Abstract
Kinetically assembled nanoparticles are fabricated from an advanced class of bioactive macromolecules that have potential utility in counteracting atherosclerotic plaque development via receptor-level blockage of inflammatory cells. In contrast to micellar analogs that exhibit poor potency and structural integrity under physiologic conditions, these kinetic nanoparticle assemblies maintain structural stability and demonstrate superior bioactivity in mediating oxidized low-density lipoprotein (oxLDL) uptake in inflammatory cells.
Keywords: Nanomedicine, Cardiovascular Disease, Macrophages, Atherosclerosis, Flash NanoPrecipitation, Biomedical Applications
Rapid progress in nanotechnology has allowed development of novel therapeutic vehicles that provide enhanced pharmacological properties of bioactives.[1–3] Of these vehicles, polymeric micelles, including polymer-lipid conjugates, have received considerable attention due to the ease in synthesizing reactive-functional macromolecules and the ability to sequester aqueous insoluble therapeutics.[4–6] These assemblies, although attractive, can be labile due to equilibrium between micelles and unimeric chains (Figure 1A).[7, 8] This dynamic equilibrium raises uncertainty about micelle integrity when introduced into a physiological environment.[8] In fact, researchers have observed both poor drug retention and micelle dissolution in vitro as well as in vivo, ascribed to unimer or drug partitioning into neighboring plasma proteins or lipophilic sinks.[1, 4, 8–14] Premature micelle dissolution leads to suboptimal therapeutic efficacy and biodistribution.
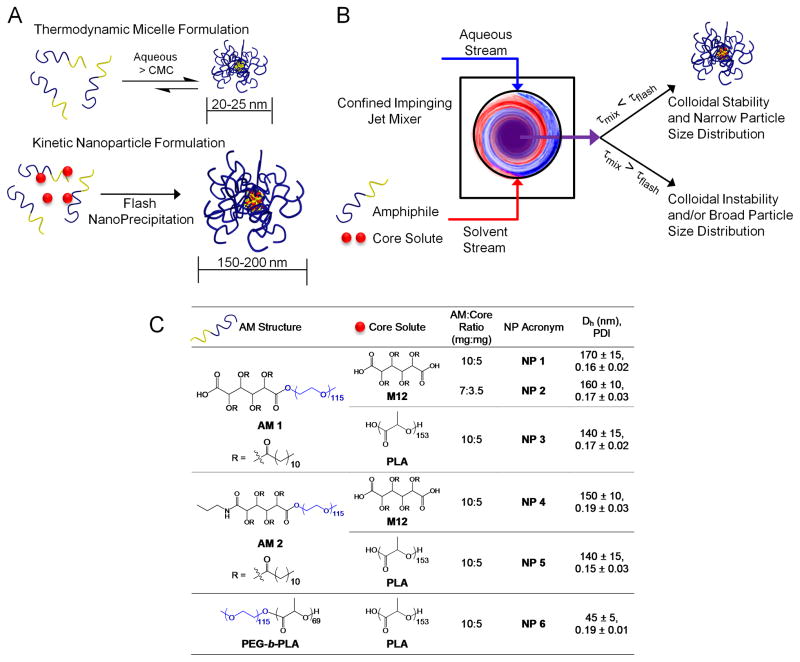
Figure 1.
Kinetically assembled nanoparticles (NPs) of bioactive macromolecules fabricated via Flash NanoPrecipitation. A) Schematic contrasts differences between thermodynamic micelle assemblies by aqueous dissolution above the critical micelle concentration (top) and kinetic assembled NPs formulated by Flash NanoPrecipitation (bottom). B) Basic components of the Flash NanoPrecipitation process requisite to the formation of colloidal stable NPs. τmix represents the time required for complete and homogenous mixing of the aqueous and solvent stream, while τflash is the precipitation or NP formation time. C) Table listing the chemical structures of amphiphilic macromolecules (AMs) and the hydrophobic core solutes; AM-to-core solute weight ratios investigated; NP acronyms; and hydrodynamic diameters (Dh) and polydispersity indices (PDI). Data are from an n = 3–5 conducted in triplicate with error representing ± S.E.M.
In contrast to thermodynamically assembled micellar systems of macromolecules, we propose a new approach to stabilize biological active macromolecules around hydrophobic cores, resulting in stable nanoparticles (NPs), which are resistant to thermodynamic disruption into unimers. The organic nanoparticles were fabricated by adapting flash nanoprecipitation principles,[15–20] as depicted in Figure 1A. The process highlighted in Figure 1B shows the rapid mixing of an aqueous stream with a water-miscible organic solvent stream containing selected hydrophobic solutes and amphiphilic macromolecules (AMs). Rapid mixing causes supersaturation of the hydrophobic solute in the aqueous stream, which induces nucleation and NP growth.[21, 22] To fabricate stabilized nanoparticles of bioactive macromolecules we employed core-forming solutes that mirror the hydrophobic domains of the AMs.
