SUMMARY
The Saccharomyces cerevisiae monopolin complex directs proper chromosome segregation in meiosis I by mediating co-orientation of sister kinetochores on the meiosis I spindle. The monopolin subunits Csm1 and Lrs4 form a V-shaped complex that may directly crosslink sister kinetochores. We report here biochemical characterization of the monopolin complex subunits Mam1 and Hrr25 and of the complete four-protein monopolin complex. By purifying monopolin subcomplexes with different subunit combinations, we have determined the stoichiometry and overall architecture of the full monopolin complex. We have determined the crystal structure of Csm1 bound to a Mam1 fragment, showing how Mam1 wraps around the Csm1 dimer and alters the stoichiometry of kinetochore-protein binding by Csm1. We further show that the kinase activity of Hrr25 is altered by Mam1 binding, and we identify Hrr25 phosphorylation sites on Mam1 that may affect monopolin complex stability and/or kinetochore binding in meiosis.
INTRODUCTION
Faithful transmission of the genome through cell division is critical for survival. In sexually reproducing eukaryotes, the specialized meiotic cell division gives rise to gametes or spores with exactly half the chromosome complement of the mother cell (Lee and Amon, 2001; Nasmyth, 2001; Marston and Amon, 2004). This reduction in ploidy is accomplished by two rounds of chromosome segregation without an intervening DNA replication step: In meiosis I, each chromosome aligns with and segregates from its homolog; and in meiosis II, replicated sister chromatids segregate as in mitosis. Homologous chromosome segregation in meiosis I is enabled by physical links between homologs called crossovers or chiasmata, and by the meiosis I-specific co-orientation, or attachment to the same spindle pole, of each pair of sister chromatids.
The mechanism of meiosis I sister chromatid co-orientation in different eukaryotes depends on their centromere and kinetochore architecture. In most eukaryotes, the kinetochore assembles on a “regional” centromere that spans tens of kb to multiple Mb; each kinetochore can then attach to multiple microtubules (Przewloka and Glover, 2009; Verdaasdonk and Bloom, 2011). Proper kinetochore-microtubule attachment depends on the heterochromatin surrounding centromeres, and sister chromatid co-orientation in meiosis I further depends on the pattern of cohesin complex deposition near centromeres as well as a cohesin-complex binding protein, Moa1 (Yokobayashi and Watanabe, 2005; Sakuno et al., 2009; Tada et al., 2011). In contrast, the budding yeast Saccharomyces cerevisiae and its close relatives have a minimal ~150 base pair (bp), sequence-defined “point” centromere, on which assembles a kinetochore that binds a single microtubule (Westermann et al., 2007). In these organisms, sister kinetochore co-orientation in meiosis I depends on the Aurora B/Ipl1 kinase, as well as on a specialized four-protein complex known as monopolin (Tóth et al., 2000; Rabitsch et al., 2003; Petronczki et al., 2006; Monje-Casas et al., 2007). The monopolin complex has been proposed to link sister kinetochores, effectively fusing them into a single microtubule-binding site to achieve co-orientation (Rabitsch et al., 2003; Corbett et al., 2010).
The monopolin complex consists of four proteins, Mam1, Csm1, Lrs4, and Hrr25, which are interdependent for kinetochore localization in meiosis I, and are all necessary for sister kinetochore co-orientation (Tóth et al., 2000; Rabitsch et al., 2003; Petronczki et al., 2006). We have shown previously that Csm1 and Lrs4 form a V-shaped complex with two globular “heads” spaced ~10 nm apart (Corbett et al., 2010). Each head comprises a dimer of Csm1 C-terminal domains, and each of these domains can bind the kinetochore proteins Dsn1 and Mif2. Thus, the full complex has two pairs of kinetochore-binding sites separated by ~10 nm. We proposed that the Csm1:Lrs4 complex is a kinetochore crosslinker, with each head binding one sister kinetochore. Two additional observations support a direct interaction of Csm1:Lrs4 with kinetochores. First, the Csm1:Lrs4 subcomplex localizes to kinetochores independently of Mam1 in mitotic anaphase, where it appears to increase chromosome segregation fidelity (Brito et al., 2010a). Second, orthologs of Csm1 and Lrs4, but not Mam1, are also present in regional-centromere fungi including Schizosaccharomyces pombe (Pcs1 and Mde4, respectively), where they bind kinetochores in mitosis and inhibit merotelic attachments (Rabitsch et al., 2003; Tanaka et al., 2009; Corbett et al., 2010; Rumpf et al., 2010; Tada et al., 2011).
In S. cerevisiae, Csm1 and Lrs4 relocalize from the nucleolus to kinetochores in response to Lrs4 phosphorylation during early prophase of meiosis I (Rabitsch et al., 2003; Katis et al., 2004; Huang et al., 2006; Katis et al., 2010), but they are not effectively retained at kinetochores without the meiosis-specific protein Mam1, which binds Csm1 (Rabitsch et al., 2003). Mam1, in turn, does not localize to kinetochores without both Csm1:Lrs4 and Hrr25, a ubiquitous casein kinase 1 δ/ε (Petronczki et al., 2006). Hrr25 has diverse functions in the cell but appears to play two distinct roles in meiosis I. First, it partners with Cdc7-Dbf4 kinase in phosphorylating the meiosis-specific cohesin subunit Rec8, allowing Rec8 cleavage along chromosome arms at the onset of anaphase I (Katis et al., 2010). Second, it supports sister chromatid co-orientation, which requires both Mam1-dependent kinetochore localization and its kinase activity (Petronczki et al., 2006). Hrr25 phosphorylates Mam1 at sites not yet determined (Petronczki et al., 2006).
