Abstract
Background
Troponin I (TNNI3) is the inhibitory subunit of the thin filament regulatory complex Troponin, which confers calcium-sensitivity to striated muscle actomyosin ATPase activity. Mutations (2-7%) in this gene had been reported in hypertrophic cardiomyopathy patients (HCM). However, the frequencies of mutations and associated clinical presentation have not been established in cardiomyopathy patients of Indian origin, hence we have undertaken this study.
Methods
We have sequenced all the exons, including the exon-intron boundaries of TNNI3 gene in 101 hypertrophic cardiomyopathy patients (HCM), along with 160 healthy controls, inhabited in the same geographical region of southern India.
Results
Our study revealed a total of 16 mutations. Interestingly, we have observed Arginine to Glutamine (R to Q) mutation at 3 positions 98, 141 and 162, exclusively in HCM patients with family history of sudden cardiac death. The novel R98Q was observed in a severe hypertrophic obstructive cardiomyopathy patient (HOCM). The R141Q mutation was observed in two familial cases of severe asymmetric septal hypertrophy (ASH++). The R162Q mutation was observed in a ASH++ patient with mean septal thickness of 29 mm, and have also consists of allelic heterogeneity by means of having one more synonymous (E179E) mutation at g.4797: G → A: in the same exon 7, which replaces a very frequent codon (GAG: 85%) with a rare codon (GAA: 14%). Screening for R162Q mutation in all the available family members revealed its presence in 9 individuals, including 7 with allelic heterogeneity (R162Q and E179E) of which 4 were severely affected. We also found 2 novel SNPs, (g.2653; G → A and g.4003 C → T) exclusively in HCM, and in silico analysis of these SNPs have predicted to cause defect in recognition/binding sites for proteins responsible for proper splicing.
Conclusion
Our study has provided valuable information regarding the prevalence of TNNI3 mutations in Indian HCM patients and its risk assessment, these will help in genetic counseling and to adopt appropriate treatment strategies.
Keywords: TNNI3-Troponin I, Cardiomyopathy, SNPs, HCM, Indians, Mutations
Background
Inherited cardiomyopathy is a disorder of ‘cardiac muscle’ associated with abnormalities of cardiac wall thickness, chamber size, contraction, relaxation, conduction and rhythm, were found to be the major cause of heart failure. It is also different from other heart disease, as it frequently affects all the age groups, including young children, adults and competitive athletes [1]. Over the last two decades, a large number of mutations have been identified in sarcomeric genes as a cause of hypertrophic and dilated cardiomyopathy. The sarcomere composed of thick and thin filaments, the thick filament is constituted mainly of myosin, and the thin filament is composed of actin, tropomyosin and troponin complex. Cardiac troponin I (TNNI3), the inhibitory sub-unit of troponin complex, is 6.2 kb in size and comprised of 8 exons that encode for 210 amino acids, prevents the contraction of muscle in the absence of calcium and troponin C, and expressed exclusively in cardiac tissue [2]. Troponin I has three binding sites, one for troponin T (residue 61-112), another for troponin C (residue 113-164) and the third one for actin-tropomyosin (residue 130-148; 173-181) of the thin filaments [3]. During the functioning of the contractile apparatus depolarization of muscle leads to intracellular release of calcium, which binds with troponin C. A conformational change occurs in troponin-tropomyosin complex in such a way that actin molecules can interact with myosin, resulting in muscle contraction. Several mutations leading to familial hypertrophic cardiomyopathy (FHC) have been identified in this gene [4-6]. Analysis of cardiac beta myosin heavy chain (MYH7) and myosin binding protein C (MyBPC3) in Indian HCM and DCM patients revealed few genetic variants associated with the disease [7,8]. Since the Indian populations are culturally and geographically highly heterogeneous; genetically we expected that they might exhibit unique set of mutations. As there is no comprehensive study on Indian population, we have analysed all the exons and the exon-intron boundries of TNNI3 gene of Indian cardiomyopathy patients to assign its role in the etiology of cardiomyopathy among Indian populations.
