Abstract
Decapping scavenger (DcpS) enzymes catalyze the cleavage of a residual cap structure following 3′→5′ mRNA decay. Some previous studies suggested that both m7GpppG and m7GDP were substrates for DcpS hydrolysis. Herein, we show that mononucleoside diphosphates, m7GDP (7-methylguanosine diphosphate) and m32,2,7GDP (2,2,7-trimethylguanosine diphosphate), resulting from mRNA decapping by the Dcp1/2 complex in the 5′→3′ mRNA decay, are not degraded by recombinant DcpS proteins (human, nematode and yeast). Furthermore, whereas mononucleoside diphosphates (m7GDP and m32,2,7GDP) are not hydrolyzed by DcpS, mononucleoside triphosphates (m7GTP and m32,2,7GTP) are, demonstrating the importance of a triphosphate chain for DcpS hydrolytic activity. m7GTP and m32,2,7GTP are cleaved at a slower rate than their corresponding dinucleotides (m7GpppG and m32,2,7GpppG, respectively), indicating an involvement of the second nucleoside for efficient DcpS-mediated digestion. Although DcpS enzymes cannot hydrolyze m7GDP, they have a high binding affinity for m7GDP and m7GDP potently inhibits DcpS hydrolysis of m7GpppG, suggesting that m7GDP may function as an efficient DcpS inhibitor. Our data have important implications for the regulatory role of m7GDP in mRNA metabolic pathways, due to its possible interactions with different cap-binding proteins, such as DcpS or eIF4E.
mRNA degradation plays a significant role in the post-transcriptional regulation of gene expression.1 Understanding the control of gene expression requires defining the molecular and cellular basis of mRNA turnover. There are two major pathways that are utilized for degradation of mRNA in eukaryotic cells, both initiated by the removal of the poly(A) tail.2 In the 5′→3′ decay pathway, the cap is hydrolyzed exposing the mRNA 5′ end to the exoribonuclease activity.3 In the 3′→5′ decay pathway, exosome-mediated degradation of mRNA releases a cap containing dinucleotide or a short capped oligonucleotide.4 Decapping plays an important role in mRNA turnover and decapping rates can differ significantly for different mRNAs.5
Eukaryotic cells contain different classes of enzymes responsible for the cleavage of mono- and trimethylated cap struct ures. Dcp2 is an RNA decapping enzyme generating m7GDP from an mRNA chain and enables 5′→3′ mRNA decay. In the 3′→5′ decay of mRNAs, 5′ capped dinucleotides or short capped oligonucleotides result from the mRNA degradation. The DcpS (Decapping Scavenger) enzyme acts on them releasing m7GMP. DcpS can only hydrolyze the cap on mRNA oligonucleotides up to 10 nucleotides in mammals and 3 nucleotides in nematodes.6–8 Nematode mRNAs have either monomethylguanosine (MMG) or trimethylguanosine (TMG) caps, and DcpS from C. elegans and A. suum recognize both caps whereas human DcpS can degrade only the monomethylated cap structure.7,8
A detailed characterization of the DcpS binding affinity and catalytic properties in different eukaryotic organisms, as well as the knowledge of its biological functions in cellular processes, are essential for the design and synthesis of novel cap analogs and improvement of their therapeutic properties. Infections of humans and animals caused by parasitic nematodes are great medical and economic problems (~ 3 billion people infected)9 and there is a need for drugs that will specifically block parasite gene expression, without influencing the metabolism of mammalian cells. Nematode DcpS has the unique ability to efficiently hydrolyze the TMG cap whereas the human enzyme cannot.8
An important therapeutic aspect of DcpS studies is the regulation of the enzyme activity in human cells in spinal muscular atrophy (SMA).10 Biochemical and structural data indicate that DcpS enzyme could be a molecular target for SMA treatment. It has been demonstrated that the therapeutic effect in this disease might be based on the inhibition of this enzyme by C5-substituted quinazolines, that hold human DcpS in a catalytically inactive conformation.10
Several studies have suggested that DcpS in cell extracts (human, yeast and Xenopus) and recombinant human and yeast decapping scavengers were able to efficiently hydrolyze m7GDP, the product of Dcp2 activity.11,12 In this context, DcpS proteins were proposed to function not only in the 3′→5′ mRNA decay pathway, but also to be involved in hydrolysing m7GDP generated from cleavage of the capped mRNA in 5′→3′ decay pathway. In this scheme, m7GMP would be the final product of both degradation pathways, and a general indicator of eukaryotic mRNA degradation process. However, a detailed analysis of the function of DcpS enzymes in this pathway has not been carefully examined. Moreover, in independent experiments performed by Cohen et al., efficient hydrolysis of m7GDP to m7GMP was not observed, using either recombinant human and C. elegans nematode DcpS proteins or A. suum nematode whole-cell embryo extracts.8 In A. suum extracts, a very small percentage of m7GDP was converted to m7GMP, but the activity responsible for this conversion was attributed to non-specific enzymes, not to DcpS. Liu et al.6 also reported the hydrolytic stability of m7GDP in enzymatic assays with the human decapping scavenger. Thus, data regarding the role of DcpS in the hydrolysis of m7GDP are contradictory. In this paper, we re-examined the hydrolytic activity of four recombinant DcpS enzymes from multiple organisms: human (HsDcpS), A. suum nematode (AsDcpS), C. elegans nematode (CeDcpS) and S. cerevisiae yeast Dcs1 homodimer (ScDcpS), previously evaluated by different research groups.6,8,11,12 We carried out enzymatic assays using conditions in which an efficient cleavage of m7GDP was previously reported by van Dijk et al.11, as well as conditions optimal for HsDcpS binding studies13 using a highly sensitive high-performance liquid chromatography (HPLC) assay for monitoring the reaction progress and detection of degradation products. We show that recombinant DcpS enzymes have no ability to hydrolyze m7GDP, regardless of their origin. Furthermore, our DcpS binding and inhibition studies indicate that m7GDP is strongly bound by all DcpS enzymes and efficiently inhibits their hydrolytic activity towards m7GpppG. We determined the association constants (KAS) of m7GDP and m32,2,7GDP, and also the inhibition parameters, the half maximal inhibitory concentrations (IC50) and the inhibition constants (Ki) of m7GDP. Overall, the resistance of m7GDP to DcpS-mediated hydrolysis and its strong binding and inhibitory properties towards decapping scavengers suggest that m7GDP may play a regulatory role for cap-binding proteins associated with several mRNA pathways. We thus propose an amended model of mRNA degradation.
MATERIALS AND METHODS
Synthesis of cap analogs
Cap analogs investigated in this paper (m7GMP, m7GDP, m7GTP, m7GpppG, m7Gpppm7G, m32,2,7GMP, m32,2,7GDP, m32,2,7GTP, m32,2,7GpppG, GpppG) were synthesized according to previously described methods.14–16 The concentrations of compounds were determined on the basis of their absorption coefficients (εmax) determined in 0.1 M phosphate buffer: ε258(m7GMP, m7GDP, m7GTP, m32,2,7GMP, m32,2,7GDP, m32,2,7GTP) = 11.4 × 103 M−1 cm−1 at pH 6.0, ε251(GpppG) = 25.5 × 103 M−1 cm−1, ε255(m7GpppG) = 22.6 × 103 M−1 cm−1 and ε259(m7Gpppm7G) = 16.0 × 103 M−1 cm−1 at pH 7.0,17 and ε258(m32,2,7GpppG) = 26.3 × 103 M−1 cm−1 at pH 7.0.18 Absorption spectra were recorded on Lambda 20UV/VIS spectrophotometer (Perkin-Elmer, Co., Waltham, MA, USA) at 20°C.