We outline here the molecular architecture of the monopolin complex from S. cerevisiae, focusing on Mam1 and Hrr25. We show that Mam1 binds Csm1 and Hrr25 through distinct domains that are likely to be flexibly linked and that Mam1 binding modulates the kinase activity of Hrr25. We have determined the X-ray crystal structure of the Mam1-Csm1 complex, which shows that a single copy of Mam1 binds a Csm1 dimer and occludes one of its two kinetochore-protein binding sites. Finally, we have prepared the intact, four-protein monopolin complex by coexpression of its components, allowing us to examine its subunit stoichiometry and to sketch a preliminary molecular picture of this complex.
RESULTS
Mam1 Interactions with Csm1 and Hrr25
Csm1 and Lrs4, which are found throughout fungi, form the conserved structural core of the S. cerevisiae monopolin complex (Rabitsch et al., 2003; Gregan et al., 2007; Corbett et al., 2010). Mam1 is present only in point-centromere yeast, which include S. cerevisiae and its close relatives. Within the 302-residue protein, there are two well-conserved regions encompassing residues 92–190 and 224–260. All Mam1 constructs we tested were insoluble when expressed alone in Escherichia coli, suggesting that the protein requires binding partners for solubility. Hrr25 is a member of the ubiquitous casein kinase 1 δ/ε family, with the kinase domain comprising residues 1–293. Following the kinase domain is a central domain (residues 294–394), well conserved in point-centromere yeasts, and a poorly conserved proline/glutamine (P/Q)-rich region (residues 395–494). While the full-length protein is poorly behaved on its own, a construct spanning both conserved regions (residues 1–394) can be solubly expressed at high levels in E. coli. Hrr251–394 is an active kinase, as measured by extensive nonspecific auto-phosphorylation: Mass spectrometry indicates that over one third of Ser and Thr residues in the purified protein are phosphorylated to some degree (Figure S1A and Table S1). Size exclusion chromatography and multiangle light-scattering analysis (SEC-MALS) on both wild-type Hrr251–394 and a kinase-dead mutant (Lys38 to Arg; K38R) shows that this construct is monomeric in solution (Table S2).
To isolate Mam1 regions that interact with Csm1 and Hrr25 (Rabitsch et al., 2003; Petronczki et al., 2006), we expressed in vitro a series of Mam1 constructs truncated in 30-residue steps from both termini and tested their association with His6-tagged Csm1 and Hrr251–394. We found Mam1’s C-terminal region (residues 211–302, further refined to 221–290) to be sufficient for Csm1 binding (Figures 1A, S2A, and S2C). The Mam1221–290 fragment formed stable complexes with both full-length Csm1 and the isolated Csm1 C-terminal domain (residues 69–181) when coexpressed in E. coli. A smaller construct, Mam1221–270, also bound Csm1, but the complex was unstable over the course of several days (data not shown). Equilibrium analytical ultracentrifugation analysis of the Csm1:Mam1221–290 complex showed that it contains a dimer of Csm1 and one copy of Mam1 (Table S2).
Figure 1. Monopolin Complex Interactions.
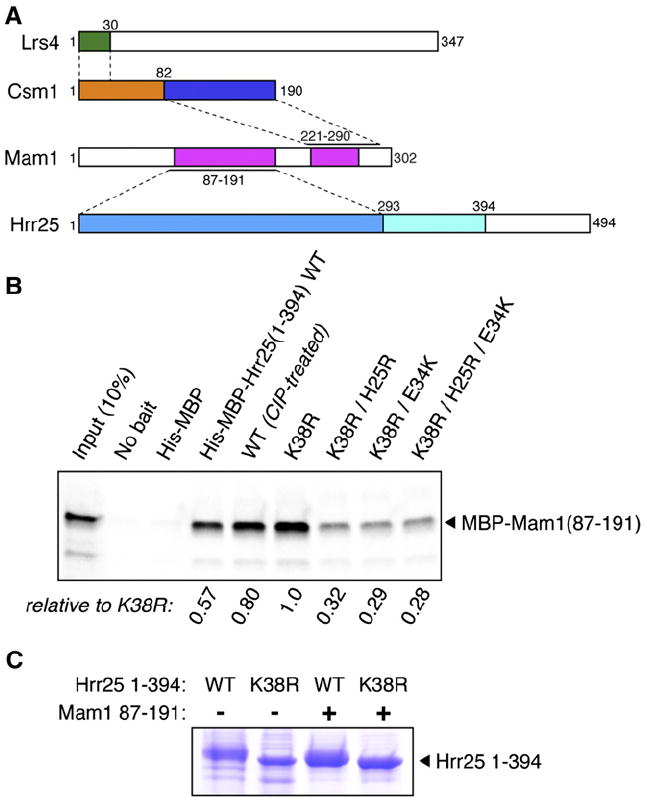
(A) Subunit interactions. Lrs4 is shown in green (Csm1-interacting region) and white (C-terminal uncharacterized region), Csm1 is shown in yellow (coiled coil) and blue (globular domain), Mam conserved regions are shown in magenta, and Hrr25 is shown in light blue (kinase domain), cyan (conserved extension in point-centromere fungi), and white (P/Q-rich domain). Interacting regions (from Rabitsch et al., 2003; Petronczki et al., 2006; Corbett et al., 2010) and the present work) are indicated by dashed lines; see Figures S2A–S2D for details.