Results
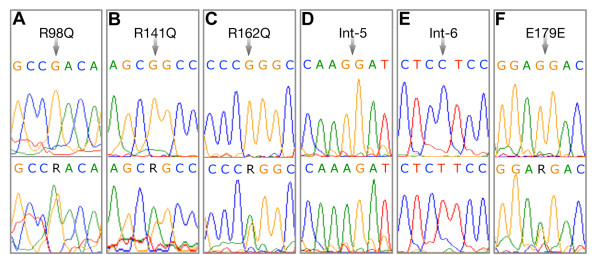
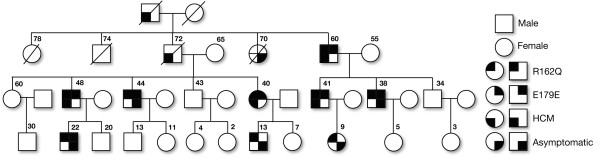
Screening of all the exons including the exon-intron boundaries of the TNNI3 gene in 101 individuals with HCM (Table1) along with 160 healthy controls from India revealed a total of 16 mutations, including 15 SNPs, and a 4 bp deletion/insertion polymorphism (Table2). Of the 15 SNPs, 7 were exonic (one novel, 3 reported non-synonymous and 3 synonymous mutations), and 9 were intronic mutations (Table2). Interestingly, we found three heterozygous arginine to glutamine (R to Q) substitution at 3 positions 98, 141 and 162 in TNNI3 Figure1A, 1B, 1C, of these R98Q in exon 6 of TNNI3 gene is novel (Figure1A) observed in a 28 years old severe hypertrophic obstructive cardiomyopathy patient (HOCM) with interventricular septum (IVS) thickness of 25 mm and his 5 years old asymptomatic son, with the family history of sudden cardiac deaths (Figure2). The dominant R141Q mutation in exon 7 of TNNI3 gene (Figure1B), lies within the “minimum inhibitory sequence” (residues 137 to 148) region, was observed in two individuals with severe familial asymmetric septal hypertrophy (ASH+), with interventricular septum (IVS) thickness of 25 and 28 mm, respectively. The dominant R162Q mutation (Figure1C) in exon 7 of TNNI3 gene was observed in an individual with severe asymmetric septal hypertrophy (ASH) with mean thickness of 29 mm had abnormal echocardiogram/ECG. Screening for this mutation (R162Q) in all the available family members (Figure3) revealed its presence in 9 individuals (Figure1C). Seven out of 9 individuals with R162Q mutation showed allelic heterogeneity with having a synonymous mutation at g.4797: G → A: E179E (Figure1F) in exon 7, which replaces a very frequent codon (GAG: 85%) with rare codon (GAA: 14%) (Table3). Four out of 7 Individuals with allelic heterogeneity (R162Q and E179E) (Figure1C, 1F) had presented with severe septal hypertrophy (ASH++) with the mean thickness of 27, 28, 29, 32 mm and the ECGs were abnormal in all the four individuals. History of sudden cardiac death was also been recorded in this family (Figure3).
Table 1.
Clinical features exhibited by hypertrophic cardiomyopathy (HCM) patients
| Baseline characteristics | (n = 101) |
|---|---|
| Age, years |
49 ± 10 |
| Sex, males,% |
62 |
| NYHA class III, IV (%) |
29 |
| Dyspnea,% |
65 |
| Angina Pectoris,% |
54 |
| Syncope,% |
33 |
| LVESD, mm |
20.3 ± 3.7 |
| LVEDD, mm |
36 ± 6.8 |
| Septum, mm |
21.2 ± 4.2 |
| Abnormal ECG,% |
62 |
| Family history of HCM,% |
37 |
| Family history of SCD,% |
32 |
| Left ventricular outflow obstruction | 49 |
NYHA - New York Heart Association; LVESD - Left ventricular end systolic dimension; LVEDD - Left ventricular end diastolic dimension; ECG - Electrocardiogram; HCM - Hypertrophic cardiomyopathy; SCD – Sudden cardiac death.
Table 2.
