Abstract
In this work [14C]spermidine binding to total proteins solubilized from plasma membrane purified from zucchini (Cucurbita pepo L.) hypocotyls was investigated. Proteins were solubilized using octyl glucoside as a detergent. Specific polyamine binding was thermolabile, reversible, pH dependent with an optimum at pH 8.0, and had a Kd value of 5 μm, as determined by glass-fiber-filter assays. Sephadex G-25 M gel-filtration assays confirmed the presence of a spermidine-protein(s) complex with a specific binding activity. By sodium dodecyl sulfate-polyacrylamide gel electrophoresis and native polyacrylamide gel electrophoresis of collected fractions having the highest specific spermidine-binding activity, several protein bands (113, 75, 66, and 44 kD) were identified. The specificity of spermidine binding was examined by gel-filtration competition experiments performed using other polyamines and compounds structurally related to spermidine. Partial purification on Sephadex G-200 led to the identification of 66- and 44-kD protein bands, which may represent the putative spermidine-binding protein(s) on the plasmalemma.
Polyamines are biologically ubiquitous compounds that are implicated in many aspects of growth and development in a wide range of organisms (microorganisms, animals, and plants) (Tabor and Tabor, 1985; Bagni, 1989; Persson et al., 1996), although their specific mechanism of action is not well understood. By virtue of their cationic nature at physiological pH, they can interact with several molecules and thereby affect their structure and function. Hydrogen binding, ionic and covalent linkages, and hydrophobic interactions may occur that involve the charged functional amino and imino groups and the tetra- and trimethylene chains (Feuerstein and Marton, 1989; Schuber, 1989; Serafini-Fracassini et al., 1995).
One possible mechanism by which polyamines might act as growth substances involves their binding to specific regulatory proteins, as has been observed in many organisms. In both animal and plant systems it has been postulated that these compounds may play a role in the posttranslational modifications of proteins (Äschlimann and Paulsson, 1994; Serafini-Fracassini et al., 1995), as well as in the modulation of many enzyme activities such as protein kinases, phosphatases (Datta et al., 1986; Friedman, 1986), and 1,3-β-glucan synthase (Kauss and Jeblick, 1985). Despite the large amount of information on the covalent interactions between polyamines and proteins, little is known about the noncovalent binding of polyamines to PM proteins, even though this may represent one of the first steps in their action at the cellular level.
In Escherichia coli two periplasmic polyamine-binding proteins (PotD and PotF) have been isolated as part of two membrane transport systems, pPT104 and pPT79, respectively (Kashiwagi et al., 1993; Pistocchi et al., 1993b). In particular, the crystal structure of PotD, which strongly binds spermidine, was recently determined (Sugiyama et al., 1996). This crystallographic study led to the elucidation of the mechanism of spermidine recognition and the characteristics of the main chain folding at the binding site.
Nevertheless, no information is available on polyamine-binding proteins of plant membranes. In a previous paper, the main characteristics of spermidine binding to purified zucchini (Cucurbita pepo L.) PM vesicles were described (Tassoni et al., 1996). Results indicated that the specific interaction was saturable, reversible, heat labile, and detergent- and pronase-sensitive, thus suggesting that the binding occurred with a protein component of the PM.
In this paper we have defined the features of specific spermidine binding to solubilized PM proteins. This represents a necessary step before purification of the specific polyamine-binding protein(s). Moreover, a partial purification of the specific spermidine-binding protein(s) was performed to identify the putative receptor(s) and/or transporter(s) on the plant plasmalemma.
MATERIALS AND METHODS
Seeds of zucchini (Cucurbita pepo L. hybrid Strorr's Green, Asgrow Co., Lodi, Italy) were planted in moist vermiculite and grown at 22 ± 1°C in total darkness for 7 d. Etiolated hypocotyls (6–12 cm long) were excised and stored at −20°C until use.
Isolation of PM Vesicles, Solubilization of Membrane Proteins, and Polyamine Analysis
A very pure PM-enriched fraction was obtained after the aqueous phase-partitioning procedure described by Tassoni et al. (1996). Buffer A, containing 50 mm Tris-HCl, pH 7.5, 2.0 mm EDTA, 250 mm Suc, and 10 mm 2-mercaptoethanol, was slightly modified by adding a mixture of protease inhibitors, namely 0.2 mm PMSF, 5 mm 6-amino-n-hexanoic acid (Gray, 1982), 1 mm benzamidine, 10 μg mL−1 N-α-p-tosyl-l-Arg methyl ester, and 10 μg mL−1 N-α-benzoyl-l-Arg methyl ester (Miller et al., 1995) (all from Sigma). PM vesicles were resuspended in buffer A at pH 8.0 and stored at −80°C until use. For protein solubilization, 1% (w/v) OG was added to purified PM vesicles (final protein concentration 2 mg mL−1). The solution was sonicated twice for 30 s by means of an ultrasonic disintegrator (20 kHz, amplitude 10 μm peak to peak, model 150-W, Measuring and Scientific Equipment Ltd., Crawley, UK), left on ice for 30 min, and then centrifuged at 20,000g for 30 min at 4°C. About 95% of the total PM proteins was recovered in a soluble form. Protein content was determined by the method of Lowry et al. (1951) with BSA as a standard.
