Abstract
Arrestin-1 (visual arrestin) binds to light-activated phosphorylated rhodopsin (P-Rh*) to terminate G-protein signaling. To map conformational changes upon binding to the receptor, pairs of spin labels were introduced in arrestin-1 and double electron–electron resonance was used to monitor interspin distance changes upon P-Rh* binding. The results indicate that the relative position of the N and C domains remains largely unchanged, contrary to expectations of a “clam-shell” model. A loop implicated in P-Rh* binding that connects β-strands V and VI (the “finger loop,” residues 67–79) moves toward the expected location of P-Rh* in the complex, but does not assume a fully extended conformation. A striking and unexpected movement of a loop containing residue 139 away from the adjacent finger loop is observed, which appears to facilitate P-Rh* binding. This change is accompanied by smaller movements of distal loops containing residues 157 and 344 at the tips of the N and C domains, which correspond to “plastic” regions of arrestin-1 that have distinct conformations in monomers of the crystal tetramer. Remarkably, the loops containing residues 139, 157, and 344 appear to have high flexibility in both free arrestin-1 and the P-Rh*complex.
Keywords: EPR, signal transduction, site-directed spin labeling
Arrestin was first discovered in the visual system as a protein that blocks (“arrests”) the signaling of the prototypical G protein-coupled receptor (GPCR) rhodopsin (Rh) via specific binding to the phosphorylated activated form P-Rh* (1). Mammals express four arrestin subtypes: Arrestin-1 and -4 are specific for the visual system, whereas arrestin-2 and -3 are ubiquitous (2). [We use systematic names of arrestins: arrestin-1 (historic names S-antigen, 48-kDa protein, or visual or rod arrestin), arrestin-2 (β-arrestin or β-arrestin1), arrestin-3 (β-arrestin2 or hTHY-ARRX), and arrestin-4 (cone or X-arrestin).] The discovery of nonvisual arrestins (3) showed that phosphorylation followed by arrestin binding is a common mechanism of GPCR regulation. Crystal structures of all four arrestin subtypes in their basal state revealed similar topology: two cup-like domains linked by an interdomain hinge (Fig. 1) (4–7). Arrestin-1 was proposed to undergo a conformational rearrangement during the P-Rh* interaction that results in the release of the C-terminal sequence (C tail) (8, 9) but does not involve major secondary structure changes (8, 10). Recent site-directed spin labeling (SDSL) studies identified specific parts of arrestin-1 engaged by different functional forms of rhodopsin and provided direct evidence of binding-induced conformational changes (11, 12). A conformational change in the so-called finger loop (Fig. 1) implicated in P-Rh* recognition was also observed using NMR and fluorescence quenching (13, 14). Arrestin-1 shows a remarkable selectivity for P-Rh*. Observed binding to inactive phosphorylated (P-Rh) or active unphosphorylated rhodopsin (Rh*) is usually less than 10% of the binding to P-Rh*, whereas its binding to inactive unphosphorylated rhodopsin (Rh) is barely detectable (15). A sequential multisite binding model was proposed to explain arrestin-1 selectivity (16). This model suggests that arrestin-1 elements that recognize rhodopsin-attached phosphates and active rhodopsin conformation serve as sensors. Simultaneous engagement of both sensors, which only P-Rh* can achieve, triggers a global conformational change, allowing arrestin-1 transition to a high-affinity receptor-binding state. The activation mechanism appears to be conserved in nonvisual arrestins (17, 18) that initiate a second round of signaling upon receptor binding. Thus, the arrestin–receptor complex serves as a nucleus of a signalosome (19), where the conformation of the receptor-bound arrestin apparently determines its interactions with multiple signaling proteins (20).
Fig. 1.
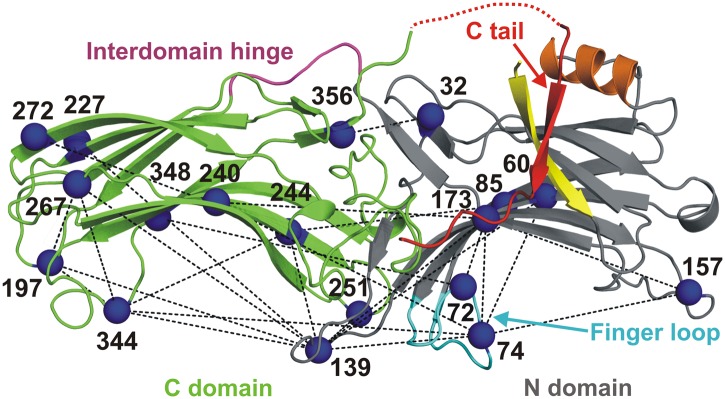
Ribbon model of arrestin-1 [Protein Data Bank (PDB) ID code 1CF1, chain D] (4). The N and C domains are gray and green, respectively; the interdomain hinge is magenta and the finger loop is cyan. The flexible C tail (red) forms a strong intramolecular interaction with the adjacent α-helix I (orange) and β-strand I (yellow), which is crucial for the stability of the basal state. A red dotted line represents the loop connecting the C domain to the C tail, which was not resolved in the crystal structure. Spin-labeled sites are shown as blue spheres at their α-carbons, and black dotted lines represent 25 interspin distances measured using DEER. Sites 60, 85, 173, 227, 240, 244, 267, 272, and 348 are considered stable (nonplastic) reference sites for detecting intradomain movements.
To obtain a global picture of receptor-induced conformational changes in arrestin-1, SDSL and double electron–electron resonance (DEER) were used to measure long-range (∼17–60 Å) intramolecular distances in arrestin-1 in the solution tetramer (the “basal” state) and bound to P-Rh* (the “active” state). DEER is a powerful tool capable of monitoring structural changes of a protein via distance measurements between attached spin labels, and for resolving discrete conformations and/or structural heterogeneity as inferred from the distance distributions (21–23). For these experiments, pairs of nitroxide side chains (R1) were introduced into arrestin-1 (11), targeting four loops on the receptor-binding surface that were expected to be flexible based on the crystal structure, and at positions in the rigid β-strand sandwich cores of both domains to serve as reference points (see legend to Fig. 1 for identity of reference sites). A total of 25 distances was measured in the absence and presence of P-Rh*. The data reveal receptor binding-induced movements of multiple arrestin-1 elements. A Rosetta model based on these data provides the description of the active receptor-bound arrestin.