(B) Pulldown showing interactions between wild-type (WT), kinase-dead (K38R), and H25R/E34K mutant Hrr251–394 and Mam1 (CIP, calf intestinal phosphatase).
(C) Hrr251–394 autophosphorylation. His6-Hrr251–394 wild-type (WT) or kinase-dead (K38R) was expressed in E. coli, alone or with Mam187–191, and purified by nickel-affinity pulldown. In agreement with mass spectrometry (see Figure S1A), Hrr25 WT appears less phosphorylated when coexpressed with Mam1.
See also Figures S1 and S2 and Tables S1 and S2.
Using the same series of Mam1 truncations, we identified a central region (residues 91–210) as necessary for Hrr25 binding and subsequently identified a fragment containing residues 87–191 that stably bound Hrr251-394 when coexpressed in E. coli (Figures 1A, S2B, and S2D). Using the Mam187–191 fragment, we tested the importance for Mam1 binding of Hrr25 residues His25 and Glu34, probable surface-exposed residues implicated in Mam1 association in vivo (Petronczki et al., 2006). The H25R, E34K, and H25R+E34K mutations of Hrr25 all strongly impaired Mam187–191 binding (Figure 1B). We also found that Mam187–191 binds more tightly to the kinase-dead K38R mutant than to wild-type Hrr25; this difference is due to nonspecific autophosphorylation of the wild-type protein, which probably occludes the Mam1-binding interface (Figure 1B; cf. binding of native vs. phosphatase-treated Hrr25).
When coexpressing Mam187–191 with Hrr251–394, we noticed that the mobility of wild-type Hrr25 coexpressed with Mam1 on SDS-PAGE is between that of the wild-type protein expressed in isolation and of the kinase-dead (K38R) protein (Figure 1C). Mass spectrometry showed that coexpression with Mam187–191 eliminated detectable autophosphorylation at 10 of the 28 sites identified with Hrr251–394 alone and tended to reduce phosphorylation levels at other sites (Figure S1A and Table S1). Thus, Mam1 binding appears to lead to reduced Hrr25 kinase activity and/or increased specificity. Using the same coexpressed samples, we also identified Hrr25 phosphorylation sites on Mam1. Ser214, located between the Csm1- and Hrr25-binding regions of Mam1, was strongly phosphorylated when coexpressed with Hrr25, and a Ser/Thr-rich stretch within the Csm1-binding region (residues 257–264: TSENPFSS) was also phosphorylated (Figure S1B).
We reconstituted monopolin subcomplexes containing Hrr25 and Mam1. Ultracentrifugation and multiangle light scattering showed that the Hrr251–394 K38R:Mam187–191 complex has a 1:1 stoichiometry. While we were unable to express full-length Hrr25 on its own, we could coexpress Hrr25 full-length K38R with Mam187–191 and show that this complex also has a 1:1 stoichiometry (Table S2). When we coexpressed all four monopolin complex proteins using a Mam1 construct encompassing both the Csm1 and Hrr25 binding sites (Hrr251–394 K38R:Mam187–302: Csm1:Lrs41–102), the mass of the resulting complex, as measured by analytical ultracentrifugation, was consistent with a stoichiometry of 2 Hrr25:2 Mam1:4 Csm1:2 Lrs4; this stoichiometry also agrees with all of our measurements on monopolin subcomplexes (Table S2). The ultracentrifugation data for the four-protein complex did, however, show some evidence of aggregation or self-association, leaving open the possibility that the fully assembled monopolin complex could form higher order structures under certain conditions (see the Discussion section). Thus, our present data lead to the following picture for the monopolin complex: one V-shaped 4 Csm1:2 Lrs4 complex (Corbett et al., 2010) with a 1 Mam1:1 Hrr25 complex associated with each of the two Csm1 globular heads. The poor conservation and predicted disorder of the Mam1 region between its Csm1- and Hrr25-binding regions (residues ~192–220) indicate that Hrr25 is likely to be flexibly tethered to the rest of the complex, allowing it significant positional freedom.