Bioactive AMs have been proposed as therapeutic ligands for inflammatory cells with potential to mitigate atherosclerosis,[23–26] a condition involving the accumulation of lipid-rich plaques and chronic inflammation of the vascular wall.[27] Plaque deposits initiate when low-density lipoproteins (LDL) accumulate in the artery wall and subsequently undergo oxidative modification (oxLDL). OxLDL stimulates endothelial inflammation triggering monocyte recruitment and subsequent monocyte differentiation to macrophages. Intimal macrophages exhibit unregulated uptake of oxLDL, via scavenger receptors, leading to the formation and accumulation of lipid-laden macrophages called foam cells, which in turn drives plaque development. The design of new bioactives that can inhibit macrophage internalization of oxLDL may hold promise in managing atherosclerosis.[28] Notably, a new class of synthetically derived bioactive AMs, displaying various structural features (i.e. ionic charge, charge placement, hydrophilic/lipophilic balance, and rotational mobility of the lipophile), has been recently designed and investigated for mediating oxLDL uptake by macrophage-like THP-1 cells.[23, 25, 26] Computer modeling and in vitro experimentation[25, 29] suggest that these bioactive AMs can inhibit oxLDL uptake by competing for or occupying cell surface scavenger receptors, the main binding sites and cellular entry points for oxLDL. More recently, localized delivery of these bioactive AMs, carrying a cholesterol efflux drug, to injured carotid arteries of Sprague Dawley rats fed a high fat diet led to decreased cholesterol accumulation in the intima and inhibited retention of macrophages.[29] To date, micellar assemblies of these bioactive AMs have been explored, however, these systems display decreased efficacy in serum containing media and are susceptible to the aforementioned drawbacks of micellar constructs under physiologic conditions such as those in circulation in vivo.
The key hypothesis of this report is that kinetically assembled macromolecular bioactive NPs offer a more stable alternative to the thermodynamically assembled micellar systems, and can potentially exhibit enhanced biological activity in the presence of serum proteins. To test this hypothesis, a new class of bioactive NPs was fabricated to examine the NP potential for enhanced inhibition of human macrophage uptake of oxLDL and foam cell formation under physiologically relevant conditions. The Flash NanoPrecipitation process enables the fabrication of NP constructs with various shell components based on amphiphilic macromolecular chains and core components containing derivative hydrophobic domains (Figure 1C). Colloidal stability was investigated by dynamic light scattering (DLS) and in vitro studies were carried out utilizing human monocyte-derived macrophages (HMDMs) isolated from peripheral blood mononuclear cells (PBMCs). The ability of the NP system to inhibit oxLDL uptake in HMDMs and prevent formation of the foam cell phenotype was studied in the presence of serum proteins and directly compared to HMDMs treated with an analogous micellar system. NP interactions with macrophage scavenger receptors, which mediate the uptake of modified cholesterols, were also investigated.
The compositions for stabilizing AMs and the organic solutes employed in Flash NanoPrecipitation are listed in Figure 1C. Based on previous findings that electrostatic charge and structural features affect bioactivity, the bioactive AMs chosen for the study are comprised of a mucic acid modified with lauroyl groups, 5 kDa poly(ethylene glycol) (PEG), and either a carboxylic acid (AM 1) or propyl amide (AM 2) terminal functionality.[23, 26] NPs formulated with PEG-block-poly(lactic acid) (PEG-b-PLA) copolymer, where each block has a molecular weight of 5 kDa, served as a non-active control for in vitro studies, as PEG and PLA are cytocompatible, but do not exhibit inherent biological activity towards managing atherosclerosis. The hydrophobic solutes (Figure 1C), which constitute the core of the NP and provide an anchoring site for the AM, were chosen to be either mucic acid modified with lauroyl groups, 2,3,4,5-tetrakis-dodecanoyloxy-hexanedioic acid abbreviated as M12, or an 11 kDa PLA. Both anionically terminated macromolecules (AM 1) and uncharged macromolecules (AM 2) were packaged into NPs by Flash NanoPrecipitation as depicted in Figure 1. The rapid solvent exchange of this process leads to kinetically trapped NPs while maintaining control of core compositions.[14, 16] Rapid micromixing is produced by novel impinging jet micromixers as previously described.[15, 21, 22] To ensure uniform kinetics and thus a homogenous NP size distribution, the time of mixing both streams, τmix, was prescribed to be less than the NP formation time, τflash, or in other words the induction times of both AM aggregation and precipitation/nucleation of the core solute. In this study, two streams were used: a THF solvent stream containing bioactive AM and the core molecules M12 or PLA (2:1 by weight) and an anti-solvent stream of water. Mixing produces high supersaturation with drives rapid nucleation and growth. After mixing, the final H2O:THF ratio was 9:1 v/v and the newly formed NPs were immediately dialyzed against MilliQ H2O to remove the residual solvent THF. Uniform particle size distributions were observed by DLS and hydrodynamic diameters (Dh) of NPs ranged from 150 to 200 nm, with the exception of the NP 6 system, ~ 45 nm. Dh and polydispersities (PDI) for the NPs are listed in Figure 1C. The number of AM 1 chains per NP 1 was determined to be between 3300 and 5600, while the number of M12 molecules per NP 1 was between 10000 and 17000 (see Supporting Information for estimated values using an approach presented previously [30] and based on the volume of a typical NP with average diameter of 150 ± 10 nm).