Polyamines noncovalently (PCA-soluble fraction) and covalently bound to solubilized proteins (about 2 mg mL−1) were determined by HPLC analysis after dansylation, as described by Scaramagli et al. (1995). In particular, polyamines covalently bound (PCA-insoluble fraction) were determined after hydrolysis at 110°C for 20 h with 6 n HCl in flame-sealed vials.
Binding and Dissociation Assay
The standard binding assay (1 mL final volume) contained 0.2 mg of protein in buffer A, pH 8.0, and 5.55 kBq [14C]spermidine (specific activity 4.07 GBq mmol−1; Amersham) with or without 1 mm unlabeled spermidine to give total and nonspecific binding (samples A and B, respectively). Samples were incubated on ice for 5 min and then directly filtered through glass-fiber filters (Whatman GF/B) previously soaked for about 2 h in 0.3% (v/v) polyethylenimine (Sigma) (Bruns et al., 1983), and placed in a Büchner funnel connected to a vacuum pump. After rapid filtration, filters were rinsed with 5 mL of buffer A, pH 8.0, to wash out the unbound spermidine, and then placed overnight in 2 mL of scintillation cocktail (Ultima Gold, Canberra Packard, Groningen, The Netherlands) before radioactivity was determined in a scintillation counter (model LS 1800, Beckman). Specific binding was obtained by subtracting the radioactivity of sample B (nonspecific binding) from that of sample A (total binding). All binding experiments were repeated at least twice with triplicate samples.
To study the dissociation rate, total and nonspecific samples were incubated until a steady-state level of occupancy was achieved, then 4 mm unlabeled spermidine was added and samples were filtered as described above after different dissociation times.
Gel-Filtration Assays
Solubilized proteins (about 1 mg mL−1) were incubated for 5 min on ice with 7.4 kBq [14C]spermidine in the absence or presence of 1.7 mm unlabeled spermidine for total and nonspecific binding assays, respectively (0.6 mL final volume). To verify that a good separation of bound ligand from free ligand occurred, blank assays without solubilized proteins were performed. The mixtures were then applied to Sephadex G-25 medium prepacked columns (9-mL bed volume, PD-10, Pharmacia) previously equilibrated with about 25 mL of buffer A, pH 8.0, to which 0.01% (w/v) OG was added to prevent the detergent concentration from falling below the critical micellar concentration (Hulme, 1992). Column elution was carried out with buffer A and 20 fractions, each of about 0.4 mL, were recovered. Radioactivity in a 0.2-mL aliquot of each fraction was determined by liquid-scintillation counting, whereas the remainder of the fraction was used for A280 measurement. Specific binding activity was calculated as the difference between total and nonspecific values. Fractions showing the highest specific spermidine-binding activity were then analyzed by 12% (w/v) SDS-PAGE (Laemmli, 1970) and native PAGE (Hendrick and Smith, 1968). Gels were stained with silver salts (Sammons et al., 1981) and then analyzed by densitometry using Molecular Analyst/PC Image Analysis software (version 1.1.1, 1992–1994, model GS-670, Bio-Rad).
Sephadex G-25 gel-filtration competition experiments were performed between labeled spermidine and the following analog and polyamines (20 μm final concentration): MGBG, putrescine, spermidine, and spermine. Sephadex G-25 gel-filtration fractions (3.0 mL, approximately 0.4 mg mL−1 protein) with the highest specific spermidine-binding activity were incubated for 5 min on ice with 18.5 kBq [14C]spermidine. They were applied to a column (40 cm × 1.6 cm) of Sephadex G-200 equilibrated in buffer A with 0.01% (w/v) OG and maintained at 3.5°C. Elution was carried out with the same buffer. Fractions (0.4 mL) were collected and analyzed for radioactivity and A280 as described above for Sephadex G-25 fractions. SDS-PAGE (12%, w/v) and silver staining were performed on fractions that showed the highest spermidine-binding activity.
