Abstract
X-ray free-electron lasers (X-FELs) produce X-ray pulses with extremely brilliant peak intensity and ultrashort pulse duration. It has been proposed that radiation damage can be “outrun” by using an ultra intense and short X-FEL pulse that passes a biological sample before the onset of significant radiation damage. The concept of “diffraction-before-destruction” has been demonstrated recently at the Linac Coherent Light Source, the first operational hard X-ray FEL, for protein nanocrystals and giant virus particles. The continuous diffraction patterns from single particles allow solving the classical “phase problem” by the oversampling method with iterative algorithms. If enough data are collected from many identical copies of a (biological) particle, its three-dimensional structure can be reconstructed. We review the current status and future prospects of serial femtosecond crystallography (SFX) and single-particle coherent diffraction imaging (CDI) with X-FELs.
Introduction
The past decade has witnessed several important developments that potentially have significant impact on structural biology. First, the development of X-FELs with extremely high peak intensity and ultrashort pulse duration allows data collection of biological samples with significantly reduced radiation damage. Radiation damage is presently a major factor limiting the attainable resolution in the imaging of biological material, in particular when using X-rays [1]. Initial radiation damage by X-rays in the energy range up to about 20 keV is dominated by the photoelectric effect. Upon absorption of an X-ray photon, an electron is ejected, typically from an inner shell. The resulting core hole is filled by an outer shell electron. For biologically relevant elements such as C, N, O, S and P, the transition energy is transmitted primarily to another outer shell electron, which is then ejected as an Auger electron within a few to tens of femtoseconds (fs). Still on a femtosecond timescale, secondary electron ionization cascades ensue through electron-impact ionization. The removal of electrons and the formation of radicals leads to long timescale local electronic [2,3] and chemical changes [4,5] and will ultimately decrease the overall diffraction power of the sample. Cooling samples during data collection slows the long timescale damage since it reduces the mobility of the generated radicals. Protein crystals cooled to the liquid nitrogen temperature can tolerate a dose of ~30 MGy [6]. Solem predicted in 1986 that this dose can be significantly increased when using very intense X-ray pulses [7]. In this case, diffraction-limited resolution diffraction patterns can be captured before the obliteration of the sample by radiation damage [7]. With the emergence of X-FELs this concept was taken up by Hajdu and coworkers who calculated the pulse length dependence of the ionization induced atomic displacements and their effect on data quality using molecular dynamics simulations, suggesting that useable diffraction data can be obtained before explosion of the sample for very short FEL pulse lengths (“diffraction-before-destruction”) [•• 8].
Second, while X-ray crystallography is presently the primary methodology used for 3D structure determination of protein molecules at medium to atomic resolution, many biological systems are difficult to crystallize. In 1999, the methodology of X-ray crystallography was extended to allow structure determination of non-crystalline specimen and nanocrystals [•• 9]. Compared to a regular crystal, the diffraction intensity from a non-crystalline specimens or nanocrystal is weak due to the loss or reduction of the amplification from the large number of unit cells in a sizable crystal. The diffraction pattern of a non-crystalline specimen or a nanocrystal can be sampled at an interval finer than the Bragg peak frequency, which in principle allows the direct phase determination by the oversampling method with iterative algorithms [10]. The combination of this methodology with X-FELs thus potentially opens a door to study biological systems without the requirement of sizable crystals.
X-ray free electron lasers
X-ray free electron lasers are a new generation of X-ray sources that produce X-ray pulses with a peak brilliance a billion times higher than synchrotron radiation and a pulse duration ranging from a few to hundreds of femtoseconds [11]. An X-FEL consists of a linear accelerator and a long undulator magnet. Low-emittance and intense electron bunches are first accelerated to several to tens of gigaelectron volt (GeV) by using the linear accelerator. The high energy electron bunches pass through the undulator with a sinusoidal magnetic field. When the electron bunches move along a sinusoidal, oscillating trajectory, they emit electromagnetic radiation. Initially because the size of each electron bunch is larger than the wavelength of the radiation, the radiation is incoherent and the intensity of the radiation is linear proportional to the number of electrons in the electron bunch. If the undulator magnet is precisely tuned to match the phase and wavelength of the radiation, the radiation interacts with the electron bunch and the interaction accelerates part of the electrons and de-accelerates the others, such that the electrons form micro-bunches. The light emitted from these micro-bunches interferes constructively, resulting in an intensity proportional to ~N2, with N denoting the number of electrons inside the micro-bunches. The increased intensity leads to an amplification of the micro-bunching process, which includes more and more electrons. This self-amplified process results in an exponential growing intensity of the emitted light in the undulator until all electrons are part of a micro-bunch. At the end of the undulator, the electron beam is dumped and the intense radiation produces X-FEL pulses with full transverse coherence, high peak intensity (~1012 photons/pulse), and short pulse duration. The principle of this process is termed self-amplified stimulated emission (SASE) [12]. A more thorough review of the SASE X-FEL can be found elsewhere [11].
Presently, several soft and hard X-FELs are in operation worldwide, including the Free electron LASer in Hamburg (FLASH) [13], the Linac Coherent Light Source (LCLS) [14] at the SLAC National Accelerator Laboratory and the SPring-8 Angstrom Compact free electron Laser (SACLA) at the RIKEN Harima Institute in Japan [15]. In addition, the European X-ray free electron laser (XFEL) in Hamburg and the SwissFEL at the Paul Scherrer Institute in Switzerland are under construction and are expected to operate in 2016 and 2017, respectively. Furthermore, X-FEL facilities are under construction or planned in Italy, China and South Korea. Although SASE X-FELs are currently under rapid development worldwide, there are several issues associated with this kind of coherent X-ray sources. First, a SASE X-FEL is not a true laser due to the lack of temporal coherence [11]. Second, SASE starts with noise and is a stochastic process, which results in energy, intensity and position jitters of the X-ray pulses. Although these issues will be resolved or alleviated by using different types of seeding technologies in the future [11], the present X-FELs with extremely intense and ultrashort X-ray pulses already open exciting new opportunities in structural biology.
Serial femtosecond crystallography
X-ray crystallography is the oldest and most successful method to determine high-resolution 3D structures of molecules of any size. Accordingly, the method’s impact in biology and other areas of science is immense. A key feature of crystallography is the coherent addition of diffracted intensities of the molecules in the crystal into Bragg reflections with the absorbed dose being distributed between them. Thus, the resolution possible in a reconstructed image of the periodically averaged molecule in a crystal, for a given dose per molecule below the radiation damage threshold, depends on the number of molecules in the crystal [16]. This is the reason for the demand of large crystals for the collection of high-resolution data. For many systems, however, growing large, well ordered crystals is very difficult if not impossible. It is here that X-FELs are expected to make an impact in crystallography by allowing using tiny1 crystals for data collection, beyond the conventional radiation damage limits [•• 17].
Indeed, in a pioneering experiment, it has been demonstrated recently that tiny crystals of photosystem I, a megadalton membrane protein complex, yield diffraction data that are useful for structure analysis [•• 17] (reviewed last year [18] in this journal). The experiment was performed at the LCLS at the AMO beamline [19] using the CAMP instrument [• 20] that houses two pairs of fast-readout large-area pnCCD detectors, positioned to capture both high- and low-angle diffraction. A gas-focused liquid microjet [21] was used to inject randomly oriented photosystem I micro- and nanocrystals into the X-FEL interaction region. There, they intersected with 300 fs long X-ray pulses with 1.8 keV photon energy. Since the crystals were destroyed upon exposure to a single femtosecond FEL pulse, the data were collected in a serial fashion, with the liquid jet providing a fast, gentle, and convenient means of replenishment as shown in Fig. 1(a). Serial femtosecond crystallography (SFX) data were integrated using a Monte Carlo-like approach [22,23] which resulted in interpretable electron density of the photosystem I data, using the known structure for phasing. The resolution was limited to 8.5 Å by the wavelength of 6.9 Å and the geometry of the experimental setup [•• 17].
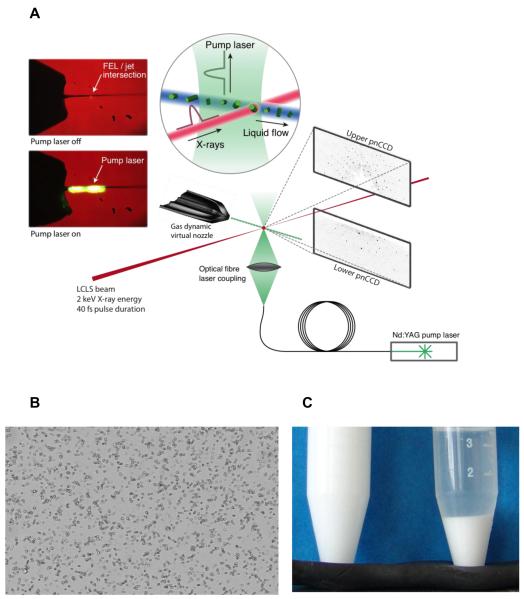
Figure 1.