Results
Fig. 1 shows the sites where the R1 side chain was introduced for distance measurement. The 25 selected pairs yield three classes of distances: 12 interdomain, 8 within the N domain, and 5 within the C domain; the pairs are identified in Fig. 1 and listed in Table S1. All spin-labeled arrestins retained the ability to bind P-Rh* (Fig. S1). DEER data were collected for each pair in the solution tetramer and upon binding to P-Rh* in native disk membranes (Fig. S2). Representative distance distributions derived from fits to the data are shown in Fig. 2A; the remaining distributions are given in Fig. S3A. From the distributions, the most probable and median distances were obtained (Fig. S4 and Table S1). The median distances were used as geometric constraints to predict possible conformations of arrestin-1 in solution tetramer and bound to P-Rh* using the RosettaEPR tool (24, 25). Typically, the most probable and median were close in value, but in the case of multimodal and strongly asymmetric distributions the median best represents the flexible structure. The RosettaEPR tool generates a set of protein conformations based on EPR distance restraints as evaluated by a knowledge-based potential. The ensemble of models that fulfill the distance restraints reveals the conformational space consistent with experiment. It should be noted that independent of modeling, the dramatic distance changes observed in the transition from the solution tetramer to the P-Rh*–bound state clearly identify the nature of the movements; the models incorporate all distance data to provide a global picture of possible structural changes involving the regions sampled experimentally.
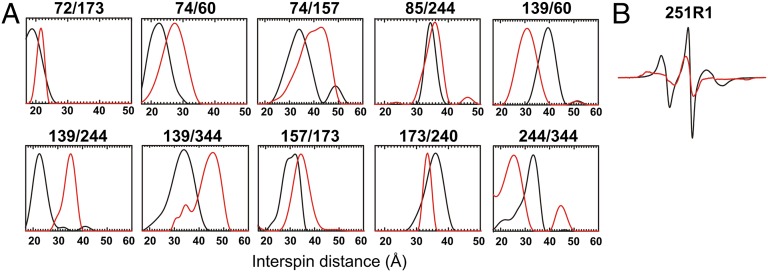
Fig. 2.
(A) Representative DEER data of doubly labeled arrestin-1 mutants in the solution tetramer (black traces) and P-Rh*–bound forms (red traces); remaining data are shown in Fig. S3. (B) CW EPR spectrum of arrestin-1 251R1 in the solution tetramer (black) and P-Rh*–bound (red) forms. Other unpublished CW spectra are shown in Fig. S7.
The main conclusions of the study are based on changes in median distances between nitroxide spin labels upon arrestin-1 binding to P-Rh*. However, the width and shape of a DEER-determined distance distribution contain additional information on structural heterogeneity of the protein but can also have contributions from rotameric equilibria of the R1 side chain. Detailed interpretation of the distribution width and modality is beyond the scope of the present report. Nevertheless, simple qualitative conclusions are possible and will be mentioned. For example, distance distributions with effective widths substantially greater than those characteristic of R1 pairs in rigid proteins [∼5–7 Å full width at half-height (22, 26)] presumably reflect structural heterogeneity of the protein. On the other hand, a narrow distribution width does not necessarily imply structural homogeneity, because a distribution of states wherein the interspin distance is constant is possible.
Conformation of Arrestin-1 in the Solution Tetramer.
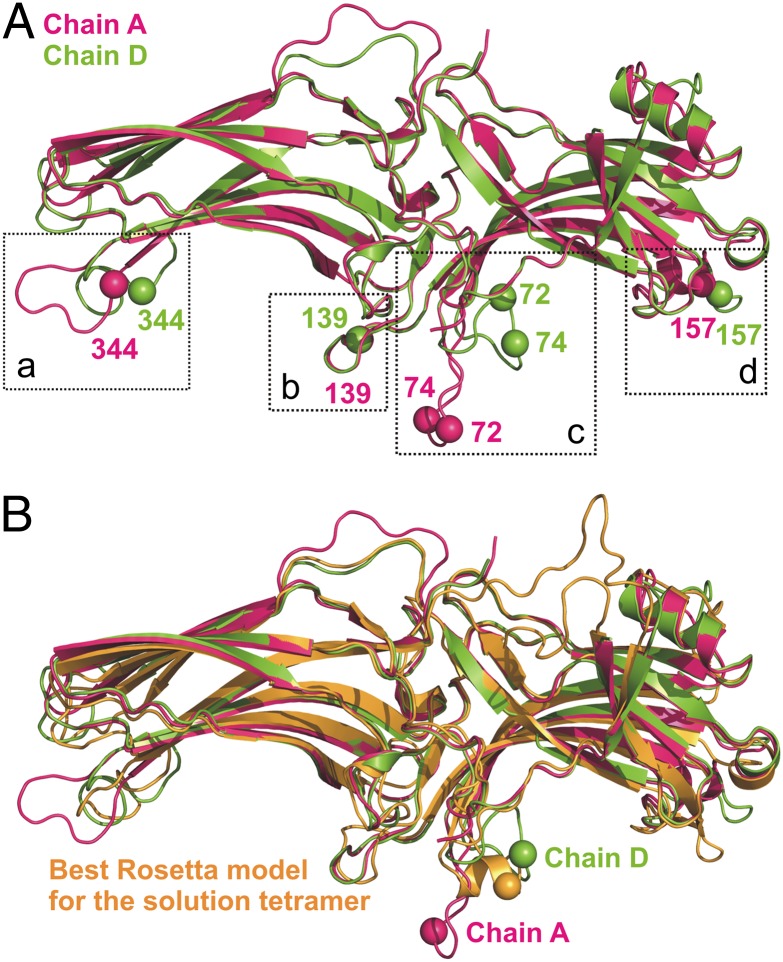
Arrestin-1 is known to self-associate, existing in a monomer–dimer–tetramer equilibrium (27). At physiological concentrations [>2 mM in the cell body (28)], as well as under DEER conditions (∼200 μM), the tetramer is the prevalent form. Not surprisingly, the crystal structure of arrestin-1 was solved as a tetramer (4). Interestingly, it contains four conformationally distinct monomers (chains A–D) showing the natural plasticity of the arrestin-1 molecule. For example, the “finger loop” (residues 67–79), which is highly conserved in the arrestin family (29) and has been identified as a key receptor-binding element in two subtypes (11, 30), has two distinct conformations in the crystal structure, bent (chain D) and extended (chain A) (Fig. 3A). Recently, a model for the solution tetramer of arrestin-l was determined (31) by Rosetta docking based on SDSL and light-scattering data. In this model, the monomers of the tetramer are conformationally equivalent, with each monomer having the conformation of chain A of the crystal structure. In the present study, additional DEER distance constraints and RosettaEPR suggest a refinement of the model.
Fig. 3.
(A) Overlaid chain A (magenta) and chain D (green) from the crystal structure of arrestin-1 (PDB ID code 1CF1). The Cα positions of residues studied in this work in four plastic loops (a–d) on the concave side of the molecule are shown as spheres and reveal the conformational differences between the two chains. The spheres are color-coded according to their backbone ribbon color. In particular, the finger loop (c) assumes two distinct conformations: extended (chain A) and bent (chain D). (B) Superposition of the crystal structures in A with the best arrestin-1 Rosetta model for the solution tetramer based on DEER data (orange). The Cα positions of residue 74 are shown as spheres for comparison.