Solution stability of the fabricated NPs upon storage and co-incubation with fetal bovine serum (FBS) was monitored by DLS (Figure 2). Colloidal stability of the bioactive AM NP, NP 1, is displayed in Figure 2A. No significant changes in the intensity distributions, Dh, or count rate relative to time zero were detected during the 37 °C 4 week incubation period, indicating extended aqueous stability of the colloids. Colloidal stabilities of other NP systems upon storage are elaborated in the Supporting Information, Figure S1. To mimic physiologic conditions, colloidal stability was additionally monitored in the presence of serum proteins. The intensity distribution of the NP 1 at 0.6 mg mL−1 in RPMI media containing up to 20 % FBS was measured initially and after 24 h incubation (Figure 2B). For comparison, NPs were also incubated at 37 °C under serum-free conditions. No appreciable difference in the NP Dh under serum-free or serum containing media was observed. The difference in Dh between NPs dispersed in deionized water (Figure 2A; ~160 nm) versus salt- and serum-containing RPMI media (Figure 2B; ~190 nm) potentially arises from the shielding of the anionic charges of the M12 core and the AM 1 bioactive AM by counterions. Because DLS operates on Brownian motion and determines Dh from particle diffusion coefficients, counterion diffusion in DI water and the electrostatic coupling between the counterions and macro-ion can lead to a faster macro-ion diffusivity and, therefore, apparent smaller hydrodynamic size.[31] Electrostatic screening by the buffer is evident in zeta potential measurements where NP charge in deionized water was −30 mV versus −6 mV in phosphate buffer. The small distribution peaks in Figure 2B under 100 nm can be attributed to serum proteins as reported previously.[32] As shown in Figure 2B, the particle size was unaffected by the addition of serum proteins even after 24 hours of incubation. These results indicate enhanced stability against aggregation in serum media, which is an important requisite for in vivo drug delivery, and has been problematic for many NP formulations.[33, 34]
Figure 2.
Kinetically assembled nanoparticles exhibit colloidal stability when stored for extended time (4 weeks) or incubated in the presence of serum proteins. A) Intensity distribution of NP 1 stored at 37 °C for 4 weeks. Hydrodynamic diameters (Dh) and polydispersity indices (PDI) are listed next to the respective sample. B) Intensity distribution of NP 1 incubated with 20% fetal bovine serum (FBS) at time zero and 24h. NP incubation under serum-free conditions is plotted for reference. C) The release of AM 1 from kinetically fabricated NPs (black symbols) is significantly lower than that from thermodynamically assembled micelles (gray symbols) when dialyzed under sink conditions against PBS (open symbols) or PBS + 10% FBS (closed symbols). The release of fluorescein labeled AM 1 (20% by weight) was monitored via UV-vis spectroscopy (error bars ± S.D.).
As previously described, micelle formulations can suffer from premature release or exhibit unimer partitioning into neighboring hydrophobic/lipophilic environments.[1, 4, 8–14, 35] In order to demonstrate the enhanced physiological stability of kinetically fabricated NPs versus micelles, the percent release of AM 1, under sink conditions, was monitored via dialysis against both PBS and PBS containing 10% FBS (Figure 2C). The fractional release of AM 1 from both systems was accomplished by fluorescently labeling the hydrophilic PEG chains with a reactive fluorescein molecule and measuring absorbance via UV-vis spectroscopy. Fluorescently labeled AM 1 was incorporated at 20 wt% for both systems with the remainder comprising of unlabeled AM 1. Incorporation of fluorescently labeled AM 1 had no effect on the Dh of the fluorescently labeled systems relative to the unlabeled systems. As expected, in both micellar and NP systems, the release of AM 1 was greater in the presence of FBS vs PBS. However, the quantitative level of retention of AM 1 by the NP system versus micellar system in both PBS and PBS containing 10% FBS was markedly superior. These findings indicate that kinetic fabrication of these bioactive AMs into NPs, rather than thermodynamically assembled micelles, provides a drug delivery formulation with enhanced structural integrity in physiologic conditions. This data also validates our premise that the kinetic trapping process anchors the AM within the NP core, thus, obviating the dynamic equilibrium between unimers and micelles as seen in thermodynamic assemblies.[15]
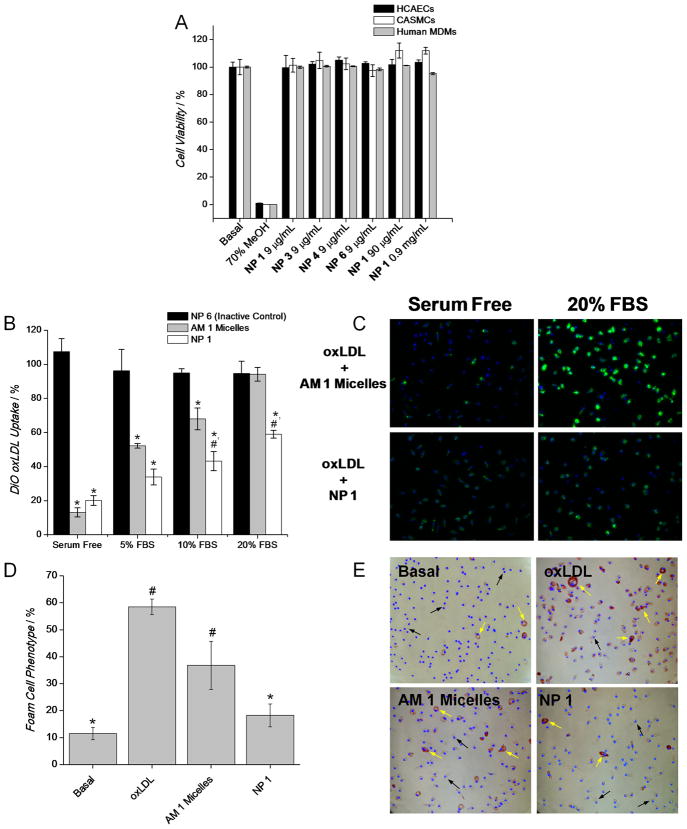
To confirm that the new NP configurations did not alter the intrinsic cytocompatibility of macromolecules, we compared the viability of cells incubated with various NP systems using a LIVE/DEAD fluorometric assay. Figure 3A shows the percent cell viability data for several NP systems in three different cell types that are relevant to vascular remodeling leading to atherosclerotic lesions: human monocyte-derived macrophages (HMDMs), human coronary artery endothelial cells (HCAECs) and human coronary artery smooth muscle cells (HCASMCs). As seen in Figure 3A, all tested NPs demonstrated undetectable levels of cytotoxicity over the range of concentrations studied. In addition, cytocompatibility for the NP 1 system was determined at NP concentrations ranging from 9 to 900 μg mL−1, which corresponds to a bulk AM 1 concentration of 10−6 to 10−4 M, respectively.