(a) Experimental setup for serial femtosecond crystallography. Tiny crystals are injected into an X-FEL beam using a liquid microjet. This setup is very convenient for pump-probe experiments using laser excitation. Shown is an experiment on the photo-induced dissociation of photosystem I-ferredoxin cocrystals, taking place on a microsecond time-scale (from ref [43]). (b) Microcrystalline sample for serial femtosecond crystallography data collection. Lysozyme crystals (≤1 × 1 × 3 μm3) as seen through a conventional light microscope, (c) Microcrystalline lysozyme suspended in solution (left) and after settling (right) (from ref.[42]). It is important to note that the quantities depicted here are by no means excessive; several milliliters of very dense crystalline slurry are currently required for SFX experiments.
High-resolution SFX data collection became feasible in February 2011 with the commissioning of the Coherent X-ray Imaging (CXI) instrument [24] at LCLS, providing access to 10-keV photons (1st harmonic). The prospects of serial femtosecond crystallography for high-resolution data collection and structure analysis were explored using hen egg-white lysozyme as a well-characterized model system. SFX data were collected to 1.9 Å resolution of micrometer-sized lysozyme crystals (Fig. 1(b)) using 40 fs and 5 fs X-ray pulses of 9.3-keV photons [•• 25]. Both SFX data sets scale reasonably with very low dose synchrotron data collected of crystals kept at room temperature. The SFX data yield clear molecular replacement solutions, the derived electron density maps show the expected features and the refinement statistics are similar to the synchrotron data. This establishes SFX as a viable and valuable complement to existing macromolecular crystallography techniques for the analysis of tiny crystals [•• 25].
Molecular replacement [26] has been used for phasing the SFX data [25,27]. So far, no de-novo phasing has been demonstrated. Recently, the MAD method [28,29] has been theoretically generalized for phasing SFX data at high X-ray intensity by taking into account the detailed electronic damage dynamics of heavy atoms [• 30]. Although experimental results are needed to confirm the feasibility of this proposed generalization, in particular since the calculation only included the ionization of the heavy atoms, it may potentially offer an ab initio structure determination method for SFX with X-FELs.
Growing, detecting, and testing tiny crystals
Given the limitations imposed by radiation damage combined with signal-to-noise considerations, the emphasis has been on optimizing crystallization conditions that yield only very few large crystals instead of showers of small ones. Although finding initial crystallization conditions is largely driven by trial and error, i.e. testing many different conditions, often using commercially available crystallization screens, once crystallization parameters have been identified, crystallization can be rationalized by a phase diagram that shows where liquid, crystalline or amorphous precipitate states are stable [31]. Crystallization involves two steps, nucleation and crystal growth. Nucleation takes place in a region of moderate supersaturation, while crystal growth occurs under conditions of lower supersaturation just below the nucleation zone where crystals are stable. Thus, for conventional large crystal growth, conditions are sought that lead the system into the so-called metastable zone where the concentration of the protein in the solution drops once nuclei have formed, resulting in crystal growth without the formation of further nuclei (see for example www.emeraldbiosystems.com/blog/post/protein-crystallization-phase-diagrams.aspx). Therefore, to grow large amounts of tiny crystals, a higher degree of supersaturation has to be achieved, resulting in the formation of many nuclei in the “labile zone” [32]. The most convenient way to achieve this is batch crystallization because of its simplicity, high reproducibility and scalability from a few microliters to hundreds of milliliters, allowing both screening and bulk preparation for the SFX experiments. Rapid and thorough mixing of precipitant and protein solutions can be essential for obtaining uniform crystal size distributions. Batch methods can also be used for lipidic sponge phase crystallization of membrane proteins for SFX as has been demonstrated recently for the photosynthetic reaction center of B. viridis [• 27].
Many macromolecules crystallize spontaneously if sufficiently concentrated. This can be achieved for example by centrifugation using centricons or by dialysis using appropriate molecular weight cut-offs. High local macromolecular concentrations can also occur in cells; indeed, microcrystals can be observed in vivo, providing eukaryotes and prokaryotes with a convenient and efficient way for storage or disposal [33]. Insect cells are interesting in this respect since many protein and virus crystals have been observed in-vivo. For example, polyhedrin forms a crystalline matrix coating the nuclear polyhedrosis baculovirus. Heterologous expression in baculovirus Sf9 expression system under the polyhedrin promoter can be used for in-vivo crystallization of proteins as has been shown for calcineurin [34] and recently for cathepsin from Trypanosoma brucei (TbCatB) and inosine monophosphate dehydrogenase from Trypanosoma brucei [• 35]. In the case of TbCatB ~5 × 105 crystals were purified from 106 cells kept for 8 days in suspension culture. Since the crystals form inside the cell, they are limited in their size (10–15 μm long and 0.5–1 μm wide) and are very stable in regular buffers (pH> 4) and water. They were used for SFX experiments at the AMO beamline at the LCLS and diffracted to 8 Å resolution, limited by the geometry of the experimental setup and the available photon energy [• 35]. The advantages of in vivo crystallization are i) production of post-translationally modified proteins of interest, ii) high stability and easy isolation by centrifuging the crystals after cell lysis, and iii) a narrow crystal size distribution.
The identification of tiny crystals by light microscopy can be challenging, in particular when they occur as showers or in optically dense lipidic phases. In situ analysis of the crystallization setups exploiting UV fluorescence or birefringence may help in the identification of crystalline order as does the emerging application of Second-Order Non-linear optical Imaging of Chiral Crystals (SONICC) [36,37], which yields a strongly enhanced signal in the presence of chromophores. The latter two approaches, however, depend on the symmetry of the crystal lattice and the orientation of the crystals.
Electron microscopy is an excellent way to identify nanocrystals since it allows both imaging and diffraction studies, yielding information on both the size and crystallinity of the particles. However, sample transfer, in particular when using very small volumes, washing, blotting and, if desired, staining can be challenging especially when using crystallization solutions with high viscosity or high salt concentrations. Nevertheless, high throughput screening of 2D crystallization trials has been demonstrated recently [38] and there seems no reason that this cannot be transported to 3D crystals.
In situ (powder) diffraction is an option for screening tiny crystals [39], but the diffraction signal is likely not sufficient for nanoliter setups using standard beamlines. Nevertheless, powder diffraction does offer a convenient way of testing tiny crystals: crystalline slurries can be filled into quartz capillaries or mylar sleeves which are subsequently attached to conventional magnetic bases and stored in the associated plastic caps. Upon settling of the crystals, the pellet can be analyzed at room temperature by powder diffraction. Although the resulting diffraction pattern is unlikely to represent the diffraction limit of a single crystal at an X-FEL, it demonstrates the crystallinity of the sample. Moreover, this approach can be used for optimization of crystallization conditions or of crystal storage solutions, similar to testing crystals at a home source before high-resolution data collection at a synchrotron. An important tool for improving the diffraction properties of large crystals is dehydration. At this point it is unclear whether similar approaches are applicable to tiny crystals.
Practical aspects of serial femtosecond crystallography
Crystals of macromolecules require maintaining a certain hydration level during data collection. The liquid microjet [21] developed at Arizona State University allows keeping the sample in solution during injection and data collection which takes place in a vacuum. The crystalline liquid slurry is contained in a relatively large diameter inner capillary surrounded by a focusing sheath gas [• 40]. This design of a gas dynamic virtual nozzle (GDVN) provides both a means for reducing the probability of clogging of the capillaries while keeping the diameter of the liquid jet small (submicrometer to a few micrometer) and preventing of freezing of the jet [• 40].
The GDNV provides a stable liquid jet for a wide range of solution compositions before breaking up in droplets. Stable jets require typically sample flow rates of 5-25 μl/min depending on the GDVN and solution composition, with the jet flowing at linear velocities of around 10 m/s. To achieve a high hit rate, defined as the fraction of detector frames collected containing a diffraction pattern of a crystal, while reducing the occurrence of double hits, the crystal concentration should be adjusted such that up to ~63% of the X-ray interaction volume contains a crystal. With an FEL focus of 1 μm2, a diameter of the liquid jet of 5 μm2 this corresponds to a crystal densities of ~1011/ml. For micrometer protein crystals with a density of 1.35 g/cm3 and 50% solvent content, this is equivalent to 130 mg/ml of protein. With a flow rate of 10 μl/min, and an FEL repetition rate of 120 Hz (LCLS) or 10/30 Hz (SACLA), one out of 440,000 (LCLS) or 5.3 million/ 1.8 million (SACLA) crystals is hit by the FEL beam, emphasizing the need to develop methods that reduce sample consumption and high-lighting the importance of high repetition X-FELs, such as the European XFEL. Possible solutions to decrease “unproductive” sample consumption include approaches to slow the jet to an extent that the delivery rate of fresh crystals to the FEL interaction zone approaches the FEL repetition rate or to use fixed targets [41].