ATPase, ADC, and ODC Activities
Vanadate-sensitive ATPase activity in PM vesicles, solubilized proteins, and fractions having maximum spermidine-binding activity was determined as described by Tassoni et al. (1996). ADC and ODC activities were assayed as described by Altamura et al. (1993): for a 2 mm final concentration, unlabeled Arg and Orn, respectively, were added to the assay mixture.
RESULTS AND DISCUSSION
Solubilization of Membrane Proteins and Polyamine Content
Three detergents (Chaps, Triton X-100, and OG) at 0.3 and 1% concentration were tested on purified PM vesicles to establish the best condition for solubilizing zucchini PM proteins. Chaps and OG were more effective than Triton X-100 at all concentrations tested (data not shown). Treatment with 0.3% (w/v) Chaps or 1% (w/v) OG led to the recovery of more than 95% of total PM proteins in the supernatant, whereas with 0.3% (w/v) OG or 0.3 and 1% (v/v) Triton X-100, the yield of proteins was less than 90%. OG (1%, w/v) was chosen for subsequent work because of the greater stability of many membrane proteins in nonionic detergents (Thomas and McNamee, 1990), and because this detergent has been successfully used to solubilize other plant PM proteins in an active form (de Boer et al., 1989).
Polyamines noncovalently (PCA-soluble fraction) and covalently (PCA-insoluble fraction) bound to solubilized proteins were determined (Table I). All three polyamines (putrescine, spermidine, and spermine) were more abundant in the noncovalently bound form, and the spermidine content was 2- and 16-fold higher than putrescine and spermine, respectively. This reflects the pattern of polyamine content previously reported for zucchini PM vesicles (Tassoni et al., 1996), and suggests that very few polyamines are bound to acidic PM phospholipids. The content of noncovalently bound spermidine was about 22-fold higher than that of covalently bound spermidine (Table I).
Table I.
Noncovalently and covalently bound polyamine content in proteins solubilized from zucchini PM vesicles
| Polyamine | Noncovalently Bound (PCA-soluble fraction) | Covalently Bound (PCA-insoluble fraction) |
|---|---|---|
| nmol mg−1 protein | ||
| Putrescine | 1.53 ± 0.39 | 0.21 ± 0.07 |
| Spermidine | 3.27 ± 1.11 | 0.15 ± 0.07 |
| Spermine | 0.21 ± 0.09 | 0.04 ± 0.02 |
The values are means ± sd of four samples of two replicate experiments.
Characteristics of [14C]Spermidine Binding to Solubilized Proteins
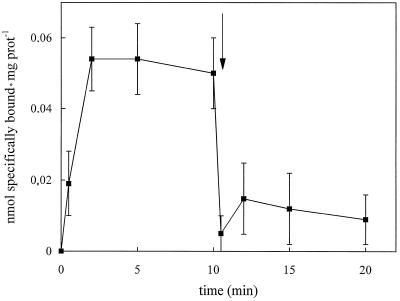
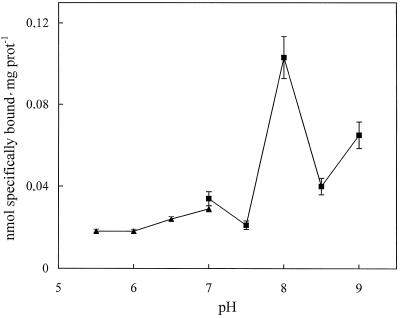
Preliminary results indicated that OG in the range between 0.8% and 1% (w/v), which coincides with the detergent concentration used for protein solubilization, gave the maximum specific spermidine-binding activity (data not shown). Thus, 1% (w/v) OG was used in subsequent binding assays. Figure 1 shows the association of [14C]spermidine to total solubilized proteins. Steady-state binding was reached after 2 min and, upon addition of an excess of unlabeled spermidine (Fig. 1, arrow), the labeled polyamine rapidly dissociated and thereafter specific binding remained constant. This showed that the specific interaction between spermidine and PM proteins is reversible. In Figure 2 the effect of pH on spermidine binding, using Tris-Mes buffer in the range from pH 5.5 to 7.0 and Tris-HCl buffer from pH 7.0 to 9.0, is reported. Specific spermidine binding to solubilized proteins is pH dependent, with an optimum at pH 8.0.
Figure 1.
Kinetics of specific [14C]spermidine binding to total proteins solubilized from zucchini PM vesicles. The assay was performed at pH 8.0. The dissociation of specifically bound spermidine was started by adding 4 mm unlabeled spermidine (arrow). The values are means ± sd of triplicate samples from two replicate experiments. prot, Protein.
Figure 2.