Protein expression and purification
HsDcpS, AsDcpS, CeDcpS and ScDcpS were expressed in Escherichia coli according to the previously described procedures19,20 and were purified as His-tagged proteins by affinity chromatography using Ni-NTA agarose under native conditions. To obtain homogeneous fractions of enzymes, all recombinant decapping scavengers were further purified by gel filtration at 4°C through a Pharmacia Superdex-200 gel filtration column (GE Healthcare Bio-Science AB, Uppsala, Sweden), using an ÄKTA FPLC system (Pharmacia-Biotech, Uppsala, Sweden) and 20 mM Tris HCl buffer containing 50 mM KCl, 0.2 mM EDTA, 20% glycerol (final pH 7.6). The peaks corresponding to the respective decapping scavenger proteins were identified by comparison of their FPLC profiles with reference proteins (BSA, ovalbumin and lysozyme) and stored at −80 °C in 20 mM Tris HCl buffer containing 50 mM KCl, 0.2 mM EDTA, 20% glycerol (final pH 7.6) and 1 mM DTT at ~50 μM. The concentration of decapping scavenger enzymes was estimated by the Bradford assay21 and spectrophotometrically from the enzymes’ molar absorption coefficient (ε280 = 30.4 × 103 M−1cm−1 for HsDcpS, ε280 = 40.3 × 103 M−1cm−1 for AsDcpS, ε280 = 38.9 × 103 M−1cm−1 for CeDcpS and ε280 = 62.8 × 103 M−1cm−1 for ScDcpS) calculated from amino acid composition of a monomer using Clustal 2.1 algorithm on the ExPASy Server.
HPLC analysis of reaction progress
The hydrolytic activity of recombinant DcpS was assayed using two experimental conditions: (1) 45 mM Tris HCl containing 9 mM MgAc and 27 mM (NH4)2SO4 (final pH 8.0), at 37 °C; (2) 50 mM Tris HCl containing 200 mM KCl, 0.5 mM EDTA and 1 mM DTT (final pH 7.6), at 20 °C. Initial substrate concentration for DcpS hydrolysis assays was 15 μM. Before each experiment, 1 mL of buffer solution containing the investigated cap analog was incubated at 20 or 37 °C for 10 min. The hydrolysis process was started by the addition of recombinant DcpS (final concentration either 0.02 or 0.2 μM). At 5, 10, 30, 60 min and after 24 h of the hydrolysis, 200 μL aliquots of the reaction mixture were withdrawn and the reaction was stopped by heat inactivation of the enzyme (2.5 min at 97 °C). The samples were then subjected to analytical HPLC (Agilent Technologies 1200 Series, Santa Clara, CA, USA) using a reverse-phase Supelcosil LC-18-T column (4.6 mm × 250 mm, 5 μm) with UV/VIS and fluorescence detectors. Substrate and products were eluted at room temperature with a linear gradient of methanol from 0% to 25% in aqueous 0.1 M KH2PO4 over 15 min at a flow rate of 1.3 mL/min. The fluorescence at 337 nm (excitation at 280 nm) and absorbance at 260 nm were monitored continuously during the analysis. Hydrolysis products were identified by comparison of their retention times with those of the reference standards.
Fluorescence titration analysis of binding affinity
DcpS-cap binding affinity was determined by monitoring the quenching of intrinsic DcpS Trp fluorescence. The time-synchronized-titration method (TST) was applied for this study.22 The experiments were performed on LS-50B spectrofluorometer (Perkin-Elmer Co., Waltham, MA, USA) in a quartz cuvette (Hellma, Müllhaim, Germany) with an optical path length of 4 mm for absorption and 10 mm for emission. All measurements were performed as previously described for HsDcpS: 20 °C, in 50 mM Tris HCl buffer containing 200 mM KCl, 0.5 mM EDTA and 1 mM DTT (final pH 7.6): experimental condition (2).13 One μL aliquots of cap analog solutions of increasing concentration (from 5 μM to 2 mM) were added to 1.4 mL of DcpS solution (initial concentration 0.2 μM). The fluorescence intensity was monitored at 340 nm with a 4 nm bandwidth (excitation at 280 nm with a 2.5 nm bandwidth) and corrected for sample dilution and inner filter effects.23 The equilibrium association constants for single titrations (KAS) were determined by fitting the theoretical dependence of the fluorescence intensity on the total concentration of the cap analog to the experimental data points using the previously described equation.22 The final KAS were calculated as weighted averages of 3–5 independent titrations, with the weights taken as the reciprocal standard deviations squared. The numerical least-squares non-linear regression analysis was performed using ORIGIN 8.0 (Microcal Software Inc., USA).
Determination of inhibition parameters for m7GDP
The inhibitory properties of m7GDP were examined on recombinant DcpS enzymes in decapping assays using m7GpppG as the substrate. Experiments were performed in the binding affinity buffer (50 mM Tris HCl containing 200 mM KCl, 0.5 mM EDTA and 1 mM DTT, final pH 7.6), at 20 °C: experimental condition (2). The inhibition of the rate of m7GpppG (15 μM) hydrolysis in the presence of 0.1, 0.2, 0.5, 1, 2, 5, 10, 20 and 50 μM m7GDP was monitored by HPLC. One mL of buffer solution containing the substrate and inhibitor was incubated at 20°C for 10 min and the reaction initiated by the addition of recombinant DcpS. After 10 min, the reaction was stopped by heat inactivation of the enzyme at 97 °C for 2.5 min (200 μL aliquots of the reaction mixture were withdrawn and placed into eppendorf tube preincubated at 97 °C). Reaction products were analyzed by HPLC as described above, with a slightly different linear gradient of methanol (from 0% to 15% in aqueous 0.1 M KH2PO4 over 15 min). The concentration of DcpS (0.02 μM for CeDcpS, 0.02 μM for AsDcpS, 0.10 μM for ScDcpS and 0.12 μM for HsDcpS) was optimized to obtain 50% substrate conversion within 10 min of hydrolysis without the inhibitor. IC50 values were determined by plotting substrate conversion versus inhibitor concentration (IC50 is defined as the concentration of inhibitor causing the decrease of substrate conversion by 50% under fixed enzyme concentration and different substrate concentrations). For calculation of Ki values we used the equation for tight binding competitive inhibitors:24
where Ki is the inhibition constant, [S] is the substrate concentration, Km is the Michaelis constant, and [E] is the enzyme concentration.
RESULTS
Resistance of m7GDP and m32,2,7GDP to hydrolysis catalyzed by DcpS enzymes
Naturally occurring mononucleoside diphosphates, m7GDP (all eukaryotes) and m32,2,7GDP (nematodes), are products of Dcp2 decapping activity on mRNA. Previous reports described conflicting data on the ability of DcpS to convert m7GDP to m7GMP6,8,11,12. To re-evaluate whether scavenger decapping enzymes are able to efficiently hydrolyze m7GDP, as well as to examine m32,2,7GDP susceptibility to DcpS digestion, highly purified human, nematode (A. suum and C. elegans) and yeast (S. cerevisiae Dcs1) recombinant DcpS proteins (Figure 1) were examined using two assay conditions: (1) those described by van Dijk et al.11 for investigation of m7GDP hydrolysis by DcpS proteins (37 °C, 45 mM Tris HCl containing 9 mM MgAc and 27 mM (NH4)2SO4, final pH 8.0) and (2) those proposed by Darzynkiewicz et al.13 for analysis of human DcpS binding affinity (20 °C, 50 mM Tris HCl containing 200 mM KCl, 0.5 mM EDTA and 1 mM DTT, final pH 7.6). The first condition was used to replicate van Dijk’s experiments and the second one was developed to assay DcpS hydrolytic and binding characterization. In both cases, two concentrations of DcpS were examined, 0.02 μM and 0.2 μM. The lower concentration was optimal for evaluation of cap analog degradation during 60 min as described by van Dijk et al.11 The 10-fold higher enzyme concentration was used to examine the hydrolytic stability of mononucleoside diphosphates towards DcpS. The hydrolytic activity of the decapping scavengers for the m7GpppG cap analog was also examined.