A high-performance liquid chromatography (HPLC) system can be used to supply the liquid pressure to deliver the sample into the nozzle. Such a setup also allows rapid switching between water and sample in case of clogging or problems with data collection, which can save the sample. Recently, a propeller-mounted syringe pump was developed that minimizes sample settling (Fig. 1(c)) during data collection by gently rotating a custom-made, temperature-controlled container storing the crystalline slurry [42]. The system is rated to 400 bar, allowing injection of very viscous samples, including various lipidic phases (e.g. [• 27]) used for membrane protein crystallization.
Crystals of macromolecules are characterized by a network of large solvent channels (solvent content typically 50 %) allowing diffusion of small molecules to their binding sites in the crystallized macromolecule to induce chemical reactions or structural changes. Since diffusion times are significantly reduced for micron to submicron-sized crystals, they are very attractive for time-resolved mechanistic mixing experiments, using the liquid microjet. Due to their relatively low absorption, tiny crystals are also well suited for the study of light-induced reactions. A recent example is a pump probe experiment to study the light-induced dissociation of a ferredoxin-photosystem I complex on a microsecond timescale [43] (Fig. 1a).
Data evaluation in serial femtosecond crystallography
In SFX, the crystals are intersected by an X-FEL beam only fleetingly, and each exposure corresponds to a still image, representing a thin slice of the rocking curve of the Bragg reflections. The CrystFEL software suite [44] has been developed to analyze SFX data, using established procedures for indexing and a Monte Carlo like integration averaging out variations in crystal size and orientation and incident beam intensity when the sampling of the reflections is high enough [• 22]. Depending on the space group of the crystals, the independent indexing of the collected diffraction patterns may result in an equal probability of choosing one of multiple indexing possibilities, for example in the case of polar space groups [45]. Therefore, the resulting dataset will appear to have higher symmetry than the actual crystal lattice and display perfect “twinning”. This feature complicates structure determination. The possible ambiguity in indexing is an open issue in data evaluation. The inclusion of profile fitting and post refinement is currently pursued by several groups. It will not only improve data quality but also likely reduce the number of diffraction patterns required for the convergence of the integrated reflection intensities, and thus the amount of sample required to collect high quality data sets.
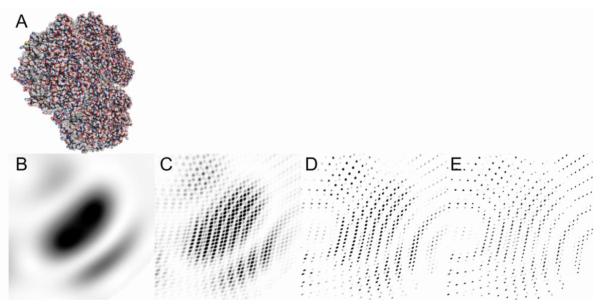
For very small crystals, the assumption of an infinite lattice is no longer valid. This has direct consequences for the observed diffraction pattern which corresponds to the Fourier transform of the convolution of the electron density distribution with the crystal lattice. In case of a finite lattice the intensities between Bragg peaks do not cancel out. Instead, each Bragg peak contains the Fourier transform of the crystal shape (shape transform) [17,46-48] and N-2 interference fringes are present between adjacent Bragg peaks with N denoting the number of unit cells along the particular lattice direction (Fig. 2). Currently, none of these finite-lattice aspects are taken into account in SFX data evaluation, but once they can be included accurately to reconstruct the 3D reciprocal lattice features, they bear the potential for direct phase determination [46,48].
Figure 2.
Simulated diffraction patterns of photosystem I (a). (b) The molecular transform of the molecule is continuous. The interference of several molecules arranged in a lattice of 2×2×2 (c), 4×4×4 (c) and 8×8×8 (e) results in the appearance of Bragg peaks and the fringes between the Bragg peaks.
So far, 2D crystals have been within the realm of electron diffraction but this may change with the advent of X-FELs. Indeed, SFX diffraction data have been collected at the LCLS of beef catalase crystals which are typically used to calibrate transmission electron microscopes. The expected diffraction features were observed, with rod-like diffraction in one direction.
Radiation damage analysis in serial femtosecond crystallography
The SFX experiment on photosystem I was performed using X-ray pulses of up to 300 fs duration. Strong Bragg peaks were observed at the edge of the detector, corresponding to 8 Å resolution [•• 17]. Even when taken into account that the liquid jet surrounding the crystals may act as a tamper [49], this may seem at odds with predictions based on plasma models and hydrodynamics codes that predict 5 Å movements of the generated ions in less than 100 fs. Thus, follow-up studies were performed on microcrystals of photosystem I [• 50] and lysozyme [• 51] to analyze the pulse-length dependence of radiation damage. For lysozyme, diffraction data were collected using nominal pulse durations of 70, 78, 85, 100, 150, 200, 250, 300 and 400 fs, with absorbed dose up to 3GGy, two orders of magnitude higher than the conventional damage limit for protein crystals kept at cryogenic temperature during data collection [6]. To examine the quality of the X-FEL data, virtual powder patterns were computed by summing up the individual peaks and compared with conventional synchrotron powder patterns of identically prepared crystals. The correlation coefficients between intensities extracted by curve fitting from the X-FEL data and conventional powder data were >90 % for all pulse lengths investigated, and all dataset can be used for structure determination by molecular replacement.
Radiation damage in SFX differs substantially from that observed in regular crystallography due to the difference in time-scales. The high irradiance of an FEL pulse results in very rapid formation of a hot electron gas by impact ionization, and impulse rearrangement of atoms and ions. The Coulomb repulsion of the ions and the rapid rise in electron temperature of the system causes displacement of both the atoms and ions during the pulse [52]. This heating leads to a high pressure that drives the explosion of the sample. The atomic ionization and rearrangement of atoms, ions and electrons manifests itself by degradation of Bragg peak intensity and increase of diffuse scattering [53]. To analyze the effects of the X-ray pulse length on the data quality, all lysozyme SFX data sets were compared with the 70 fs one, which is assumed to be the least effected by radiation damage. A decrease of the intensities at high scattering angles was observed for pulse lengths longer than 100 fs. Given the stochastic nature of the experiment and the approximately Gaussian beam intensity profile which is significantly larger than the crystals, some crystals will be in the center of the X-ray beam when exposed to pulses with high fluence, resulting in strong diffraction, whereas others will just be grazed, or exposed to low fluence pulses, resulting in weaker diffraction. Since the lysozyme crystals had a uniform size distribution, diffraction intensity and dose can be correlated. By sorting the X-FEL data according to their relative intensity, it became apparent that the highest-intensity data shows the strongest decrease of Bragg-intensity as a function of resolution with increasing pulse duration. Both dose and dose-rate dependent damage were observed even in the 70 fs data set.
When an X-FEL pulse interacts with a protein crystal, both time-dependent losses of electrons due to photoionization, Auger decay and impact ionization, and time-dependent uncorrelated disorder in the crystalline lattice take place. This has been modeled for events distributed evenly in the unit cell and a correction factor has been deduced [• 50]. In order to understand whether non-uniform modifications of the electron density in the unit cell take place that cannot be corrected, further higher resolution studies are necessary [• 51].
Phase determination by the oversampling method
X-ray crystallography can nowadays be routinely used to determine the 3D structure of macromolecules, as long as sizable, well-ordered crystals are obtained. This remarkable achievement is partially attributed to the availability of powerful phase determination techniques such as the direct methods [54], isomorphous replacement [55], multiple wavelength anomalous dispersion (MAD) [28,29] and molecular replacement [26]. However, when a crystal becomes very small or only has one unit cell (i.e. no crystallinity), a different class of phasing techniques exist, denoted the oversampling method. Partially based on the observation made by Boyes-Watson, Davidson and Perutz on haemoglobin [56], Sayre suggested in 1952 in his half-page paper that measuring the diffraction intensities at as well as between the Bragg peaks may provide the phase information [57]. Following Sayre’s idea, Bates proposed an explanation to the oversampling method in 1982 [58]. Based on the argument that the autocorrelation function of any function is exactly twice the size of the object itself, he concluded that the phase information can be recovered if and only if the diffraction intensities are sampled at least twice finer than the Bragg peak frequency.