Effect of pH on specific [14C]spermidine binding to total proteins solubilized from zucchini PM vesicles. The assay was performed with Tris-Mes buffer from pH 5.5 to 7.0 (▴) and with Tris-HCl buffer from pH 7.0 to 9.0 (▪). The values are means ± sd of triplicate samples from two replicate experiments. prot, Protein.
Both the reversibility and the pH dependence of specific spermidine binding reflect properties previously found in PM-enriched fractions (Tassoni et al., 1996). Heat lability was also similar in solubilized proteins, specific binding activity being reduced to one-half in assays performed at 40°C compared with those carried out on ice (data not shown). Similar pH dependence has been observed with respect to putrescine and spermidine uptake in several systems, including sugar beet seedlings (Christ et al., 1989), carrot cell cultures, African violet petals (Pistocchi et al., 1993a), the lichen Evernia prunastri (Escribano and Legaz, 1985), and the seaweed Ulva rigida (Badini et al., 1994). This suggests that binding proteins in zucchini may be part of a polyamine-transport system in the plasmalemma that consists of different proteins. It may be analogous to the system in Escherichia coli, in which PotD protein, a primary receptor of the spermidine preferential transport system pPT104, has been characterized and crystallized (Sugiyama et al., 1996).
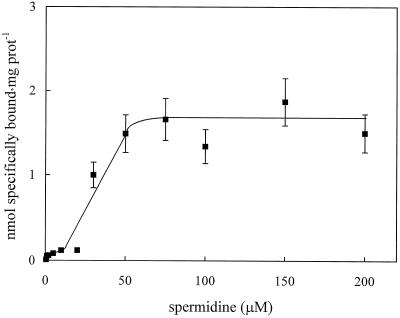
The concentration dependence of spermidine binding to solubilized proteins (Fig. 3) showed that saturation was reached at 50 μm polyamine concentration with an apparent Kd of 5 μm (EBDA program, version 3.0, 1983, GA McPherson [Elsevier-Biosoft, Cambridge, UK]). This value is 1 order of magnitude higher in affinity than that found in PM fractions (Kd approximately 40 μm; Tassoni et al., 1996), but is similar to the Kd value observed for the PotD protein in E. coli (Kashiwagi et al., 1993).
Figure 3.
Concentration dependence of specific [14C]spermidine binding to total proteins solubilized from zucchini PM vesicles. Different amounts of unlabeled spermidine were added to 5.55 kBq [14C]spermidine to give the final spermidine concentrations shown. The values are means ± sd of multiple samples from three replicate experiments. prot, Protein.
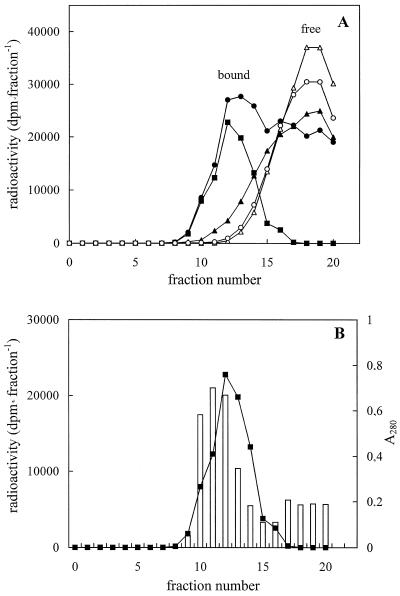
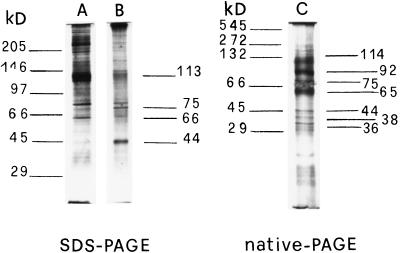
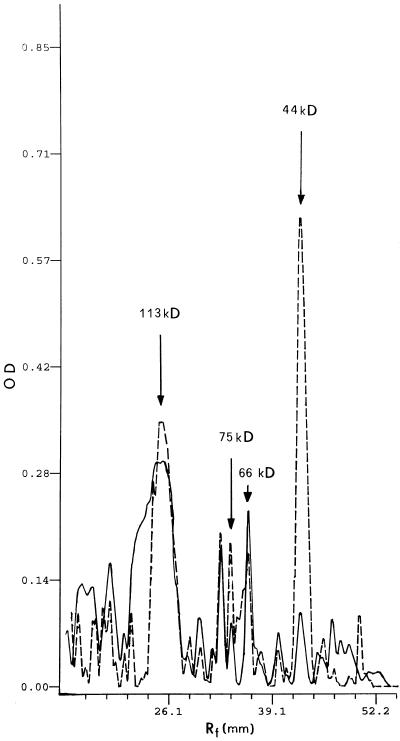
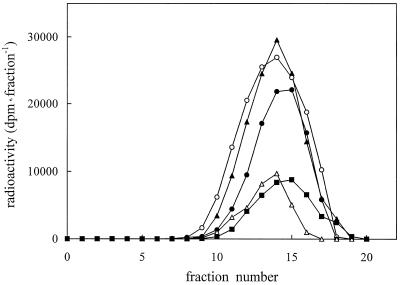
Another approach to the study of spermidine-binding sites on solubilized proteins involves a gel-filtration technique using prepacked columns (PD-10, Pharmacia). In Figure 4A, the elution pattern of the gel filtration of the [14C]spermidine-binding assay is shown. A good separation of bound from free [14C]spermidine was obtained, indicating that the gel-filtration technique represents a suitable method for polyamine-binding studies. In addition, the specific-to-nonspecific binding ratio was higher than that obtained by filtration with glass-fiber filters. Fractions 10 to 14 had the highest specific spermidine-binding activity (Fig. 4B). Total solubilized proteins and gel-filtration active fractions were analyzed by SDS-PAGE, stained with silver salts, and compared (Fig. 5, lanes A and B). Active fractions (Fig. 5, lane B) exhibited bands of 