Structure of Csm1 Bound to a Mam1 Fragment
We crystallized the complex of full-length Csm1 with Mam1221–290, determined its structure at a resolution of 3.05 Å and refined the structure to R/Rfree values of 21.3%/26.0% (Table S3). The asymmetric unit contains a dimer of Csm1 and one copy of Mam1, in agreement with measured stoichiometry in solution. We located the Csm1 dimer by molecular replacement, modeled residues 223–263 of Mam1, and confirmed the sequence assignment with simulated annealing omit maps, and from the anomalous scattering of a sulfur atom in a complex with Mam1 Phe243 mutated to Met (Figures 2 and S3). In an effort to validate the sequence assignment, we mutated five conserved residues in Mam1 and 14 in Csm1 and tested their effects on the Csm1:Mam1 interaction; none of these single-residue mutations had a discernible effect on binding. This behavior, while unexpected, is presumably due to the large extended Csm1:Mam1 interface (discussed later). The Mam1 region encompassing residues 271–290 is completely disordered in our crystals, despite its contribution to tight Csm1 binding in solution. This region is strongly positively charged (pI = 10.5), and it may therefore contribute to Csm1 binding through nonspecific ionic interactions with the negatively charged Csm1 C-terminal domain (pI = 5.0 for Csm169–190).
Figure 2. Structure of the Csm1:Mam1 Complex.
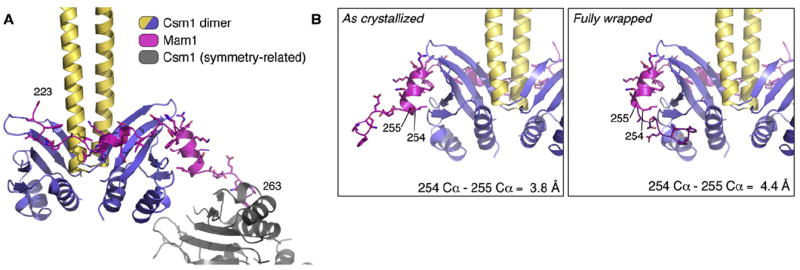
(A) Overall structure of the 2 Csm1: 1 Mam1 complex, with Csm1 colored yellow and blue, and Mam1 indicated in purple. A symmetry-related copy of Csm1 that the Mam1 C-terminal region contacts is in gray. The N- and C-terminal residues of the ordered segment of Mam1 (residues 223 and 263) are labeled.
(B) A 180° rotated view of the Mam1 C-terminal segment conformation as crystallized (left) and translocated based on crystallographic and Csm1-dimer symmetry onto Csm1 chain B to generate a fully wrapped model (right).
See also Figure S3 and Table S3.
Csm1 is a homodimer with an N-terminal coiled coil (residues 3–82 of 190) and a C-terminal globular domain (residues 83–190) and is structurally related to several kinetochore complexes: Spc24/Spc25, Ctf19/Mcm21, and Mad1 (Wei et al., 2006; Corbett et al., 2010; Kim et al., 2012; Schmitzberger and Harrison, 2012). Mam1 binds to the C-terminal domain of Csm1, adopting an extended conformation that wraps around the Csm1 dimer (Figure 2A). Mam1 makes distinct contacts with each of the two Csm1 monomers, denoted here as “A” and “B”. The N-terminal portion of the Mam1 fragment contacts Csm1 monomer A, the central part of the peptide wraps across monomer B, and the C-terminal portion (residues 255–263) extends away from the dimer to bind a crystallographic symmetry-related copy of monomer A. This arrangement suggests that Mam1 could induce oligomerization of Csm1 dimers, but we observed no evidence of higher order complexes in solution. The surface of Csm1 that binds the Mam1 C-terminal segment is, in fact, adjacent to the surface that binds the preceding Mam1 segment, but it is buried in Csm1 monomer B by crystal packing interactions. We could therefore model a fully wrapped 2 Csm1:1 Mam1 complex by translocating the bound conformation of the Mam1 C-terminal segment (residues 255–263) back on to Csm1 monomer B: The configuration of the translocated C terminus places the Cα of residue 255 4.4 Å from the Cα of the untranslocated residue 254 (canonical Cα-Cα distance: 3.8 Å), showing that a minor backbone reconfiguration could accommodate this fully wrapped conformation (Figure 2B). As there is no evidence that Mam1 causes Csm1 oligomerization in solution, we conclude that the fully wrapped conformation probably represents the physiologically relevant form of the Csm1:Mam1 complex.
The Csm1 globular domain shares a fold with several other kinetochore proteins; this fold appears to be a conserved protein-protein interaction module that is used repeatedly within the kinetochore. The structure of the Csm1:Mam1 complex represents a picture of how this module interacts with a binding partner. We speculate that other members of this fold family, including Spc24/25 and Mad1, may similarly interact with their binding partners at the kinetochore through binding of short, flexible peptide segments across these domains’ surfaces.
Mam1 Binding Affects Csm1-Kinetochore Protein Interactions
We previously showed that S. cerevisiae Csm1 binds the kinetochore proteins Dsn1 and Mif2 through a conserved hydrophobic surface patch on its C-terminal globular domain. We confirmed these findings in the case of Dsn1 using purified, intact MIND complex (consisting of Dsn1, Mtw1, Nsl1, and Nnf1). We also showed that the S. pombe Csm1 ortholog (Pcs1) binds orthologs of Dsn1 (Mis13) and Mif2 (Cnp3) through the same conserved surface (Corbett et al., 2010). In the Csm1:Mam1221–290 structure described here, the C-terminal segment of Mam1 interacts with this conserved hydrophobic surface, with the side chain of Phe262 inserted into a hydrophobic pocket formed by several aromatic residues and contacting residues Tyr156 and Leu161, whose mutation affects Dsn1/Mif2 binding (Figure 3A). Thus, Mam1 appears to occlude one of the two kinetochore-protein binding sites on a Csm1 dimer.