Mutations observed in troponin I (TNNI3) gene of the cases/controls
|
S.NO |
Position |
Location |
AA Change |
Major/Minor Allele |
Mutations observed |
Reported/Novel |
|
|---|---|---|---|---|---|---|---|
| Control | HCM | ||||||
| 1 |
g.1389 |
Intron 1 |
--- |
(T/C) |
Nil |
2 |
rs11667847 |
| 2 |
g.1403 |
Intron 1 |
--- |
(A1/G) |
Nil |
1 |
rs11671293 |
| 3 |
g.1215 |
Intron 1 |
C/A |
|
Nil |
1 |
rs3729707 |
| 4 |
g.1486-90 |
Intron 1 |
4b p del/In |
ACAG |
P |
P |
poly |
| 5 |
g.1698 |
Intron 2 |
--- |
(T/C) |
Nil |
4 |
rs3729836 |
| 6 |
g.1810 |
Intron 3 |
--- |
(G/A) |
Nil |
3 |
rs3729837 |
| 7 |
g.1897 |
Intron 3 |
--- |
(G/A) |
Nil |
22 |
rs3729838 |
| 8 |
g.2560 |
Exon 5 |
R68R |
(G/T) |
Nil |
4 |
rs3729711 |
| 9 |
g.2563 |
Exon 5 |
R69R |
(C/A) |
Nil |
2 |
Reported |
| 10 |
g.2601 |
Exon 5 |
P82R |
(C/G) |
1 |
2 |
Reported |
| 11 |
g.2653 |
Intron 5 |
--- |
(G/A) |
Nil |
1 |
Novel |
| 12 |
g.4003 |
Intron 6 |
--- |
(C/T) |
Nil |
2 |
Novel |
| 13 |
g.4019 |
Exon 6 |
R98Q |
G/A) |
Nil |
+1(S) |
Novel |
| 14 |
g.4682 |
Exon 7 |
R141Q |
(G/A) |
Nil |
2 |
Reported to be associated with HCM |
| 15 |
g.4745 |
Exon 7 |
R162Q |
(G/A) |
Nil |
1+8 (FM) |
Reported to be associated with HCM |
| 16 | g.4797 | Exon 7 | E179E | (G/A) | Nil | 1+6 (FM) | rs3729841 |
AA- Amino acid, HCM-Hypertrophic cardiomyopathy, P-Polymorphic, SNP-Single nucleotide polymorphism, Nil- No mutation observed, S-Son, FM-family members.
Figure 1.
(A - F). Sequence electropherograms of TNNI3 gene. Upper panel representing the control sequences, whereas the lower panel showing the mutations observed in the HCM patients. The mutations sites are shown with arrows. A. Novel missense heterozygous mutation at the nucleotide position g.4019 (G > A) that changes the amino acid Arginine (CGA) to Glutamine (CAA) at the residue 98. B. A heterozygous mutation at the nucleotide position g.4682 (G > A) that changes the amino acid Arginine (CGG) to Glutamine (CAG) at residue 141. C. A heterozygous mutation at the nucleotide position g.4745 (G > A) that changes the amino acid Arginine (CGG) to Glutamine (CAG) at residue 162. D. A novel homozygous splice acceptor site SNP at the nucleotide position g.2653 (AA) in intron 5. E. A novel homozygous SNP at the nucleotide position g.4003 (TT) in Intron 6. F. A heterozygous silent mutation (E179E), at the nucleotide position g.4797 (G > A) in exon 7.
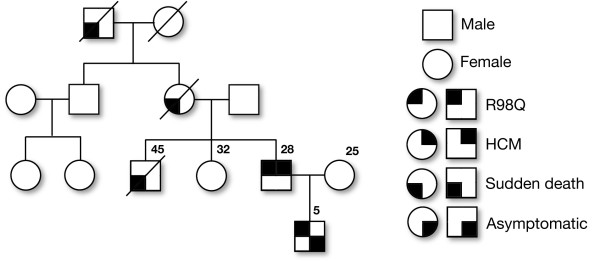
Figure 2.
A pedigree of a hypertrophic cardiomyopathy (HCM) family with R98Q mutation in the exon 6 of cardiac troponin ITNNI3gene is depicted.
Figure 3.
A pedigree of a four-generation hypertrophic cardiomyopathy (HCM) family with R162Q and E179E mutations in the exon 7 of cardiac (TNNI3)Troponin I gene.
Table 3.
