Clones that confer the production of N-acyl amino acids to model cultured bacterial hosts are frequently identified in activity-based screens of soil DNA libraries. The N-acyl amino acids encoded by these clones are synthesized from acyl-(acyl-carrier-protein)s (acyl-ACPs, ACP) and amino acids using a diverse group of bacterial enzymes referred to as N-acyl amino acid synthases (NASs).[1, 2] The acyl-ACP substrates of NAS enzymes are common intermediates in the de novo synthesis of fatty acids by the type II fatty acid synthase system (FAS II).[3] In addition to N-acyl amino acids, FAS II-derived acyl-ACPs also serve as acyl-donors in the production of N-acyl homoserine lactone autoinducers, the cofactor lipoic acid and several lipopeptide antibiotics (Figure 1A, compounds 1–4).[4–9] During a recent screen of soil metagenomic libraries for clones exhibiting antimicrobial activity, we uncovered an NAS-containing clone (clone EC5) that confers the production of long-chain N-acyl-phenylalanines and N-acyl-tryptophans to multiple Gram-negative host species.[10] The NAS-encoding gene found on clone EC5, nasA, is the first gene in a predicted two gene operon, nasAB (GenBank Accession No. GQ869383). The second gene in this operon, nasB, encodes for a putative 1118 residue protein that contains six well-characterized Pfam homology domains (Figure 1B).[11] Based on generic functional predictions for these domains, we hypothesized that this enzyme was likely to be involved in the formation of ACP-linked fatty acids similar to those produced by bacterial FAS II systems. Here we report results from a series of heterologous expression experiments in Burkholderia graminis C4D1M, which show that nasB functions to provide branched-chain acyl-ACPs for nasA. Many bacteria, particularly Gram-negative species, do not natively produce branched-chain fatty acids.[3] NasB and its homologs are likely used by bacteria with straight-chain specific FAS II systems for generating branched-chain acyl-ACP substrates and should be useful as tools for synthesizing these substrates in model heterologous expression systems in which they are not natively produced (e.g. E. coli).
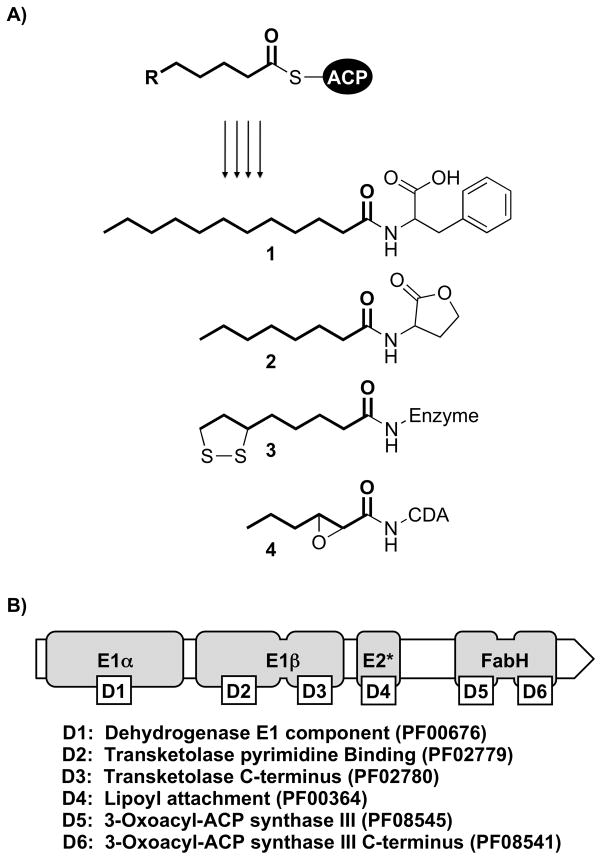
Figure 1.
A) Incorporation of FAS II intermediates (acyl-ACPs) into secondary metabolites and cofactors: N-dodecanoyl-phenylalanine (1), N-octanoyl homoserine lactone (2), enzyme-bound lipoic acid (3), and calcium-dependent antibiotic (CDA) (4). B) The domain architecture of nasB includes all three Pfam homology domains found in the E1α/E1β subuntis of 2-oxo acid dehydrogenase complexes, as well as both of the Pfam homology domains found in the E. coli FabH protein (β-ketoacyl-ACP synthase III). Separating these two sets of domains is an E2-like (E2*) lipoyl-attachment site.