Figure 3. Mam1 Affects Csm1-Kinetochore Interactions.
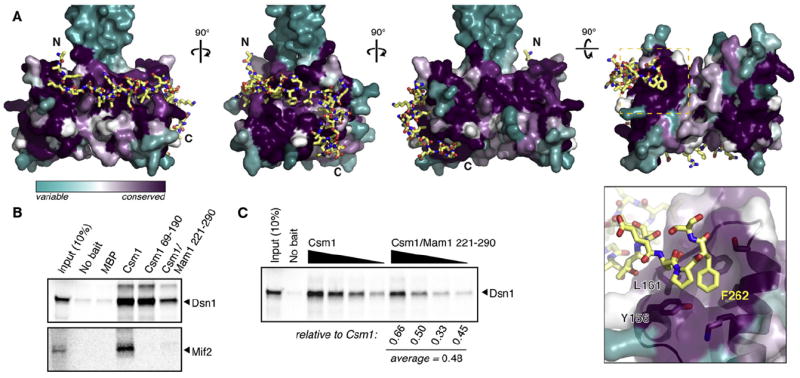
(A) Four views of the Mam1 C-terminal segment (yellow sticks) binding Csm1 (colored cyan-to-purple, based on conservation within point-centromere fungi). Close-up panel: The interaction with Csm1’s kinetochore protein-binding patch (including residues Y156 and L161; Corbett et al., 2010) is anchored by Mam1 Phe262.
(B) Pulldown of in vitro-expressed (35)S-methione-labeled Dsn1 or Mif2 with Csm1 and the Csm1-Mam1 complex.
(C) Pulldown of Dsn1 using equimolar amounts of Csm1 (left four lanes: 10, 5, 2, and 1 μg Csm1) and Csm1:Mam1221–290 (right four lanes: matched molar amounts of Csm1:Mam1221–290), showing that Csm1:Mam1221–290 binds roughly half as much Dsn1 as does Csm1 alone.
To test whether Mam1 affects interactions of Csm1 with its kinetochore binding partners, we examined the binding of both Dsn1 and Mif2 to Csm1 and the Csm1:Mam1221–290 complex using an in vitro pulldown assay (Figure 3B). While Dsn1 bound robustly to both Csm1 alone and the Csm1:Mam1221–290 complex, Mif2 bound only weakly to Csm1 and did not detectably bind to Csm1:Mam1221–290. Thus, the previously described binding of Csm1 to Mif2 may not be functionally relevant, at least in S. cerevisiae (see the Discussion section). When we compared Dsn1 binding by equivalent molar amounts of full-length Csm1 and Csm1:Mam1221–290, the Csm1:Mam1 complex bound about half as much Dsn1 as did Csm1 alone (Figure 3C), which is consistent with the idea that Mam1 occludes one of the two kinetochore-protein binding sites on the Csm1 dimer.
DISCUSSION
Sister kinetochore co-orientation in S. cerevisiae meiosis I depends on the monopolin complex, which now appears to have multiple functions at kinetochores. First, structural and biochemical data have suggested that the Csm1 and Lrs4 subunits crosslink sister kinetochores directly (Corbett et al., 2010). Second, these proteins associate with the condensin complex and may recruit condensins to the kinetochore in both S. cerevisiae meiosis I and S. pombe mitosis (Brito et al., 2010b; Tada et al., 2011). Finally, sister co-orientation also requires recruitment of the Hrr25 kinase to kinetochores in S. cerevisiae meiosis I, although the roles of the kinase at the kinetochore are currently unclear (Petronczki et al., 2006).
When combined with our earlier data, the work presented here allows us to construct a molecular model for the bulk of the four-protein monopolin complex (Figure 4). The complex contains two dimers of Csm1 and two copies of Lrs4, as reported previously (Corbett et al., 2010). Bound to each Csm1 dimer head is one copy of Mam1, which, in turn, recruits one copy of the Hrr25 kinase. While we have not observed any convincing evidence of higher order monopolin complex assembly beyond what is shown in Figure 4, Petronczki et al. (2006) have reported that Hrr25 self-associates specifically in meiosis I. This potential Hrr25 self-association could, in turn, promote oligomerization of the monopolin complex, which may play a role in sister kinetochore crosslinking by the complex.
Figure 4. Model of the Intact Monopolin Complex, with Proteins Colored as in Figure 1.