The codon usage in human cTnI (GenBank No. NM_000363) gene
| Nt. Site | SNP Reference | Loction | Nt. change | Codon Site | Type | Codons | aa | fraction | frequency | codon usage |
|---|---|---|---|---|---|---|---|---|---|---|
| g.2560 |
rs3729711 |
Exon 5 |
G/T |
68 |
Wild |
CGG |
(R) |
0.24 |
28.436 |
6 |
| Mutant |
CGT |
(R) |
0.04 |
4.739 |
1 |
|||||
| g.2563 |
Novel |
Exon 5 |
C/A |
69 |
Wild |
CGC |
(R) |
0.36 |
42.654 |
9 |
| Mutant |
CGA |
(R) |
0.12 |
14.218 |
3 |
|||||
| g.4797 |
rs3729841 |
Exon 7 |
G/A |
179 |
Wild |
GAG |
(E) |
0.857 |
85.308 |
10 |
| Mutant | GAA | (E) | 0.143 | 14.218 | 3 |
C → T transition resulting in the replacement of proline with arginine (P82R) in exon 5, lies within the troponin T binding domain (61-112), was observed in 2 HCM patients and a control individual. A total of three synonymous mutations (g.2560; G > T; g.2563; C > A; g.4797; G > A) were observed exclusively in HCM patients (Tables 2, 3). The G → T mutation at g.2560 (R68R), observed in 4 HCM patients, replaces a frequent codon (CGG: 24%) with a less frequent one (CGT: 4%) (Table3). The C → A mutation at g.2563 (R69R), observed in 2 HCM patients, replaces the frequent codon (CGC: 36%) with rare codon (CGA: 12%) (Table3). The 4 bp deletion/insertion polymorphism in TNNI3 gene was observed almost in equal frequency in both the patients and the controls, suggesting that it was not associated with HCM.
In silico analysis of 2 novel SNPs using Splicing Rainbow tool predicted abnormal splicing pattern (Table4) by removing or creating binding sites for hnRNPs or the SR proteins (http://www.ebi.ac.uk/asd-srv/wb.cgi?method). A novel splice acceptor site mutation at g.2653; G → A (Table4; Figure1D), was found exclusively in a HCM patient, had been predicted to abort a binding site for hnRNP.U1U2 and one more novel mutation at g.4003 C → T (Table4; Figure1E), was observed exclusively in the two severe HCM patients, revealed a drastic change in the binding sites for hnRNPs and SR proteins (2 sites in hnRNPs and 3 sites in SR proteins). The disturbed binding sites due to g.4003 C → T mutation were hnRNP-E1E2, hnRNP-I1I2, SRp20, SC35, U2AF65 (Table4; Figure1E), further emphasize its regulatory role, however it needs further investigation. In addition to the novel mutations, we have also observed SNPs reported elsewhere; rs11667847, rs3729836, rs3729837, rs11671293, rs3729838, rs3729711, rs3729841. The allele frequencies of the SNPs were comparable to the HapMap populations (http://www.HapMap.org).
Table 4.
The hn RNPs and SR proteins binding site sequences in normal and mutant as predicted by “Splicing Rainbow” tool
|
S:NO |
SNP |
LOCATION |
Splicing Rainbow & the binding site sequences |
||
|---|---|---|---|---|---|
| Normal | Mutant | ||||
|
1 |
g.2653:G > A |
Intron 5 |
GGATGCGAGG |
hnRNP.U1U2 |
(Site destroyed) |
|
2 |
g.4003:C > T |
Intron 6 |
TCCTCCTCCA |
hnRNP.E1E2 |
(Site destroyed) |
| |
|
|
(No site) |
hnRNP.I1 I2 |
CCACGTTCCTCTTCCAG (New site) |
| |
|
|
CCTCCTCC |
SRp20 |
CCTCTTCCA |
| |
|
|
CTCCTCCA |
SC 35 |
(Site destroyed) |
| (No site) | U2AF65 | TCTT (New site) | |||
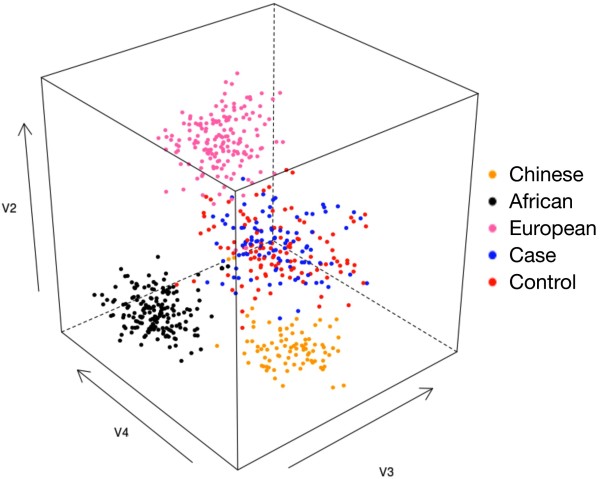
Our concern is that the difference in frequencies of the alleles observed between the cases and controls are associated with disease or a difference unrelated to disease arising from underlying genetic differences between the populations from which the case and control samples were drawn: ‘population stratification’. In order to maximize our chances detecting such stratification, we genotyped cases and control samples using a panel of 50 ancestry-informative markers (AIMs) for inferring ancestry [7] and performed a principal components analysis on the data together with HapMap samples of Chinese, European, Yoruban [9], and found no significant difference in ancestry between cases and controls (Figure3). On the other hand, the Chinese, European, Yoruban, the cases/controls were making clusters among themselves, confirming that the 50 AIMs are sufficient for detecting whether or not ancestry differences along this axis are present. Thus, the population stratification along the axis was ruled out as the cause of the disease association.