Comparison of the best-scoring Rosetta model with each chain of the crystal structure showed that chain D, wherein the finger loop is in a bent rather than extended configuration, is in best agreement with the DEER data (Fig. 3B). This finding is consistent with previous studies that examined the finger-loop conformation of arrestin-1 using site-specific cysteine mutagenesis and intramolecular fluorescence quenching (13). In addition, the top Rosetta model shows that the finger loop can adopt an α-helical conformation in the solution tetramer, in contrast to the crystal, where it is disordered or unresolved (4). Previously reported EPR spectra of R1 in positions 72 and 74 (11) in the finger loop show at least two components, suggesting a possibility of at least two distinct conformational states. Taken together, these results suggest that the finger loop may exist in multiple conformations in equilibrium. In this regard, the widths of the distance distributions between 72R1 and 74R1 and reference sites 173R1 and 348R1 are sufficiently broad in the basal state (8∼16 Å) to accommodate this possibility.
Conformational Changes in Arrestin-1 upon Binding to P-Rh*.
The N and C domains in arrestin-1 are connected by a 12-residue “hinge” (Fig. 1). The addition of extra residues to the hinge does not affect P-Rh* binding, whereas increasing deletions progressively reduces the ability of arrestin-1 to bind P-Rh* (32). This suggested that the transition of arrestin-1 to the active receptor-bound state requires an extended hinge, which led to the idea that the domains move relative to each other, closing in on the receptor (the “clam-shell” model) (16). To test this model and to explore the global conformation of receptor-bound arrestin-1, the same 25 interspin distances measured in the solution tetramer were determined for arrestin-1 bound to P-Rh* (Table S1).
Three interdomain distances (32R1/356R1, 85R1/244R1, 173R1/240R1) that link the rigid β-sandwich domains revealed only small changes (<3 Å) upon binding to P-Rh* (Fig. 2A and Table S1); the distance between 32R1 and 356R1 was too short to measure with DEER and was examined by continuous-wave (CW) line broadening (Fig. S3B). Each result rules out large domain movement and therefore rejects the clam-shell model.
Conformational changes induced by P-Rh* binding in the finger loop were investigated using distances between R1 residues in the loop (at sites 72 and 74) and at various reference points in the β-sheet core of both domains (Fig. 1 and Table S1). All pairs involving the finger loop show distance changes of up to 8.5 Å (Fig. 2A, Figs. S3 and S5, and Table S1), indicating dislocation of the finger loop in complex with P-Rh*. Upon P-Rh* binding, the widths of the distance distributions between residues 72R1 and 74R1 in the loop and reference site 173R1 become narrower, as does that between 72R1 and reference site 348R1 (Fig. 2A and Fig. S3), consistent with a more restricted motion for this loop at a direct receptor interaction site (11).
The most striking observed movement was that of the loop containing residue 139, which is adjacent to the finger loop (Fig. 1). Upon P-Rh* binding, interspin distances involving position 139 show remarkably large changes, up to ∼17 Å (Fig. 2A, Figs. S3 and S5, and Table S1). EPR spectra of 139R1 suggested that this loop has high mobility in both the basal and active states (11), consistent with the relatively broad distributions for distances between 139R1 and R1 at reference sites 60, 173, 251, and 267, both in the absence and presence of P-Rh*. All distance changes are consistent with a movement of the 139 loop away from the finger loop, which apparently resides at the receptor-binding surface (11). We propose that the observed large movement of the 139 loop facilitates receptor binding involving the finger loop and adjacent elements. One of these adjacent elements is apparently the loop containing 251R1, which is strongly immobilized by receptor contact in the complex (Fig. 2B), along with R1 residues in the finger loop (11).
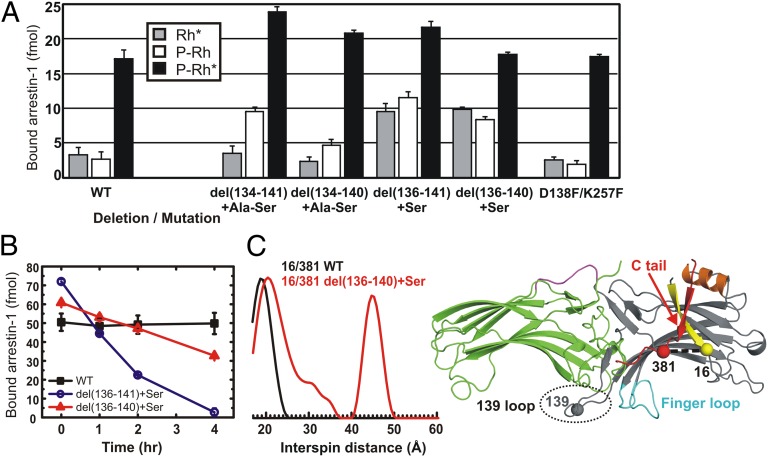
The role of the 139 loop was further studied by mutagenesis (Fig. 4). Deletions in this loop increase P-Rh* binding (Fig. 4A). More importantly, they reduce arrestin-1 selectivity for P-Rh*, enhancing its binding to inactive P-Rh and unphosphorylated Rh*. Moreover, they reduce the thermal stability of the arrestin-1 molecule (Fig. 4B) in a manner inversely correlated with the ability to bind nonpreferred forms of rhodopsin, P-Rh and Rh*. This behavior is reminiscent of constitutively active or preactivated forms of arrestin-1 (15), and was further investigated using a 16R1/381R1 double mutant that monitors the position of the C tail (Fig. 4C). β-Strand I and the C tail form a strong intramolecular interaction in the basal state of arrestin-1, and the C tail is released upon binding to P-Rh*, as shown by an increase of the interspin distance between two residues (11, 12). Deletions in the 139 loop disrupt this interaction, resulting in the release of the C terminus even in the absence of P-Rh* (Fig. 4C). Collectively, the data suggest that the 139 loop stabilizes the basal conformation of arrestin-1 and serves as a gatekeeper, preventing its binding to dark P-Rh and Rh*, thereby increasing arrestin-1 selectivity for P-Rh*.
Fig. 4.