Figure 3.
Nanoparticle cytocompatibility (A) and in vitro comparison of kinetic assembled NPs versus thermodynamic assembled micelles for inhibiting oxLDL uptake (B–C) and foam cell formation (D–E), a critical requisite for atherosclerotic therapies, in HMDMs. A) Cell viability of HMDMs, HCAECs and HCASMCs after incubation with NPs for 24 h and 10 % FBS. Experiment repeated in triplicate, error bars = ± S.D. B) Quantification of DiO oxLDL (5 μg mL−1) uptake by HMDMs after 24 h incubation with AM 1 micelles or NP 1 under 0 to 20% FBS, [AM 1] = 10−6 M. NP 6 serve as a non-active control and all treatments are normalized to cells incubated exclusively with DiO oxLDL. C) Representative fluorescent images contrasting the level of oxLDL uptake between HMDMs co-incubated with DiO oxLDL (green) and either AM 1 micelles (top) or NP 1 (bottom) under serum-free or 20 % FBS for 24 h. Nuclei were counterstained with Hoechst 33342. D) Quantification and E) bright field images of foam cell phenotype after 48 h incubation of HMDMs with no oxLDL (Basal), oxLDL, AM 1 micelles + oxLDL, or NP 1 + oxLDL with 10% FBS ([oxLDL] = 50 μg mL−1, [AM 1] = 10−5 M). Foam cells were quantified using OilRed O (red) staining and marking cell nuclei with Hoechst 33342. Examples of native HMDM phenotype and foam cell phenotype are highlighted by black and yellow arrows, respectively. Data for Figure 3B and D are from an n = 3 conducted in triplicate (error bars = ± S.E.M.). Figure 3B, statistical significance was evaluated at p < 0.05, * indicates significance versus NP 6 and # versus AM 1 micelles. Figure 3D, statistical significance was evaluated at p < 0.05, * indicates significance versus oxLDL and # versus Basal conditions.
The key biological efficacy of the kinetically assembled macromolecules was evaluated in terms of the degree of inhibition of oxLDL uptake in HMDMs by AM NPs versus analogous micellar systems (Figure 3 B–E). Although the bioactivity of other NP systems was evaluated (See Supporting Information), the remainder of this account will focus on the bioactivity and cell specificity of the NP 1 system, which is comprised of AM 1 as the stabilizing NP shell and M12 as the core solute at an AM: core solute weight ratio of 10 mg:5 mg. Human PBMC monocytes isolated from whole blood were first differentiated into macrophages using macrophage colony stimulating factor for one week prior to initiating experiments. HMDMs were co-incubated with 5 μg mL−1 of 3,3′-dioctadecyloxacarbocyanine (DiO)-labeled oxLDL and either NPs or micelles for 24 h in the presence of 0 (serum-free), 5, 10 or 20% FBS. Uptake of oxLDL in HMDMs was determined by densitometry analysis via ImageJ of a series of digitized images (9 fields for each condition, 3 per well, were captured at 20X magnification per biological replicate, n = 3). The key results for NP 1 and the AM 1 micelle systems are shown in Figure 3B & C, where all conditions were normalized to HMDMs only receiving DiO-labeled oxLDL. In addition, NP 4 versus AM 2 micellar systems were investigated and the oxLDL uptake results are available in the Supporting Information, Figure S2.