The N-terminal region of nasB contains the same set of protein domains that are found in the E1α and E1β subunits of 2-oxo acid dehydrogenase complexes (e.g. the pyruvate dehydrogenase complex, which converts pyruvate to acetyl-CoA).[12] Following these domains is an E2-like (E2*) lipoyl-attachment site. Although lipoyl-attachment sites are found in the E2 subunits of 2-oxo acid dehydrogenase complexes, nasB lacks the catalytic lipoamide acyltransferase domain found at the C-terminus of canonical E2 subunit proteins (Pfam PF00198; exchanges the acyl-lipoate thioester with CoA, yielding a soluble acyl-CoA).[13] Instead, the C-terminal portion of nasB contains both of the protein domains that are found in the E. coli FabH protein, a β-ketoacyl-ACP synthase III (KASIII)-type enzyme that catalyzes the initial condensation reaction of FAS II (acetyl-CoA and malonyl-ACP, forming acetoacetyl-ACP).[14]
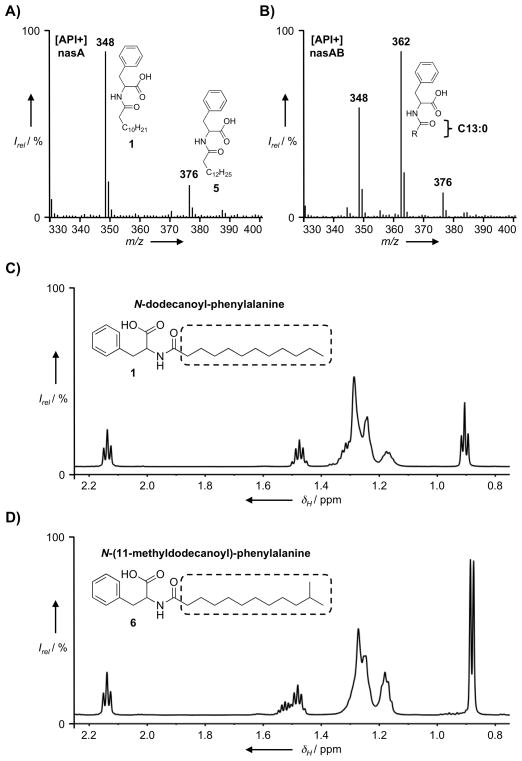
To test our original hypothesis that nasB provides substrates for the biosynthesis of N-acyl amino acids, the nasA gene was subcloned alone and in combination with nasB (i.e. the nasAB operon) into the broad host-range vector pJWC1, and these constructs were then electroporated into B. graminis for heterologous expression studies.[10] Ethyl acetate extracts from cultures of B. graminis were examined by HPLC-MS for differences in the N-acyl amino acids synthesized by each construct. Extracts of both cultures contained mixtures of N-acyl-phenylalanines, including significant amounts of the 12-carbon and 14-carbon saturated N-acyl derivatives of phenylalanine [m/z (M+H)+ = 348 and 376, compounds 1 and 5, respectively] (Figure 2A–B). Interestingly, the most abundant mass peak in the nasAB extract was from a compound with m/z (M+H)+ = 362 (Figure 2B), which was notably absent from the nasA-only extract (Figure 2A). The presence of this additional mass peak suggested that nasB afforded the production of a new 13-carbon saturated N-acyl-phenylalanine derivative.
Figure 2.
API-positive mode ionization data from the HPLC-MS analysis of extracts of B. graminis cultures transformed with A) pJWC1-nasA and B) pJWC1-nasAB. Comparison of the fatty acid region of the 1D 1H NMR spectrum of C) N-dodecanoyl-phenylalanine (1) with that of D) N-(11-methyldodecanoyl)-phenylalanine (6).