The effects of 139-loop deletions on the thermal stability and selectivity of arrestin-1. (A) Direct binding (SI Materials and Methods) of the WT and four 139 deletion mutants, (i) del(134–141)+Ala-Ser, (ii) del(134–140)+Ala-Ser, (iii) del(136–141)+Ser, and (iv) del(136–140)+Ser, and one double mutant (D138F/K257F), which replaces the salt bridge with potential hydrophobic interaction to hold the 139 loop in its basal position. One or two Ala or Ser residues were added to the deletion mutants to relieve strain. (B) Thermal stability was measured for WT and two deletion mutants as a function of time of incubation at 39 °C. The results in A and B are presented as the mean of three and two independent experiments, each performed in triplicate and duplicate, respectively. The error bars represent standard deviations. (C) (Left) Distance distribution of 16R1/381R1 of full-length (black) and 139-loop deletion mutant del(136–140)+Ser (red) arrestin-1. (Right) Arrestin-1 crystal structure showing the location of the 139 loop with respect to the finger loop. The locations of positions 16, 139, and 381 are indicated by spheres at the Cα carbons.
The differences in conformation between the four monomers in the crystallographic tetramer of arrestin-1 (4) are most prominent in three loops: residues (i) 68–79, (ii) 155–165, and (iii) 337–347. Such plastic and/or unstructured regions of a protein are often involved in interaction with other binding partners (33). Not surprisingly, region i is the finger loop, discussed above, which plays a key role in receptor recognition (15). To investigate the movement of loops ii and iii during complex formation, distances between R1 residues in these distal regions (157 and 344 in Fig. 1) and one or more fixed reference sites in the β-sheets of the molecule (173, 244, and 267 in Fig. 1) were monitored. The results indicate that 157R1 and 344R1 move away from and toward the central crest of the molecule, respectively (Fig. 5A, Fig. S3, and Table S1). To study correlated motion between plastic regions of arrestin-1, additional distances between positions 157 and 344 and such regions were monitored; these distance changes were used in global modeling.
Fig. 5.
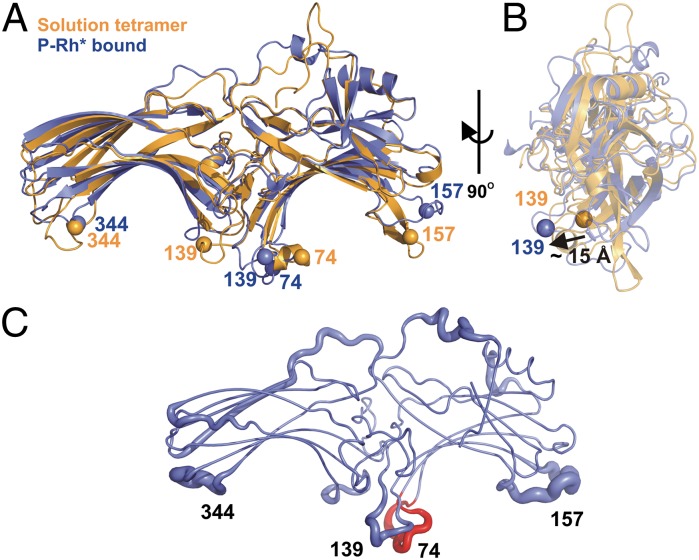
Models of arrestin-1 in the solution tetramer (orange) and P-Rh*–bound forms (blue) generated by RosettaEPR using DEER distance constraints. The C tail is not shown for clarity. (A) Front view. Cα locations of four residues of interest are shown as spheres to show the direction of conformational change of each loop. The spheres are color-coded according to their backbone ribbon color. (B) Side view. The 139 loop moves out of the arrestin-1 plane by ∼15 Å upon binding to P-Rh*. (C) The 10 best models of P-Rh*–bound arrestin-1 suggest plasticity of the four loops containing the indicated residues. The finger loop is shown in red. The thickness of the tube representation reflects the average rmsd (SI Materials and Methods) per residue among the top 10 models (Table S2). Residues 1–10 and 382–404 are omitted for clarity.
Based on all measured distances in arrestin-1 bound to P-Rh*, 4,037 models of the bound state were generated with RosettaEPR (24, 25) and scored based on global agreement with the distance constraints (Table S2). The best-scoring model (blue trace) compared with that of the monomer in the solution tetramer (orange trace) is shown in Figs. 5 A and B. In this model, the finger loop is displaced toward the expected binding interface with P-Rh*. Interestingly, 7 out of the 10 best models show a propensity of the finger loop to form an α-helix upon P-Rh* binding. In addition, the top-scoring Rosetta models show variable positions of sites 344R1 and 157R1, suggesting that the associated elements may retain their plasticity within the complex (Fig. 5C). This is consistent with an earlier finding that 157R1 and 344R1 have high mobility in both the basal state and in the receptor-bound state (11), indicating that they are not involved in direct receptor contact. The broad distributions of distances involving 344R1 to reference sites 244R1 and 267R1 and 157R1 to reference site 173R1 highlight the apparent flexibility of these loops even in the complex with the receptor.
The relatively large (∼70 Å) long axis of all arrestins in the basal state (4), compared with a more compact cytoplasmic surface of the receptors (35∼40 Å) (34), along with evidence that the concave sides of both arrestin domains are engaged by the receptor (11, 15, 35, 36), led to the idea that a single arrestin molecule could bind two receptors in a dimer (37, 38). However, it has been reported that arrestin-1 saturates rhodopsin at a 1:1 molar ratio (39) and binds the P-Rh* monomer in nanodiscs with physiological affinity and stoichiometry (40, 41), supporting a one-to-one binding model. In contrast, two recent studies showed that the apparent binding stoichiometry depends on the percentage of active receptors in native disk membranes, and that arrestin-1 binding at high levels of activated rhodopsin is best-described by a combination of 1:1 and 1:2 interactions (42, 43). The present study was done in high activation density with excess P-Rh* in native disk membranes. To test whether our distance measurements exclusively reflect multiple receptor binding, interdomain (173R1/240R1), intra-N domain (139R1/60R1), and intra-C domain (267R1/344R1) distances were compared for P-Rh*–bound arrestin-1 obtained in native disk membranes and nanodiscs containing monomeric P-Rh* (41). The results show that the distance changes are very similar in both cases (Fig. S6 and SI Materials and Methods). Therefore, we conclude that the DEER measurements apply to a high-affinity one-to-one arrestin–P-Rh* complex, consistent with the finding that regardless of the binding stoichiometry, arrestin-1 stabilizes the active state (metarhodopsin II) of only one rhodopsin (42). However, the data do not eliminate the possibility of complexes with other stoichiometry in the native disk membrane that do not alter arrestin conformation.