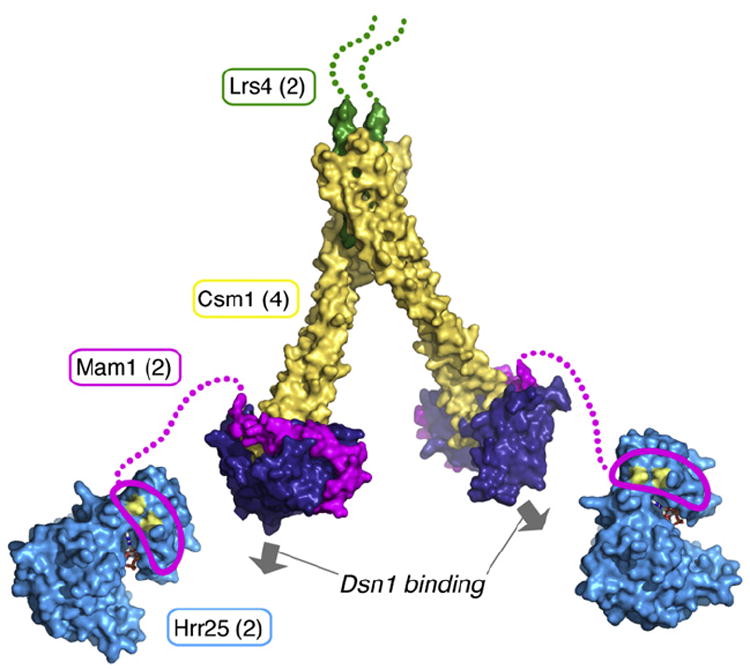
Hrr25 is represented by the structure of S. pombe casein kinase I (Xu et al., 1995), with residues H25 and E34 shown in yellow and bound ATP shown as sticks. The central domain of Mam1 (magenta outline) is shown bound to the N-terminal lobe of Hrr25 (Petronczki et al., 2006), with the linker region (residues 192–220) shown as a dotted magenta line. The copy number of each protein in the complex is indicated in parentheses, and the two available Dsn1-binding sites are indicated by arrows.
Our data clarify earlier ambiguity concerning the kinetochore target of S. cerevisiae monopolin. In previous work, we tested both Dsn1 and Mif2 for Csm1 binding, because prior two-hybrid screening with S. cerevisiae proteins had implicated Dsn1 as a Csm1 binding partner (Wong et al., 2007) while data showing that the S. pombe Csm1 and Mif2 orthologs Pcs1 and Cnp3 interact had, by inference, implicated Mif2 as a possible binding partner (Tanaka et al., 2009). We found that both proteins bound Csm1 in vitro but that the interaction of Csm1 with Mif2 was less robust (Corbett et al., 2010). Our present data show that Dsn1 binds tightly to both Csm1 alone and the Csm1:Mam1 complex, while Mif2 does not bind the latter at all. This result suggests that the Csm1-Mif2 interaction we detected previously was probably nonspecific and functionally irrelevant in S. cerevisiae. In support of this conclusion, we have isolated small regions of both Dsn1 and Tof2 (the Csm1 binding partner at the rDNA locus; Corbett et al., 2010) that robustly bind Csm1 (data not shown), but we have not been able to identify any truncated Mif2 constructs that bind. Our data leave open the question of why the S. pombe and S. cerevisiae monopolin complexes appear to bind different proteins at the kinetochore. The differences might correlate with the distinct functions of these complexes: S. pombe monopolin inhibits merotelic attachments in mitosis, while the S. cerevisiae complex mediates sister kinetochore co-orientation in meiosis I. Further work will be required to address this question.
Our findings raise interesting questions about the roles of Mam1 and Hrr25 in regulating the stoichiometry and affinity of kinetochore binding by the monopolin complex. Hrr25 is needed in meiosis I to phosphorylate the meiotic cohesin subunit Rec8 along chromosome arms to promote its cleavage (Katis et al., 2010); it is also independently required at kinetochores as a component of monopolin, where it phosphorylates Mam1 (Petronczki et al., 2006). We have identified Hrr25 phosphorylation sites on Mam1 in the linker between its Hrr25- and Csm1-binding regions (Ser214) and in a short stretch (residues 257–264) that associates with one of the two conserved kinetochore-binding surfaces on the Csm1 dimer. Our structural and biochemical data show that unphosphorylated Mam1 stably binds Csm1 and occludes one kinetochore-binding interface on the dimer, reducing by half Csm1’s stoichiometry of binding to the kinetochore protein Dsn1. It is currently unclear, however, how Mam1 phosphorylation might affect these interactions. Phosphorylation of Mam1 at residues 257–264 could partially release Mam1 from Csm1, opening up the previously occluded second kinetochore-binding patch and potentially strengthening the Csm1-kinetochore interaction. On the other hand, we have also observed that Hrr25’s kinase activity adversely affects the stability of the monopolin complex itself and, specifically, that of the Mam1-Csm1 interaction. When we coexpress Mam1, Csm1, and Lrs4 with the kinase-dead K38R mutant of Hrr25, we obtain a very stable complex. In contrast, when we coexpress monopolin subunits with wild-type Hrr25, the resulting complex can be purified but separates over the course of 1–2 days into Mam1:Hrr25 and Csm1:Lrs4 subcomplexes (data not shown). Moreover, published data show that the Csm1-Lrs4 and Mam1-Hrr25 interactions are much more readily detected in cells than the Csm1-Mam1 interaction (Petronczki et al., 2006). Thus, it is possible that Mam1 phosphorylation by Hrr25 at Ser214, residues 257–264, or another unidentified site may destabilize the monopolin complex.