To determine the nature of the presumed 13-carbon acyl-chain, the metabolite corresponding to m/z = 362 (6) was isolated from the ethyl acetate extract of B. graminis [nasAB] culture broth. Analysis of the 1D 1H and 13C NMR spectra of 6 suggested this compound was structurally similar to previously reported N-acyl-phenylalanines (see Supplementary Materials).[10, 15] The presence of phenylalanine was also apparent from the fragments observed in the LRESI MS spectrum (m/z 166 and 120). In the 1H NMR spectrum of 6, signals arising from the fatty acyl-chain are similar to those observed in the spectra of straight-chain N-acyl-phenylalanines isolated from E. coli, except that the 3-proton triplet arising from the terminal fatty acid methyl-group is replaced by a 6-proton doublet (δH = 0.88 ppm, J (H,H) = 6.6 Hz) (Figure 2C–D). In the 1H-1H COSY spectrum of 6, these methyl group protons are coupled to a single methine proton (m, δH = 1.52 ppm) that is in turn coupled to the methylene envelope of the fatty acyl-chain. Collectively, this data indicates that compound 6 is the structurally novel N-acyl amino acid N-(11-methyldodecanoyl)-phenylalanine, which contains an iso-branched 13-carbon saturated acyl-chain (iso-C13:0). The fatty acid profile of wild-type B. graminis does not contain branched-chain fatty acids of any type, indicating that nasB functions to provide a source of branched-chain acyl-ACP substrates for nasA.[16]
The production of branched-chain fatty acids has been most extensively studied in the model Gram-positive bacterium Bacillus subtilis, where the FAS II system is responsible for producing both iso- and anteiso-branched fatty acids.[17] The most important difference between the FAS II system of B. subtilis and that of E. coli, a model for straight-chain fatty-acid biosynthesis, is the substrate specificity of FabH, the β-ketoacyl-ACP synthase that performs the initial condensation reaction in FAS II-mediated fatty acid biosynthesis.[18] In B. subtilis, but not in E. coli, FabH efficiently utilizes branched-chain acyl-CoA primers. These primers are produced from α-keto acids derived from the transamination of the proteinogenic branched-chain amino acids.[19, 20] The enzymatic machinery that carries out this function is a specialized α-keto acid decarboxylase referred to as the branched-chain α-keto acid dehydrogenase complex (BCKAD).[21] BCKAD contains the standard repertoire of protein domains that are typically found in the E1α, E1β, E2 and E3 subunits of other 2-oxo acid dehydrogenase complexes.
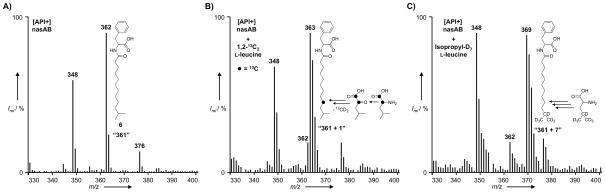
The N-terminal domains of nasB are the same as those found in the E1α/E1β subunits of BCKAD, suggesting that nasB also uses α-keto acid substrates derived from the transamination of amino acids. The odd length and terminal branching pattern of the iso-C13:0 acyl-chain substructure of 6 suggested that L-leucine (7) was the likely source of the preferred α-keto acid substrate of nasB. To test this hypothesis and evaluate the α-keto acid decarboxylase activity of nasB, we fed 1,2-13C2-labelled L-leucine to a culture of B. graminis [nasAB]. The most abundant mass peak in the organic extract of this culture was m/z (M+H)+ = 363, one mass unit greater than that of unlabelled 6. This result is consistent with the conversion of 1,2-13C2 L-leucine to 1,2-13C2 2-oxoisocaproic acid by endogenous transaminase and the subsequent decarboxylation of this substrate by nasB to form a C1 isotopically-enriched isovaleryl-intermediate that feeds into the biosynthesis of 6 (Figure 3B; compare with Figure 3A). The mass peaks corresponding to linear-chain N-acyl-phenylalanines 1 and 5 (m/z (M+H)+ = 348 and 376) were unaltered. To further confirm that the branched portion of the iso-C13:0 acy-chain of 6 originated from the branched side-chain of L-leucine, we also fed isopropyl-D7-labelled L-leucine to a culture of B. graminis [nasAB]. As expected, this resulted in the appearance of a prominent mass peak with m/z (M+H)+ = 369, corresponding to the incorporation of the leucine-derived isopropyl-D7 label into 6 (Figure 3C). Again, the mass peaks corresponding to linear-chain N-acyl-phenylalanines were unaltered. In B. graminis, branched-chain amino acid aminotransferase (BCAT; GenBank Accession No. EDT09763) likely performs the initial transamination reaction that provides the α-keto acid 8 used by nasB.