113, 75, 66, and 44 kD, two of which (75 and 44 kD) were enriched compared with total solubilized proteins (Fig. 5, lane A). Additional bands at 92, 38, and 36 kD were observed on native PAGE (Fig. 5, lane C). The densitometric analysis of lanes A and B of SDS-PAGE (Fig. 6) revealed that, changing from total solubilized proteins to gel-filtration fractions, the 44- and 75-kD protein bands increased in intensity by about 90% and 50%, respectively, whereas, according to the computer calculation, the 113-kD band did not significantly vary in intensity and the 66-kD band decreased by about 40%.
Figure 4.
Sephadex G-25 M gel-filtration assay of [14C]spermidine binding to proteins solubilized from zucchini PM vesicles. A, Blank and standard binding assay elution patterns. B, Specific binding elution pattern and A280 protein elution profile. Proteins were eluted with buffer A, pH 8.0, containing 0.01% (w/v) OG and fractionated in 0.4-mL aliquots. Open symbols, Blank assay; closed symbols, standard binding assay; circles, total binding; squares, specific binding; and triangles, nonspecific binding. The values are means ± sd of triplicate samples of three replicate experiments. The sd values are contained within the symbols.
Figure 5.
SDS-PAGE of total solubilized proteins (lane A) and gel-filtration fractions 10 to 14 (lane B), and native PAGE of gel-filtration fractions (lane C). Protein bands were stained with silver.
Figure 6.
Densitometric profile of SDS-PAGE of total solubilized proteins (Fig. 5, lane A) and gel-filtration fractions (Fig. 5, lane B). Continuous line, Total solubilized proteins; broken line, gel-filtration fractions. OD, Optical density.
Gel-filtration competition experiments were carried out between [14C]spermidine and other polyamines and a spermidine analog. As shown in Figure 7, MGBG, a competitive inhibitor that resembles spermidine with regard to the distance between the positively charged amino-terminal groups (Seiler and Dezeure, 1990), competed for [14C]spermidine binding as effectively as unlabeled spermidine. Spermine exerted a lower but still noticeable inhibitory effect, whereas putrescine did not compete. These results demonstrate that spermidine-binding protein(s) exhibit considerable specificity, perhaps attributable to the presence of negatively charged groups in the binding site, the distance of which strictly resembles that of positively charged amino-terminal groups of spermidine. In fact, in PM vesicles, putrescine, in which the two positively charged amino groups are closer than in spermidine, failed to compete for specific binding (Tassoni et al., 1996), although spermine competition was greater then that found with solubilized proteins. Moreover, transport studies performed on carrot protoplasts confirmed the higher capacity of spermidine to bind to plasmalemma with respect to putrescine (Pistocchi et al., 1988).
Figure 7.
Effect of different competitors on specific [14C]spermidine binding to proteins solubilized from zucchini PM vesicles. The various antagonists were present at 20 μm final concentration. ▴, No antagonist; ○, putrescine; •, spermine; ▵, MGBG; and ▪, spermidine. The values are means ± sd of double samples of two replicate experiments. The sd values are contained within the symbols.