Discussion
About 2–7% mutations in TNNI3 have been reported to be associated with hypertrophic cardiomyopathy patients from various populations [3,10]. As the data are often population specific, we investigated the prevalence of TNNI3 mutations in 101 hypertrophic cardiomyopathy patients from southern India. We have identified R → Q mutation at 3 positions (R98Q, R141Q, R162Q) (Figure1A, 1B, 1C), of which a novel R98Q missense mutation was identified exclusively in a HOCM patient (1%) with early onset of clinical manifestation. This mutation was present at a functionally significant domain that lies within the Troponin T binding domain (61-112) (Figure1A), and was identified in a 28 year old proband and his 5 year old asymptomatic son. There were 3 premature sudden deaths (Grand father, mother and brother) in the family (Figure2), suggesting that the R98Q substitution is significantly associated with an adverse phenotype. The R141Q, R162Q mutations (Figure1B, 1C) reported to be associated with HCM in large tertiary referral center population [11] were also observed in our study. Both these mutations (R141Q, R162Q) were located in the carboxy terminal part of troponin I and the first binding site for troponin C domain and changes the net charge from +1 to 0. All the three R → Q mutations at 98, 141 and 162 were exclusively observed in the hypertrophic cardiomyopathy patients, these regions were reported to be the functionally significant domains. A dominant R141Q was observed in two HCM (2%) patients, (Figure1B). Screening of dominant R162Q and E179E mutations in exon 7 of TNNI3 gene with the available family members revealed the presence of R162Q mutation in 9 individuals and the allelic heterogeneity (R162Q and E179E) in 7 out of 9 individuals (Figure1C, 1F). It is known that the non-random use of synonymous codons creates codon usage bias [12], and the translational speed and co-translational folding are the main factors that affect the correlation between synonymous codon usage and protein structure [13,14]. Though the significance of synonymous codon is still debatable, the 7 out of 9 individuals, who showed allelic heterogeneity (R162Q and E172E), four (out of 7) were affected more severely in the family (Figure3) therefore the role of second synonymous mutation (E172E) (Figure1F) could not be ignored.
Interestingly, two novel SNPs, one at the nt position g.2653; G → A and another one at the nt position g.4003 C → T, which were exclusively present in cases, were estimated using Splicing Rainbow tool (http://www.ebi.ac.uk/asd-srv/wb.cgi?method). In our study a splice acceptor site mutation at g.2653; G → A was found to remove a binding site for hnRNP.U1U2 and disturb the splicing (Table4; Figure1D). The g.4003 C → T mutation had revealed drastic changes at the binding sites for hnRNPs and SR proteins (hnRNP-E1E2, hnRNP-I1I2, SRp20, SC35, U2AF65), (Table3; Figure1E), thus predicted to disturb the splicing significantly. It has already been suggested that the alignment of the sequences present in the vicinity of the splice junctions has led to consensus sequences for both the splice donor and splice acceptor sites [15,16]. A panel of 50 ancestry-informative markers (AIMs) [6] for inferring ancestry was used and performed a principal components analysis on the data together with HapMap samples [9] had ruled out that these allelic differences between the cases and the controls was the cause of the disease association (Figure4).
Figure 4.
Principal components analysis of Indian case/control samples together with Chinese, African and European HapMap samples, based on a panel of 50 ancestry-informative SNP markers (AIMs).