Figure 3.
API-positive mode ionization data from the HPLC-MS analysis of extracts of B. graminis [nasAB] cultures fed with either A) nothing, B) 1,2-13C2-labelled L-leucine or C) isopropyl-D7-labelled L-leucine.
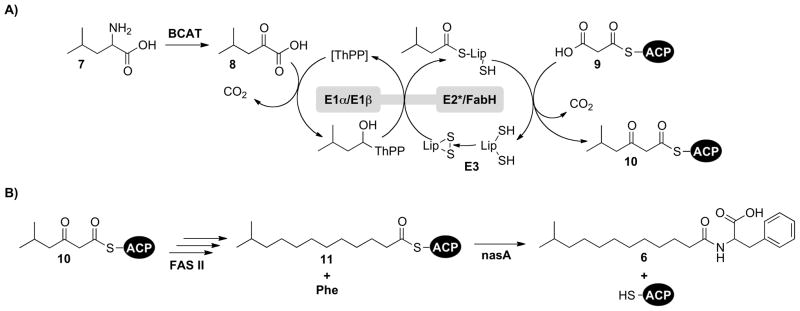
These feeding experiments support a model in which the nasB enzyme preferentially converts the product of leucine transamination, 2-oxoisocaproic acid (8), to 5-methyl-3-oxohexanoyl-ACP (10) through an integrated series of biochemical reactions (Scheme 1A). In this model, the E1α/E1β-like components of nasB first decarboxylate 2-oxoisocaproic acid (8), forming a covalent thiamine pyrophosphate (ThPP) adduct that is subsequently transferred to the bound lipoic acid cofactor. As previously noted, nasB lacks the catalytic E2 lipoamide acyltransferase domain that would normally transfer this covalently-bound intermediate from the lipoic acid cofactor to CoA. The absence of this catalytic domain instead suggests that the FabH-like component of nasB condenses malonyl-ACP (9) directly with the enzyme-bound isovaleryl-lipoate thioester, forming 5-methyl-3-oxohexanoyl-ACP (10). This FAS II intermediate can then undergo 3 rounds of canonical fatty acid chain elongation, yielding 11-methyldodecanoyl-ACP (11), which along with L-phenylalanine, is used by nasA for the production of N-(11-methyldodecanoyl)-phenylalanine (6) (Scheme 1B).
Scheme 1.
A) The proposed condensation of 2-oxoisocaproic acid (8) and malonyl-ACP (9) by nasB to form 5-methyl-3-oxohexanoyl-ACP (10). B) Scheme for the biosynthesis of N-(11-methyldodecanoyl)-phenylalanine (6), beginning with the FAS II-mediated chain elongation of 10, yielding 11-methyldodecanoyl-ACP (11).
The nasAB operon structure found on clone EC5 suggests that the branched-chain acyl-ACPs produced by nasB are specifically intended for use by nasA. Branched acyl-chains may therefore represent a characteristic feature used to distinguish the N-acyl amino acids produced by the uncultured EC5 bacterium from the more common straight-chain varieties isolated from numerous other environmental DNA clones. While the precise role(s) of N-acyl amino acids remains to be determined, it has been proposed that they may function as signaling molecules in a manner similar to that of several other N-acylated small molecules, most notably the N-acyl homoserine lactones (e.g. compound 2).[1] Variations in the acyl-chain length, degree of unsaturation and C-3 oxidation state of N-acyl homoserine lactones are the primary structural determinants used to target specific populations of receptor proteins.[22, 23] Although less common by comparison, the incorporation of branched acyl-chains into N-acyl homoserine lactones has also been reported.[24] The above examples highlight how acyl-chain diversification strategies can allow individual bacteria to increase the number of structurally and functionally unique metabolites they produce.