In summary, the data presented here support the idea that Mam1 and Hrr25 regulate the kinetochore-binding affinity and/or stoichiometry of the monopolin complex. They also suggest that Hrr25’s kinase activity may affect the stability of the monopolin complex itself through Mam1 phosphorylation. The functional consequences of Mam1 phosphorylation at each identified site, and the details of how Hrr25 is regulated by Mam1 binding, remain important questions for understanding how the monopolin complex ensures sister kinetochore co-orientation in meiosis I.
EXPERIMENTAL PROCEDURES
Proteins were cloned into vectors with N-terminal, TEV-protease cleavable His6 tags and purified using Ni2+ affinity, ion-exchange, and gel filtration chromatography. For pulldown assays, bait proteins were expressed in vitro by coupled transcription/translation, and assays were performed as described elsewhere (Corbett et al., 2010). Csm1:Mam1221–290 was crystallized in 100 mM HEPES, pH 7.5, 100–150 mM MgCl2, and 6%–9% PEG 4000, cryo-protected in 25% PEG 400, and flash-frozen in liquid nitrogen; and diffraction data were collected at NE-CAT beamlines 24ID-C and 24ID-E at the Advanced Photon Source at Argonne National Laboratory. The structure was determined by molecular replacement using the structure of Csm1, and Mam1 residues 223–263 were manually built into density-modified difference maps and refined, B-factor sharpened 2Fo-Fc maps (see Extended Experimental Procedures for details; see Table S3 for data collection and refinement statistics).
Supplementary Material
Acknowledgments
We thank the staff at the Advanced Photon Source NE-CAT beamlines, especially Kay Perry, for advice and assistance with data collection and interpretation; James Whittle for assistance with diffraction data collection; Ross Tomaino for assistance with mass spectrometry; Qiaozhen Ye for technical assistance; and members of the Corbett and Harrison laboratories and Angelika Amon for helpful discussions. K.D.C. acknowledges prior support from the Charles A. King Trust Postdoctoral Fellowship Program (Bank of America, N.A., Co-Trustee) and current salary support from the Ludwig Institute for Cancer Research; S.C.H. is an investigator in the Howard Hughes Medical Institute.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes Extended Experimental Procedures, three figures, and three tables and can be found with this article online at doi:10.1016/j.celrep.2012.05.012.
ACCESSION NUMBERS
Coordinates and structure factors for the Csm1: Mam1221–290 structure have been deposited into the Protein Data Bank (PDB) under accession code 4EMC.
Publisher's Disclaimer: LICENSING INFORMATION
This is an open-access article distributed under the terms of the Creative Commons Attribution-Noncommercial-No Derivative Works 3.0 Unported License (CC-BY-NC-ND; http://creativecommons.org/licenses/by-nc-nd/3.0/legalcode).
References
- Brito IL, Monje-Casas F, Amon A. The Lrs4-Csm1 monopolin complex associates with kinetochores during anaphase and is required for accurate chromosome segregation. Cell Cycle. 2010a;9:3611–3618. doi: 10.4161/cc.9.17.12885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brito IL, Yu H-G, Amon A. Condensins promote coorientation of sister chromatids during meiosis I in budding yeast. Genetics. 2010b;185:55–64. doi: 10.1534/genetics.110.115139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett KD, Yip CK, Ee L-SS, Walz T, Amon A, Harrison SC. The monopolin complex crosslinks kinetochore components to regulate chromosome-microtubule attachments. Cell. 2010;142:556–567. doi: 10.1016/j.cell.2010.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregan J, Riedel CG, Pidoux AL, Katou Y, Rumpf C, Schleiffer A, Kearsey SE, Shirahige K, Allshire RC, Nasmyth K. The kinetochore proteins Pcs1 and Mde4 and heterochromatin are required to prevent merotelic orientation. Curr Biol. 2007;17:1190–1200. doi: 10.1016/j.cub.2007.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Brito IL, Villén J, Gygi SP, Amon A, Moazed D. Inhibition of homologous recombination by a cohesin-associated clamp complex recruited to the rDNA recombination enhancer. Genes Dev. 2006;20:2887–2901. doi: 10.1101/gad.1472706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katis VL, Matos J, Mori S, Shirahige K, Zachariae W, Nasmyth K. Spo13 facilitates monopolin recruitment to kinetochores and regulates maintenance of centromeric cohesion during yeast meiosis. Curr Biol. 2004;14:2183–2196. doi: 10.1016/j.cub.2004.12.020. [DOI] [PubMed] [Google Scholar]