In 1998, Miao et al. proposed a different explanation to the oversampling method and concluded that Bates’ statement is inaccurate [59]. In their explanation, solving the phase problem is equivalent to solving the unknown electron density from a set of non-linear equations where each diffraction intensity point is considered as a non-linear equation. When the diffraction intensities are sampled at the Bragg peak frequency, there are exactly twice the unknown variables than the number of independent equations where the independent equations are defined as those diffraction intensity points without any crystallographic or Friedel symmetry. When the diffraction intensities are sampled at an interval finer than the Bragg peak frequency, the number of independent equation is increased but the number of unknown variables remains the same. Equivalently, sampling the diffraction intensities finer than the Bragg peak frequency corresponds to surrounding the electron density with a “no-density” region. The larger the sampling frequency, the larger the no-density region. To quantify the sampling degree, an oversampling ratio was introduced [59], defined as
| (1) |
When σ ≥ 2, the number of independent non-linear equations is larger than or equal to the number of unknown variables, and the phases can, in principle, be recovered from the diffraction intensities. Having the number of independent equations more than the number of unknown variables is a necessity, but not a guarantee to a unique solution. By using the theory of polynomials, it can be shown that, while a finite number of multiple solutions may exist in one dimension, they are pathologically rare in two and three dimensions [60]. Physically, oversampling the diffraction intensities requires better spatial and temporal coherence of the incident beam than the Bragg peak sampling [61,62]. This is because the higher the oversampling frequency, the finer the recording of the features in the diffraction pattern has to be. This explanation was confirmed by both numerical simulation and experiments [59,62].
Oversampling the diffraction intensities with σ ≥ 2, in principle, makes the phases uniquely encoded in the intensities. However, it remains a challenging task to solve a large number of non-linear equations. A very effective way to find the solution (i.e. global minimum) is to use Fourier-based iterative algorithms. In 1972, Gerchberg and Saxton developed an iterative algorithm for retrieving the phases in electron microscopy [63]. The algorithm iterates back and forth between real and reciprocal space in which an electron micrograph in real space and its diffraction pattern in reciprocal space are used as constraints. In 1978, Fienup further improved the iterative algorithm by using the finite size of an object (i.e. support) and positivity as constraints in real space [64]. Emphasis should be made that oversampling and support are two related, but different concepts. When the diffraction intensities are sampled at an interval slightly finer than the Bragg peak frequency, a support exists in real space, but the phases are not unique. The unique phases exist only when the sampling frequency is sufficiently fine with σ ≥ 2. To date, several Fourier-based iterative algorithms have been developed, including error reduction [64], hybrid input-output [64], difference map [65], shrink-wrap [66], guided hybrid input-output [67], and relaxed averaged alternating reflections [68]. Each iteration in the Fourier-based iterative algorithms typically consists of the following five steps.
i). The measured Fourier magnitudes are combined with the phases obtained from previous iteration. In the first iteration, a random phase set is usually used.
ii). Applying the inverse fast Fourier transform (FFT), a new electron density map is calculated.
iii). A support is defined based on the oversampling ratio [59]. The electron density outside the support and the negative electron density are slowly pushed close to zero.
iv). Applying the FFT to the modified electron density map, a new Fourier transform is calculated and its phases are used for the next iteration.
v). An error metric is defined based on either the difference between the measured and calculated Fourier magnitudes or the electron density outside the support divided by that inside the support.
While the oversampling method with iterative algorithms works well for simulated data, the experimental realization of this method faced several obstacles: i) the high dynamic range of the diffraction intensities (typically spanning a few orders), ii) the missing data problem (i.e. the low frequency data cannot usually be measured due to the use of a beamstop, and iii) the presence of different sources of noise in the diffraction pattern. It was not until 1999 that Miao and colleagues performed the first demonstration experiment and established the coherent diffraction imaging (CDI) method [•• 9]. CDI has since been applied to a broad range of biological samples and inorganic materials [10,47,69,70]. The application of the oversampling method to phasing nanocrystals has also been pursued both theoretically [46,48] and experimentally [47,71]. By measuring intensities around the Bragg peaks, Robinson and colleagues were able to determine the 3D shape and the strain field of nanocrystals [71]. Although the oversampling method has been widely applied to determine the structure of non-crystalline specimens and the strain field of inorganic nanocrystals, it has not been demonstrated experimentally for ab initio phasing of a 3D structure.
Single-particle coherent diffraction imaging
One of the most challenging applications of X-FELs is arguably the potential of imaging single large protein complexes in three dimensions. Unlike X-ray crystallography, single-particle CDI faces several obstacles. First, the X-ray diffraction signal from a single biomolecule is extremely weak which makes data acquisition a challenging task. Second, in order to enhance the signal-to-noise ratio and to obtain 3D structure information, the diffraction patterns of a large number of “identical” differently oriented particles have to be measured. The heterogeneity of biomolecules poses a challenge. Third, compared to electrons, the interaction cross section of X-rays with atoms is about five to six orders lower. This explains why cryo electron microscopy (cryo-EM) has been widely used to image the 3D structure of large protein complexes and virus particles [72]. However, compared to electrons, X-rays have a much higher penetration depth and are not limited in their intensity by charging effects. Furthermore, X-FELs can provide intense X-ray pulses with ~1012 photons per pulse. When the X-ray pulses are shorter than tens of femtosecond, the diffraction pattern of a biomolecule can, in principle, be measured before the molecule is destroyed [•• 8]. Follow-up studies suggest that in order to limit average atomic ionization in a biomolecule, X-FEL pulses may have to be shorter than 2-4 fs [73].
Over the past few years, a number of numerical simulations have been performed to explore the feasibility of single-particle CDI. It was first demonstrated through numerical simulations in 2001 that a molecular diffraction pattern at 2.5 Å resolution accumulated from 3×105 copies of single rubisco biomolecules each generated by a femtosecond-level X-FEL pulse can in principle be successfully phased and transformed into an electron density map comparable to that obtained by more conventional methods [• 74]. In this study, however, the molecular orientation was assumed to be determined by other methods such as those developed in single-particle cryo-EM [72]. In subsequent years, several groups have developed more advanced methods to classify the weak diffraction patterns and then determine the molecular orientations based on the diffraction patterns [75-81]. This is an important step as the diffraction intensity of a single biomolecule from a single X-FEL pulse is very weak, and the signal to noise ratio of most detector pixels is likely much smaller than 1. After the classification and orientation determination, the 2D diffraction patterns can then be aligned, averaged and assembled to obtain a 3D diffraction volume, which can be directly phased to obtain the 3D electron density of the molecule using the oversampling method with iterative algorithms [10].
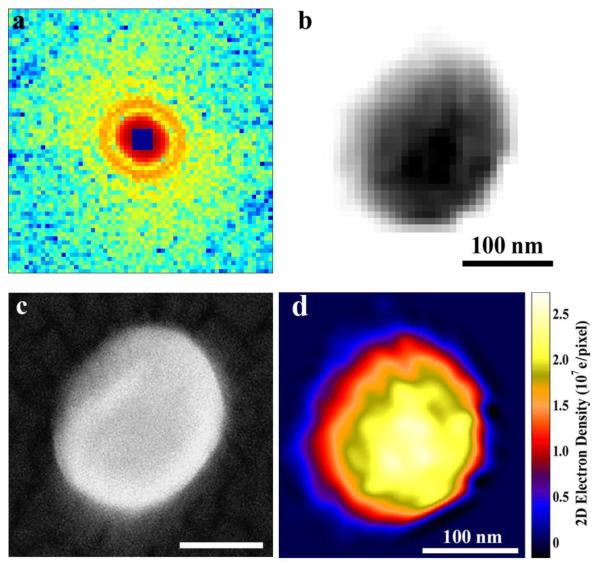
To perform a single-particle CDI experiment, two different schemes have been implemented: one with fixed targets and the other with particle injectors. The fixed target experiment was first conducted on single murine herpesvirus-68 virions at SPring-8 [•• 82]. A herpesvirus virion has an asymmetric tegument and envelope outside of the icosahedrally symmetric capsid composed of defined numbers of subunits. The unstained herpesvirus virions were chemically fixed by 3% glutaraldahyde and supported on 30 nm thick silicon nitride membranes. Individual virions were illuminated by a coherent X-ray beam of 5 keV photon energy, and the diffraction patterns were captured by a CCD detector. Fig. 3(a) shows a coherent diffraction pattern of a single, unstained herpesvirus virion; no effects of radiation damage were observed at 22 nm resolution. The square at the center of the diffraction pattern represents the missing center problem, which is confined within the centro-speckle and allows for consistent phase retrieval [83]. By using the oversampling method with iterative algorithms, the diffraction pattern was directly phased to obtain an image with a resolution of ~22 nm (Fig. 3(b)) which is in good agreement with the scanning electron microscope image of the same virion (Fig. 3(c)). By measuring the incident and diffracted X-ray intensities, the electron density of the reconstruction can be quantified. Fig. 3(d) shows the quantitative electron density of the herpesvirus virion, where the yellow region represents the viral capsid. Although this experiment was conducted on a synchrotron radiation source, the fixed target scheme of single particle CDI can be directly transferred to the X-FELs.
Figure 3.