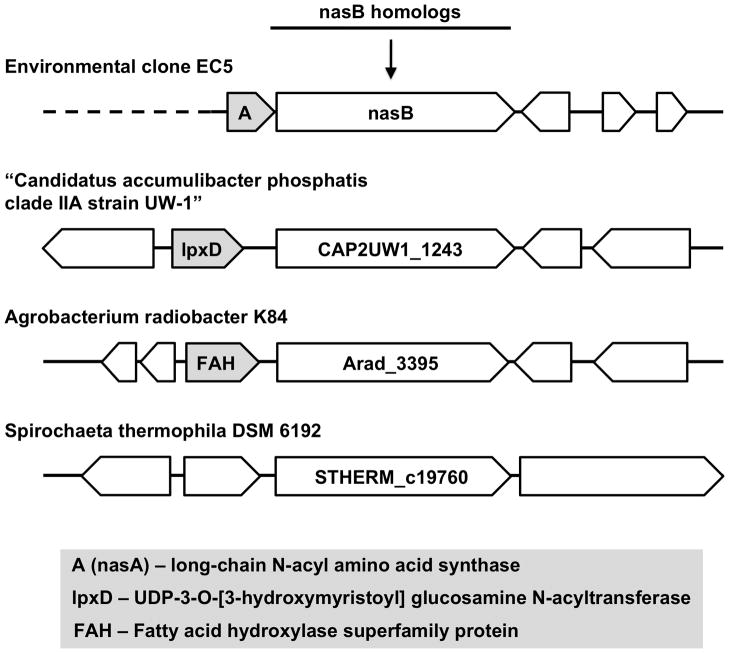
The genomes of three different species of bacteria are predicted to encode homologs of nasB containing the same putative domain architecture. These nasB homolgs do not form operon structures with nasA homologs, but are instead found within divergent genetic contexts (Figure 4). The nasB homolog from “Cand. A. phosphatis” is adjacent to a homolog of the N-acyltransferase lpxD, which transfers the acyl group from 3-hydroxytetradecanoyl-ACP to UDP-3-O-(3-hydroxytetradecanoyl)-D-glucosamine during lipopolysaccharide biosynthesis (GenBank Accession No. ACV34570; Pfam PF04613). The nasB homolog from A. radiobacter is adjacent to a putative fatty acid hydroxylase superfamily enzyme predicted to be involved in the oxidative modification of lipids and sterols (GenBank Accession No. ACM27362; Pfam PF04116). Although the biosynthetic abilities of the known nasB homologs have not been investigated, the diversity of genes to which they are linked suggests that nasB homologs are involved in multiple different metabolic pathways.
Figure 4.
Putative nasB homolgs from sequenced bacterial genomes are found in a variety of genetic contexts. The labels for nasB homologs are taken from the corresponding KEGG database entries. The GenBank Accession Numbers for CAP2UW1_1243, Arad_3395 and STHERM_c19760 are ACV34571, ACM27363 and ADN02911, respectively.
The aggregation of distinct enzymatic domains enhances the catalysis of multi-step reactions.[25] This is particularly true for multi-enzyme complexes that utilize covalent transfer as a means of channeling substrates through successive active sites and for selecting against competing metabolic pathways. In branched-chain fatty acid producing bacteria such as B. subtilis, BCKAD carries out the conversion of α-keto acids to the corresponding acyl-CoAs, which then become substrates for FabH. As a covalent alternative to the CoA-based strategy used by most bacteria, the FabH-like component of nasB may instead utilize the bound isovaleryl-lipoate thioester, eliminating the need for diffusible acyl-CoA intermediates along with the potential for these intermediates to be consumed by competing catabolic pathways. Although speculative, this proposed mechanism is supported by functional inferences for each of the Pfam homology domains found in nasB, as well as by the absence of the catalytic E2 lipoamide acyltransferase domain.