The comparative uptake inhibition of oxLDL was done using equivalent levels of AM 1 concentration of 10−6 M, which corresponds to a NP concentration of 9 μg mL−1. This concentration was chosen to be an effective range of bioactivity in the micellar form and because it is sufficiently above the critical micelle concentration of AM 1, ~10−7 M.[23, 24] As shown in Figure 3B, NP and micelle vehicles exhibited similar inhibition of oxLDL uptake when HMDMs were treated under serum-free conditions. However, as FBS concentration was increased to 20%, the efficacy of the micelles decreased to 6 % while NP 1 maintained more than 40 % inhibition of oxLDL uptake. Retention of bioactivity for the NP system can be attributed to a slower rate of partitioning to surrounding serum proteins as the bioactive AM is kinetically anchored to the NP core through favorable hydrophobic interactions. In contrast, the micelle system loses bioactivity in the presence of serum proteins likely due to the disruption of the micelle organization and premature release of the bioactive AM.[4, 11] A significant difference in oxLDL uptake between NP 1 and the traditional micelle system, as determined by one way ANOVA and subsequent post-hoc analysis, is evident at conditions incubated with 10% FBS (p < .05) and 20% (p < .01). Visual evidence is provided in Figure 3C, which clearly shows the level of fluorescence in each system is similar under serum-free conditions, but is significantly weaker (i.e. the AM 1 is still bioactive) when cells are treated with NP 1 20 % FBS. Fluorescent images for DiO oxLDL uptake alone and other serum-containing conditions for AM 1 micelles and corresponding NPs are available in the Supporting Information (Figure S3). In initial in vitro studies, utilizing human macrophage cell lines (i.e.; differentiated THP-1s), the hydrophobic core M12 had minimal-to-no therapeutic efficacy towards inhibiting oxLDL uptake as determined by co-incubating macrophages with PEG-b-PLA NPs loaded with M12 and DIO oxLDL (data not shown). Due to this observation PEG-b-PLA/M12 NPs were not further investigated in human primary cells. Further, the inhibition of DiO oxLDL uptake under serum-free conditions (Figure 3B) is similar for both the NP 1 and AM 1 micelle systems suggesting that M12 of the NP core does not, by itself, additionally contribute toward enhanced bioactivity. While the hydrophobic moiety of AM 1 is nearly identical to that of M12, it is hypothesized that the PEG group of AM 1 significantly increases the partition coefficient between the NP core and the serum phase, which allows AM 1 to become bioavailable. Clearly the NP system retains bioactivity in the presence of serum protein. However, there are two possible hypotheses for the mechanism of action. First, the NPs are stabilizing the bioactive AM and it remains bioactive as is the case in micelle form, or second, M12 is the bioactive and the NP formulation allows delivery of M12. In a previous report, our laboratories demonstrated that minor structural changes in the bioactive AM, such as charge type, charge number and rotational mobility have a marked effect on the inhibition of oxLDL uptake.[26] Perhaps, the absence of PEG or an additional carboxylic acid (M12 versus AM 1), affect how M12 interacts with the cell surface or macrophage scavenger receptors responsible for oxLDL uptake. Whether the binding event itself or the PEG chain attached to the bound receptor blocks oxLDL uptake is unresolved; a more detailed studied is required to clarify this observation. Importantly, the non-active NP with the PLA core, NP 6, displays no inhibitory effects.
Excessive accumulation of cholesterol through intimal macrophage uptake of modified LDLs alters the macrophage phenotype.[36, 37] Excess cholesterol leads to the formation of numerous intracellular lipid droplets, thus giving the cell a “foam-like” appearance.[38] As foam cell formation is associated with the development of atherosclerotic plaque, therapeutics that can retard this change in macrophage phenotype hold promise. Therefore, we examined foam cell formation in HMDMs while in the presence of the NP 1 (Figure 3D & E). Foam cell phenotype was induced by incubating HMDMs with 50 μg mL−1 of oxLDL and either NPs or micelles for 48 h in the presence of 10% FBS. To maintain the AM 1 to oxLDL concentration ratio used in the DiO oxLDL uptake studies above, a AM 1 concentration of 10−5 M, equivalent to 90 μg/mL for the NP, was used. The intracellular lipids were visualized by staining with Oil Red O after cell fixation (Figure 3E). After staining, foam cells were labeled red and subsequently counted via image analysis software. The percentage of foam cells for the oxLDL control condition was near 60 ± 3 %, while HMDMs incubated with NP 1 reduced foam cell formation close to the basal level with statistical significance (p < .05), 18 ± 4 %, versus 12 ± 2 %. The low level of foam cell formation seen in the basal condition is attributed to the presence of serum lipids in FBS. Notably, foam cell formation in HMDMs receiving treatment with AM 1 micelles did not display statistical significance from HMDMs incubated with oxLDL. This result further corroborates the DiO oxLDL studies and demonstrates that the methods used to package and deliver the bioactive AMs, micelles versus kinetically formed NPs, can have a significant effect on inhibiting oxLDL uptake in the presence of serum proteins.