Discussion
G protein-mediated signaling by the majority of GPCRs is terminated by binding of arrestin to the phosphorylated form of the activated receptor (3, 44). Indirect evidence suggesting that arrestin-1 binding to P-Rh* involves a significant conformational change has accumulated for more than 20 y (9, 10, 32, 45). For example, the release of the arrestin-1 C tail upon receptor binding was suggested by an increased accessibility of this element in the P-Rh*–associated form (9, 45) and by the higher mobility of C-terminal residues in the presence of mimics of phosphorylated receptor (46). However, direct proof that the C tail moves away from β-strand I and α-helix I, with which it interacts in the basal conformation, was obtained only recently with SDSL by measuring P-Rh* binding-induced changes in distances between C-tail residues and the body of the molecule (11, 12). Here we report direct evidence of conformational changes accompanying arrestin-1 binding to P-Rh* in other parts of the molecule, using intramolecular distance measurements in the solution tetramer and P-Rh*–bound arrestin-1.
Twenty-five pairs of spin labels were used to systematically study multiple regions in arrestin-1. In view of strong experimental support for one-to-one arrestin–receptor interaction, binding-induced conformational changes in arrestin were expected to make the receptor-binding surface more compact, largely by the proposed movement of the two arrestin domains (47). This model was supported by the findings that progressive deletions in the interdomain hinge reduced the ability of all arrestins to bind receptors (32, 48). However, the data presented here reveal only subtle changes in the interdomain distances, ruling out a large clam shell-like relative motion of the N and C domains (16), but a small rotation of one domain relative to the other about the long axis could occur. Hinge deletions could affect such rotations or act indirectly, possibly through shifts in the conformational ensemble of the flexible arrestin molecule (49). It should be noted that the idea of a misfit between the large receptor-binding surface of arrestins and smaller arrestin-binding surface of GPCRs is purely speculative. Many residues on the concave side of the N domain of arrestin-1 directly bind receptor-attached phosphates (7, 50). Receptor elements that are phosphorylated are either unresolved or partially resolved [the C termini of rhodopsin (34) and β2-adrenoceptor (51)] or simply deleted [the third cytoplasmic loop of M2 muscarinic receptor (52)] even in the best GPCR crystal structures. Hence, the location of these elements with respect to the rest of the GPCR structure is unknown. Thus, it is possible that phosphorylated parts of the receptor are 20∼30 Å away from other arrestin-binding elements, so that receptor-attached phosphates bind the N domain whereas other parts of the receptor interact with the finger loop and the C domain covering a large surface of arrestin. In this case, a significant domain movement does not seem to be necessary. On the other hand, it may be that the N domain and finger-loop region are the primary sites of interaction with P-Rh* and that the C domain has an accessory function, including the possibility of interaction with another receptor molecule in an inactive state (42, 43), the membrane surface, or other signaling proteins.
Among the four vertebrate subtypes, arrestin-1 shows the highest receptor specificity and selectivity for P-Rh* (50). However, the mechanism of arrestin activation by GPCRs is conserved in all subtypes (17, 18, 53), suggesting that receptor binding induces similarly small domain movement in nonvisual arrestins, which leaves a large portion of the molecule essentially unchanged. This can explain why many nonreceptor signaling proteins bind comparably to free and GPCR-associated arrestins (53).
Two rearrangements in the central “crest” on the receptor-binding side of arrestin-1 were detected upon complex formation. The first involves the finger loop, wherein the sites monitored (residues 72 and 74) move only slightly toward the expected location of the receptor-binding surface (11); no evidence was found for a transition of the finger loop from folded to a fully extended conformation (13). Recently, it has been shown by NMR that an arrestin-1 peptide corresponding to the finger loop becomes α-helical upon binding to P-Rh* (14). Preventing this change by disulfide formation inhibits arrestin-1 binding to P-Rh*, indicating that conformational flexibility is required for arrestin-1 transition to the P-Rh*–bound state. Modeling based on EPR restraints suggests a propensity for the finger loop to form a helix in both free and bound states. Interestingly, members of three protein families that preferentially bind active GPCRs, namely G proteins (54, 55), G protein-coupled receptor kinases (56–58), and arrestins (14), all have flexible sequences that transition to a helical conformation when bound to GPCRs. In each case, the helical segment is proposed to bind in or near the central cavity of the activated receptor.
The second central crest motion is the large distance change of the 139 loop upon binding to P-Rh*. This loop is located next to the finger loop (Fig. 1); both loops are conserved in all arrestins, indicative of their biological importance. Collectively, the SDSL data are consistent with a dramatic displacement of the loop, with its tip swinging by ∼15 Å (Fig. 5B), away from the receptor-binding surface defined by the finger loop (11). This explains previous findings by SDSL that the highly mobile residue 139R1 is constrained by receptor contact in complex with inactive P-Rh but reverts to high mobility upon binding to P-Rh*, in contrast to the behavior of R1 in the finger loop, which becomes strongly immobilized in the complex with P-Rh* (11). The similarly strong immobilization of 251R1 in a loop adjacent to the finger loop further implicates the central crest of arrestin in receptor binding. The high mobility of 139R1 in both the solution tetramer and complex indicate that the 139 loop is not directly engaged by P-Rh* but instead that its large movement facilitates receptor binding. More intriguingly, we found that this loop plays an important role biasing the arrestin-1 population of conformers toward the basal state. Without the 139 loop, the population is shifted toward an active-like state, resulting in the release of C tail even before binding to P-Rh* (Fig. 4C). This result suggests allosteric communication between the 139 loop and α-helix I and/or β-strand I that controls the position of the C tail.
Residues 157 and 344 are located on distal tips of the N and C domains, respectively (Fig. 1). The insertion of a 10-amino acid myc tag into these two regions resulted in a decrease in P-Rh* binding similar to that caused by myc insertion into the finger loop (59). However, we previously found that spin labels at positions 157 and 344 remain mobile upon P-Rh* binding (11), suggesting that they do not contact the receptor directly. The DEER data reveal broad distributions for distances between 344R1 and reference sites when bound to P-Rh*, suggesting conformational heterogeneity of the 344 loop in the complex. This is also reflected in large rmsd deviations among the top 10 bound Rosetta models for the loops containing 157 as well as 344 (Fig. 5C).
In summary, the available data indicate that the three plastic domains of arrestin-1 identified in the crystal structure are in fact structurally heterogeneous in the solution tetramer, judged by both EPR spectral analysis (11) and the DEER distance mapping presented above. Upon complex formation with P-Rh*, the finger loop and the 157 and 344 loops are moved; the finger loop becomes ordered while the distal 157 and 344 loops remain flexible; and a clam shell-like relative motion of the N and C domains is not observed. Distance mapping revealed an unexpected large-scale movement of a flexible loop containing residue 139, which is located in proximity to key elements of the central crest involved in receptor binding. Mutagenesis results suggest that this loop may act as a gatekeeper that determines binding selectivity. The relative position of the N- and C-terminal domains remains essentially the same as in the solution tetramer. The set of experimental distance constraints has been used with RosettaEPR to provide a plausible model for the conformation of the arrestin-activated (P-Rh*–bound) state. Having a model for a conformation of the receptor-bound arrestin provides a structural basis for mechanistic studies of arrestin-mediated signaling. The data presented here identify arrestin elements that change upon receptor binding but which are not directly involved in the binding interaction. Thus, it is possible that these elements determine preferential interactions of receptor-bound or free arrestin with certain partners (20). If this is the case, disruption of the binding sites for individual partners could be used to rechannel arrestin-mediated signaling to desired pathways for therapeutic purposes (60). Although the crystal structure of an arrestin–P-Rh* complex would yield higher resolution, EPR and NMR can provide dynamic information that cannot be supplied by crystallography, which is particularly important in view of the high plasticity of arrestin.