Single particle coherent diffraction imaging of herpesvirus virions. (a) Coherent X-ray diffraction pattern acquired from a single, herpesvirus unstained virion. (b) The diffraction pattern was directly phased to obtain an image with a resolution of ~22 nm. (c) SEM image of the same virion. (d) Quantitative characterization of the reconstructed electron density map of the herpesvirus virion. [From ref. 82]
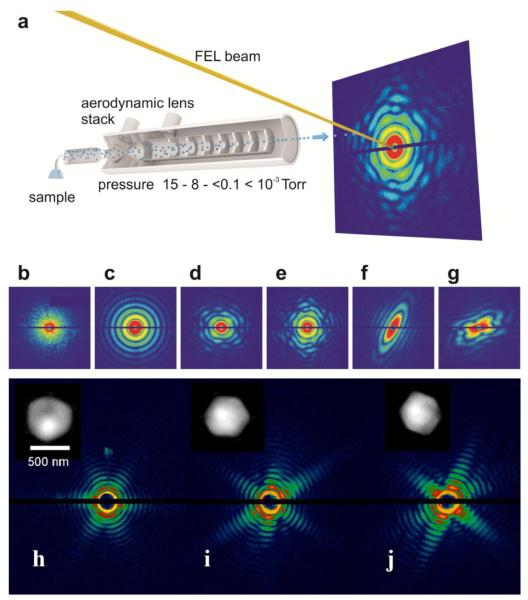
The other scheme is to directly inject single particles into an X-FEL beam with an aerodynamic lens stack. Fig. 4(a) shows a schematic layout of aerosolized particles injected into the X-FEL beam in random orientations [84]. The injected particles are intercepted by X-FEL pulses and the diffractions patterns are measured by a set of detectors. Figs. 4(b-f) show coherent X-ray diffraction patterns of a large aggregate, a water droplet, single T4 phage particles, a nanorice grain and two nanograins, measured with the LCLS pulses [84]. For virus particles smaller than 200 – 300 nm in diameter, it has been observed that the size distribution of the particles estimated from the recorded diffraction patterns is larger than the actual particle size. This size increase is likely caused by aggregation of protein fragments, salts or a residual shell of ammonium acetate buffer around the virus particles. For larger virus particles such as miniviruses, this effect has not been observed [•• 85], presumably because they have a similar size as the initial aerosol droplets. Figs. 4(h-j) show three representative coherent X-ray diffraction patterns measured from single mimivirus particles using LCLS pulses [•• 85]. The LCLS pulses with photon energy of 1.2 keV and 8×1011 photons per pulse were focused to a spot of ~3 μm in diameter. The diffraction patterns exhibit symmetries reflecting the pseudoicosahedral shape of the mimiviruses (Fig. 4(h-j)). By using the oversampling method with iterative algorithms, the diffraction patterns were directly phased to obtain images with a resolution of ~32 nm (insets in Fig. 4(h-j)) [•• 85,86]. In order to obtain the 3D structure, a series of diffraction patterns has to be measured from multiple identical copies of the particles. To date, a 3D reconstruction has been performed on nanorice particles in which 56 randomly oriented diffraction patterns were used [87,88].
Figure 4.
(a) Schematic layout of aerosolized particles injected into the X-FEL beam in random orientations. The injected particles are intercepted by the X-FEL pulses and the diffractions patterns are measured by a set of pnCCD detectors. Coherent X-ray diffraction patterns of a large aggregate (b) a water droplet (c), single T4 phage particles (d, e), a nanorice grain (f), and two nanogrrains (g), measured with the LCLS pulses. (h-j) Three representative coherent X-ray diffraction patterns measured from single mimivirus particles using LCLS pulses. The diffraction patterns were directly phased to obtain images with a resolution of ~32 nm (insets). [From refs. 84 and 86]
Another potentially important direction is a newly developed CDI technique, denoted ankylography, which under certain circumstances can reconstruct the 3D structure of a small object (or a large object at low resolution) from its diffraction pattern sampled on the Ewald sphere [• 89-91]. Due to the size and resolution limitation, ankylography may be only applicable to certain classes of samples, but its advantage is the possibility of obtaining 3D structure without the requirement of averaging. The first ankylography paper has generated a huge debate in the scientific community. For those who are interested in ankylography, several ankylographic source codes have been posted on a public website and can be freely downloaded and tested (http://www.physics.ucla.edu/research/imaging/Ankylography). Finally, the Coherent X-ray Imaging Data Bank has recently been established (www.CXIDB.org) to make the single-particle CDI data and the reconstruction algorithms publicly available.
Conclusions
X-FELs constitute a novel type of light source that produces X-ray pulses with unprecedented peak flux and ultrashort pulse durations. The application of X-FEL radiation to structural biology has just begun, including the recent measurements of X-ray diffraction patterns from micron-sized protein crystals at near atomic resolution [•• 25]. These experimental results suggest that the conventional radiation damage barrier in structure determination of biological systems can be significantly alleviated by using the ultra intense and short X-FEL pulses. Presently, however, several issues have to be resolved before serial femtosecond crystallography can be applied to routinely determine 3D structures using tiny crystals. First, sample consumption in serial femtosecond crystallography is currently a major bottleneck. It is thus an urgent need to develop sample delivery systems to reduce the sample amount required in the experiment. Second, ab initio phase determination methods need to be developed to phase the diffraction patterns in serial femtosecond crystallography. Third, crystal indexing, post refinement and profile fitting have to be improved to reduce the number of hits required for structural determination. With the hard X-rays now available at LCLS and SACLA, great impact can be anticipated if increasingly smaller crystals can be used for structure determination, relaxing the requirements on crystal size. Smaller crystals will however demand for higher X-FEL flux to achieve sufficient signal levels and will be accompanied by increased doses. This will bring the question whether radiation damage can be “outrun” on the atomic level into focus.
The other potentially important direction is the imaging of large protein complexes with X-FELs without the need of crystallization. The methodology of X-ray crystallography has been extended to allow imaging of non-crystalline specimens [•• 9]. The continuous diffraction patterns from non-crystalline specimens allow the direct phase determination by the oversampling method with iterative algorithms [10]. With X-FELs diffraction patterns of single virus particles have been measured and then directly phased to obtain images [•• 85,86]. In order to obtain high resolution 3D structure information of single large protein complexes, there are several challenges that need to be overcome. First, the pulse fluence of X-FELs is not high enough to allow the measurement of high resolution diffraction signal from single large protein complexes. This can, in principle, be overcome by improving the X-FEL peak intensity and using better focusing optics. Second, the dynamic range of the detectors presently used for single-particle imaging with X-FELs is ~103. This has to be increased by at least 1 – 2 orders. Third, due to the weak signal from single particles, the background scattering has to be further reduced. Finally, sample preparation is critical for single particle imaging with X-FELs. It not only requires a very large quantity of identical particles, but also should enable to inject identical particles into an X-FEL beam without aggregation of protein fragments, salts or a residual shell of the buffer around the particles. As structural biology with X-FELs is a new, rapidly developing field, it is not surprising that many issues remain to be resolved and more developments need to be made. As more X-FELs worldwide start to operate within the next few years, it is safe to predict that the future of structural biology with X-FELs will be exciting, bright and also challenging.
Highlights.
X-ray free electron lasers produce X-ray pulses with extremely brilliant peak intensity and ultrashort pulse duration.
The concept of “diffraction-before-destruction” has been verified on biological samples at the Linac Coherent Light Source.
The continuous diffraction patterns from single biological particles allow solving the classical “phase problem”.
Serial femtosecond crystallography has bee used to determine the 3D structure of micron-sized protein crystals.
Coherent X-ray diffraction imaging of single virus particles has been experimentally demonstrated.
Acknowledgements
We are grateful to Thomas R.M. Barends, Lukas Lomb and Yunfei Zou for help with the manuscript. J.M. thanks the support by the National Institutes of Health (grant # GM081409-01A1).
Footnotes
“Tiny” is used here as smaller than a few microns.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Howells MR, Beetz T, Chapman HN, Cui C, Holton JM, Jacobsen CJ, Kirz J, Lima E, Marchesini S, Miao H, et al. An assessment of the resolution limitation due to radiation-damage in x-ray diffraction microscopy. J Electron Spectrosc Relat Phenom. 2009;170:4–12. doi: 10.1016/j.elspec.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schlichting I, Berendzen J, Chu K, Stock AM, Maves SA, Benson DE, Sweet RM, Ringe D, Petsko GA, Sligar SG. The catalytic pathway of cytochrome p450cam at atomic resolution. Science. 2000;287:1615–1622. doi: 10.1126/science.287.5458.1615. [DOI] [PubMed] [Google Scholar]
- 3.Berglund GI, Carlsson GH, Smith AT, Szoke H, Henriksen A, Hajdu J. The catalytic pathway of horseradish peroxidase at high resolution. Nature. 2002;417:463–468. doi: 10.1038/417463a. [DOI] [PubMed] [Google Scholar]
- 4.Burmeister WP. Structural changes in a cryo-cooled protein crystal owing to radiation damage. Acta Crystallogr D. 2000;56:328–341. doi: 10.1107/s0907444999016261. [DOI] [PubMed] [Google Scholar]
- 5.Ravelli RBG, McSweeney SM. The ‘fingerprint’ that X-rays can leave on structures. Structure. 2000;8:315–328. doi: 10.1016/s0969-2126(00)00109-x. [DOI] [PubMed] [Google Scholar]
- 6.Owen RL, Rudino-Pinera E, Garman EF. Experimental determination of the radiation dose limit for cryocooled protein crystals. Proc Natl Acad Sci USA. 2006;103:4912–4917. doi: 10.1073/pnas.0600973103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Solem JC. Imaging biological specimens with high-intensity soft x rays. J. Opt. Soc. Am. B. 1986;3:1551–1565. [Google Scholar]
- 8.Neutze R, Wouts R, van der Spoel D, Weckert E, Hajdu J. Potential for biomolecular imaging with femtosecond X-ray pulses. Nature. 2000;406:752–757. doi: 10.1038/35021099. •• In this paper, Neutze et al. investigate the theoretical feasibility of collecting data from a single protein molecule using molecular dynamics simulations and find that while a molecule exposed to an FEL pulse will disintegrate in a matter of tens of femtoseconds, sufficient time remains in which its structure remains unchanged, thus allowing the collection of useful diffraction data with ultrashort pulses.