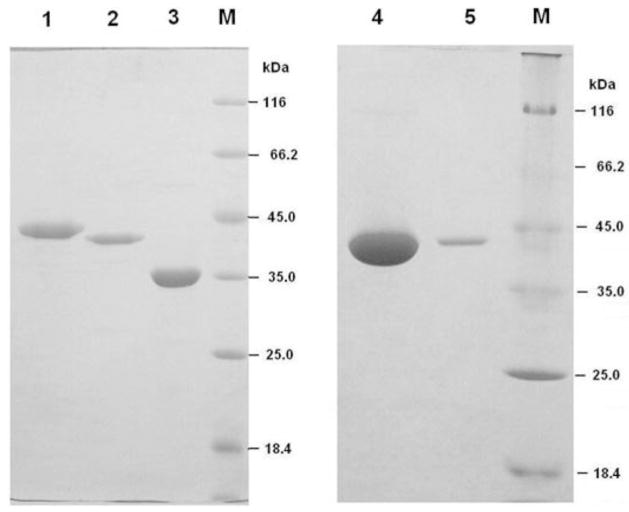
Figure 1.
SDS-PAGE of purified recombinant human, nematode and yeast DcpS proteins. His-tagged decapping scavengers were expressed in E. coli and purified using Ni-NTA agarose affinity and gel filtration chromatography. HsDcpS, AsDcpS and CeDcpS are presented in lanes 1, 2 and 3, respectively. ScDcpS (S. cerevisiae Dcs1) is presented in lanes 4 and 5. Protein markers are in lane M. The amount of protein loaded on the gel is 5 μg of HsDcpS, 4 μg of AsDcpS, 7 μg of CeDcpS, 25 μg and 2 μg of ScDcpS (lane 4 and 5, respectively). DcpS Coomassie Blue-staining of 15% SDS-PAGE gels indicates a high purity of these recombinant proteins.
The rate of hydrolysis of m7GpppG for all the DcpS proteins was very fast at 37 °C under assay condition (1), with the reaction products, m7GMP and GDP, appearing immediately following enzyme addition. In contrast, m7GDP and m32,2,7GDP were not degraded by the DcpS proteins, as indicated from the HPLC analysis, regardless of the enzyme concentration or incubation time (up to 24 h).
As illustrated in Figure 2, using the alternate assay condition (2), m7GpppG was very efficiently converted to products, within less than 3 minutes, whereas both mononucleoside diphosphates, m7GDP and m32,2,7GDP, were resistant to cleavage by all the DcpS enzymes. As shown in Figure 2B,D, the intensity of chromatographic peaks corresponding to the input amounts of m7GDP and m32,2,7GDP remains unchanged even after long incubation period (24 h).
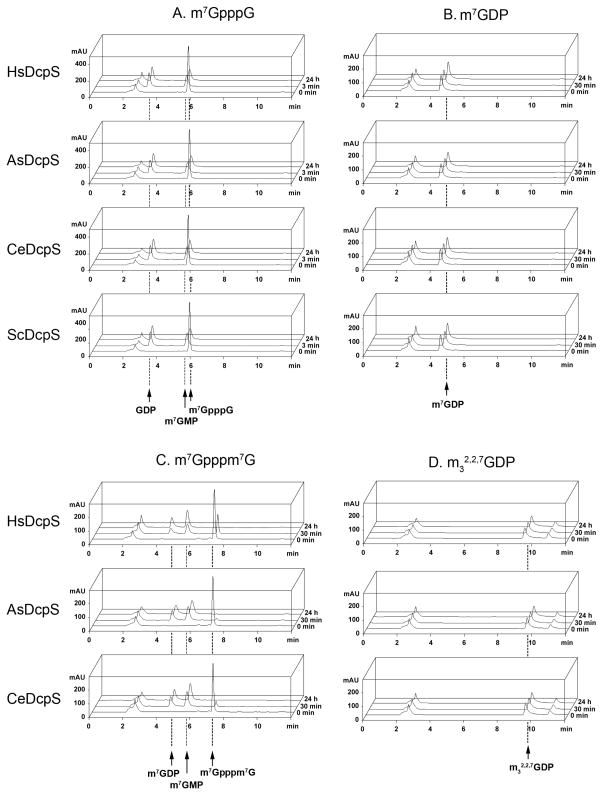
Figure 2.
HPLC analysis of hydrolytic susceptibility of m7GpppG, m7Gpppm7G, 7-methylguanosine diphosphate (m7GDP), and 2,2,7-trimethylguanosine diphosphate (m32,2,7GDP) to recombinant DcpS. All experiments were performed using condition set (2): 20 °C in 50 mM Tris HCl buffer containing 200 mM KCl, 0.5 mM EDTA and 1 mM DTT (final pH 7.6) with 0.2 μM DcpS. Initial concentrations of cap analogs were 15 μM. The HPLC peak with a retention time of 2.5 min corresponds to the reaction buffer.
(A) DcpS enzymes efficiently hydrolyze m7GpppG. Complete conversion of the substrate to m7GMP and GDP products occurs within less than 3 minutes.
(B) and (D) m7GDP and m32,2,7GDP, respectively, are not hydrolyzed following 24 h of incubation with DcpS. The HPLC peak with a retention time of 10.8 min in the m32,2,7GDP profile corresponds to a substrate contaminant.
(C) Hydrolysis of m7Gpppm7G is significantly slower than that of m7GpppG. After 30 min, a small amount of the substrate is still observed in all cases. Chromatographic peaks corresponding to the products, m7GDP and m7GMP, retain their intensity after the complete degradation of m7Gpppm7G (24 h of investigation), confirming the resistance of m7GDP to enzymatic cleavage.
Taken together, none of the four DcpS enzymes examined, from human, S. cerevisiae, A. suum and C. elegans, hydrolyzes m7GDP or m32,2,7GDP, even at high enzyme concentration (0.2 μM) or under different assay conditions. We conclude that the mononucleoside diphosphates, m7GDP and m32,2,7GDP, are not hydrolyzed by recombinant DcpS.
Stability of m7GDP confirmed by m7Gpppm7G incubation with DcpS
To further confirm the stability of m7GDP towards DcpS proteins, we additionally examined the hydrolysis of m7Gpppm7G. As shown in Figure 2C, hydrolysis of m7Gpppm7G by DcpS proteins resulted in the simultaneous appearance of only two reaction products, m7GMP and m7GDP, and the latter was not further converted to any products. The ratio of the chromatographic peak areas corresponding to reaction products (m7GDP:m7GMP) remains constant, even after a long time of incubation (24 h). This is a further demonstration that the mononucleoside diphosphate m7GDP is not hydrolyzed by DcpS to m7GMP.
Activity of human and nematode DcpS on m7GpppG, m32,2,7GpppG and GpppG
Recombinant DcpS enzyme from A. suum, a parasitic nematode of pigs, has not been previously examined. We previously carried out some kinetic analysis of the hydrolysis of m7GpppG and m32,2,7GpppG by C. elegans DcpS.20 Here, we performed a direct comparison of hydrolysis rates of AsDcpS with human and C. elegans DspS enzymes towards the two cap structures, both existing at the 5′ end of nematode mRNAs, as well as towards the unmethylated dinucleotide GpppG. Experiments were performed using assay condition (2), which facilitated simultaneous characterization of DcpS-cap binding affinity and hydrolysis (Figure 3). A low enzyme concentration (0.02 μM) was chosen to be able to analyze reaction progress at several time points with complete degradation of m7GpppG within 60 min (Figure 3).
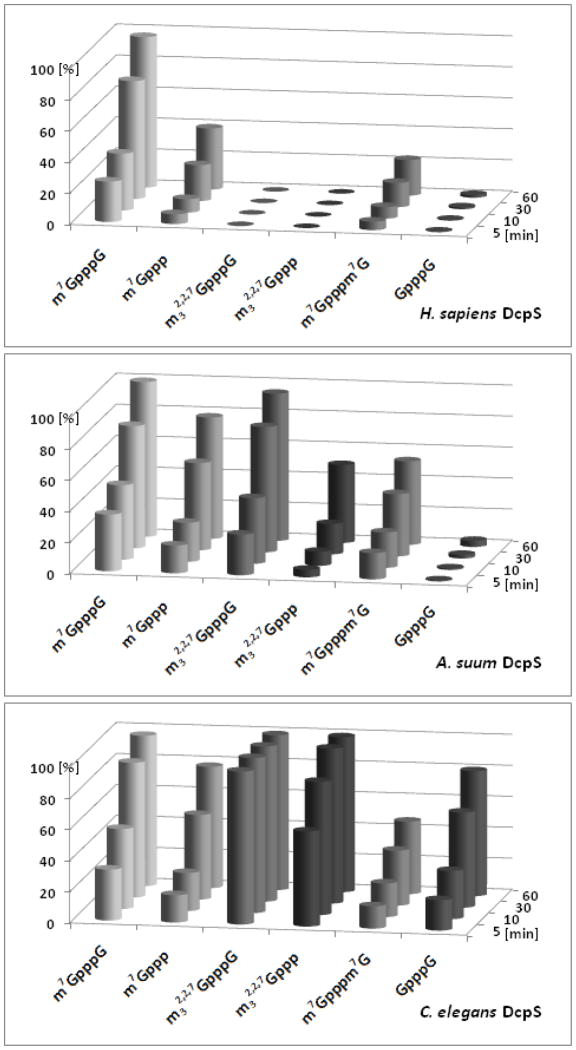
Figure 3.