Materials and Methods
Preparation of Arrestin-1 Double Mutants and Phosphorylated Rhodopsin.
Site-directed mutagenesis, expression, and purification of arrestin-1 were performed as previously described (61). All mutations were introduced on the fully functional cysteine-less base mutant VSV-CL (C63V, C128S, and C143V) (11). P-Rh in the disk membranes was prepared as described in SI Materials and Methods.
Spin Labeling and Sample Preparation.
For spin labeling, arrestin-1 cysteine mutants in 50 mM 3-(N-morpholino)propanesulfonic acid and 100 mM NaCl (pH 7.2) buffer were mixed with a 10-fold molar excess of 1-oxyl-2,2,5,5-tetramethyl-3-pyrroline-3-methyl methanethiosulfonate spin label (a generous gift of Kalman Hideg, University of Pécs, Pécs, Hungary) overnight at 4 °C. Removal of excess spin label and concentration were performed using an Amicon ultracentrifugal filter device (Millipore). The final sample contained 20% glycerol as a cryoprotectant. For the solution tetramer arrestin-1 samples, doubly labeled arrestin-1 mutants were mixed with cysteine-less arrestin-1 WT (unlabeled) at a ratio of 1:3 before concentration to eliminate undesired intermolecular distances within the solution tetramer. For measurements with P-Rh*, a two- to fourfold molar excess of P-Rh in native disk membranes was pelleted at 100,000 × g for 10 min and then resuspended in the dark with the doubly labeled arrestin-1 mutants in the buffer. P-Rh was light-activated by illumination for 1 min at room temperature before DEER measurement. Continuous-wave EPR spectra were collected for each pair in free and P-Rh*–bound forms to check binding, and unpublished CW spectra for four single sites are shown (Fig. 2B and Fig. S7).
DEER Spectroscopy and Data Analysis.
For DEER measurements, 15 μL of sample (∼200 μM) was loaded into sealed quartz capillaries (1.5 mm i.d. × 1.8 mm o.d.) and flash-frozen using liquid nitrogen. Data were collected at 80 K on a Bruker Elexys 580 spectrometer fitted with an MS-2 split-ring resonator as previously described (62). Data analysis to obtain the dipolar evolution functions (Fig. S2) and distance distributions using Tikhonov regularization was performed using DeerAnalysis2011 software (63). To estimate the median distances, the distance distributions were integrated and normalized to the maximum amplitude. The median distance was estimated as that corresponding to 0.5 of the integrated intensity (Fig. S4).
RosettaEPR Protein Modeling Based on DEER Distance Restraints.
The basis of RosettaEPR modeling has been published (24, 25). In the present implementation, a crystal structure of arrestin-1 (4) was used as the template for comparative modeling. The crystal structure contains four copies of the protein in the asymmetric unit (chains A–D). The four copies display structural plasticity in loop regions involving residues 67–79, 132–143, 152–169, and 335–345. Modeling was focused on these regions due to the presumed flexibility in solution and hence potential functional importance in protein–protein interactions. Details of the modeling are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. Christian Altenbach for his help with distance data analysis. This study was supported in part by National Institutes of Health (NIH) Grants EY011500, GM077561, and GM081756 (to V.V.G.), NIH/National Eye Institute Grants EY005216 and EY00331, and The Jules Stein Professorship Endowment (W.L.H.). Work in the Meiler laboratory is supported through the NIH (GM080403 and MH090192) and the National Science Foundation (0742762). N.S.A. was supported by National Research Service Award MH086222. O.P.E. holds The Anne and Max Tanenbaum Chair in Neuroscience at the University of Toronto and is supported by the Canada Excellence Research Chair program.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1216304109/-/DCSupplemental.
References
- 1.Wilden U, Hall SW, Kühn H. Phosphodiesterase activation by photoexcited rhodopsin is quenched when rhodopsin is phosphorylated and binds the intrinsic 48-kDa protein of rod outer segments. Proc Natl Acad Sci USA. 1986;83(5):1174–1178. doi: 10.1073/pnas.83.5.1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gurevich VV, Gurevich EV, Cleghorn WM. Arrestins as multi-functional signaling adaptors. Handbook of Experimental Pharmacology. 2008 doi: 10.1007/978-3-540-72843-6_2. Protein-Protein Interactions as New Drug Targets, eds Klussmann E, Scott J (Springer, Berlin), Vol 186, pp 15–37. [DOI] [PubMed] [Google Scholar]
- 3.Lohse MJ, Benovic JL, Codina J, Caron MG, Lefkowitz RJ. Beta-arrestin: A protein that regulates beta-adrenergic receptor function. Science. 1990;248(4962):1547–1550. doi: 10.1126/science.2163110. [DOI] [PubMed] [Google Scholar]
- 4.Hirsch JA, Schubert C, Gurevich VV, Sigler PB. A model for arrestin’s regulation: The 2.8 Å crystal structure of visual arrestin. Cell. 1999;97(2):257–269. doi: 10.1016/s0092-8674(00)80735-7. [DOI] [PubMed] [Google Scholar]
- 5.Han M, Gurevich VV, Vishnivetskiy SA, Sigler PB, Schubert C. Crystal structure of beta-arrestin at 1.9 Å. Structure. 2001;9(9):869–880. doi: 10.1016/s0969-2126(01)00644-x. [DOI] [PubMed] [Google Scholar]
- 6.Zhan X, Gimenez LE, Gurevich VV, Spiller BW. Crystal structure of arrestin-3 reveals the basis of the difference in receptor binding between two non-visual subtypes. J Mol Biol. 2011;406(3):467–478. doi: 10.1016/j.jmb.2010.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sutton RB, et al. Crystal structure of cone arrestin at 2.3 Å: Evolution of receptor specificity. J Mol Biol. 2005;354(5):1069–1080. doi: 10.1016/j.jmb.2005.10.023. [DOI] [PubMed] [Google Scholar]
- 8.Palczewski K, Riazance-Lawrence JH, Johnson WC., Jr Structural properties of arrestin studied by chemical modification and circular dichroism. Biochemistry. 1992;31(16):3902–3906. doi: 10.1021/bi00131a003. [DOI] [PubMed] [Google Scholar]