- 9.Miao J, Charalambous P, Kirz J, Sayre D. Extending the methodology of X-ray crystallography to allow imaging of micrometre-sized non-crystalline specimens. Nature. 1999;400:342–344. •• This paper represents the first experimental demonstration of coherent diffraction imaging (CDI), in which the coherent diffraction pattern of a non-crystalline specimen is first measured and then directly phased to obtain an image by using the oversampling method with an iterative algorithm.
- 10.Miao J, Chapman HN, Kirz J, Sayre D, Hodgson KO. Taking X-ray Diffraction to the Limit: Macromolecular Structures from Femtosecond X-ray Pulses and Diffraction Microscopy of Cells with Synchrotron Radiation. Annu Rev Biophys Biomol Struct. 2004;33:157–176. doi: 10.1146/annurev.biophys.33.110502.140405. [DOI] [PubMed] [Google Scholar]
- 11.McNeil Brian W. J., Thompson Neil R. X-ray free-electron lasers. Nat Photon. 2011;4:814–821. [Google Scholar]
- 12.Bonifacio R, Pellegrini C, Narducci LM. Collective Instabilities and High Gain Regime in a Free-Electron Lasers. Opt Commun. 1985;50:373. [Google Scholar]
- 13.Ayvazyan V, Baboi N, Bähr J, Balandin V, Beutner B, Brandt A, Bohnet I, Bolzmann A, Brinkmann R, Brovko OI, et al. First operation of a free-electron laser generating GW power radiation at 32 nm wavelength. Eur Phys J D. 2006;37:297–303. [Google Scholar]
- 14.Emma P, Akre R, Arthur J, Bionta R, Bostedt C, Bozek J, Brachmann A, Bucksbaum P, Coffee R, Decker F-J, et al. First lasing and operation of an Ångstrom-wavelength free-electron laser. Nat Photon. 2010;4:641–647. [Google Scholar]
- 15.Shintake T, Tanaka H, Hara T, Tanaka T, Togawa K, Yabashi M, Otake Y, Asano Y, Bizen T, Fukui T, et al. A compact free-electron laser for generating coherent radiation in the extreme ultraviolet region. Nat Photon. 2008;2:555–559. [Google Scholar]
- 16.Holton JM, Frankel KA. The minimum crystal size needed for a complete diffraction data set. Acta Crystallogr D. 2010;66:393–408. doi: 10.1107/S0907444910007262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chapman HN, Fromme P, Barty A, White TA, Kirian RA, Aquila A, Hunter MS, Schulz J, DePonte DP, Weierstall U, et al. Femtosecond X-ray protein nanocrystallography. Nature. 2011;470:73–77. doi: 10.1038/nature09750. •• The first demonstration of SFX, this paper reports data collection on a stream of nanocrystals of the membrane protein complex Photosystem I, resulting in an interpretable 8.5 Å resolution electron density map.
- 18.Fromme P, Spence JC. Femtosecond nanocrystallography using X-ray lasers for membrane protein structure determination. Curr Opin Struct Biol. 2011;21:509–516. doi: 10.1016/j.sbi.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bozek JD. AMO instrumentation for the LCLS X-ray FEL. Eur Phys J Special Topics. 2009;169:129–132. [Google Scholar]
- 20.Strüder L, Sascha E, Daniel R, Robert H, Peter H, Gerhard L, Heike S, Rouven E, Christian R, Klaus H, et al. Large-format, high-speed, X-ray pnCCDs combined with electron and ion imaging spectrometers in a multipurpose chamber for experiments at 4th generation light sources. Nucl Instru Methods A. 2010;614:483–496. •The design of the CAMP instrument is presented including the fast-readout, large-area pnCCD detectors. Many pioneering imaging experiments carried out at the LCLS [•• 17, 27,35, • 50, • 51,53,,84, •• 85,86] were performed in the CAMP instrument.
- 21.Weierstall U, Spence JCH, Doak RB. Injector for scattering measurements on fully solvated species Rev. Sci Instr. 2012;83:035108. doi: 10.1063/1.3693040. [DOI] [PubMed] [Google Scholar]
- 22.Kirian RA, Wang Xiaoyu, Weierstall Uwe, Schmidt Kevin E., Spence John C. H., Hunter Mark, Fromme Petra, White Thomas, Chapman Henry N., et al. Femtosecond protein nanocrystallography—data analysis methods. Opt Express. 2011;18:5713–5723. doi: 10.1364/OE.18.005713. • This paper demonstrates that SFX data can be processed with Monte-Carlo integration methods.
- 23.Kirian RA, White TA, Holton JM, Chapman HN, Fromme P, Barty A, Lomb L, Aquila A, Maia FR, Martin AV, et al. Structure-factor analysis of femtosecond microdiffraction patterns from protein nanocrystals. Acta Crystallogr A. 2011;67:131–140. doi: 10.1107/S0108767310050981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boutet S, Williams GJ. The Coherent X-ray Imaging (CXI) instrument at the Linac Coherent Light Source (LCLS) New J Phys. 2010;12:035024. [Google Scholar]
- 25.Boutet S, Lomb L, Williams GJ, Barends TRM, Aquila A, Doak RB, Weierstall U, Deponte D, Steinbrener J, Shoeman RL, et al. High-Resolution Protein Structure Determination by Serial Femtosecond Crystallography. Science. 2012;337:362–364. doi: 10.1126/science.1217737. •• The first publication of high-resolution SFX experiments with the model protein lysozyme. 1.9 Å resolution electron density maps were obtained with 40 fs and 5 fs FEL pulse lengths, proving the applicability of XFELs to high-resolution structural biology.
- 26.Argos P, Rossmann MG. Molecular replacement method. In: Ladd MFC, Palmer RA, editors. Theory and Practice of Direct Methods in Crystallography. Plenum; New York: 1980. pp. 361–417. [Google Scholar]
- 27.Johansson LC, Arnlund D, White TA, Katona G, DePonte DP, Weierstall U, Doak RB, Shoeman RL, Lomb L, Malmerberg E, et al. Lipidic phase membrane protein serial femtosecond crystallography. Nat Methods. 2012;9:263–265. doi: 10.1038/nmeth.1867. • This manuscript shows that the liquid injector [21] can also be used to deliver micrometer-sized lipidic sponge phase crystals of membrane proteins, such as Blastochloris viridis photosynthetic reaction center, into the FEL interaction region.
- 28.Phillips JC, Wlodawer A, Goodfellow JM, Watenpaugh KD, Sieker LC, Jensen LH, Hodgson KO. Applications of Synchrotron Radiation to Protein Crystallography. II. Anomalous Scattering, Absolute Intensity, and Polarization. Acta Crystallogr A. 1977;33:445–55. [Google Scholar]
- 29.Hendrickson WA, Smith JL, Sheriff S. Direct phase determination based on anomalous scattering. Meth Enzymol. 1985;115:41–55. doi: 10.1016/0076-6879(85)15006-8. [DOI] [PubMed] [Google Scholar]
- 30.Sang-Kil Son, Henry N. Chapman and Robin Santra: Multiwavelength Anomalous Diffraction at High X-Ray Intensity. Phys Rev Lett. 2011;107:218102. doi: 10.1103/PhysRevLett.107.218102. • Detailed electronic damage calculations on isolated heavy atoms exposed to FEL pulses indicte the existence of a Karle-Hendrickson-type equation in the high-intensity regime.