The frequencies of observed polymorphism in the present study were also comparable with HapMap populations. The high prevalence of R → Q mutations (4%) at three positions (R98Q, R141Q, R162Q) and the absence of remaining reported mutations observed in other populations (2-7%), is clearly elucidating the unique origin of Indian populations. These mutations (R98Q, R141Q and R162Q) can be used as markers for screening HCMs in this region. However, this study needs to be extended to know true spectrum and the prevalence of mutations present in the remaining part of the India.
Conclusion
We have identified three R → Q mutations (R98Q, R141Q, R162Q) accounting for about 4% of HCM, of these R98Q was novel. Interestingly, two more novel SNPs, predicted to disturb the splicing significantly were also been identified in this study. Principal components analysis of the data generated by a panel of 50 ancestry-informative markers (AIMs) [6] with cases and controls for inferring ancestry together with HapMap samples had ruled out that the mutations (R98Q, R141Q, R162Q and the 2 novel SNPs (g.2653; G → A and g.4003 C → T) observed in the HCM patients was the cause of the disease association. The overall findings have provided valuable information for risk assessment, genetic counseling and adopt treatment strategies for hypertrophic cardiomyopathy among Indian patients.
Methods
Ethical statement
All of the DNA samples analyzed in the present study were derived from blood samples that were collected with the informed written consent of the donors. The Institutional Ethics Committee of Care Hospitals, Hyderabad and the Centre for Cellular and Molecular Biology (CSIR), Hyderabad, India, have approved the study. This study conforms to the principles outlined in the Declaration of Helsinki (WMA World Medical Association Declaration of Helsinki). The study subjects were all South Indian patients with HCM, diagnosed based on the NYHA (The Criteria Committee of the New York Heart Association, 1994), WHO (http://www.who.int/cardiovascular_diseases) guidelines for HCM; the same was applied as appropriate to rule out HCM.
Case and control samples
Blood (about 10.0 ml) samples were collected from 101 hypertrophic cardiomyopathy patients from South India (Table1, Additional file 1: Table S1). The patients underwent physical/clinical examinations, such as; 12 lead ECGs, and trans-thoracic two-dimensional echocardiography, and Doppler studies (Table1, Additional file 1: Table S1). One hundred and sixty healthy individuals from the same ethnic background, without hypertension and cardiomyopathy, based on the electrocardiograph and echocardiograph measurements, were also recruited for the study as controls.
DNA isolation
DNA was isolated from the blood samples using the following protocol: Erythrocytes were lysed with 15.0 ml of erythrocyte lysis buffer (10 mm Tris pH 8.0, 320 M sucrose, 5 M MgCl2, 1% Triton X-100; Sigma Chemical Company, St Louis, MO, USA) for 5 min. After complete lysis of erythrocytes, leucocytes were pelleted by centrifugation at 500 g for 5 min. The leucocyte pellet was dissolved in 8.0 ml of leucocyte lysis buffer (400 mM Tris, 60 mM EDTA, 150 M NaCl, and 1% SDS; Sigma) and mixed thoroughly. To this, 2.0 ml of 5.0 M sodium perchlorate (E. Merck, Darmstadt, Germany) was added and mixed thoroughly for 2–3 min. DNA was precipitated with absolute alcohol after extracting once with phenol:chloroform (1 : 1) and once with chloroform. DNA was washed once with 70% ethanol and dissolved in TE buffer (10 mM Tris pH 8.0, 1 mM EDTA).
Genetic analysis
Primer sequences covering the exons, exon-intron boundaries of TNNI3 gene, were obtained from the website (http://genepath.med.harvard.edu/~seidman/cg3/genes/TNNI3exons.html). Primers were synthesized using an ABI 392 oligo synthesizer (Perkin–Elmer, Foster City, CA, USA), and the PCRs (polymerase chain reactions) were carried out under standard conditions, containing 50 ng of genomic DNA, 5 pM of each primer, 200 mM dNTPs, 10X PCR buffer containing 1.5 mM MgCl2, and 1 unit of AmpliTaq Gold (Perkin–Elmer). Amplification was carried out in a thermal cycler (MJ Research, Waltham, MA, USA) using the following cycling conditions: 94°C for 5 min, 35 cycles at 94°C for 1 min, 55–60°C for 1 min and 72°C for 1 min, followed by a final extension at 72°C for 10 min. Amplicons were purified by treating them with ExoSAP-IT, [composed of Exonuclease 1 and Shrimp alkaline phosphatase (USB Corporation, 26, 111 Miles Road, Cleveland, Ohio 44128, USA)], according to the manufacturer’s instructions. The purified PCR products were bi-directionally sequenced using the ABI BigDye Terminator cycle sequencing kit (Perkin–Elmer, Foster City, CA, USA) and analyzed using on ABI 3730 DNA Analyzer (Applied Biosystems, Foster City, CA, USA) [17]. Sequences were edited and compared with the reference sequence (TNNI3) using AutoAssembler software (Applied Biosystems, Foster City, CA, USA).