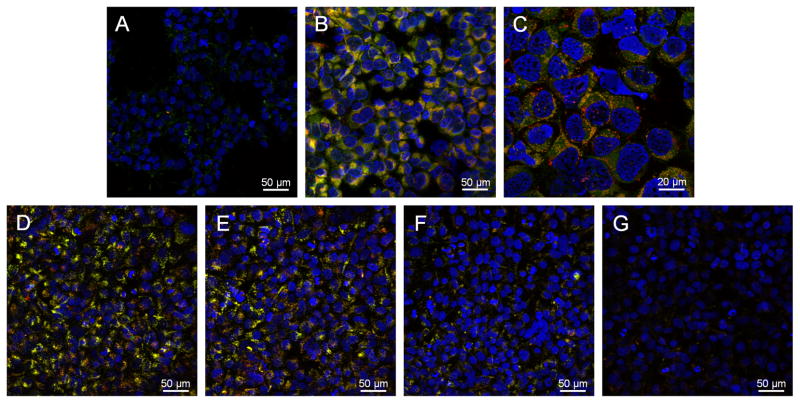
To further confirm the molecular cell targeting potential of kinetically assembled macromolecules, we examined the scavenger receptor interactions of the NP 1. Scavenger receptors (SRs) have been identified and implicated in the unregulated uptake of oxLDL by a number of inflammatory and vascular cell types.[37] Class A and B scavenger receptors account for 75 to 90% uptake of modified LDLs.[39] Therefore, blocking or binding of these receptors would be advantageous for limiting oxLDL internalization in developing atherosclerotic lesions. Previous reports by our laboratories demonstrated that bioactive AMs exhibit enhanced affinity of binding towards scavenger receptor A1 (SR-A1).[25] We used this established model system to demonstrate the binding potential of NP 1 with SRs in cells engineered to express SR-A1 (HEK-SRA). Dual fluorescent NPs were prepared via Flash NanoPrecipitation utilizing fluorescein-labeled AM 1, incorporated at 20 wt% of total AM 1 weight, and by co-precipitating a highly hydrophobic red fluorophore, 2,2,10,10-tetraethyl-6,14-bis-(triisopropylsilylethynyl)-1,3,9,11-tetraoxa-dicyclopenta[b,m]pentacene (EtTP-5), with M12. Human embryonic kidney (HEK) cells, genetically engineered with or without inducible SR-A1 expression, were incubated with fluorescent NP 1 for 24 h. Confocal images in Figure 4A–C clearly show the level of NP associated fluorescence is significantly increased in SR-A1 expressing cells (Figure 4B & C) relative to non-expressing cells (Figure 4A). Further, NP specificity towards SR-A1 was confirmed via receptor blocking experiments (Figure 4D–G). SR-A1 expressing cells incubated with fluorescent NPs (Figure 4D) resulted in a high level of cell associated fluorescence; however, when cells were pre-incubated with either primary SR-A1 monoclonal mouse antihuman antibody (mAb; Figure 4F) or polyinosinic acid (PIA; Figure 4G), a known SR-A1 ligand, cell associated fluorescence was noticeably reduced. To discern non-specific from specific binding, SR-A1 monoclonal antibody was replaced with purified mouse IgG1 (Figure 4E) as an isotype control. Importantly, when SR-A1 was blocked with antibody, cell associated fluorescence decreased ~ 50%, as evidenced from fluorescence microscopy, in comparison to the isotype control (Figure 4E versus 4F). In addition, blocking SR-A1 with PIA reduced cell associated NP fluorescence ~ 95% (Figure 4D vs. 4G). These results provide strong visual evidence that the NP 1 system displays affinity towards SR-A1 and highlight the possibility of developing NPs that associate with other cell surface receptors responsible for oxLDL uptake. Interaction of these macromolecular bioactive NPs with not only SR-A1, but other SRs is currently being investigated.
Figure 4.
Fluorescent NPs, labeled at the PEG termini with fluorescein (green) and loaded with the hydrophobic fluorophore ETTP5 (red), exhibit affinity towards scavenger receptor A1, a key mediator of oxLDL accumulation in atherosclerotic lesions. NPs appear yellow in fields supported by stable colocalization of core-shell fluorophores. HEK cells engineered to express SR-A1 were incubated overnight (A–C) or for 6 h (D–E) with fluorescent NP 1, [AM 1] = 10−5 M. Confocal images of NP binding/uptake in basal cells with low SR-A1 expression (A) and following SR-A1 expression (B) indicate substantial NP interaction with SR-A1. C) Higher magnification image of cells with positive SR-A1 expression clearly showing NP association/internalization. Primary SR-A1 mAb (mouse antihuman) or polyinosinic acid (PIA), a well known ligand for SR-A1, blocking experiments (D–G) confirms NP affinity towards SR-A1. D) NPs incubated with SR-A1 expressing cells and following incubation with E) 10 μg mL−1 isotype control antibody (IgG1), F) 10 μg mL−1 primary anti-SR-A1 monoclonal antibody and G) 10 μg mL−1 PIA for 6h. Yellow signal indicates co-localization of both fluorophores associated with the NP. Cell nuclei were counterstained with Hoechst 33342.
