- 6.Desnues C, Raoult D. Virophages question the existence of satellites. Nat Rev Microbiol. 2012;10(3):234–, author reply 234. doi: 10.1038/nrmicro2676-c3. [DOI] [PubMed] [Google Scholar]
- 7.Fischer MG, Suttle CA. A virophage at the origin of large DNA transposons. Science. 2011;332(6026):231–234. doi: 10.1126/science.1199412. [DOI] [PubMed] [Google Scholar]
- 8.Yau S, et al. Virophage control of antarctic algal host-virus dynamics. Proc Natl Acad Sci USA. 2011;108(15):6163–6168. doi: 10.1073/pnas.1018221108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun S, et al. Structural studies of the Sputnik virophage. J Virol. 2010;84(2):894–897. doi: 10.1128/JVI.01957-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rossmann MG, Johnson JE. Icosahedral RNA virus structure. Annu Rev Biochem. 1989;58:533–573. doi: 10.1146/annurev.bi.58.070189.002533. [DOI] [PubMed] [Google Scholar]
- 11.Harrison SC, Olson AJ, Schutt CE, Winkler FK, Bricogne G. Tomato bushy stunt virus at 2.9 A resolution. Nature. 1978;276(5686):368–373. doi: 10.1038/276368a0. [DOI] [PubMed] [Google Scholar]
- 12.Abad-Zapatero C, et al. Structure of southern bean mosaic virus at 2.8 A resolution. Nature. 1980;286(5768):33–39. doi: 10.1038/286033a0. [DOI] [PubMed] [Google Scholar]
- 13.Rossmann MG, et al. Structure of a human common cold virus and functional relationship to other picornaviruses. Nature. 1985;317(6033):145–153. doi: 10.1038/317145a0. [DOI] [PubMed] [Google Scholar]
- 14.Hogle JM, Chow M, Filman DJ. Three-dimensional structure of poliovirus at 2.9 A resolution. Science. 1985;229(4720):1358–1365. doi: 10.1126/science.2994218. [DOI] [PubMed] [Google Scholar]
- 15.Tsao J, et al. The three-dimensional structure of canine parvovirus and its functional implications. Science. 1991;251(5000):1456–1464. doi: 10.1126/science.2006420. [DOI] [PubMed] [Google Scholar]
- 16.McKenna R, et al. Atomic structure of single-stranded DNA bacteriophage phi X174 and its functional implications. Nature. 1992;355(6356):137–143. doi: 10.1038/355137a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roberts MM, White JL, Grütter MG, Burnett RM. Three-dimensional structure of the adenovirus major coat protein hexon. Science. 1986;232(4754):1148–1151. doi: 10.1126/science.3704642. [DOI] [PubMed] [Google Scholar]
- 18.Lin T, et al. The refined crystal structure of cowpea mosaic virus at 2.8 A resolution. Virology. 1999;265(1):20–34. doi: 10.1006/viro.1999.0038. [DOI] [PubMed] [Google Scholar]
- 19.Nandhagopal N, et al. The structure and evolution of the major capsid protein of a large, lipid-containing DNA virus. Proc Natl Acad Sci USA. 2002;99(23):14758–14763. doi: 10.1073/pnas.232580699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Benson SD, Bamford JKH, Bamford DH, Burnett RM. Viral evolution revealed by bacteriophage PRD1 and human adenovirus coat protein structures. Cell. 1999;98(6):825–833. doi: 10.1016/s0092-8674(00)81516-0. [DOI] [PubMed] [Google Scholar]
- 21.Huiskonen JT, Kivelä HM, Bamford DH, Butcher SJ. The PM2 virion has a novel organization with an internal membrane and pentameric receptor binding spikes. Nat Struct Mol Biol. 2004;11(9):850–856. doi: 10.1038/nsmb807. [DOI] [PubMed] [Google Scholar]
- 22.Abrescia NG, et al. Insights into assembly from structural analysis of bacteriophage PRD1. Nature. 2004;432(7013):68–74. doi: 10.1038/nature03056. [DOI] [PubMed] [Google Scholar]
- 23.Khayat R, et al. Structure of an archaeal virus capsid protein reveals a common ancestry to eukaryotic and bacterial viruses. Proc Natl Acad Sci USA. 2005;102(52):18944–18949. doi: 10.1073/pnas.0506383102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khayat R, Fu CY, Ortmann AC, Young MJ, Johnson JE. The architecture and chemical stability of the archaeal Sulfolobus turreted icosahedral virus. J Virol. 2010;84(18):9575–9583. doi: 10.1128/JVI.00708-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bahar MW, Graham SC, Stuart DI, Grimes JM. Insights into the evolution of a complex virus from the crystal structure of vaccinia virus D13. Structure. 2011;19(7):1011–1020. doi: 10.1016/j.str.2011.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hyun JK, et al. Membrane remodeling by the double-barrel scaffolding protein of poxvirus. PLoS Pathog. 2011;7(9):e1002239. doi: 10.1371/journal.ppat.1002239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murshudov GN, Dodson EJ, Vagiin AA. Application of Maximum Likelihood Methods for Macromolecular Refinement. Macromolecular Refinement. CLRC Daresbury Laboratory; 1996. pp. 93–104. [Google Scholar]