Comparison of the hydrolytic activity of HsDcpS, AsDcpS and CeDcpS. The extent of decapping (x) was determined as the % of hydrolyzed substrate measured by HPLC system. The standard deviation of the mean x value is below 5%. All experiments were performed at 20 °C in 50 mM Tris HCl buffer containing 200 mM KCl, 0.5 mM EDTA and 1 mM DTT (final pH 7.6) with 0.02 μM DcpS. Initial concentrations of cap analogs were 15 μM.
As presented in Figure 3, m7GpppG hydrolysis occurs very efficiently and at similar rates for all three DcpS proteins, with 95% of the substrate hydrolyzed after 60 min. The comparison of the m32,2,7GpppG hydrolysis catalyzed by human and nematode decapping scavengers demonstrated that A. suum and C. elegans DcpS readily hydrolyze a trimethylated dinucleotide, but the human enzyme does not (Figure 3). Digestion of m32,2,7GpppG was not observed after 60 min or 24 h incubation with the human enzyme. The TMG cap is hydrolyzed by C. elegans DcpS at a significantly higher rate than that observed for m7GpppG hydrolysis. In contrast, AsDcpS hydrolysis of MMG is slightly greater than that observed for m32,2,7GpppG. This is the first demonstration of a difference in specificity for a trimethylated cap between nematode DcpS enzymes from different species. The hydrolysis of unmethylated dinucleotide GpppG is much slower as compared to m7GpppG for CeDcpS and does not occur with either AsDcpS or HsDcpS.
Activity of human and nematode DcpS on m7GTP and m32,2,7GTP
Since m7GDP and m32,2,7GDP were found to be resistant to DcpS enzymatic hydrolysis, we next examined DcpS hydrolytic properties towards m7GTP and m32,2,7GTP to gain insight into the contribution of either a second nucleoside or a length of the cap’s phosphate chain on the activity of the DcpS enzymes. We found that 7-methylguanosine triphosphates are efficiently hydrolyzed by the human and nematode DcpS proteins, whereas only the nematode decapping scavengers have activity on the m32,2,7GTP (Figure 3). The rate of m7GTP degradation is identical for AsDcpS and CeDcpS, but 2-fold slower for HsDcpS. A significant difference is observed for m32,2,7GTP hydrolysis by the two nematode decapping scavengers. m32,2,7GTP is a much better substrate for CeDcpS than for AsDcpS, as indicated by a higher hydrolysis rate (Figure 3). This difference is similar to the one observed for m32,2,7GpppG, which confirms that AsDcpS and CeDcpS differ in their activity for the two additional CH3 groups at N2 position of 7-methylguanosine.
The rate of mononucleoside triphosphates hydrolysis is slower in comparison with the respective dinucleotides (Figure 3), demonstrating the importance of the second nucleoside on the efficiency of the hydrolysis. We conclude that the second nucleoside is not absolutely required for DcpS activity, however it enhances DcpS hydrolysis. Furthermore, direct comparison of enzymatic cleavage of mononucleoside di- and triphosphates illustrates the requirement of a triphosphate bridge for efficient hydrolysis by decapping scavengers.
Binding affinity of human and nematode DcpS for mononucleotide cap analogs
Our data demonstrate that m7GDP and m32,2,7GDP are resistant to enzymatic cleavage by DcpS. We next examined whether DcpS proteins were able to bind these nucleotides. To determine the association constants (KAS) for the complexes of DcpS with these mononucleotide cap analogs, we performed fluorescence titration assays (Figure 4). Assay condition (2) were previously determined and optimized to measure cap interaction with human DcpS using quenching of intrinsic Trp fluorescence upon ligand binding.13
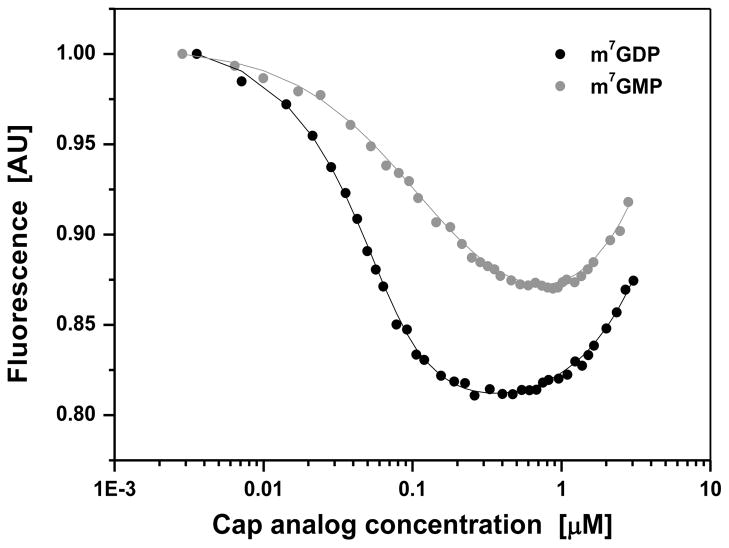
Figure 4.
Fluorescence titration curves for binding of mononucleotide cap analogs to C. elegans DcpS. Titration experiments were carried out at 20 °C, in 50 mM Tris HCl containing 200 mM KCl, 0.5 mM EDTA and 1 mM DTT (final pH 7.6). Protein fluorescence was excited at 280 nm and observed at 340 nm. The intensity of fluorescence is represented as relative values (AU: arbitrary units). The observed increasing fluorescence signal at the higher concentration of cap analogs originates from their emission in free forms present in solutions. The shift of the titration curve for m7GMP towards the higher concentrations, compared to the curve for m7GDP, indicates weaker binding of m7GMP by C. elegans DcpS.
To further determine the role of the phosphate groups in the interactions with DcpS proteins, we compared the binding affinities of m7GDP and m32,2,7GDP with mononucleoside monophosphates, m7GMP and m32,2,7GMP (Figure 4 and Table 1). We obtained that all DcpS proteins have a high binding affinity for m7GDP and significant differences were observed towards mono- and diphosphate mononucleosides. Human DcpS has a 6-fold higher affinity for m7GDP (KAS = 114 ± 8 μM−1) compared to m7GMP (KAS = 19 ± 1 μM−1).13 An even larger difference between the binding affinities of these two mononucleotides is observed for CeDcpS, m7GDP (KAS = 97.04 ± 12.52 μM−1) versus m7GMP (KAS = 8.00 ± 0.65 μM−1). AsDcpS has a higher affinity for m7GMP (KAS = 26.35 ± 1.84 μM−1) than HsDcpS and CeDcpS, however it is one order of magnitude lower than for m7GDP (KAS = 185.13 ± 16.48 μM−1). A significantly higher affinity for m7GDP than for m7GMP indicates the importance of the second phosphate group for cap recognition by DcpS proteins. The resistance of m7GDP to enzymatic hydrolysis and its strong binding to human and nematode (A. suum and C. elegans) decapping scavengers suggested that m7GDP might be a potential DcpS inhibitor.
Table 1.