In summary, a new approach for fabrication of macromolecular bioactive NPs that retain bioactivity and colloidal stability in the presence of serum proteins has been presented. Core solutes and bioactive AMs possessing comparable hydrophobicities were chosen and assembled into aqueous stable NPs via Flash NanoPrecipitation. The formulated NPs, in contrast to analogous bioactive AM micelles, revealed an improved bioactivity in the presence of serum proteins, as evidenced by inhibition of oxLDL uptake and foam cell formation, thus demonstrating potential utility in the management of atherosclerosis. Furthermore, NP affinity towards cell surface receptors that are active participants in the development and progression of atherosclerotic lesions was demonstrated. In contrast to traditional delivery methodologies, kinetic fabrication provides high therapeutic loading capacities and NPs with tunable hydrodynamic diameters and drug partitioning rates, thus improving the pharmacological properties of the packaged therapeutic. These findings highlight the utility of kinetically formed NPs versus thermodynamically formed micelles for managing atherosclerosis. The observation of increased stability and bioavailabity of these kinetically assembled nanoparticles in serum media indicates the potential to enhance the delivery and efficacy of a wide range of drugs that have been studied in labile polymeric micelle formulations. These findings provide impetus for further applications of similar nanoparticles to different pathologies including targeting atherosclerotic plaques and lesions and various cancerous tissues. The nanoparticle systems can also be used for fundamental studies related to the partitioning of bioactive macromolecules between nanoparticle cores, protein and lipid components, and cellular membranes during cellular uptake and delivery.[40–42]
Experimental Section
Materials and experimental procedures are provided in the Supporting Information.
Supplementary Material
Acknowledgments
The authors would like to thank the National Institute of Biomedical Imaging and Bioengineering (T32 EB005583), National Heart, Lung and Blood Institute (R21HL93753, R01HL107913) and the American Heart Association (AHA 0756036T) for financial support. We would like to thank Dr. Howard Bowman of Surmodics Pharmaceuticals (Birmingham, AL) for the synthesis of the PEG-b-PLA polymers.
Footnotes
Supporting Information is available online from Wiley InterScience or from the author.
Contributor Information
Dr. Adam W. York, Department of Biomedical Engineering, Rutgers University, 599 Taylor Road, Piscataway, NJ 08854, USA
Kyle R. Zablocki, Department of Biomedical Engineering, Rutgers University, 599 Taylor Road, Piscataway, NJ 08854, USA
Daniel R. Lewis, Department of Chemical and Biochemical Engineering, Rutgers University, 98 Brett Road, Piscataway, NJ 08854, USA
Li Gu, Department of Chemistry and Chemical Biology, Rutgers University, 610 Taylor Road, Piscataway, NJ 08854, USA.
Prof. Kathryn E. Uhrich, Department of Chemistry and Chemical Biology, Rutgers University, 610 Taylor Road, Piscataway, NJ 08854, USA
Prof. Robert K. Prud’homme, Department of Chemical and Biological Engineering, Princeton University, Princeton, NJ 08544, USA
Prof. Prabhas V. Moghe, Email: moghe@rci.rutgers.edu, Department of Biomedical Engineering, Rutgers University, 599 Taylor Road, Piscataway, NJ 08854, USA. Department of Chemical and Biochemical Engineering, Rutgers University, 98 Brett Road, Piscataway, NJ 08854, USA.
References
- 1.Allen TM, Cullis PR. Science. 2004;303:1818. doi: 10.1126/science.1095833. [DOI] [PubMed] [Google Scholar]
- 2.Koo OM, Rubinstein I, Onyuksel H. Nanomed Nanotechnol Biol Med. 2005;1:193. doi: 10.1016/j.nano.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 3.Farokhzad OC, Langer R. ACS Nano. 2009;3:16. doi: 10.1021/nn900002m. [DOI] [PubMed] [Google Scholar]
- 4.Gaucher G, Dufresne M-H, Sant VP, Kang N, Maysinger D, Leroux J-C. J Controlled Release. 2005;109:169. doi: 10.1016/j.jconrel.2005.09.034. [DOI] [PubMed] [Google Scholar]
- 5.Nishiyama N, Kataoka K. Pharmacol Therapeut. 2006;112 doi: 10.1016/j.pharmthera.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 6.York AW, Kirkland SE, McCormick CL. Adv Drug Deliv Rev. 2008;60 doi: 10.1016/j.addr.2008.02.006. [DOI] [PubMed] [Google Scholar]
- 7.Torchilin VP. Current Drug Delivery. 2005;2 [Google Scholar]
- 8.Bae YH, Yin H. J Control Release. 2008;131:2. doi: 10.1016/j.jconrel.2008.06.015. [DOI] [PubMed] [Google Scholar]