Vanadate-sensitive ATPase, a PM marker, and ADC and ODC, two polyamine biosynthetic enzymes, were assayed in PM vesicles, total solubilized proteins, and gel-filtration fractions showing the highest spermidine-binding activity. In E. coli specific transport systems for spermidine and putrescine (pPT104 and pPT79) contain, in addition to periplasmic substrate-binding proteins (PotD and PotF, respectively), PotA and PotG proteins that have ATPase activity (Kashiwagi et al., 1993, 1995; Pistocchi et al., 1993b). Moreover, the latter two proteins have molecular masses of 43 and 45 kD, similar to our 44-kD protein band found in the SDS-PAGE and native PAGE of active fractions (Fig. 5, lanes B and C). Because it is known that the loading through Sephadex gels does not generally interfere with this enzyme activity (Penefsky, 1977), vanadate-sensitive ATPase activity was also tested in gel-filtration fractions to determine if this activity is related to specific spermidine binding. We detected vanadate-sensitive ATPase activity in PM vesicles (27.11 ± 2.70 nmol min−1 mg−1 protein) and total solubilized proteins (10.73 ± 1.49 nmol min−1 mg−1 protein), but none in gel-filtration fractions. This result shows that ATPase activity, although present in total solubilized proteins, does not co-elute during Sephadex G-25 gel filtration with the putative spermidine-binding proteins. However, the result does not exclude the possibility that a similar enzyme activity could be correlated with specific spermidine binding at the plasmalemma level. ADC and ODC activities were undetectable in PM vesicles, total solubilized proteins, and gel-filtration fractions, consistent with their reported subcellular localization in nuclei, mitochondria, and chloroplasts (Panagiotidis et al., 1982; Torrigiani et al., 1985; Borrell et al., 1995).
Specific spermidine-binding activity was investigated by gel-filtration assay of solubilized plasmalemma proteins stored overnight at different temperatures (4°C, −20°C, and −80°C, respectively) to identify the optimal storage temperature for subsequent multistep purification procedures. Storage at −20°C resulted in recovery of the highest binding activity (80% of fresh control), whereas proteins stored at 4°C and −80°C lost about 50% and 90%, respectively, of their binding capacity (data not shown).
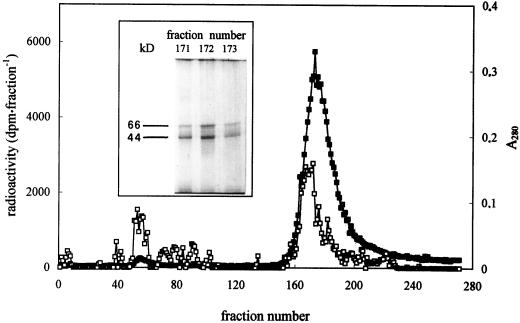
To achieve partial purification of spermidine-binding proteins, Sephadex G-25 gel-filtration fractions with the highest specific spermidine-binding activity were applied to a Sephadex G-200 column. The elution profile (Fig. 8) of [14C]spermidine binding shows the presence of a radioactivity peak coinciding with the major protein peak (A280). SDS-PAGE analysis of the most active spermidine-binding fractions showed two protein bands of 66 and 44 kD (Fig. 8, inset), suggesting that the 113- and 75-kD bands previously found in Sephadex G-25 gel filtration are not involved in spermidine binding.
Figure 8.
Sephadex G-200 gel-filtration elution profile. Sephadex G-25 gel-filtration fractions showing the highest specific spermidine-binding activity were applied to a Sephadex G-200 column after incubation for 5 min at 5°C with 18.5 kBq [14C]spermidine. Proteins were eluted by using buffer A, pH 8.0, with 0.01% (w/v) OG and 0.4-mL aliquots were collected. Closed symbols, radioactivity (dpm fraction−1) elution profile; open symbols, A280 protein elution profile. Inset, Silver-stained 12% (w/v) SDS-PAGE of the most-active spermidine-binding fractions (fractions 171, 172, and 173).
In conclusion, the characteristics of spermidine binding to total solubilized proteins from plant plasmalemma turned out to be similar to those previously found in PM vesicles. This confirms the hypothesis that the specific polyamine interaction between spermidine and PM occurs with the protein component of the membrane. This interaction could represent one of the first steps by which polyamines act as growth regulators. If the spermidine-binding protein(s) is a polyamine receptor, this specific binding could activate the reaction chain responsible for the range of polyamine effects at the cellular level. However, if the binding protein is part of a PM carrier(s), polyamine entry would stimulate the physiological response once inside the cell. One or both of the two proteins (66 and 44 kD) found after partial purification with Sephadex G-200 gel filtration are probably involved in specific spermidine binding to the PM. Work is in progress with the aim of identifying and characterizing the polyamine-binding protein(s).
ACKNOWLEDGMENTS
We are very grateful to Prof. M.A. Venis (Horticulture Research International, Wellesbourne, UK) for useful scientific discussion and help in preparing the manuscript, and to Prof. Patrizia Aducci (University of Rome “Tor Vergata,” Italy) for her most valuable suggestions.