- 28.Xiang Y, et al. Structural changes of bacteriophage ϕ29 upon DNA packaging and release. EMBO J. 2006;25(21):5229–5239. doi: 10.1038/sj.emboj.7601386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang W, et al. Structure of epsilon15 bacteriophage reveals genome organization and DNA packaging/injection apparatus. Nature. 2006;439(7076):612–616. doi: 10.1038/nature04487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Holm L, Rosenström P. Dali server: Conservation mapping in 3D. Nucleic Acids Res. 2010;38(Web Server issue):W545-9. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zubieta C, Schoehn G, Chroboczek J, Cusack S. The structure of the human adenovirus 2 penton. Mol Cell. 2005;17(1):121–135. doi: 10.1016/j.molcel.2004.11.041. [DOI] [PubMed] [Google Scholar]
- 32.Stewart PL, et al. Cryo-EM visualization of an exposed RGD epitope on adenovirus that escapes antibody neutralization. EMBO J. 1997;16(6):1189–1198. doi: 10.1093/emboj/16.6.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chiu CY, Mathias P, Nemerow GR, Stewart PL. Structure of adenovirus complexed with its internalization receptor, alphavbeta5 integrin. J Virol. 1999;73(8):6759–6768. doi: 10.1128/jvi.73.8.6759-6768.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martín CS, et al. Combined EM/X-ray imaging yields a quasi-atomic model of the adenovirus-related bacteriophage PRD1 and shows key capsid and membrane interactions. Structure. 2001;9(10):917–930. doi: 10.1016/s0969-2126(01)00642-6. [DOI] [PubMed] [Google Scholar]
- 35.Zauberman N, et al. Distinct DNA exit and packaging portals in the virus Acanthamoeba polyphaga mimivirus. PLoS Biol. 2008;6(5):e114. doi: 10.1371/journal.pbio.0060114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Greber UF, Willetts M, Webster P, Helenius A. Stepwise dismantling of adenovirus 2 during entry into cells. Cell. 1993;75(3):477–486. doi: 10.1016/0092-8674(93)90382-z. [DOI] [PubMed] [Google Scholar]
- 37.Caspar DLD, Klug A. Physical principles in the construction of regular viruses. Cold Spring Harb Symp Quant Biol. 1962;27:1–24. doi: 10.1101/sqb.1962.027.001.005. [DOI] [PubMed] [Google Scholar]
- 38.Simpson AA, Nandhagopal N, Van Etten JL, Rossmann MG. Structural analyses of Phycodnaviridae and Iridoviridae. Acta Crystallogr D Biol Crystallogr. 2003;59(Pt 12):2053–2059. doi: 10.1107/s090744490302225x. [DOI] [PubMed] [Google Scholar]
- 39.Zubieta C, Blanchoin L, Cusack S. Structural and biochemical characterization of a human adenovirus 2/12 penton base chimera. FEBS J. 2006;273(18):4336–4345. doi: 10.1111/j.1742-4658.2006.05430.x. [DOI] [PubMed] [Google Scholar]
- 40.Sokolova A, et al. Solution structure of bacteriophage PRD1 vertex complex. J Biol Chem. 2001;276(49):46187–46195. doi: 10.1074/jbc.M106848200. [DOI] [PubMed] [Google Scholar]
- 41.Wickham TJ, Mathias P, Cheresh DA, Nemerow GR. Integrins αvβ3 and αvβ5 promote adenovirus internalization but not virus attachment. Cell. 1993;73(2):309–319. doi: 10.1016/0092-8674(93)90231-e. [DOI] [PubMed] [Google Scholar]
- 42.Rosenthal PB, Henderson R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J Mol Biol. 2003;333(4):721–745. doi: 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- 43.Tang G, et al. EMAN2: An extensible image processing suite for electron microscopy. J Struct Biol. 2007;157(1):38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 44.Ludtke SJ, Baldwin PR, Chiu W. EMAN: Semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128(1):82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 45.Grigorieff N. FREALIGN: High-resolution refinement of single particle structures. J Struct Biol. 2007;157(1):117–125. doi: 10.1016/j.jsb.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 46.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brünger AT. Version 1.2 of the Crystallography and NMR system. Nat Protoc. 2007;2(11):2728–2733. doi: 10.1038/nprot.2007.406. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.