- 31.Asherie N. Protein Crystallization and Phase Diagrams. Methods. 2004;34:266–272. doi: 10.1016/j.ymeth.2004.03.028. [DOI] [PubMed] [Google Scholar]
- 32.Chayen NE. Methods for separating nucleation and growth in protein crystallisation. Prog Biophys Mol Bio. 2005;88:329–337. doi: 10.1016/j.pbiomolbio.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 33.Doye JPK, Poon WCK. Protein crystallization in vivo. Curr Opin Colloid In. 2006;11:40–46. [Google Scholar]
- 34.Fan GY, Maldonado F, Zhang Y, Kincaid R, Ellisman MH, Gastinel LN. In vivo calcineurin crystals formed using the baculovirus expression system. Microsc Res Tech. 1996;34:77–86. doi: 10.1002/(SICI)1097-0029(19960501)34:1<77::AID-JEMT11>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 35.Koopmann R, Cupelli K, Redecke L, Nass K, DePonte DP, White TA, Stellato F, Rehders D, Liang M, Andreasson J, et al. In vivo protein crystallization opens new routes in structural biology. Nat Methods. 2012;9:259–262. doi: 10.1038/nmeth.1859. • In vivo crystallization of polyhedrin-free, glycosylated cathepsin from Trypanosoma brucei overexpressed in Sf9 insect cells is described. The tiny crystals are ideally suited for SFX experiments. Preliminary diffraction studies at LCLS are reported.
- 36.Kissick DJ, Wanapun D, Simpson GJ. Second-order nonlinear optical imaging of chiral crystals. Annu Rev Anal Chem. 2011;4:419–437. doi: 10.1146/annurev.anchem.111808.073722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wampler RD, Kissick DJ, Dehen CJ, Gualtieri EJ, Grey JL, Wang HF, Thompson DH, Cheng JX, Simpson GJ. Selective detection of protein crystals by second harmonic microscopy. J Am Chem Soc. 2008;130:14076–14077. doi: 10.1021/ja805983b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Coudray N, Hermann G, Caujolle-Bert D, Karathanou A, Erne-Brand F, Buessler JL, Daum P, Plitzko JM, Chami M, Mueller U, et al. Automated screening of 2D crystallization trials using transmission electron microscopy: a high-throughput tool-chain for sample preparation and microscopic analysis. J Struct Biol. 2011;173:365–374. doi: 10.1016/j.jsb.2010.09.019. [DOI] [PubMed] [Google Scholar]
- 39.Bingel-Erlenmeyer R, Olieric V, Grimshaw JPA, Gabadinho J, Wang X, Ebner SG, Isenegger A, Schneider R, Schneider J, Glettig W, et al. SLS crystallization platform at Beamline X06DA-A fully automated pipeline enabling in situ X-ray diffraction screening. Cryst Growth Des. 2011;11:916–923. [Google Scholar]
- 40.DePonte DP, Weierstall U, Schmidt K, Warner J, Starodub D, Spence JCH, Doak RB. Gas dynamic virtual nozzle for generation of microscopic droplet streams. J Phys D: Appl Phys. 2008;41:195505. • A concentric, dual capillary setup is described that uses an outer gas stream for focusing of the relatively thick inner liquid jet column. This greatly reduces the risk of clogging of the inner capillary.
- 41.Zarrine-Afsar A, Barends TRM, Müller C, Fuchs MR, Lomb L, Schlichting I, Miller RJD. Crystallography on a chip. Acta Cryst. D. 2012;68:321–323. doi: 10.1107/S0907444911055296. [DOI] [PubMed] [Google Scholar]
- 42.Lomb L, Steinbrener J, Beisel D, Berndt D, Kieser C, Lukat M, Neef N, Shoeman RL. An Anti-Settling Sample Delivery Instrument for Serial Femtosecond Crystallography. J Appl Crys. 2012;45:674–678. [Google Scholar]
- 43.Aquila A, Hunter MS, Doak RB, Kirian RA, Fromme P, White TA, Andreasson J, Arnlund D, Bajt S, Barends TR, et al. Time-resolved protein nanocrystallography using an X-ray free-electron laser. Opt Express. 2012;20:2706–2716. doi: 10.1364/OE.20.002706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.White TA, Kirian RA, Martin AV, Aquila A, Nass K, Barty A, Chapman HN. CrystFEL: a software suite for snapshot serial crystallography. J Appl Cryst. 2012;45:335–341. [Google Scholar]
- 45.White TA, Barty A, Stellato F, Holton J, Kirian RA, Chapman HN. Crystallographic data processing for free-electron laser sources. J. Appl Cryst. 2012 doi: 10.1107/S0907444913013620. Submitted to. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miao J, Sayre D. On possible extensions of X-ray crystallography through diffraction pattern oversampling. Acta Crystallogr A. 2000;56:596–605. doi: 10.1107/s010876730001031x. [DOI] [PubMed] [Google Scholar]
- 47.Robinson I, Harder R. Coherent X-ray diffraction imaging of strain at the nanoscale. Nat Mater. 2009;8:291–298. doi: 10.1038/nmat2400. [DOI] [PubMed] [Google Scholar]
- 48.Spence JCH, Kirian RA, Wang X, Weierstall U, Schmidt KE, White T, Barty A, Chapman HN, Marchesini S, Holton J. Phasing of coherent femtosecond X-ray diffraction from size-varying nanocrystals. Opt Express. 2011;19:2866–2873. doi: 10.1364/OE.19.002866. [DOI] [PubMed] [Google Scholar]
- 49.Hau-Riege SP, London RA, Chapman HN, Szoke A, Timneanu N. Encapsulation and diffraction-pattern-correction methods to reduce the effect of damage in x-ray diffraction imaging of single biological molecules. Phys Rev Lett. 2007;98:198302. doi: 10.1103/PhysRevLett.98.198302. [DOI] [PubMed] [Google Scholar]
- 50.Barty A, Caleman C, Aquila A, Timneanu N, Lomb L, White TA, Andreasson J, Arnlund D, Bajt S, Barends TRM, et al. Self-terminating diffraction gates femtosecond X-ray nanocrystallography measurements. Nat Photon. 2012;6:35–40. doi: 10.1038/nphoton.2011.297. • Based on general considerations on crystals containing molecules with homogeneously distributed elements Barty et al show that the diffracted X-rays are gated by a rapid loss of crystalline periodicity, producing apparent pulse lengths significantly shorter than the duration of the incident pulse.
- 51.Lomb L, Barends TRM, Kassemeyer S, Aquila A, Epp SW, Erk B, Foucar L, Hartmann R, Rudek B, et al. Radiation damage in protein serial femtosecond crystallography using an x-ray free-electron laser. Phys Rev B. 2011;84:214111. doi: 10.1103/PhysRevB.84.214111. • This publication evaluates the effect of radiation damage on SFX diffraction patterns, finding both dose and dose-rate dependent effects.