Equilibrium association constants (KAS) for the complexes of mononucleotide cap analogs with the human and nematode DcpS, obtained from analysis of steady-state fluorescence titration at 20 °C, in 50 mM Tris HCl buffer containing 200 mM KCl, 0.5 mM EDTA and 1 mM DTT (final pH 7.6); nd – not determined.
| Cap analog |
KAS [μM−1]
|
||
|---|---|---|---|
| HsDcpS | AsDcpS | CeDcpS | |
| m7GMP | 19 ± 1 | 26.35 ± 1.84 | 8.00 ± 0.65 |
| m7GDP | 114 ± 8 | 185.13 ± 16.48 | 97.04 ± 12.52 |
| m32,2,7GMP | nd | 0.28 ± 0.27 | 2.88 ± 0.67 |
| m32,2,7GDP | nd | 0.53 ± 0.16 | 1.44 ± 0.26 |
Inhibitory properties of m7GDP in DcpS-mediated hydrolysis of m7GpppG
To characterize the inhibitory potency of m7GDP on DcpS proteins hydrolysis of m7GpppG, IC50 values were determined. The results, 3.95 ± 0.23 μM for yeast DcpS, 2.17 ± 0.13 μM for C. elegans, 1.44 ± 0.11 μM for A. suum and 4.17 ± 0.50 μM for human enzyme, indicate similar inhibitory properties of m7GDP towards all the decapping scavengers. To calculate inhibition constants Ki from IC50, we first determined the type of inhibition by comparing DcpS reaction velocities without inhibitor and in the presence of increasing inhibitor concentrations. The observed increasing Km value without affecting Vmax indicated m7GDP is a competitive inhibitor of the decapping scavengers. Inhibition constants were calculated for CeDcpS and ScDcpS using the IC50 values determined here and Km values obtained in the previous studies (1.17 ± 0.14 μM and 0.14 μM, respectively).12,20 The inhibition constants (Ki = 0.156 ± 0.020 μM for CeDcpS and 0.036 ± 0.002 μM for ScDcpS) demonstrate that m7GDP is a strong inhibitor of these enzymes.
DISCUSSION
Biological consequences of m7GDP resistance towards DcpS enzymes. DcpS-mediated hydrolysis of m7GDP to m7GMP described in previous studies led to the conclusion that scavenger decapping enzymes play roles in both eukaryotic mRNA decay pathways, converting m7GpppN to m7GMP in the 3′ → 5′ pathway and m7GDP to m7GMP in the 5′ → 3′ pathway.11,12 In contrast, the data from Cohen et al.8 and Liu et al.6 indicated that human and nematode DcpS enzymes are not able to hydrolyze m7GDP to m7GMP. Here, we reexamined these contradictory results through a careful analysis of m7GDP hydrolysis using highly purified recombinant decapping scavengers from human, A. suum, C. elegans and S. cerevisiae, and a highly sensitive analytical HPLC assay to monitor reaction progress and hydrolysis products.
All these DcpS enzymes very efficiently hydrolyze their natural substrate, m7GpppG (Figure 3). The m7GDP hydrolysis tests were performed under the same assay conditions as previously chosen by van Dijk et al.11 and also under conditions adopted for HsDcpS binding studies. Under both conditions, m7GDP and its trimethylated derivative, m32,2,7GDP, are resistant to DcpS hydrolysis, regardless of DcpS origin, as shown by HPLC analysis. Additional experiments using m7Gpppm7G (that is converted to m7GDP and m7GMP) further support that these scavenger decapping enzymes are not able to hydrolyze m7GDP. Based on the analysis of the four DcpS enzymes from different organisms (mammals, nematodes, fungi) we suggest that this is likely a general feature of scavenger decapping enzymes. Therefore, we conclude that DcpS enzymes generate m7GMP only from dinucleotide caps in the 3′ → 5′ decay pathway.
The reason of conflicting results obtained by various research groups with respect to the DcpS ability to hydrolyze m7GDP (Liu et al.6 and Cohen et al.8 versus van Dijk et al. 11 and Malys and McCarthy12) has not been discussed until now. The definite explanation for this disagreement is not possible, since some details in the description of experimental procedures are lacking in the previous studies. However, we can presume the most likely possible reasons for the differences observed in m7GDP susceptibility to DcpS mediated degradation. (1) The concentrations of substrates are not described by van Dijk et al.11 and Malys and McCarthy12, thus, the assays and kinetics of the hydrolysis in these cases might over estimate DcpS activity. (2) Our experiments are based on N-terminally His-tagged DcpS proteins compared to C-terminally His-tagged scavengers used by groups that observed the opposite results. Due to the C-terminal localization of the catalytic HIT motif it is possible that the C-terminally tagged proteins display an altered scavenger decapping activity. (3) The purity of the protein samples from different preparations may not be the same. Gel filtration procedure applied in our studies is an additional (to SDS-PAGE) control for the homogeneity of DcpS enzymes, not performed in the previously reported procedures.6,8,11,12 Moreover, we tested in comparative studies four DcpS orthologs (human, C. elegans, A. suum and S. cerevisiae) under a variety of conditions, including those used by the other groups, clearly obtaining m7GDP resistance to DcpS mediated hydrolysis, regardless of the protein source.
DcpS enzymes strongly bind m7GDP
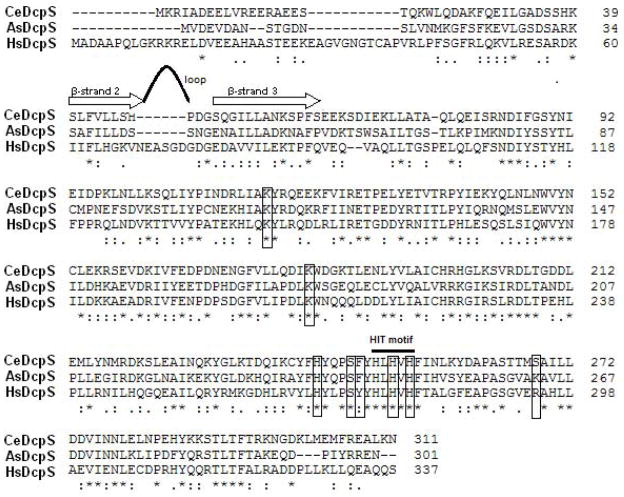
Another important finding from the current study is the strong binding of human and nematode DcpS enzymes for m7GDP. The association constants for the decapping scavengers are 97–185 μM−1. Such high KAS values lead us to conclude that m7GDP may efficiently bind DcpS proteins, and therefore competitively inhibit their hydrolytic activity. A sequence alignment of human and the two nematode DcpS enzymes (Figure 5), and the analysis of the crystallographic structure of HsDcpS-m7GDP complex25 suggests that the binding mode of m7GDP is likely the same, with respect to the interactions of the two phosphate groups with the conserved amino acids of the HIT fold domain.
Figure 5.
Amino acid alignment of human and nematode A. suum and C. elegans DcpS obtained using the Clustal 2.0.12 program illustrating conserved residues involved in the cap phosphate recognition and hydrolysis. The amino acids of each decapping scavenger are numbered on the right. Identical residues are indicated by stars below the lines. Conserved substitutions are indicated by (.) and semi-conserved by (:). Gaps in the alignment are illustrated as (−). Decapping scavenger enzymes constitute their own branch within the superfamily of pyrophosphatases containing a highly conserved HIT motif (His-ϕ-His-ϕ-His-ϕ, where ϕ is a hydrophobic residue) utilized to cleave the substrate’s phosphate chain. Hydrolysis is performed by a nucleophilic attack on the γ phosphate group and the cleavage of the β-γ phosphate bond as proposed by Lima et al.32 The DcpS amino acids involved in a binding of the cap’s phosphate chain, according to the resolved structure of HsDcpS,19 are given in frames. Most of the residues interacting with the triphosphate bridge in the human DcpS-m7GpppG complex are strictly conserved in CeDcpS and AsDcpS. The two non-conserved amino acids are Tyr273 (replaced by Phe in both nematode enzymes) and Arg294 (replaced by Lys in A. suum DcpS and Ser in C. elegans DcpS). Note that mutation of Tyr273 to Phe in human DcpS results in a protein with 120% of the wild-type enzyme activity,25 while mutation of Arg294 to Lys increases the activity to 114%,19 which may explain the higher activity of nematode DcpS enzymes comparing to the human one. Corresponding protein sequence between β-strand 2 and β-strand 3 of HsDcpS19 (strands are indicated as arrows above the alignment) is shortened by 6 amino acids in C. elegans and A. suum proteins. In the structure of human DcpS with m7GpppG or m7GpppA, this region forms an extended loop.19 In the apo-form of human and mouse DcpS proteins, this loop region was not observed in the electron density map, and thus was assumed to be disordered.25
Remarkably, DcpS binding affinity for 7-methylguanosine diphosphate, m7GDP, is higher than for its monophosphate counterpart, m7GMP, as indicated by the association constants (Table 1, Figure 4). Weaker interactions of m7GMP with DcpS seem to be very important during the catalytic cycle, triggering the conformational changes of the enzyme that enable product release. In addition, our binding affinity results (Table 1) are in a good agreement with the previously reported differences for inhibition of recombinant C. elegans decapping scavenger with m7GDP and m7GMP. Cohen et al.8 noted that competitor concentration for 50% inhibition of CeDcpS hydrolytic activity was about 20-fold higher for m7GMP (2.5 μM) than for m7GDP (0.12 μM).