Sequenom iPLEX assay
We genotyped case/control samples for 50 Ancestry Informative Markers (AIM’s) using the Sequenom iPLEX assay where two markers failed to give results in all the individuals, therefore our final analysis was based upon 48 AIM’s. The detailed information about these markers was published elsewhere [7].
In silico analysis
To evaluate the novel mutations observed exclusively in the hypertrophic cardiomyopathy (HCM) patients in this study was the potential cause for the defect in splicing, we have analyzed those sites with ASD Workbench wrapper (http://www.ebi.ac.uk/asd-srv/wb.cgi) tools such as PPT analysis, BP analysis. The regulatory sequence were used to identify the presence of polypyrimidine tracks, branch point sites, binding sites for splicing factors, and exonic splicing enhancers/silencers (ESE/ESS) or intronic splicing enhancers/silencers (ISE/ISS), respectively, at the mutation sites. Splicing Rainbow tool searches for the SR proteins (serine/arginine-rich) as well as hnRNP motifs were done.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
DSR, PN, KT designed the study. DSR, PN carried out the blood samples collection and/or preparation of the samples. DSR performed the sequencing and statistical analysis and interpretation of the data. SP assisted with sequencing analysis. KT, PN cross-checked statistical analyses and interpretation of the data carried out by DSR. CN evaluated the patients and provided clinical information for the study. LS oversaw the data undertaken by DSR and provided the logistical support. DSR prepared the first draft of the manuscript. KT made the final interpretation of the data, and prepared the final manuscript. All authors read and approved the final manuscript.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Supplementary Material
Table S1. Clinical phenotype of the Hypertrophic Cardiomyopathy Patients.
Contributor Information
Deepa Selvi Rani, Email: deepa@ccmb.res.in.
Pratibha Nallari, Email: prathinallari@yahoo.com.
Singh Priyamvada, Email: priyaiitr@gmail.com.
Calambur Narasimhan, Email: calambur@hotmail.com.
Lalji Singh, Email: lalji@ccmb.res.in.
Kumarasamy Thangaraj, Email: thangs@ccmb.res.in.
Acknowledgements
We thank all the patients and their family members, and normal healthy individuals who have participated in this study. We also thank S. Nizamuddin for his help in PCA. DSR and PN acknowledge the help of Department of Genetics, Osmania University, Hyderabad.
References
- Maron BJ. Hypertrophic cardiomyopathy. A systematic review. JAMA. 2002;287:1308–1320. doi: 10.1001/jama.287.10.1308. [DOI] [PubMed] [Google Scholar]
- Bhavsar PK, Brand NJ, Yacoub MH, Barton PJR. Isolation and characterization of the human cardiac troponin I gene (TNNI3) Genomics. 1996;35:11–23. doi: 10.1006/geno.1996.0317. [DOI] [PubMed] [Google Scholar]
- Mogensen J, Murphy RT, Kubo T, Bahl A, Moon JC, Klausen IC, Elliott PM, McKenna WJ. Frequency and clinical expression of cardiac troponin I mutations in 748 consecutive families with hypertrophic cardiomyopathy. J Am Coll Cardiol. 2004;44:2315–2325. doi: 10.1016/j.jacc.2004.05.088. [DOI] [PubMed] [Google Scholar]
- Kimura A, Harada H, Park JE, Nishi H, Satoh M, Takahashi M, Hiroi S, Sasaoka T, Ohbuchi N, Nakamura T, Koyanagi T, Hwang TH, Choo JA, Chung KS, Hasegawa A, Nagai R, Okazaki O, Nakamura H, Matsuzaki M, Sakamoto T, Toshima H, Koga Y, Imaizumi T, Sasazuki T. Mutations in the cardiac troponin I gene associated with hypertrophic cardiomyopathy. Nat Genet. 1997;16:379–382. doi: 10.1038/ng0897-379. [DOI] [PubMed] [Google Scholar]