Abbreviations:
- ADC
Arg decarboxylase
- Chaps
3-[(3-cholamidopropyl)-dimethyl-ammonio]-1-propane-sulfonate
- MGBG
methylglyoxal-bis(guanylhydrazone)
- ODC
Orn decarboxylase
- OG
octyl glucoside
- PCA
perchloric acid
- PM
plasma membrane
Footnotes
This work was supported in part by funds from the National Research Council of Italy, special project Ricerche Avanzate per Innovazioni nel Sistema Agricolo, subproject no. 2, and in part by funds from the Ministero dell'Università, della Ricerca Scientifica e Tecnologica. O.S. was a recipient of a grant from the Roche Research Foundation (Basel, Switzerland).
LITERATURE CITED
- Altamura MM, Torrigiani P, Falasca G, Rossini P, Bagni N. Morpho-functional gradients in superficial and deep tissues along tobacco stem: polyamine levels, biosynthesis and oxidation, and organogenesis in vitro. J Plant Physiol. 1993;142:543–551. [Google Scholar]
- Äschlimann D, Paulsson M. Transglutaminases: proteins cross-linking enzymes in tissues and body fluids. Thromb Haemostasis. 1994;71:402–415. [PubMed] [Google Scholar]
- Badini L, Pistocchi R, Bagni N. Polyamine transport in the seaweed Ulva rigida (Chlorophyta) J Phycol. 1994;30:599–605. [Google Scholar]
- Bagni N. Polyamines in plant growth and development. In: Bachrach U, Heimer YM, editors. The Physiology of Polyamines, Vol II. Boca Raton, FL: CRC Press; 1989. pp. 107–120. [Google Scholar]
- Borrell A, Culiañez-Macià FA, Altabella T, Besford RT, Flores D, Tiburcio AF. Arginine decarboxylase is localized in chloroplasts. Plant Physiol. 1995;109:771–776. doi: 10.1104/pp.109.3.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruns RF, Lawson-Wendling K, Pugsley TA. A rapid filtration assay for soluble receptors using polyethylenimine-treated filters. Anal Biochem. 1983;132:74–81. doi: 10.1016/0003-2697(83)90427-x. [DOI] [PubMed] [Google Scholar]
- Christ M, Harr J, Felix H. Transport of polyamines in sugar beet seedlings. Z Naturforsch. 1989;44c:59–63. [Google Scholar]
- Datta N, Hardison LK, Roux SJ. Polyamine stimulation of protein phosphorylation in isolated pea nuclei. Plant Physiol. 1986;82:681–684. doi: 10.1104/pp.82.3.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer AH, Watson BA, Cleland RE. Purification and identification of the fusicoccin binding protein from oat root plasma membrane. Plant Physiol. 1989;89:250–259. doi: 10.1104/pp.89.1.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escribano MI, Legaz ME. Putrescine accumulation does not affect RNA metabolism in the lichen Evernia prunastri. Endocyt Cell Res. 1985;2:239–248. [Google Scholar]
- Feuerstein BG, Marton LJ. Specificity and binding in polyamine/nucleic acid interactions. In: Bachrach U, Heimer YM, editors. The Physiology of Polyamines, Vol I. Boca Raton, FL: CRC Press; 1989. pp. 109–124. [Google Scholar]
- Friedman DL. Polyamine-activated protein phosphatase activity in HeLa cell nuclei. Biochem Biophys Res Commun. 1986;134:1372–1378. doi: 10.1016/0006-291x(86)90401-8. [DOI] [PubMed] [Google Scholar]
- Gray JC. Use of proteolytic inhibitors during isolation of plastid proteins. In: Edelman M, Hallick RB, Chua NH, editors. Methods in Chloroplasts Molecular Biology. Amsterdam: Elsevier Science Publishers; 1982. pp. 1103–1123. [Google Scholar]
- Hendrick JL, Smith AJ. Size and charge isomer separation and estimation of molecular weights of proteins by disc gel electrophoresis. Arch Biochem Biophys. 1968;126:155–164. doi: 10.1016/0003-9861(68)90569-9. [DOI] [PubMed] [Google Scholar]
- Hulme EC. Gel filtration assays for solubilized receptors. In: Hulme EC, editor. Receptor-Ligand Interactions: A Practical Approach. Oxford, UK: IRL Press; 1992. pp. 255–263. [Google Scholar]
- Kashiwagi K, Endo H, Kobayashi H, Takio K, Igarashi K. Spermidine-preferential uptake system in Escherichia coli. J Biol Chem. 1995;270:25377–25382. doi: 10.1074/jbc.270.43.25377. [DOI] [PubMed] [Google Scholar]