To function in a heterologous host like B. graminis, nasB must recognize host-derived malonyl-ACP. The use of host ACP allows the acyl-ACP products of nasB to be treated as native intermediates by the endogenous FAS II system, permitting subsequent utilization by acyl-ACP requiring enzymes that fail to discriminate between branched-chain and straight-chain substrates.[18] The introduction of nasB into bacterial strains that normally produce only straight-chain fatty acids may therefore provide access to branched-chain variants of numerous membrane lipids and acylated small molecules. Previous attempts to engineer E. coli for the production of branched-chain fatty acids have relied on the replacement of E. coli FabH with alternative FabH homologs derived from bacterial species that produce primarily branched-chain fatty acids. Such efforts have met with limited success.[26] The potential for nasB to be used as a tool for the engineering of branched-chain metabolites is intriguing and warrants additional study.
Experimental Section
Bacterial culture conditions
Cultures of Burkholderia graminis C4D1M were grown aerobically in YENB medium (7.5 g L−1 yeast extract, 8.0 g L−1 nutrient broth) at 30°C on a rotary shaker (200 rpm). Broad host-range plasmids were selected for with tetracycline (30 μg mL−1). Electrocompetent B. graminis cells were prepared according to Sharma and Schimke (1996).[27]
Analytical and Preparative HPLC-MS
Organic extracts were subjected to analytical HPLC-MS using the following conditions: (Waters XBridge C18 5 mm column [4.6 by 150 mm], 1.5 mL min−1 flow rate); 3 min at 50:50 H2O-methanol with formic acid (0.1%), followed by a linear gradient from 50:50 H2O-methanol with formic acid (0.1%) to 100% methanol with formic acid (0.1%) over 12 min, followed by 100% methanol with formic acid (0.1%) for 5 min. Preparative HPLC-MS was performed using the same solvent conditions (Waters XBridge C18 5 mm column [10 by 150 mm], 7 mL min−1 flow rate). Under these conditions, compound 6 was eluted between minutes 14.4–14.6 at a final yield of approximately 1.5 mg L−1 of culture broth.
Compound 6, N-(11-methyldodecanoyl)-phenylalanine
White crystalline powder; 1H NMR (600 MHz, [D4]methanol, 25°C, TMS):δH/(ppm) = 7.18–7.27 (m, 5H), 4.67 (dd, J (H,H) = 4.8, 9.3 Hz, 1H), 3.22 (dd, J (H,H) = 14.0, 4.7 Hz, 1H), 2.93 (dd, J (H,H) = 13.9, 9.6 Hz, 1H), 2.14 (t, J (H,H) = 7.4 Hz, 2H), 1.52 (m, 1H), 1.48 (p, J (H,H) = 7.3 Hz, 2H), 1.16–1.27 (m, 14H), 0.88 (d, J (H,H) = 6.6 Hz, 6H); 13C NMR (150 MHz, [D4]methanol, 25°C, TMS) δC/(ppm) = 176.3, 175.1, 138.8, 130.4, 129.5, 127.9, 55.1, 40.4, 38.6, 37.0, 31.2, 30.9, 30.7, 30.6, 30.3, 29.3, 28.7, 27.0, 23.2.
Acknowledgments
This work was supported by NIH grant GM077516 and NIH MSTP grant GM07739 (JWC). SFB is a Howard Hughes Medical Institute Early Career Scientist.