- Katis VL, Lipp JJ, Imre R, Bogdanova A, Okaz E, Habermann B, Mechtler K, Nasmyth K, Zachariae W. Rec8 phosphorylation by casein kinase 1 and Cdc7-Dbf4 kinase regulates cohesin cleavage by separase during meiosis. Dev Cell. 2010;18:397–409. doi: 10.1016/j.devcel.2010.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Sun H, Tomchick DR, Yu H, Luo X. Structure of human Mad1 C-terminal domain reveals its involvement in kinetochore targeting. Proc Natl Acad Sci USA. 2012;109:6549–6554. doi: 10.1073/pnas.1118210109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B, Amon A. Meiosis: how to create a specialized cell cycle. Curr Opin Cell Biol. 2001;13:770–777. doi: 10.1016/s0955-0674(00)00282-9. [DOI] [PubMed] [Google Scholar]
- Marston AL, Amon A. Meiosis: cell-cycle controls shuffle and deal. Nat Rev Mol Cell Biol. 2004;5:983–997. doi: 10.1038/nrm1526. [DOI] [PubMed] [Google Scholar]
- Monje-Casas F, Prabhu VR, Lee BH, Boselli M, Amon A. Kinetochore orientation during meiosis is controlled by Aurora B and the monopolin complex. Cell. 2007;128:477–490. doi: 10.1016/j.cell.2006.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K. Disseminating the genome: joining, resolving, and separating sister chromatids during mitosis and meiosis. Annu Rev Genet. 2001;35:673–745. doi: 10.1146/annurev.genet.35.102401.091334. [DOI] [PubMed] [Google Scholar]
- Petronczki M, Matos J, Mori S, Gregan J, Bogdanova A, Schwickart M, Mechtler K, Shirahige K, Zachariae W, Nasmyth K. Monopolar attachment of sister kinetochores at meiosis I requires casein kinase 1. Cell. 2006;126:1049–1064. doi: 10.1016/j.cell.2006.07.029. [DOI] [PubMed] [Google Scholar]
- Przewloka MR, Glover DM. The kinetochore and the centromere: a working long distance relationship. Annu Rev Genet. 2009;43:439–465. doi: 10.1146/annurev-genet-102108-134310. [DOI] [PubMed] [Google Scholar]
- Rabitsch KP, Petronczki M, Javerzat JP, Genier S, Chwalla B, Schleiffer A, Tanaka TU, Nasmyth K. Kinetochore recruitment of two nucleolar proteins is required for homolog segregation in meiosis I. Dev Cell. 2003;4:535–548. doi: 10.1016/s1534-5807(03)00086-8. [DOI] [PubMed] [Google Scholar]
- Rumpf C, Cipak L, Schleiffer A, Pidoux A, Mechtler K, Tolić-Nørrelykke IM, Gregan J. Laser microsurgery provides evidence for merotelic kinetochore attachments in fission yeast cells lacking Pcs1 or Clr4. Cell Cycle. 2010;9:3997–4004. doi: 10.4161/cc.9.19.13233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuno T, Tada K, Watanabe Y. Kinetochore geometry defined by cohesion within the centromere. Nature. 2009;458:852–858. doi: 10.1038/nature07876. [DOI] [PubMed] [Google Scholar]
- Schmitzberger F, Harrison SC. RWD domain: a recurring module in kinetochore architecture shown by a Ctf19-Mcm21 complex structure. EMBO Rep. 2012;13:216–222. doi: 10.1038/embor.2012.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada K, Susumu H, Sakuno T, Watanabe Y. Condensin association with histone H2A shapes mitotic chromosomes. Nature. 2011;474:477–483. doi: 10.1038/nature10179. [DOI] [PubMed] [Google Scholar]
- Tanaka K, Chang HL, Kagami A, Watanabe Y. CENP-C functions as a scaffold for effectors with essential kinetochore functions in mitosis and meiosis. Dev Cell. 2009;17:334–343. doi: 10.1016/j.devcel.2009.08.004. [DOI] [PubMed] [Google Scholar]
- Tóth A, Rabitsch KP, Gálová M, Schleiffer A, Buonomo SB, Nasmyth K. Functional genomics identifies monopolin: a kinetochore protein required for segregation of homologs during meiosis i. Cell. 2000;103:1155–1168. doi: 10.1016/s0092-8674(00)00217-8. [DOI] [PubMed] [Google Scholar]
- Verdaasdonk JS, Bloom K. Centromeres: unique chromatin structures that drive chromosome segregation. Nat Rev Mol Cell Biol. 2011;12:320–332. doi: 10.1038/nrm3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei RR, Schnell JR, Larsen NA, Sorger PK, Chou JJ, Harrison SC. Structure of a central component of the yeast kinetochore: the Spc24p/Spc25p globular domain. Structure. 2006;14:1003–1009. doi: 10.1016/j.str.2006.04.007. [DOI] [PubMed] [Google Scholar]
- Westermann S, Drubin DG, Barnes G. Structures and functions of yeast kinetochore complexes. Annu Rev Biochem. 2007;76:563–591. doi: 10.1146/annurev.biochem.76.052705.160607. [DOI] [PubMed] [Google Scholar]
- Wong J, Nakajima Y, Westermann S, Shang C, Kang J-S, Goodner C, Houshmand P, Fields S, Chan CSM, Drubin D, et al. A protein interaction map of the mitotic spindle. Mol Biol Cell. 2007;18:3800–3809. doi: 10.1091/mbc.E07-06-0536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu RM, Carmel G, Sweet RM, Kuret J, Cheng X. Crystal structure of casein kinase-1, a phosphate-directed protein kinase. EMBO J. 1995;14:1015–1023. doi: 10.1002/j.1460-2075.1995.tb07082.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokobayashi S, Watanabe Y. The kinetochore protein Moa1 enables cohesion-mediated monopolar attachment at meiosis I. Cell. 2005;123:803–817. doi: 10.1016/j.cell.2005.09.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