Both trimethylated mononucleotides, m32,2,7GMP and m32,2,7GDP, are bound significantly weaker by the nematode decapping scavengers in comparison with their monomethylated counterparts. Binding affinity results (Table 1) are in a good agreement with the previously reported inhibition data for recombinant CeDcpS with mono- and trimethylated cap analogs.26 Ki values for m7GDP, m7GTP and m7GpppG were in the range 2.2–3.5 μM, approximately 10-fold lower as compared with Ki = 28 μM for m32,2,7GpppG, indicating a much more efficient inhibition by monomethylated species. Comparison of binding affinity of human and nematode DcpS for monomethylated and trimethylated mononucleotides clearly demonstrates that m7GDP could be an efficient inhibitor for DcpS.
m7GDP as a potent DcpS inhibitor
To characterize quantitatively the inhibitory potency of m7GDP, the IC50 values were determined. The results obtained for different DcpS enzymes (3.95 ± 0.23 μM for yeast, 2.17 ± 0.13 μM for C. elegans, 1.44 ± 0.11 μM for A. suum and 4.17 ± 0.50 μM for human enzyme) indicated that m7GDP efficiently inhibits the activity of decapping scavengers from different species and acts as a competitive inhibitor. The calculated Ki values (0.156 ± 0.020 for μM CeDcpS and 0.036 ± 0.002 μM for ScDcpS) clearly show the high inhibiting potency of m7GDP towards decapping scavenger enzymes.
Our results are consistent with the previous data indicating the inhibitory properties of m7GDP, and expand these observations to other species in addition to C. elegans.8,26 Furthermore, the inhibition constant for m7GDP presented by Kwasnicka et al.26 was determined for C. elegans DcpS-mediated hydrolysis of a non-biological m7GpppBODIPY cap analog. Our studies examined the more biologically relevant m7GpppG substrate.
Comparative characterization of hydrolytic activity of HsDcpS, AsDcpS and CeDcpS enzymes
Our experiments on the recombinant A. suum DcpS provide an insight into the similarities and differences of the DcpS enzyme from a parasite nematode versus DcpS protein from a potential host organism (HsDcpS), as well as versus DcpS protein from another nematode – C. elegans model organism (CeDcpS). Hydrolytic properties of human and nematode (A. suum and C. elegans) decapping scavengers were compared using non-, mono- and trimethylated cap analogs, containing two or three phosphate groups (Figure 3). It has been shown previously that human DcpS is not able to hydrolyze TMG-caps, in contrast to nematode decapping scavengers.8 HsDcpS exhibits much higher substrate specificity than nematode decapping scavengers (Figure 3). It hydrolyzes only monomethylated cap analogs containing at least a triphosphate chain, with a preference towards dinucleotide m7GpppG, which is cleaved with ~2-fold higher efficiency than m7GTP. In this study, we present for the first time a significant difference in monomethylated and trimethylated cap analogs specificity between nematode enzymes. A. suum DcpS hydrolyzes more efficiently m7GpppG than m32,2,7GpppG, in contrast to its C. elegans counterpart (Figure 3). These data are supported by activities of the two enzymes on mononucleoside triphosphates, m7GTP and m32,2,7GTP (Figure 3).
The previous kinetic studies of human and C. elegans DcpS indicated that the rate of the cap hydrolysis catalyzed by DcpS does not depend on the type of the second nucleoside.20,27 The fact that the rate of m7GpppG degradation compared to m7GTP hydrolysis is only 2-fold greater for all the DcpS enzymes studied further confirms that the second nucleoside is not crucial for DcpS activity. The presence of the second nucleoside increases efficiency of the hydrolysis, but is not absolutely necessary.
The DcpS enzyme from the parasitic A. suum nematode appears much more closely related to human DcpS than the enzyme from C. elegans. This is especially illustrated with respect to the substrate specificity for the N7 methyl group of a cap analog, since GpppG is not a substrate for either HsDcpS or AsDcpS, but undergoes some hydrolysis by CeDcpS. Our result is likewise reflected by the higher sequence identity of AsDcpS and HsDcpS (42%), than for CeDcpS comparing to HsDcpS (35%) (Figure 5).
The role of a cap’s phosphate chain length in the DcpS-mediated enzymatic hydrolysis of cap analogs
Our results indicate the importance of the length of the cap’s phosphate chain for DcpS binding and hydrolysis. Notably, the DcpS binding pocket does not require a triphosphate chain for cap analog binding. From the determined KAS constants, we conclude that 7-methylguanosine diphosphate and 7-methylguanosine monophosphate are recognized by different DcpS enzymes. However, the binding affinity is significantly higher for m7GDP than for m7GMP. From our studies, it is also clear that although DcpS enzymes have a high affinity for the two phosphate groups of m7GDP, the diphosphate chain is not sufficient for mononucleotide cap hydrolysis. Consistently, DcpS does not hydrolyze m7GDP, but does degrade the triphosphate m7GTP. An important role of the triphosphate bridge of the cap for efficient hydrolysis by DcpS is also reflected in the observation that most of the residues interacting with the phosphate groups in human DcpS-m7GpppG complex (Lys142, Lys207, His268, Ser272, His277 and His279)19 are strictly conserved in CeDcpS and AsDcpS enzymes (Fig. 5) and many of them are crucial for efficient hydrolysis. The two conserved histidine residues of the HIT motif, His277 and His279, are essential for decapping activity. His277 is the nucleophile that attacks the cap’s γ phosphate in the catalytic cycle whereas His279 and His268 make direct contacts to the γ phosphate of the substrate. His268Asn mutation and also Lys207Ala substitution (Lys207 is responsible for α phosphate binding) drastically decrease catalytic activity of human decapping scavenger (to 8% and 28%, respectively), whereas both His277Asn and His279Asn substitutions completely inactivate the DcpS enzyme.19 Moreover, mutation of Tyr273 (interacting with cap’s γ-phosphate) suggests that this amino acid also plays an important role in the catalytic cycle. Tyr273 undergoes significant conformational changes upon cap binding, contributing to the formation of the closed conformation, required for the hydrolytic activity.25 Mutation of Tyr273 to Phe in human DcpS resulted in a protein with 120% of the wild-type enzyme activity, while substitution by Ala reduced the activity to 16% .25 It was suggested, that mutation of Tyr273 to Ala disrupts interactions important for the formation of the closed configuration of DcpS active site. Mutation of Tyr273 to Phe has little effect on its interactions with residues in the closed state, but enhances the interactions in the open conformation facilitating the product release. Interestingly, Tyr 273 is replaced by Phe in both nematode DcpS enzymes (Figure 5). The second non-conserved amino acid involved in binding of a triphosphate chain (β-phosphate) in human DcpS-m7GpppG complex, Arg294, is replaced by Lys in A. suum and Ser in C. elegans DcpS. Remarkably, the Arg294Lys substitution increases human DcpS activity (114%).19 These few structural differences between nematode and human enzyme’s cap triphosphate chain-binding pocket do not significantly change the substrate specificity of nematode decapping scavengers, but may explain their higher hydrolytic activity comparing to human DcpS.