References
- 1.Brady SF, Chao CJ, Clardy J. Appl Environ Microbiol. 2004;70:6865. doi: 10.1128/AEM.70.11.6865-6870.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Van Wagoner RM, Clardy J. Structure. 2006;14:1425. doi: 10.1016/j.str.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 3.Lu YJ, Zhang YM, Rock CO. Biochem Cell Biol. 2004;82:145. doi: 10.1139/o03-076. [DOI] [PubMed] [Google Scholar]
- 4.Chooi YH, Tang Y. Chem Biol. 2010;17:791. doi: 10.1016/j.chembiol.2010.08.001. [DOI] [PubMed] [Google Scholar]
- 5.Jordan SW, Cronan JE., Jr J Biol Chem. 1997;272:17903. doi: 10.1074/jbc.272.29.17903. [DOI] [PubMed] [Google Scholar]
- 6.Miller JR, Busby RW, Jordan SW, Cheek J, Henshaw TF, Ashley GW, Broderick JB, Cronan JE, Jr, Marletta MA. Biochemistry. 2000;39:15166. doi: 10.1021/bi002060n. [DOI] [PubMed] [Google Scholar]
- 7.More MI, Finger LD, Stryker JL, Fuqua C, Eberhard A, Winans SC. Science. 1996;272:1655. doi: 10.1126/science.272.5268.1655. [DOI] [PubMed] [Google Scholar]
- 8.Schaefer AL, Val DL, Hanzelka BL, Cronan JE, Jr, Greenberg EP. Proc Natl Acad Sci U S A. 1996;93:9505. doi: 10.1073/pnas.93.18.9505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Val DL, Cronan JE., Jr J Bacteriol. 1998;180:2644. doi: 10.1128/jb.180.10.2644-2651.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Craig JW, Chang FY, Kim JH, Obiajulu SC, Brady SF. Appl Environ Microbiol. 2010;76:1633. doi: 10.1128/AEM.02169-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, Holm L, Sonnhammer EL, Eddy SR, Bateman A. Nucleic Acids Res. 2010;38:D211. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perham RN. Biochemistry. 1991;30:8501. doi: 10.1021/bi00099a001. [DOI] [PubMed] [Google Scholar]
- 13.Mattevi A, Obmolova G, Kalk KH, Teplyakov A, Hol WG. Biochemistry. 1993;32:3887. doi: 10.1021/bi00066a007. [DOI] [PubMed] [Google Scholar]
- 14.Rock CO, Cronan JE., Jr . Biochemistry of Lipids and Membranes. Benjamin/Cummings Publishing Co; Menlo Park, CA: 1985. [Google Scholar]
- 15.Clardy J, Brady SF. J Bacteriol. 2007;189:6487. doi: 10.1128/JB.00457-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Viallard V, Poirier I, Cournoyer B, Haurat J, Wiebkin S, Ophel-Keller K, Balandreau J. Int J Syst Bacteriol. 1998;48(Pt 2):549. doi: 10.1099/00207713-48-2-549. [DOI] [PubMed] [Google Scholar]
- 17.Kaneda T. Microbiol Rev. 1991;55:288. doi: 10.1128/mr.55.2.288-302.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Choi KH, Heath RJ, Rock CO. J Bacteriol. 2000;182:365. doi: 10.1128/jb.182.2.365-370.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaneda T. J Biol Chem. 1963;238:1229. [PubMed] [Google Scholar]
- 20.Willecke K, Pardee AB. J Biol Chem. 1971;246:5264. [PubMed] [Google Scholar]
- 21.Oku H, Kaneda T. J Biol Chem. 1988;263:18386. [PubMed] [Google Scholar]
- 22.Dickschat JS. Nat Prod Rep. 2010;27:343. doi: 10.1039/b804469b. [DOI] [PubMed] [Google Scholar]
- 23.Winson MK, Camara M, Latifi A, Foglino M, Chhabra SR, Daykin M, Bally M, Chapon V, Salmond GP, Bycroft BW, et al. Proc Natl Acad Sci U S A. 1995;92:9427. doi: 10.1073/pnas.92.20.9427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thiel V, Kunze B, Verma P, Wagner-Dobler I, Schulz S. Chembiochem. 2009;10:1861. doi: 10.1002/cbic.200900126. [DOI] [PubMed] [Google Scholar]
- 25.Reed LJ. Acc Chem Res. 1974;7:40. [Google Scholar]
- 26.Smirnova N, Reynolds KA. J Ind Microbiol Biotechnol. 2001;27:246. doi: 10.1038/sj.jim.7000185. [DOI] [PubMed] [Google Scholar]
- 27.Sharma RC, Schimke RT. Biotechniques. 1996;20:42. doi: 10.2144/96201bm08. [DOI] [PubMed] [Google Scholar]