- Kashiwagi K, Miyamoto S, Nukui E, Kobayashi H, Igarashi K. Functions of PotA and PotD proteins in spermidine-preferential uptake system in Escherichia coli. J Biol Chem. 1993;268:19358–19363. [PubMed] [Google Scholar]
- Kauss H, Jeblick W. Activation by polyamines, polycations, and ruthenium red of the Ca2+-dependent glucan synthase from soybean cells. FEBS Lett. 1985;185:226–230. [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- Miller DD, Scordilis SP, Hepler PK. Identification and localization of three classes of myosins in pollen tubes of Lilium longiflorum and Nicotiana alata. J Cell Sci. 1995;108:2549–2653. doi: 10.1242/jcs.108.7.2549. [DOI] [PubMed] [Google Scholar]
- Panagiotidis CA, Georgatsos JG, Kyriakidis DA. Superinduction of cytosol and chromatin-bound ornithine decarboxylase activities of germinating barley seeds by actinomycin D. FEBS Lett. 1982;146:193–196. [Google Scholar]
- Penefsky HS. Reversible binding of Pi by beef heart mitochondrial adenosine triphosphatase. J Biol Chem. 1977;252:2891–2899. [PubMed] [Google Scholar]
- Persson L, Svensson F, Lövkvist Wallström E (1996) Regulation of polyamine metabolism. In K Nishioka, ed, Polyamines in Cancer: Basic Mechanisms and Clinical Approaches. Springer Verlag, New York, pp 19–43
- Pistocchi R, Antognoni F, Bagni N. Polyamine uptake and transport in different plant systems. Curr Top Plant Physiol. 1993a;1:21–29. [Google Scholar]
- Pistocchi R, Kashiwagi K, Miyamoto S, Nukui E, Sadakata Y, Kobayashi H, Igarashi K. Characteristics of the operon for a putrescine transport system that maps at 19 minutes on the Escherichia coli chromosome. J Biol Chem. 1993b;268:146–152. [PubMed] [Google Scholar]
- Pistocchi R, Keller F, Bagni N, Matile P. Transport and subcellular localization of polyamines in carrot protoplasts and vacuoles. Plant Physiol. 1988;87:514–518. doi: 10.1104/pp.87.2.514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sammons DW, Adams LD, Nishizawa EE. Ultrasensitive silver-based color staining of polypeptides in polyacrylamide gels. Electrophoresis. 1981;2:135–139. [Google Scholar]
- Scaramagli S, Bueno M, Torrigiani P, Altamura MM, Capitani F, Bagni N. Morphogenesis in cultured thin layers and pith explants of tobacco II. Early hormone-modulated polyamine biosynthesis. J Plant Physiol. 1995;147:113–117. [Google Scholar]
- Schuber F. Influence of polyamines on membrane functions. Biochem J. 1989;260:1–10. doi: 10.1042/bj2600001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiler N, Dezeure F. Polyamine transport in mammalian cells. Int J Biochem. 1990;22:211–218. doi: 10.1016/0020-711x(90)90332-w. [DOI] [PubMed] [Google Scholar]
- Serafini-Fracassini D, Del Duca S, Beninati S. Plant transglutaminases. Phytochemistry. 1995;40:355–365. doi: 10.1016/0031-9422(95)00243-z. [DOI] [PubMed] [Google Scholar]
- Sugiyama S, Vassylyev DG, Matsushima M, Kashiwagi K, Igarashi K, Morikawa K. Crystal structure of PotD, the primary receptor of the polyamine transport system in Escherichia coli. J Biol Chem. 1996;271:9519–9525. doi: 10.1074/jbc.271.16.9519. [DOI] [PubMed] [Google Scholar]
- Tabor CW, Tabor H. Polyamines in microorganisms. Microbiol Rev. 1985;49:81–99. doi: 10.1128/mr.49.1.81-99.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassoni A, Antognoni F, Bagni N. Polyamines binding to plasma membrane vesicles isolated from zucchini hypocotyls. Plant Physiol. 1996;110:817–824. doi: 10.1104/pp.110.3.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas CT, McNamee MG. Purification of membrane proteins. Methods Enzymol. 1990;182:499–520. doi: 10.1016/0076-6879(90)82040-9. [DOI] [PubMed] [Google Scholar]
- Torrigiani P, Serafini-Fracassini D, Biondi S, Bagni N. Evidence for the subcellular localization of polyamines and their biosynthetic enzymes in plant cells. J Plant Physiol. 1985;124:23–29. [Google Scholar]