- 9.Ohguro H, Palczewski K, Walsh KA, Johnson RS. Topographic study of arrestin using differential chemical modifications and hydrogen/deuterium exchange. Protein Sci. 1994;3(12):2428–2434. doi: 10.1002/pro.5560031226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schleicher A, Kühn H, Hofmann KP. Kinetics, binding constant, and activation energy of the 48-kDa protein-rhodopsin complex by extra-metarhodopsin II. Biochemistry. 1989;28(4):1770–1775. doi: 10.1021/bi00430a052. [DOI] [PubMed] [Google Scholar]
- 11.Hanson SM, et al. Differential interaction of spin-labeled arrestin with inactive and active phosphorhodopsin. Proc Natl Acad Sci USA. 2006;103(13):4900–4905. doi: 10.1073/pnas.0600733103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vishnivetskiy SA, et al. The role of arrestin alpha-helix I in receptor binding. J Mol Biol. 2010;395(1):42–54. doi: 10.1016/j.jmb.2009.10.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sommer ME, Farrens DL, McDowell JH, Weber LA, Smith WC. Dynamics of arrestin-rhodopsin interactions: Loop movement is involved in arrestin activation and receptor binding. J Biol Chem. 2007;282(35):25560–25568. doi: 10.1074/jbc.M702155200. [DOI] [PubMed] [Google Scholar]
- 14.Feuerstein SE, et al. Helix formation in arrestin accompanies recognition of photoactivated rhodopsin. Biochemistry. 2009;48(45):10733–10742. doi: 10.1021/bi900544p. [DOI] [PubMed] [Google Scholar]
- 15.Gurevich VV, Gurevich EV. The structural basis of arrestin-mediated regulation of G-protein-coupled receptors. Pharmacol Ther. 2006;110(3):465–502. doi: 10.1016/j.pharmthera.2005.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gurevich VV, Gurevich EV. The molecular acrobatics of arrestin activation. Trends Pharmacol Sci. 2004;25(2):105–111. doi: 10.1016/j.tips.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 17.Kovoor A, Celver J, Abdryashitov RI, Chavkin C, Gurevich VV. Targeted construction of phosphorylation-independent beta-arrestin mutants with constitutive activity in cells. J Biol Chem. 1999;274(11):6831–6834. doi: 10.1074/jbc.274.11.6831. [DOI] [PubMed] [Google Scholar]
- 18.Celver J, Vishnivetskiy SA, Chavkin C, Gurevich VV. Conservation of the phosphate-sensitive elements in the arrestin family of proteins. J Biol Chem. 2002;277(11):9043–9048. doi: 10.1074/jbc.M107400200. [DOI] [PubMed] [Google Scholar]
- 19.DeWire SM, Ahn S, Lefkowitz RJ, Shenoy SK. Beta-arrestins and cell signaling. Annu Rev Physiol. 2007;69:483–510. doi: 10.1146/annurev.physiol.69.022405.154749. [DOI] [PubMed] [Google Scholar]
- 20.Gurevich VV, Gurevich EV. The new face of active receptor bound arrestin attracts new partners. Structure. 2003;11(9):1037–1042. doi: 10.1016/s0969-2126(03)00184-9. [DOI] [PubMed] [Google Scholar]
- 21.Jeschke G. Distance measurements in the nanometer range by pulse EPR. ChemPhysChem. 2002;3(11):927–932. doi: 10.1002/1439-7641(20021115)3:11<927::AID-CPHC927>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 22.Altenbach C, Kusnetzow AK, Ernst OP, Hofmann KP, Hubbell WL. High-resolution distance mapping in rhodopsin reveals the pattern of helix movement due to activation. Proc Natl Acad Sci USA. 2008;105(21):7439–7444. doi: 10.1073/pnas.0802515105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schiemann O, Prisner TF. Long-range distance determinations in biomacromolecules by EPR spectroscopy. Q Rev Biophys. 2007;40(1):1–53. doi: 10.1017/S003358350700460X. [DOI] [PubMed] [Google Scholar]
- 24.Hirst SJ, Alexander N, McHaourab HS, Meiler J. RosettaEPR: An integrated tool for protein structure determination from sparse EPR data. J Struct Biol. 2011;173(3):506–514. doi: 10.1016/j.jsb.2010.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alexander N, Bortolus M, Al-Mestarihi A, Mchaourab H, Meiler J. De novo high-resolution protein structure determination from sparse spin-labeling EPR data. Structure. 2008;16(2):181–195. doi: 10.1016/j.str.2007.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.López CJ, Oga S, Hubbell WL. Mapping molecular flexibility of proteins with site-directed spin labeling: A case study of myoglobin. Biochemistry. 2012;51(33):6568–6583. doi: 10.1021/bi3005686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim M, et al. Robust self-association is a common feature of mammalian visual arrestin-1. Biochemistry. 2011;50(12):2235–2242. doi: 10.1021/bi1018607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Song X, et al. Arrestin-1 expression level in rods: Balancing functional performance and photoreceptor health. Neuroscience. 2011;174:37–49. doi: 10.1016/j.neuroscience.2010.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gurevich EV, Gurevich VV. Arrestins: Ubiquitous regulators of cellular signaling pathways. Genome Biol. 2006;7(9):236. doi: 10.1186/gb-2006-7-9-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vishnivetskiy SA, et al. Few residues within an extensive binding interface drive receptor interaction and determine the specificity of arrestin proteins. J Biol Chem. 2011;286(27):24288–24299. doi: 10.1074/jbc.M110.213835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hanson SM, et al. A model for the solution structure of the rod arrestin tetramer. Structure. 2008;16(6):924–934. doi: 10.1016/j.str.2008.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vishnivetskiy SA, Hirsch JA, Velez MG, Gurevich YV, Gurevich VV. Transition of arrestin into the active receptor-binding state requires an extended interdomain hinge. J Biol Chem. 2002;277(46):43961–43967. doi: 10.1074/jbc.M206951200. [DOI] [PubMed] [Google Scholar]
- 33.Dunker AK, Cortese MS, Romero P, Iakoucheva LM, Uversky VN. Flexible nets. The roles of intrinsic disorder in protein interaction networks. FEBS J. 2005;272(20):5129–5148. doi: 10.1111/j.1742-4658.2005.04948.x. [DOI] [PubMed] [Google Scholar]
- 34.Li J, Edwards PC, Burghammer M, Villa C, Schertler GF. Structure of bovine rhodopsin in a trigonal crystal form. J Mol Biol. 2004;343(5):1409–1438. doi: 10.1016/j.jmb.2004.08.090. [DOI] [PubMed] [Google Scholar]
- 35.Skegro D, et al. N-terminal and C-terminal domains of arrestin both contribute in binding to rhodopsin. Photochem Photobiol. 2007;83(2):385–392. doi: 10.1562/2006-08-25-RA-1014. [DOI] [PubMed] [Google Scholar]