- 9.Cabanes A, Briggs KE, Gokhale PC, Treat JA, Rahman A. Int J Oncol. 1998;12:1035. doi: 10.3892/ijo.12.5.1035. [DOI] [PubMed] [Google Scholar]
- 10.Chowdhary RH, Shariff I, Dolphin D. J Pharm Pharmaceut Sci. 2003;6:13. [PubMed] [Google Scholar]
- 11.Toncheva V, Schacht E, SY NG, Barr J, Heller J. J Drug Target. 2003;11:345. doi: 10.1080/10611860310001633839. [DOI] [PubMed] [Google Scholar]
- 12.Fahr A, van Hoogevest P, May S, Bergstand N, Leigh MLS. Eur J Pharm Sci. 2005;26:251. doi: 10.1016/j.ejps.2005.05.012. [DOI] [PubMed] [Google Scholar]
- 13.Liu J, Zeng F, Allen C. J Controlled Release. 2005;103:481. doi: 10.1016/j.jconrel.2004.12.013. [DOI] [PubMed] [Google Scholar]
- 14.Ansell SM, Johnstone SA, Tardi PG, Lo L, Xie S, Shu Y, Harasym TO, Harasym NL, Williams L, Bermudes D, Liboiron BD, Saad W, Prud’homme RK, Mayer LD. J Med Chem. 2008;51:3288. doi: 10.1021/jm800002y. [DOI] [PubMed] [Google Scholar]
- 15.Johnson BK, Prud’homme RK. Aust J Chem. 2003;56:1021. [Google Scholar]
- 16.Gindy ME, Panagiotopoulos AZ, Prud’homme RK. Langmuir. 2008;24:83. doi: 10.1021/la702902b. [DOI] [PubMed] [Google Scholar]
- 17.Kumar V, Adamson DH, Prud’homme RK. Small. 2010;6:2907. doi: 10.1002/smll.201001199. [DOI] [PubMed] [Google Scholar]
- 18.Gindy ME, Ji S, Hoye TR, Panagiotopoulos AZ, Prud’homme RK. Biomacromolecules. 2008;9:2705. doi: 10.1021/bm8002013. [DOI] [PubMed] [Google Scholar]
- 19.Gindy ME, Prud’homme RK. Expert Opin Drug Deliv. 2009;6:865. doi: 10.1517/17425240902932908. [DOI] [PubMed] [Google Scholar]
- 20.Kumar V, Hong SY, Maciag AE, Saavedra JE, Adamson DH, Prud’homme RK, Keefer LK, Chakrapani H. Mol Pharm. 2009;7:291. doi: 10.1021/mp900245h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson BK, Prud’homme RK. AIChE Journal. 2003;49:2264. [Google Scholar]
- 22.Liu Y, Cheng C, Liu Y, Prud’homme RK, Fox RO. Chem Eng Sci. 2008;63:2829. [Google Scholar]
- 23.Tian L, Yam L, Zhou N, Tat H, Uhrich KE. Macromolecules. 2004;37:538. [Google Scholar]
- 24.Chnari E, Nikitczuk JS, Uhrich KE, Moghe PV. Biomacromolecules. 2006;7:597. doi: 10.1021/bm0506905. [DOI] [PubMed] [Google Scholar]
- 25.Plourde NM, Kortagere S, Welsh W, Moghe PV. Biomacromolecules. 2009;10:1381. doi: 10.1021/bm8014522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iverson NM, Sparks SM, Demirdirek B, Uhrich KE, Moghe PV. Acta Biomater. 2010;6:3081. doi: 10.1016/j.actbio.2010.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Libby P. Nature. 2002;420:868. doi: 10.1038/nature01323. [DOI] [PubMed] [Google Scholar]
- 28.Glass CK, Witztum JL. Cell. 2001;104:503. doi: 10.1016/s0092-8674(01)00238-0. [DOI] [PubMed] [Google Scholar]
- 29.Iverson NM, Plourde NM, Sparks SM, Wang J, Patel EN, Shah PS, Lewis DR, Zablocki KR, Nackman GB, Uhrich KE, Moghe PV. Biomaterials. 2011;32:8319. doi: 10.1016/j.biomaterials.2011.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Budijono SJ, Russ B, Saad W, Adamson DH, Prud’homme RK. Colloids Surf, A. 2010;360:105. [Google Scholar]
- 31.Tivant P, Turq P, Drifford M, Magdelenat H, Menez R. Biopolymers. 1983;22:643. [Google Scholar]
- 32.Budijono SJ, Shan J, Yao N, Miura Y, Hoye TR, Austin RH, Ju Y, Prud’homme RK. Chem Mater. 2009;22:311. [Google Scholar]
- 33.Alexis F, Pridgen E, Molnar LK, Farokhzad OC. Mol Pharmaceutics. 2008;5:505. doi: 10.1021/mp800051m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li S-D, Huang L. Mol Pharmaceutics. 2008;5:496. doi: 10.1021/mp800049w. [DOI] [PubMed] [Google Scholar]
- 35.Stam Jv, Creutz S, De Schryver FC, Jerome R. Macromolecules. 2000;33:6388. [Google Scholar]
- 36.Brown MS, Goldstein JL. Ann Rev Biochem. 1983;52:223. doi: 10.1146/annurev.bi.52.070183.001255. [DOI] [PubMed] [Google Scholar]
- 37.Tontonoz P, Nagy L, Alvarez JGA, Thomazy VA, Evans RM. Cell. 1998;93:241. doi: 10.1016/s0092-8674(00)81575-5. [DOI] [PubMed] [Google Scholar]
- 38.Brown MS, Ho YK, Goldstein JL. J Biol Chem. 1980;266:9344. [PubMed] [Google Scholar]
- 39.Shashkin P, Dragulev B, Ley K. Curr Pharm Des. 2005;11:3061. doi: 10.2174/1381612054865064. [DOI] [PubMed] [Google Scholar]
- 40.Petros RA, DeSimone JM. Nat Rev Drug Discov. 2010;9:615. doi: 10.1038/nrd2591. [DOI] [PubMed] [Google Scholar]
- 41.Faraji AH, Wipf P. Bioorg Med Chem. 2009;17:2950. doi: 10.1016/j.bmc.2009.02.043. [DOI] [PubMed] [Google Scholar]
- 42.Peer D, Karp JM, Hong SY, Farokhzad OC, Margalit R, Langer R. Nat Nanotechnol. 2007;2:751. doi: 10.1038/nnano.2007.387. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.