Degradation pathways of mRNA
Efficient conversion of m7GDP to m7GMP has been previously demonstrated in yeast, Xenopus and human cell extracts, and decapping scavengers were suggested as the enzymes catalyzing this reaction.11,28 Furthermore, data have also been described suggesting that recombinant DcpS proteins are also able to hydrolyze m7GDP.11,12 These observations led to a model of eukaryotic mRNA degradation in which DcpS acts in both major mRNA decay pathways, generating m7GMP from m7GpppN in the 3′ → 5′ pathway and from m7GDP in the 5′ → 3′ pathway.11
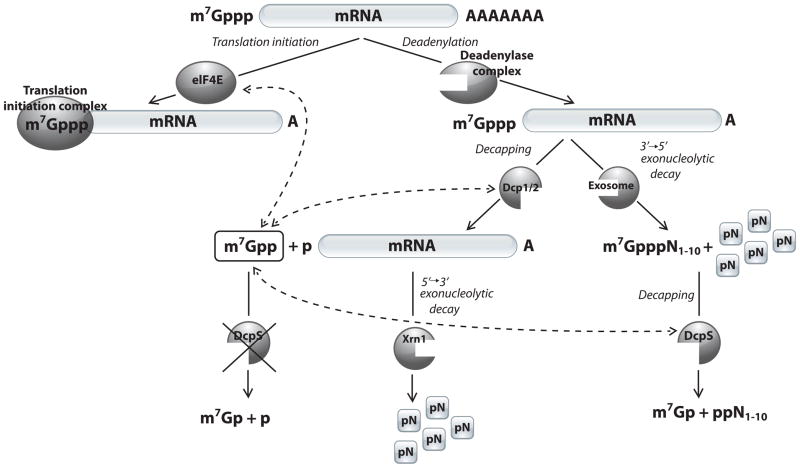
Our data demonstrate that m7GDP is resistant to DcpS hydrolysis, regardless of the DcpS origin. Moreover, m7GDP was found to be an efficient competitive inhibitor of human, nematode and yeast decapping scavengers. We therefore propose an update to the role of DcpS in mRNA degradation pathways. According to our model (Figure 6), m7GDP, the decapping product of the 5′ → 3′ mRNA decay, is not hydrolyzed by DcpS enzymes. Decapping scavengers produce m7GMP only from short capped oligonucleotides generated from 3′ → 5′ mRNA decay. As DcpS cannot hydrolyze m7GDP, there must be another yet to be identified enzymatic activity to protect cells from accumulation of this mononucleotide. The alternate possibility of m7GDP turnover could potentially be a phosphorylation of m7GDP to m7GTP (e.g. by nucleoside diphosphate kinase) and the subsequent DcpS mediated m7GTP to m7GDP, as it was shown herein.29,30
Figure 6.
An amended model of mRNA degradation pathways. mRNAs in all eukaryotes posses the m7GpppN cap at their 5′ ends. In nematodes, a significant portion of the mRNAs possesses a trimethylated cap (m32,2,7GpppG). mRNA is degraded either at 5′ → 3′ or 3′ → 5′ pathway, both initiated by the deadenylation. The 5′ → 3′ pathway is initiated by Dcp2 pyrophosphatase cleavage of the 5′ mRNA cap. In 3′ → 5′ pathway, mRNA is progressively cleaved by the exosome resulting in residual short capped oligonucleotides that are subsequently degraded by DcpS, producing m7GMP. In the present study, we show that neither m7GDP nor m32,2,7GDP are substrates for DcpS. However, m7GDP may interact with cap-binding proteins, as indicated by the dashed lines with arrowheads. The high binding affinity and strong inhibitory properties of m7GDP towards DcpS make this mononucleoside diphosphate a potential regulator of cap-dependent cellular processes. We suggest that m7GDP may inhibit DcpS and consequently affect the function of other cap-binding proteins, such as Dcp233 or eIF4E34, through the increased levels of the free cap species.
Although DcpS enzymes cannot hydrolyze m7GDP, they have a high binding affinity for m7GDP as indicated by the association constants described here (Table 1). The binding affinity of DcpS enzymes from different organisms for m7GDP is very similar. We have also shown that the m7GDP inhibitory effect on DcpS depends on the m7GDP concentration and that m7GDP is a competitive inhibitor of DcpS at the low micromolar level. The cellular concentration of m7GDP remain to be determined. To quantify the absolute level of this mononucleotide in a cell, numerous measurements would need to be carried out under a variety of developmental, environmental and genetic conditions. However, the overall levels of the extractable nucleotides may not provide insight into the local concentrations of the nucleotides in the context of proteins they may interact with. We note that current models of methylated nucleotide inhibition of eIF4E have been widely discussed and presented without discrete data on the nucleotide concentrations.28,31 Our data suggests the potential importance of m7GDP as an inhibitor of DcpS, that m7GDP is not degraded by DcpS, and that this data and its implications alone provide new insight into mRNA turnover.
Acknowledgments
Funding
This work was supported by grants from the Polish Ministry of Science and Higher Education (N N301 096339 to E.D.; N N204 089438 to J.J.), the National Institute of Health (AI49558 to R.E.D.) and co-financed with the European Union funds by the European Social Fund.
We thank Martin Bisaillon for advice and helpful discussions. The authors are grateful to Katarzyna Wnek, Anna Nowicka, Dominika Strzelecka, Aleksandra Roguska and Magdalena Wypijewska for their contribution to HPLC measurements and the total commitment to work.
ABBREVIATIONS
- A. suum
Ascaris suum
- BSA
bovine serum albumin
- C. elegans
Caenorhabditis elegans
- DcpS
decapping scavenger
- DTT
dithiothreitol
- EDTA
ethylenediaminetetraacetic acid
- FPLC
fast protein liquid chromatography
- HPLC
high performance liquid chromatography
- IC50
concentration of inhibitor causing the decrease of substrate conversion of 50%
- IPTG
isopropyl β-D-1-thiogalactopyranoside
- m7GDP
7-methylguanosine diphosphate
- m32,2,7GDP
2,2,7-trimethylguanosine diphosphate
- MMG
monomethylguanosine
- S. cerevisiae
Saccharomyces cerevisiae
- SMA
spinal muscular atrophy
- TMG
trimethylguanosine
- TST
time synchronized titration
References
- 1.Meyer S, Temme C, Wahle E. Messenger RNA turnover in eukaryotes: pathways and enzymes. Crit Rev Biochem Mol Biol. 2004;39:197–216. doi: 10.1080/10409230490513991. [DOI] [PubMed] [Google Scholar]
- 2.Couttet P, Fromont-Racine M, Steel D, Pictet R, Grane T. Messenger RNA deadenylation precedes decapping in mammalian cells. Proc Natl Sci Acad USA. 1997;94:5628–5633. doi: 10.1073/pnas.94.11.5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parker R, Song H. The enzymes and control of eukaryotic mRNA turnover. Nat Struct Mol Biol. 2004;11:121–127. doi: 10.1038/nsmb724. [DOI] [PubMed] [Google Scholar]
- 4.Wang Z, Kiledjian M. Functional link between the mammalian exosome and mRNA decapping. Cell. 2001;107:751–762. doi: 10.1016/s0092-8674(01)00592-x. [DOI] [PubMed] [Google Scholar]
- 5.Cougot N, Babajko S, Seraphin B. Cap-tabolism. Trends Biochem Sci. 2004;29:436–444. doi: 10.1016/j.tibs.2004.06.008. [DOI] [PubMed] [Google Scholar]
- 6.Liu H, Rodgers ND, Jiao X, Kiledjian M. The scavenger mRNA decapping enzyme DcpS is a member of the HIT family of pyrophosphatases. EMBO J. 2002;21:4699–4708. doi: 10.1093/emboj/cdf448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu SW, Jiao X, Liu H, Gu M, Lima CD, Kiledjian M. Functional analysis of mRNA scavenger decapping enzyme. RNA. 2004;10:1412–1422. doi: 10.1261/rna.7660804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cohen LS, Mikhli C, Friedman C, Jankowska-Anyszka M, Stepinski J, Darzynkiewicz E, Davis RE. Namatode m7GpppG and m32,2,7GpppG decapping: Activities in Ascaris embryos and characterization of C. elegansDcpS. RNA. 2004;10:1609–1624. doi: 10.1261/rna.7690504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crompton DWT. Ascaris and ascariasis. Adv Parasitol. 2001;48:285–375. doi: 10.1016/s0065-308x(01)48008-0. [DOI] [PubMed] [Google Scholar]