- 52.Caleman C, Bergh M, Scott HA, Spence JCH, Chapman H, Timneanu N. Simulations of radiation damage in biomolecular nanocrystals induced by femtosecond X-ray pulses. J Mod Opt. 2011;58:1486–1497. [Google Scholar]
- 53.Hau-Riege SP, Graf A, Döppner T, London RA, Krzywinski J, Fortmann C, Glenzer SH, Frank M, Sokolowski-Tinten K, Messerschmidt M, et al. Ultrafast Transitions from Solid to Liquid and Plasma States of Graphite Induced by X-Ray Free-Electron Laser Pulses. Phys Rev Lett. 2012;108:217402. doi: 10.1103/PhysRevLett.108.217402. [DOI] [PubMed] [Google Scholar]
- 54.Hauptman H. Phasing methods for protein crystallography. Curr Opin Struct Biol. 1997;7:672–680. doi: 10.1016/s0959-440x(97)80077-2. [DOI] [PubMed] [Google Scholar]
- 55.Green DW, Ingram VM, Perutz MF. The structure of haemolglobin IV. Sign determination by the isomorphous replacement method. Proc R Soc London Ser A. 1954;255:287–307. [Google Scholar]
- 56.Boyes-Watson J, Davidson E, Perutz MF. An X-Ray Study of Horse Methaemoglobin. I. Proc Roy Soc A. 1947;191:83–132. doi: 10.1098/rspa.1947.0104. [DOI] [PubMed] [Google Scholar]
- 57.Sayre D. Implications of a theorem due to Shannon. Acta Crystallogr. 1952;5:843. [Google Scholar]
- 58.Bates RHT. Fourier phase problem are uniquely soluble in more than one dimension: underlying theory. Optik. 1982;61:247–62. [Google Scholar]
- 59.Miao J, Sayre D, Chapman HN. Phase retrieval from the magnitude of the Fourier transforms of non-periodic objects. J Opt Soc Am A. 1998;15:1662–69. [Google Scholar]
- 60.Bruck YM, Sodin LG. On the ambiguity of the image reconstruction problem. Opt Commun. 1979;30:304–8. [Google Scholar]
- 61.Miao J, Ishikawa T, Johnson B, Anderson EH, Lai B, Hodgson KO. High Resolution 3D X-ray Diffraction Microscopy. Phys Rev Lett. 2002;89:088303. doi: 10.1103/PhysRevLett.89.088303. [DOI] [PubMed] [Google Scholar]
- 62.Miao J, Ishikawa T, Anderson EH, Hodgson KO. Phase retrieval of diffraction patterns from non-crystalline samples by using the oversampling method. Phys Rev B. 2003;67:174104. [Google Scholar]
- 63.Gerchberg RW, Saxton WO. A practical algorithm for the determination of phase from image and diffraction plane pictures. Optik. 1972;35:237–46. [Google Scholar]
- 64.Fienup JR. Reconstruction of an object from the modulus of its Fourier transform. Opt Lett. 1978;3:27–9. doi: 10.1364/ol.3.000027. [DOI] [PubMed] [Google Scholar]
- 65.Elser V. Phase retrieval by iterated projections. J Opt Soc Am A. 2003;20:40–55. doi: 10.1364/josaa.20.000040. [DOI] [PubMed] [Google Scholar]
- 66.Marchesini S. Invited Article: A unified evaluation of iterative projection algorithms for phase retrieval. Rev Sci Instru. 2007;78:011301. doi: 10.1063/1.2403783. [DOI] [PubMed] [Google Scholar]
- 67.Chen CC, Miao J, Wang CW, Lee TK. Application of the optimization technique to non-crystalline X-ray diffraction microscopy - Guided Hybrid Input-Output Method (GHIO) Phys Rev B. 2007;76:064113. [Google Scholar]
- 68.Luke DR. Relaxed averaged alternating reflections for diffraction imaging. Inverse Probl. 2005;21:37. [Google Scholar]
- 69.Chapman HN, Nugent KA. Coherent lensless X-ray imaging. Nat Photon. 2010;4:833–839. [Google Scholar]
- 70.Miao J, Sandberg RL, Song C. Coherent X-ray Diffraction Imaging. IEEE J Sel Top Quant. 2012;18:399–410. [Google Scholar]
- 71.Pfeifer MA, Williams GJ, Vartanyants IA, Harder R, Robinson IK. Three-dimensional mapping of a deformation field inside a nanocrystal. Nature. 2006;442:63. doi: 10.1038/nature04867. [DOI] [PubMed] [Google Scholar]
- 72.Frank J. Three-Dimensional Electron Microscopy of Macromolecular Assemblies. Oxford University Press; USA: 2006. [Google Scholar]
- 73.Hau-Riege SP, London RA, Szoke A. Dynamics of biological molecules irradiated by short x-ray pulses. Phys. Rev. E. 2004;69:051906. doi: 10.1103/PhysRevE.69.051906. [DOI] [PubMed] [Google Scholar]
- 74.Miao J, Hodgson KO, Sayre D. A New Approach to 3-D Structures of Biomolecules Utilizing Single Molecule Diffraction Images. Proc Natl Acad Sci. 2001;98:6641–6645. doi: 10.1073/pnas.111083998. • This paper reports through numerical simulations that a molecular diffraction pattern at 2.5 Å resolution accumulated from multiple copies of single rubisco biomolecules each generated by a femtosecond-level X-FEL pulse can be successfully phased and transformed into an accurate electron density map.
- 75.Huldt G, Szőke A, Hajdu J. Diffraction imaging of single particles and biomolecules. J Struct Biol. 2003;144:219. doi: 10.1016/j.jsb.2003.09.025. [DOI] [PubMed] [Google Scholar]
- 76.Spence JCH, Doak RB. Single molecule diffraction. Phys Rev Lett. 2004;92:198102. doi: 10.1103/PhysRevLett.92.198102. [DOI] [PubMed] [Google Scholar]
- 77.Fung R, Shneerson V, Saldin DK, Ourmazd A. Structure from fleeting illumination of faint spinning objects in flight. Nat Phys. 2009;5:64–67. [Google Scholar]
- 78.Loh N-T, Elser V. Reconstruction algorithm for single-particle diffraction imaging experiments. Phys Rev E. 2009;80:026705. doi: 10.1103/PhysRevE.80.026705. [DOI] [PubMed] [Google Scholar]
- 79.Ziaja B, Martin AV, Wang F, Chapman HC, Weckert E. Theoretical estimation for correlations of diffraction patterns from objects differently oriented in space. Ultramicroscopy. 2011;111:793–797. doi: 10.1016/j.ultramic.2010.12.021. [DOI] [PubMed] [Google Scholar]
- 80.Yoon CH, Schwander P, Abergel C, Andersson I, Andreasson J, Aquila A, Bajt S, Barthelmess M, Barty A, Bogan MJ, et al. Unsupervised classification of single-particle X-ray diffraction snapshots by spectral clustering. Opt Express. 2011;19:16542–16549. doi: 10.1364/OE.19.016542. [DOI] [PubMed] [Google Scholar]
- 81.Bortel G, Faigel G, Tegze M. Classification and averaging of random orientation single macromolecular diffraction patterns at atomic resolution. J. Struct Biol. 2009;166:226–233. doi: 10.1016/j.jsb.2009.01.005. [DOI] [PubMed] [Google Scholar]
- 82.Song C, Jiang H, Mancuso A, Amirbekian B, Peng L, Sun R, Shah SS, Zhou ZH, Ishikawa T, Miao J. Quantitative Imaging of Single, Unstained Viruses with Coherent X-rays. Phys Rev Lett. 2008;101:158101. doi: 10.1103/PhysRevLett.101.158101. paper reports for the first time the recording and reconstruction of coherent diffraction patterns from single, unstained herpesvirus virions using 5 keV X-rays. The structure of the viral capsid inside a virion was visualized at a resolution of ~22 nm.
- 83.Miao J, Nishino Y, Kohmura Y, Johnson B, Song C, Risbud SH, Ishikawa T. Quantitative Image Reconstruction of GaN Quantum Dots from Oversampled Diffraction Intensities Alone. Phys Rev Lett. 2005;95:085503. doi: 10.1103/PhysRevLett.95.085503. [DOI] [PubMed] [Google Scholar]
- 84.Kassemeyer S, Steinbrener J, Lomb L, Hartmann E, Aquila A, Barty A, Martin AV, Hampton CY, Bajt S, Barthelmess M, et al. Femtosecond free-electron laser x-ray diffraction data sets for algorithm development. Opt. Express. 2012;20:4149–4158. doi: 10.1364/OE.20.004149. [DOI] [PubMed] [Google Scholar]
- 85.Seibert MM, Ekeberg T, Maia FRNC, Svenda M, Andreasson J, Jönsson O, Odić D, Iwan B, Rocker A, Westphal D, et al. Single mimivirus particles intercepted and imaged with an X-ray laser. Nature. 2011;470:78–81. doi: 10.1038/nature09748. •• Seibert et al report the recoding of coherent X-ray diffraction patterns from single mimivirus particles intercepted with the LCLS pulses. By using various constraints in the reconstructions, the diffraction pattern was phased to obtain a 2D image of the mimivirus particle.
- 86.Martin AV, Aquila A, Bajt S, Barthelmess M, Barty A, Coppola N, DePonte DP, Fleckenstein H, Gumprecht L, Liang M, et al. Single particle imaging with soft x-rays at the Linac Coherent Light Source. Proc SPIE. 2011;8078:807809. [Google Scholar]
- 87.Bogan MJ, Boutet S, Barty A, Benner WH, Frank M, Lomb L, Shoeman R, Starodub D, Seibert MM, Hau-Riege SP, et al. Single-shot femtosecond x-ray diffraction from randomly oriented ellipsoidal nanoparticles. Phys Rev Spec Top-Acc Beams. 2010;13:94791. [Google Scholar]
- 88.Loh ND, Bogan MJ, Elser V, Barty A, Boutet S, Bajt S, Hajdu J, Ekeberg T, Maia FRNC, Schulz J, et al. Cryptotomography: Reconstructing 3D Fourier Intensities from Randomly Oriented Single-Shot Diffraction Patterns. Phys Rev Lett. 2010;104:225501. doi: 10.1103/PhysRevLett.104.225501. [DOI] [PubMed] [Google Scholar]
- 89.Raines KS, Salha S, Sandberg RL, Jiang H, Rodríguez JA, Fahimian BP, Kapteyn HC, Du J, Miao J. Three-dimensional structure determination from a single view. Nature. 2010;463:214–217. doi: 10.1038/nature08705. • This paper reports that, under certain circumstances, the 3D structure of a small object (or a large object at low resolution) can be reconstructed from its diffraction pattern sampled on the Ewald sphere. The source code of this technique (denoted Ankylography) has been posted on a public website and can be freely downloaded and tested (http://www.physics.ucla.edu/research/imaging/Ankylography).
- 90.Chen CC, Jiang H, Rong L, Salha S, Xu R, Mason TG, Miao J. Three-dimensional imaging of a phase object from a single sample orientation using an optical laser. Phys Rev B. 2011;84:224104. [Google Scholar]
- 91.Seaberg MD, Adams DE, Townsend EL, Raymondson DA, Schlotter WF, Liu Y, Menoni CS, Rong L, Chen CC, Miao J, et al. Ultrahigh 22 nm resolution coherent diffractive imaging using a desktop 13 nm high harmonic source. Opt. Express. 2011;19:22470–22479. doi: 10.1364/OE.19.022470. [DOI] [PubMed] [Google Scholar]