- Niimura H, Patton KK, McKenna WJ, Soults J, Maron BJ, Seidman JG, Seidman CE. Sarcomere protein gene mutations in hypertrophic cardiomyopathy of the elderly. Circul. 2002;105:446–451. doi: 10.1161/hc0402.102990. [DOI] [PubMed] [Google Scholar]
- Murphy RT, Mogensen J, Shaw A, Kubo T, Hughes S, McKenna WJ. Novel mutation in cardiac troponin I in recessive idiopathic dilated cardiomyopathy. Lancet. 2004;363:371–372. doi: 10.1016/S0140-6736(04)15468-8. [DOI] [PubMed] [Google Scholar]
- Dhandapany PS, Sadayappan S, Xue Y, Powell GT, Rani DS, Nallari P, Rai TS, Khullar M, Soares P, Bahl A, Tharkan JM, Vaideeswar P, Rathinavel A, Narasimhan C, Ayapati DR, Ayub Q, Mehdi SQ, Oppenheimer S, Richards MB, Price AL, Patterson N, Reich D, Singh L, Tyler-Smith C, Thangaraj K. A common MYBPC3 cardiac myosin binding protein C variant associated with cardiomyopathies in South Asia. Nat Genet. 2009;41:187–191. doi: 10.1038/ng.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanjore RR, Rangaraju A, Kerkar PG, Calambur N, Nallari P. MYBPC3 gene variations in hypertrophic cardiomyopathy patients in India. Can J Cardiol. 2008;24:127–130. doi: 10.1016/s0828-282x(08)70568-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International HapMap Consortium. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Wijngaard A, Volders P, van Tintelen JP, Jongbloed JDH, van den Berg MP, Lekanne Deprez RH, Mannens MMAM, Hofmann N, Slegtenhorst M, Dooijes D, Michels M, Arens Y, Jongbloed R, Smeets BJM. Recurrent and founder mutations in the Netherlands: cardiac Troponin I (TNNI3) gene mutations as a cause of severe forms of hypertrophic and restrictive cardiomyopathy. Neth Heart J. 2011;19:344–351. doi: 10.1007/s12471-011-0135-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Driest SL, Ellsworth EG, Ommen SR, Tajik AJ, Gersh BJ, Ackerman MJ. Prevalence and spectrum of thin filament mutations in an outpatient referral population with hypertrophic cardiomyopathy. Circulation. 2003;108:445–451. doi: 10.1161/01.CIR.0000080896.52003.DF. [DOI] [PubMed] [Google Scholar]
- Grantham R, Gautier C, Gouy M, Mercier R, Pave A. Codon catalog usage and the genome hypothesis. Nucleic Acids Res. 1980;8:r49–r62. doi: 10.1093/nar/8.1.197-c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Zhou T, Ma J, Sun X, Lu Z. The relationship between synonymous codon usage and protein structure in Escherichia coli and Homo sapiens. Biosystems. 2004;73:89–97. doi: 10.1016/j.biosystems.2003.10.001. [DOI] [PubMed] [Google Scholar]
- D’Onofrio G. Expression patterns and gene distribution in the human genome. Gene. 2002;300:155–160. doi: 10.1016/s0378-1119(02)01048-x. [DOI] [PubMed] [Google Scholar]
- Shapiro MB, Senapathy P. RNA splice junctions of different classes of eukaryotes: sequence statistics and functional implications in gene expression. Nucleic Acids Res. 1987;15:7155–7174. doi: 10.1093/nar/15.17.7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senapathy P, Shapiro MB, Harris NL. Splice junctions, branch point sites, and exons: sequence statistics, identification, and applications to genome project. Methods Enzymol. 1990;183:252–278. doi: 10.1016/0076-6879(90)83018-5. [DOI] [PubMed] [Google Scholar]
- Rani DS, Carlus SJ, Poongothai J, Jyothi A, Pavani K, Gupta NJ, Reddy AG, Rajan MM, Rao K, Chakravarty B, Singh L, Thangaraj K. CAG repeat variation in the mtDNA polymerase gamma is not associated with oligo-asthenozoospermia. Int J Androl. 2009;32:647–655. doi: 10.1111/j.1365-2605.2008.00919.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Clinical phenotype of the Hypertrophic Cardiomyopathy Patients.