- 10.Singh J, Salcius M, Liu S-W, Staker BL, Mishra R, Thurmod J, Michaud G, Mattoon DR, Printen J, Christensen J, et al. DcpS as a therapeutic target for Spinal Muscular Atrophy. ACS Chem Biol. 2008;3:711–722. doi: 10.1021/cb800120t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Dijk E, Le Hir H, Seraphin B. DcpS can act in the 5′ 3′ mRNA decay pathway in addition to the 3′ 5′ pathway. Proc Natl Acad Sci USA. 2003;100:12081–12086. doi: 10.1073/pnas.1635192100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Malys N, McCarthy JE. Dcs2, a novel stress-induced modulator of m7GpppX pyrophosphate activity that locates to P bodies. J Mol Biol. 2006;363:370–382. doi: 10.1016/j.jmb.2006.08.015. [DOI] [PubMed] [Google Scholar]
- 13.Darzynkiewicz ZM, Bojarska E, Kowalska J, Lewdorowicz M, Jemielity J, Kalek M, Stepinski J, Davis RE, Darzynkiewicz E. Interaction of human decapping scavenger with 5′ mRNA cap analogues: structural requirements for catalytic activity. J Phys: Condens Matter. 2007;19:285217. [Google Scholar]
- 14.Jankowska M, Stepinski J, Stolarski R, Temeriusz A, Darzynkiewicz E. Synthesis and properties of new NH2 and N7 substituted GMP and GTP 5′-mRNA cap analogues. Collect Czech Chem Commun. 1993;58:138–141. [Google Scholar]
- 15.Stepinski J, Bretner M, Jankowska M, Felczak K, Stolarski R, Wieczorek Z, Cai A-L, Rhoads RE, Temeriusz A, Haber D, Darzynkiewicz E. Synthesis and properties of P1,P2-, P1,P3- and P1,P4-dinucleoside di-, tri- and tetraphosphate mRNA 5′-cap analogues. Nucleosides & Nucleotides. 1995;14:717–721. [Google Scholar]
- 16.Niedzwiecka A, Stepinski J, Antosiewicz JM, Darzynkiewicz E, Stolarski R. Biophysical approach to studies of cap-eIF4E interaction by synthetic cap analogues. Methods Enzymol. 2007;430:209–246. doi: 10.1016/S0076-6879(07)30009-8. [DOI] [PubMed] [Google Scholar]
- 17.Cai A, Jankowska-Anyszka M, Centers A, Chlebicka L, Stepinski J, Stolarski R, Darzynkiewicz E, Rhoads R. Quantitative assessment of mRNA cap analogues as inhibitors of in vitro translation. Biochemistry. 1999;38:8538–8547. doi: 10.1021/bi9830213. [DOI] [PubMed] [Google Scholar]
- 18.Darzynkiewicz E, Stepinski J, Tahara SM, Stolarski R, Ekiel I, Haber D, Neuvonen K, Lehikoinen P, Labadi I, Lönnberg H. Synthesis, conformation and hydrolytic stability of P1,P3-dinucleoside triphosphates related to mRNA 5′-cap, and comparative kinetic studies on their nucleoside and nucleoside monophosphate analogs. Nucleosides & Nucleotides. 1990;9:599–618. [Google Scholar]
- 19.Gu MG, Fabrega C, Liu SW, Liu HD, Kiledjian M, Lima CD. Insight into the structure, mechanism, and regulation of scavenger mRNA decapping activity. Mol Cell. 2004;14:67–80. doi: 10.1016/s1097-2765(04)00180-7. [DOI] [PubMed] [Google Scholar]
- 20.Wypijewska A, Bojarska E, Stepinski J, Jankowska-Anyszka M, Jemielity J, Davis RE, Darzynkiewicz E. Structural requirements for Caenorhabditis elegans DcpS subtrates based on fluorescence and HPLC enzyme kinetic studies. FEBS Journal. 2010;277:3003–3013. doi: 10.1111/j.1742-4658.2010.07709.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 22.Niedzwiecka A, Marcotrigiano J, Stepinski J, Jankowska-Anyszka M, Wyslouch-Cieszynska A, Dadlez M, Gingras AC, Mak P, Darzynkiewicz E, Sonenberg N, Burley SK, Stolarski R. Biophysical studies of eIF4E cap-binding protein: recognition of mRNA 5′ cap structure and synthetic fragments of eIF4G and 4E-BP1 proteins. J Mol Biol. 2002;319:615–635. doi: 10.1016/S0022-2836(02)00328-5. [DOI] [PubMed] [Google Scholar]
- 23.Demchenko AP. Introduction to Fluorescence Sensing. Springer Science and Business Media B.V; 2009. pp. 66–76. [Google Scholar]
- 24.Copeland RA. A Practical Introduction to Structure, Mechanism, and Data Analysis. John Wiley & Sons, INC; 2000. Enzymes; pp. 305–317. [Google Scholar]
- 25.Chen N, Walsh MA, Liu YY, Parker R, Song HW. Crystal structures of human DcpS in ligand free and m7GDP-bound forms suggest a dynamic mechanism for scavenger mRNA decapping. J Mol Biol. 2005;347:707–718. doi: 10.1016/j.jmb.2005.01.062. [DOI] [PubMed] [Google Scholar]
- 26.Kwasnicka DA, Krakowiak A, Thacker C, Brenner C, Vincent SR. Coordinate expression of NADPH-dependent flavin reductase Free-1 and Hint related 7meGMP-directed hydrolase, DCS-1. J Biol Chem. 2003;278:39051–39058. doi: 10.1074/jbc.M306355200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Darzynkiewicz ZM, Bojarska E, Stepinski J, Jankowska-Anyszka M, Davis RE, Darzynkiewicz E. Affinity of dinucleotide cap analogues for human decapping scavenger (hDcpS) Nucleosides, Nucleotides and Nucleic Acids. 2007;26:1349–1352. doi: 10.1080/15257770701533818. [DOI] [PubMed] [Google Scholar]
- 28.Liu H, Kiledjian M. Scavenger decapping activity facilitates 5′ to 3′ mRNA decay. Mol Cell Biol. 2005;25:9764–9772. doi: 10.1128/MCB.25.22.9764-9772.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen CY, Gherzi R, Ong SE, Chan EL, Raijmakers R, Pruijn GJ, Stoecklin G, Moroni C, Mann M, Karin M. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell. 2001;107:451–464. doi: 10.1016/s0092-8674(01)00578-5. [DOI] [PubMed] [Google Scholar]
- 30.van Dijk E, Cougot N, Meyer S, Babajko S, Wahle E, Seraphin B. Human Dcp2: a catalytically active mRNA decapping enzyme located in specific cytoplasmic structures. EMBO J. 2002;21:6915–6924. doi: 10.1093/emboj/cdf678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bail S, Kiledjian M. DcpS, a general modulator of cap-binding protein-dependent processes? RNA Biology. 2008;5:216–219. doi: 10.4161/rna.7161. [DOI] [PubMed] [Google Scholar]
- 32.Lima CD, Klein MG, Hendrickson WA. Structure-based analysis of catalysis and substrate properties definition in the HIT protein family. Science. 1997;278:286–290. doi: 10.1126/science.278.5336.286. [DOI] [PubMed] [Google Scholar]
- 33.Floor SN, Jones BN, Hernandez GA, Gross JD. A split active site couples cap recognition by Dcp2 to activation. Nat Struc Mol Biol. 2010;17:1096–1101. doi: 10.1038/nsmb.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grudzien E, Kalek M, Jemielity J, Darzynkiewicz E, Rhoads RE. Differential inhibition of mRNA degradation pathways by novel cap analogs. J Biol Chem. 2006;27:1857–1867. doi: 10.1074/jbc.M509121200. [DOI] [PubMed] [Google Scholar]