- 36.Vishnivetskiy SA, Hosey MM, Benovic JL, Gurevich VV. Mapping the arrestin-receptor interface. Structural elements responsible for receptor specificity of arrestin proteins. J Biol Chem. 2004;279(2):1262–1268. doi: 10.1074/jbc.M308834200. [DOI] [PubMed] [Google Scholar]
- 37.Modzelewska A, Filipek S, Palczewski K, Park PS. Arrestin interaction with rhodopsin: Conceptual models. Cell Biochem Biophys. 2006;46(1):1–15. doi: 10.1385/CBB:46:1:1. [DOI] [PubMed] [Google Scholar]
- 38.Liang Y, et al. Organization of the G protein-coupled receptors rhodopsin and opsin in native membranes. J Biol Chem. 2003;278(24):21655–21662. doi: 10.1074/jbc.M302536200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hanson SM, et al. Each rhodopsin molecule binds its own arrestin. Proc Natl Acad Sci USA. 2007;104(9):3125–3128. doi: 10.1073/pnas.0610886104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tsukamoto H, Sinha A, DeWitt M, Farrens DL. Monomeric rhodopsin is the minimal functional unit required for arrestin binding. J Mol Biol. 2010;399(3):501–511. doi: 10.1016/j.jmb.2010.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bayburt TH, et al. Monomeric rhodopsin is sufficient for normal rhodopsin kinase (GRK1) phosphorylation and arrestin-1 binding. J Biol Chem. 2011;286(2):1420–1428. doi: 10.1074/jbc.M110.151043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sommer ME, Hofmann KP, Heck M. Arrestin-rhodopsin binding stoichiometry in isolated rod outer segment membranes depends on the percentage of activated receptors. J Biol Chem. 2011;286(9):7359–7369. doi: 10.1074/jbc.M110.204941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sommer ME, Hofmann KP, Heck M. Distinct loops in arrestin differentially regulate ligand binding within the GPCR opsin. Nat Commun. 2012;3 doi: 10.1038/ncomms2000. Article 995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carman CV, Benovic JL. G-protein-coupled receptors: Turn-ons and turn-offs. Curr Opin Neurobiol. 1998;8(3):335–344. doi: 10.1016/s0959-4388(98)80058-5. [DOI] [PubMed] [Google Scholar]
- 45.Palczewski K, Pulvermüller A, Buczyłko J, Hofmann KP. Phosphorylated rhodopsin and heparin induce similar conformational changes in arrestin. J Biol Chem. 1991;266(28):18649–18654. [PubMed] [Google Scholar]
- 46.Zhuang T, Vishnivetskiy SA, Gurevich VV, Sanders CR. Elucidation of inositol hexaphosphate and heparin interaction sites and conformational changes in arrestin-1 by solution nuclear magnetic resonance. Biochemistry. 2010;49(49):10473–10485. doi: 10.1021/bi101596g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vishnivetskiy SA, et al. An additional phosphate-binding element in arrestin molecule. Implications for the mechanism of arrestin activation. J Biol Chem. 2000;275(52):41049–41057. doi: 10.1074/jbc.M007159200. [DOI] [PubMed] [Google Scholar]
- 48.Hanson SM, et al. Arrestin mobilizes signaling proteins to the cytoskeleton and redirects their activity. J Mol Biol. 2007;368(2):375–387. doi: 10.1016/j.jmb.2007.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hilser VJ. Biochemistry. An ensemble view of allostery. Science. 2010;327(5966):653–654. doi: 10.1126/science.1186121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gurevich VV, Benovic JL. Visual arrestin binding to rhodopsin. Diverse functional roles of positively charged residues within the phosphorylation-recognition region of arrestin. J Biol Chem. 1995;270(11):6010–6016. doi: 10.1074/jbc.270.11.6010. [DOI] [PubMed] [Google Scholar]
- 51.Rasmussen SG, et al. Crystal structure of the human β2 adrenergic G-protein-coupled receptor. Nature. 2007;450(7168):383–387. doi: 10.1038/nature06325. [DOI] [PubMed] [Google Scholar]
- 52.Haga K, et al. Structure of the human M2 muscarinic acetylcholine receptor bound to an antagonist. Nature. 2012;482(7386):547–551. doi: 10.1038/nature10753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Breitman M, et al. Silent scaffolds: Inhibition of c-Jun N-terminal kinase 3 activity in cell by dominant-negative arrestin-3 mutant. J Biol Chem. 2012;287(23):19653–19664. doi: 10.1074/jbc.M112.358192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Scheerer P, et al. Crystal structure of opsin in its G-protein-interacting conformation. Nature. 2008;455(7212):497–502. doi: 10.1038/nature07330. [DOI] [PubMed] [Google Scholar]
- 55.Rasmussen SG, et al. Crystal structure of the β2 adrenergic receptor-Gs protein complex. Nature. 2011;477(7366):549–555. doi: 10.1038/nature10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pao CS, Barker BL, Benovic JL. Role of the amino terminus of G protein-coupled receptor kinase 2 in receptor phosphorylation. Biochemistry. 2009;48(30):7325–7333. doi: 10.1021/bi900408g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Boguth CA, Singh P, Huang CC, Tesmer JJ. Molecular basis for activation of G protein-coupled receptor kinases. EMBO J. 2010;29(19):3249–3259. doi: 10.1038/emboj.2010.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huang CC, Orban T, Jastrzebska B, Palczewski K, Tesmer JJ. Activation of G protein-coupled receptor kinase 1 involves interactions between its N-terminal region and its kinase domain. Biochemistry. 2011;50(11):1940–1949. doi: 10.1021/bi101606e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Smith WC, Dinculescu A, Peterson JJ, McDowell JH. The surface of visual arrestin that binds to rhodopsin. Mol Vis. 2004;10:392–398. [PubMed] [Google Scholar]
- 60.Gurevich VV, Gurevich EV. Custom-designed proteins as novel therapeutic tools? The case of arrestins. Expert Rev Mol Med. 2010;12:e13. doi: 10.1017/S1462399410001444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gurevich VV, Benovic JL. Arrestin: Mutagenesis, expression, purification, and functional characterization. Methods Enzymol. 2000;315:422–437. doi: 10.1016/s0076-6879(00)15859-8. [DOI] [PubMed] [Google Scholar]
- 62.Hanson SM, et al. Structure and function of the visual arrestin oligomer. EMBO J. 2007;26(6):1726–1736. doi: 10.1038/sj.emboj.7601614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jeschke G, et al. DeerAnalysis2006—A comprehensive software package for analyzing pulsed ELDOR data. Appl Magn Reson. 2006;30:473–498. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.